Abstract
The present study aimed to investigate potential prognostic long noncoding RNAs (lncRNAs) associated with colorectal cancer (CRC). An mRNA-seq dataset obtained from The Cancer Genome Atlas was employed to identify the differentially expressed lncRNAs (DELs) between CRC patients with good and poor prognoses. Subsequently, univariate and multivariate Cox regression analyses were conducted to analyze the prognosis-associated lncRNAs among all DELs. In addition, a risk scoring system was developed according to the expression levels of the prognostic lncRNAs, which was then applied to a training set and an independent testing set. Furthermore, the co-expressed genes of prognostic lncRNAs were screened using a Multi-Experiment Matrix online tool for construction of lncRNA-gene networks. Finally, Kyoto Encyclopedia of Genes and Genomes pathway and Gene Ontology (GO) function enrichment analyses were performed on genes in the lncRNA-gene networks using KOBAS, GOATOOLS and ClusterProfiler. The present study identified 82 DELs, of which long intergenic nonprotein coding RNA 2159, RP11-452L6.6, RP11-894P9.1 and RP11-69M1.6, and whey acidic protein four-disulfide core domain 21 (WFDC21P) were reported to be independently associated with the prognosis of patients with CRC. A 5-lncRNA signature-based risk scoring system was developed, which may be used to classify patients into low- and high-risk groups with significantly different recurrence-free survival times in the training and testing sets (P<0.05). Co-expressed genes of WFDC21P or RP11-69M1.6 were utilized to construct the lncRNA-gene networks. Genes in the networks were significantly enriched in ‘tight junction’, ‘focal adhesion’ and ‘regulation of actin cytoskeleton’ pathways, and numerous GO terms associated with ‘reactive oxygen species metabolism’ and ‘nitric oxide metabolism’. The present study proposed a 5-lncRNA signature-based risk scoring system for predicting the prognosis of patients with CRC, and revealed the associated signaling pathways and biological processes. The results of the present study may help improve prognostic evaluation in clinical practice.
Keywords: long noncoding RNA, Cox regression analysis, stratification analysis, pathway, Gene Ontology
Introduction
Colorectal cancer (CRC) is the third leading cause of cancer worldwide, with ~1.3 million new cases diagnosed every year (1). Surgery is a curative option for the majority of patients with early-stage CRC, whereas interventions are often aimed at improving quality of life and relieving symptoms of CRC at later stages. The 5-year survival rate of CRC varies from >90% in stage I cases to ~10% in stage IV cases (2). Recurrence and metastasis are two major reasons accounting for the poor outcome (3); therefore, promising molecular biomarker candidates to improve prognostic prediction and therapeutic outcome in CRC are required.
Long noncoding RNAs (lncRNAs), once considered transcriptional noise, have recently become an important field in research. LncRNAs are mRNA-like transcripts >200 nucleotides long, which do not encode proteins (4). Previous studies demonstrated that lncRNAs are involved in the tumorigenesis of numerous cancer phenotypes via interactions with DNA, RNA, protein and other cellular molecules (5,6). Expression of lncRNA metastasis-associated lung adenocarcinoma transcript 1 and prostate cancer-associated ncRNA transcripts 1 have been reported to be markedly elevated in CRC tissues compared with in paired normal tissues, and of prognostic significance in CRC (7,8). In addition, Qi et al (9) revealed that lncRNA LOC285194 expression is significantly reduced in CRC tissues, and is associated with poor disease-free survival. Recently, Ozawa et al (10) indicated that colon cancer-associated transcript 1 and 2 may be of strong prognostic value in CRC. These findings suggested the necessity to identify potential prognostic lncRNAs in CRC; however, there are few studies focusing on the identification of reliable prognostic lncRNAs in CRC.
In the present study, a systematic analysis of the lncRNA expression profiles in patients with CRC derived from The Cancer Genome Atlas (TCGA; https://gdc-portal.nci.nih.gov/) portal was initially conducted; differentially expressed lncRNAs (DELs) were screened between CRC patients with good and poor prognoses, from which, prognostic lncRNA signatures were identified using univariate and multivariate Cox regression analyses. Based on the expression of these signature lncRNAs, a risk scoring system was successfully developed and was employed to classify patients in a training set or an independent testing set into high-risk and low-risk groups. Furthermore, interactions between the co-expressed genes of these signature lncRNAs were analyzed in protein-protein interaction (PPI) networks, followed by functional analyses to determine their possible functional roles in CRC. The present study aimed to provide novel insight into the lncRNAs with promising prognostic value in CRC and to improve the survival of patients with CRC.
Materials and methods
Datasets
The present study was performed using an mRNA-seq dataset of colon adenocarcinoma samples retrieved from TCGA data portal, a publicly available repository. Samples with follow-ups of <2 years and without recurrence, or without recurrence-free survival (RFS) time information were excluded from the analysis; consequently, 233 remaining samples were defined as a training set. Additionally, the present study employed a TCGA rectal adenocarcinoma (READ) mRNA-seq dataset. Similarly, following the removal of samples with follow-ups of <2 years and without recurrence, or without RFS time record, 94 remaining samples in the READ set were included in the present study as a testing set. Clinical features of the training and the testing sets are presented in Table I.
Table I.
Clinical characteristics of the training set of COAD and the testing set of READ.
| Characteristics | COAD (n=233) | READ (n=94) | P-value |
|---|---|---|---|
| Age (years) | 68.15±12.38 | 65.58±10.29 | 0.0282a |
| Gender (male/female) | 126/107 | 54/40 | 0.6241 |
| pathologic_stage (I+II/III+IV) | 122/102 | 47/42 | 0.8027 |
| pathologic_M (M0/M1) | 165/39 | 19/16 | 0.0017a |
| pathologic_N (N0/N1/N2) | 133/53/47 | 49/21/22 | 0.5379 |
| pathologic_T (T0/T1/T2/T3/T4) | 0/7/31/164/31 | 0/6/14/64/9 | 0.2664 |
| lymphatic_invasion (yes/no) | 94/116 | 33/52 | 0.3665 |
| venous_invasion (yes/no) | 61/136 | 22/61 | 0.4778 |
| residual_tumor (R0/R1/R2) | 155/3/10 | 62/2/8 | 0.1543 |
| primary_therapy_outcome_success (SD/PD/CR/PR) | 0/6/9/2 | 0/2/7/0 | 0.6673 |
COAD, colon adenocarcinoma; CR, complete remission/response; PD, progressive disease; PR, partial remission/response; READ, rectal adenocarcinoma; SD, stable disease.
P<0.05.
GENCODE gene annotation (www.gencodegenes.org/) (11) is the largest publicly available catalogue of human lncRNAs. Genes in the training and testing sets were annotated by GENCODE (11). The resulting lncRNAs were used for further analysis.
Screening for prognosis-associated DELs
According to RFS time and cancer recurrence status, the 233 samples in the training set were classified into a good prognosis group (n=106), in which patients exhibited RFS time ≥2 years without cancer recurrence, and a poor prognosis group (n=127), in which patients experienced cancer recurrence. With an adjusted P-value (P<0.05) as a strict cutoff value, DELs between the two groups were screened using edgeR package (12–14) or DESeq2 package (15) (Bioconductor; www.bioconductor.org/) in R3.3.3 language.
Prognosis-associated DELs were identified from the overlapped DELs between the two packages using univariate Cox regression analysis (16) in survival package 2.41-3 (cran.r-project.org/web/packages/survival/index.html) in R 3.4.3, followed by log-rank test (cut-off value, P<0.01). Subsequently, these identified prognosis-associated DELs were subjected to multivariate Cox regression analysis.
Development of risk scoring system
Based on the expression of significant prognostic lncRNAs derived from multivariate Cox regression analysis, the risk score was calculated as the linear combination of expression levels of these lncRNAs, which were weighted by regression coefficients (16,17), and a risk scoring system was developed as follows:
Risk score = β1 × expr1 + β2 × expr2 + ··· + βn × exprn
Where βn stands for estimated regression coefficient of lncRNAn, and exprn stands for expression levels of lncRNAn.
The risk scoring system was used to divide patients in each set into high-risk and low-risk groups, with the median risk score of the set being the cut-off point. Kaplan-Meier survival analyses were employed to compare the RFS time of the high-risk and low-risk groups, followed by log-rank test. Furthermore, univariate and multivariate Cox regression analyses, and data stratification analysis (χ2) were conducted to evaluate whether the risk score was independent of clinical features. For these tests, P<0.05 was considered to indicate a statistically significant difference. In addition, the expression levels of these selected lncRNAs were compared between the high-risk group and the low-risk group using Student's t-test in the R stats package 3.4.3 (www.rdocumentation.org/packages/stats/versions/3.4.3) with a threshold of P<0.05.
Construction of lncRNA-gene networks and PPI networks
Co-expressed genes of these significant lncRNAs identified by multivariate Cox regression analysis were analyzed using the Multi-Experiment Matrix (MEM) web-based online tool (18,19) (http://biit.cs.ut.ee/mem/index.cgi) in Human Genome U133 Plus 2.0. These co-expressed genes were ranked according to a score of significance by the MEM tool. The top 100 genes of each lncRNA were selected for the construction of a lncRNA-gene network using Cytoscape 3.0 (20) (www.cytoscape.org/). In order to study the interactions between genes in each lncRNA-gene network, corresponding PPI networks were built using the Search Tool for the Retrieval of Interacting Genes (string-db.org/) (21) and visualized using Cytoscape 3.0 (20), with the threshold value set at median confidence=0.4.
Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses
GOATOOLS 0.8.2 (github.com/tanghaibao/goatools) is an easily accessible Python package used for annotation of genes to biological processes (BP), molecular function (MF) and cellular component (CC) in the GO database. KOBAS 3.0 (kobas.cbi.pku.edu.cn/) is a web server used to annotate input genes and identify putative pathways involved, allowing for ID mapping and cross-species sequence similarity mapping (22). GO terms and KEGG pathways that may involve these genes in the lncRNA-gene networks were investigated with GOATOOLS and KOBAS, respectively. ClusterProfiler (Bioconductor; https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) is a R package used for the classification of biological terms and enrichment analysis of gene clusters, and is characterized by biological theme comparison among gene clusters (23). ClusterProfiler software 3.6.0 was applied to perform GO function and KEGG pathway enrichment analyses on the genes in the lncRNA-gene networks.
Results
Analysis of prognostic lncRNAs
Following removal of lncRNAs with 0 expression in at least 50 samples, there were a total of 4,162 lncRNAs in the training set. As a result, 82 DELs between good prognosis and poor prognosis samples were identified using the edgeR and DESeq2 packages. Among these resulting DELs, 44 were identified as prognosis-associated lncRNAs in univariate Cox regression analysis (P<0.01). Furthermore, as presented in Table II, 5 of the 44 prognosis-associated lncRNAs were detected to be independently associated with prognosis in multivariate Cox regression analysis, including ENSG00000253417 [long intergenic nonprotein coding RNA 2159, (LINC02159)], ENSG00000280132 (RP11-452L6.6), ENSG00000261040 [whey acidic protein four-disulfide core domain 21 (WFDC21P)], ENSG00000246451 (RP11-894P9.1) and ENSG00000279865 (RP11-69M1.6).
Table II.
Multivariate analysis of the five lncRNA signatures.
| LncRNA | coef | exp(coef) | se(coef) | z-value | Pr (>|z|) |
|---|---|---|---|---|---|
| ENSG00000280132 | 0.3844 | 1.4688 | 0.1606 | 2.394 | 0.017 |
| ENSG00000253417 | −0.2810 | 0.7550 | 0.1002 | −2.805 | 0.005 |
| ENSG00000279865 | −0.4740 | 0.6225 | 0.2294 | −2.066 | 0.039 |
| ENSG00000261040 | −0.2715 | 0.7623 | 0.1200 | −2.262 | 0.024 |
| ENSG00000246451 | −0.6862 | 0.5035 | 0.3136 | −2.188 | 0.029 |
LncRNA, long noncoding RNA; se, standard error.
Development of a risk score system based on the 5-lncRNA signature
Based on the expression of the 5 lncRNAs derived from multivariate Cox regression analysis, a risk scoring system was developed as follows: Risk score = −0.219 × expr (LINC02159) + 0.354 × expr (RP11-452L6.6) −0.211 × expr (WFDC21P) + 0.143 × expr (RP11-894P9.1) −0.374 × expr (RP11-69M1.6)
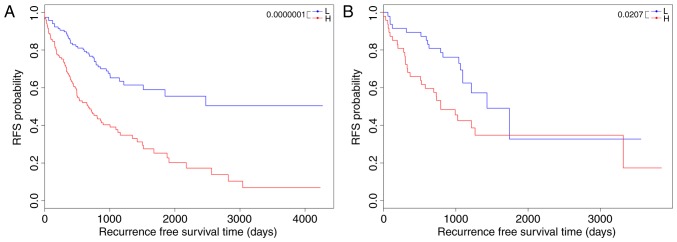
The 5-lncRNA-based risk scoring system was applied to the training set to assort patients into high-risk and low-risk groups, with the median risk score as the threshold. In the training set, the low-risk group had a markedly longer RFS time compared with the high-risk group (38.547±29.693 months vs. 26.721±25.532 months, P=1×10−7; Fig. 1A). To validate the prognostic power of the risk scoring system in the testing set, all patients in the testing set were also classified into high-risk and low-risk groups by the risk scoring system. Similar results were yielded in the testing set (34.090±18.477 months vs. 29.305±26.450 months, P=0.0207; Fig. 1B).
Figure 1.
Kaplan-Meier survival analysis of RFS time for patients classified using the 5-long noncoding RNA signature-based risk scoring system in the (A) training set and the (B) testing set. Patients in the training set and the testing set are classified into L and H groups. H, high-risk; L, low-risk; RFS, recurrence-free survival.
Expression levels of the 5 prognostic lncRNAs in the training set and the testing set
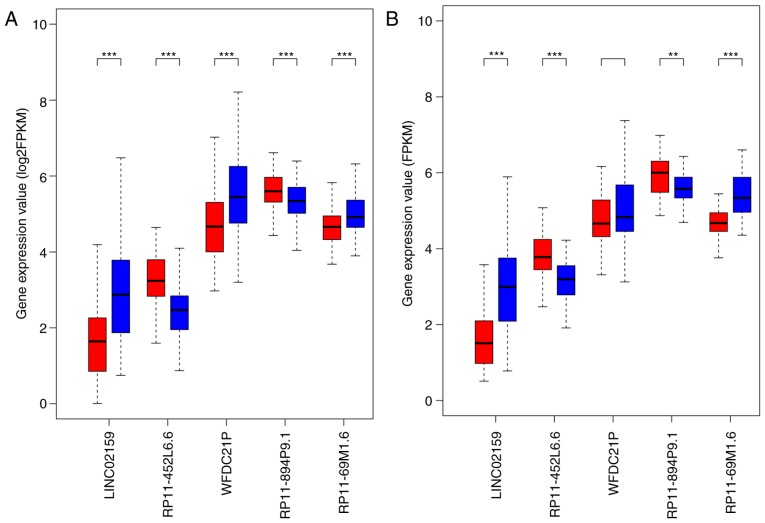
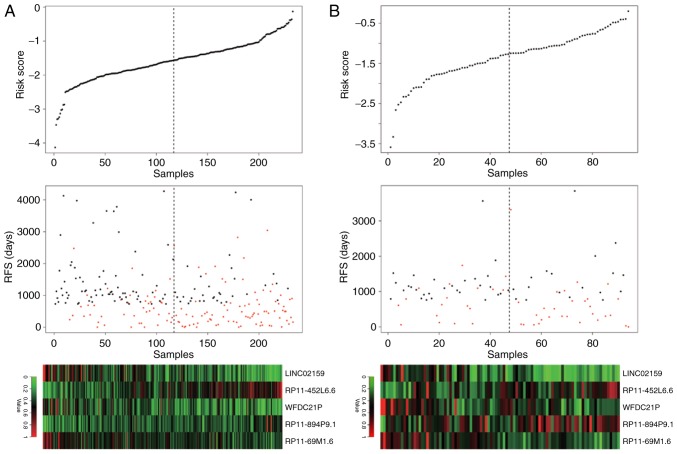
Boxplots demonstrated that in the training set, the expression levels of LINC02159, WFDC21P and RP11-69M1.6 were significantly decreased, whereas the expression levels of RP11-452L6.6 and RP11-894P9.1 were significantly elevated in the high-risk samples compared with in the low-risk samples (P<0.001; Fig. 2A), which was validated in the testing set, except for WFDC21P (Fig. 2B). WFDC21P expression appeared to be reduced in the high-risk samples compared with in low-risk samples, but the difference was not significant. The distribution of risk score, RFS time and long noncoding RNA expression in the training set and the testing set were similar, indicating the robustness of the risk scoring system based on the 5 lncRNAs (Fig. 3).
Figure 2.
Comparison of the expression of 5 lncRNAs between high- and low-risk groups in the (A) training set and the (B) testing set. Patients in the training set and the testing set were classified into low-risk and high-risk groups. Red boxplots represent expression of lncRNAs in the high-risk group; blue boxplots represent expression of lncRNAs in the low-risk group. ***P<0.001 and **P<0.01. FPKM, Fragments Per Kilobase of transcript per Million mapped read; LINC02159, long intergenic nonprotein coding RNA 2159; lncRNA, long noncoding RNA; WFDC21P, whey acidic protein four-disulfide core domain 21.
Figure 3.
Distribution of risk score, RFS time and long noncoding RNA expression in the (A) training set and the (B) testing set. The black dotted line represents the median risk score cutoff classifying patients into high- and low-risk groups. LINC02159, long intergenic nonprotein coding RNA 2159; RFS, recurrence-free survival; WFDC21P, whey acidic protein four-disulfide core domain 21.
Analysis on whether prognostic power of the 5-lncRNA signature-based risk score model is independent of clinical features
According to the results of a univariate Cox regression analysis, risk score, pathologic_stage, pathologic_M, pathologic_N, lymphatic_invasion and venous_invasion were demonstrated to be significantly associated with prognosis (Table III). Among these significant factors, risk score, pathologic_stage, pathologic_n and venous_invasion were identified to be independent predictors of prognosis according to the results of a multivariate Cox regression analysis (P<0.05; Table IV).
Table III.
Results of univariate Cox regression analysis for risk score and clinical features.
| Clinical features | coef. | Standard error | z-value | P-value | HR | Lower. 95 | Upper. 95 |
|---|---|---|---|---|---|---|---|
| Risk score | 0.9622 | 0.1882 | 5.1116 | 3.20×10−7 | 2.6174 | 1.8098 | 3.7852 |
| Age | 0.0914 | 0.1786 | 0.5115 | 0.6090 | 1.0957 | 0.7720 | 1.5550 |
| Gender | 0.1667 | 0.1798 | 0.9272 | 0.3538 | 1.1814 | 0.8306 | 1.6803 |
| pathologic_stage | 0.9726 | 0.1860 | 5.2279 | 1.71×10−7 | 2.6448 | 1.8367 | 3.8085 |
| pathologic_m | 1.4008 | 0.2137 | 6.5539 | 5.61×10−11 | 4.0583 | 2.6694 | 6.1698 |
| pathologic_n | 0.8809 | 0.1806 | 4.8763 | 1.08×10−6 | 2.4130 | 1.6935 | 3.4381 |
| lymphatic_invasion | 0.6131 | 0.1923 | 3.1876 | 0.0014 | 1.8461 | 1.2663 | 2.6913 |
| venous_invasion | 0.8316 | 0.2047 | 4.0620 | 4.87×10−5 | 2.2970 | 1.5378 | 3.4310 |
HR, hazard ratio.
Table IV.
Results of multivariate Cox regression analysis for risk score and clinical features.
| Clinical features | coef. | Standard error | z-value | P-value | HR | Lower. 95 | Upper. 95 |
|---|---|---|---|---|---|---|---|
| pathologic_stage | 1.7686 | 0.4753 | 3.721 | 0.0002a | 5.8624 | 2.3095 | 14.8814 |
| pathologic_n | −1.1287 | 0.468 | −2.412 | 0.0159a | 0.3235 | 0.1293 | 0.8094 |
| venous_invasion | 0.5074 | 0.2212 | 2.294 | 0.0218a | 1.6609 | 1.0767 | 2.5622 |
| Risk score | 0.6828 | 0.2226 | 3.068 | 0.0022a | 1.9794 | 1.2796 | 3.0620 |
HR, hazard ratio.
P<0.05.
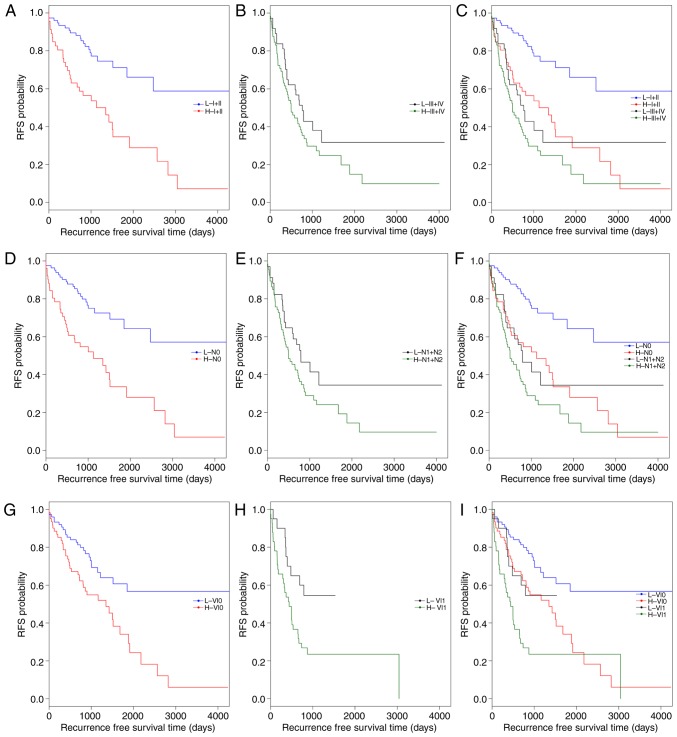
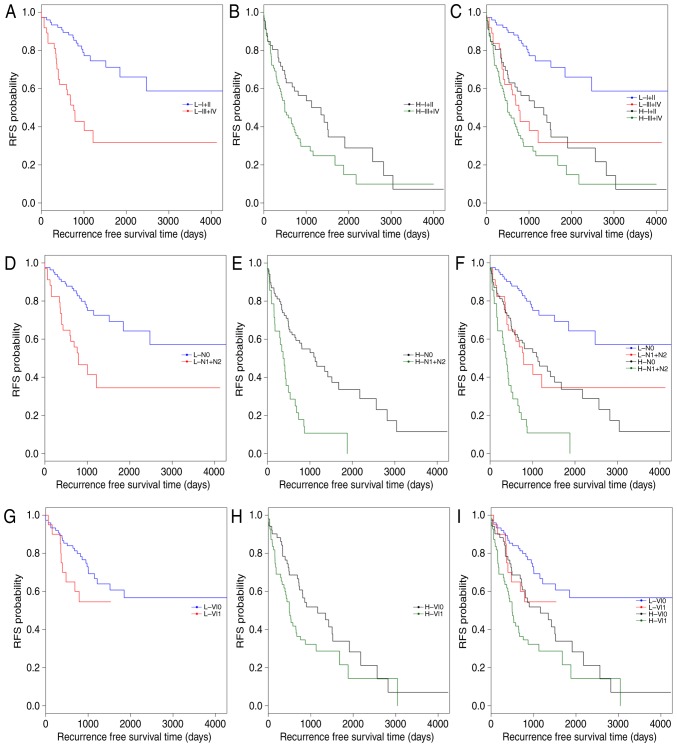
Subsequently, data stratification analysis was conducted for pathologic_stage, pathologic_n and venous_invasion (Fig. 4). All samples in the training set were stratified by pathologic_stage, pathologic_n and venous_invasion, respectively, into two groups. In each group, the 5-lncRNA based risk scoring model further split patients into a high-risk subgroup and a low-risk subgroup (Table V). Significantly different RFS time between high-risk and low-risk subgroups was observed in the pathologic_stage (I+II) group (P=4×10−5; Fig. 4A and C), the pathologic_n (N0) group (P=4×10−5; Fig. 4D and F), and patients with or without venous_invasion (P=0.0178 and P=0.001; Fig. 4G-I). However, the difference in RFS time between the high-risk and low-risk subgroups was not significant in the pathologic_stage (III+IV) group (P=0.1373; Fig. 4B) and the pathologic_n (N1+N2) group (P=0.0883; Fig. 4E). The results of multivariate Cox regression analysis and stratification analysis suggested that the risk classification power of the 5-lncRNA signature-based risk scoring system signature is independent of other clinical variables in CRC.
Figure 4.
Kaplan-Meier survival analysis for the training set of patients first stratified by clinical features and then by the risk scoring system. (A-C) All patients in the training set were categorized into pathologic stage I+II group and pathologic stage III+IV group. In either the pathologic stage (A) I+II group or (B) III+IV group, patients were further classified into H and L subgroups using the 5-lncRNA signature-based risk scoring system. (C) Combined image of A and B. (D) N0 and (E) N1+N2 patients were further classified into H and L subgroups by the 5-lncRNA signature-based risk scoring system. (F) Combined image of D and E. Patients (G) without or (H) with venous invasion were further segregated into H and L subgroups by the 5-lncRNA signature-based risk scoring system. (I) Combined image of G and H. H, high-risk; L, low-risk; RFS, recurrence free survival; VI0, patients without venous invasion; VI1, patients with venous invasion.
Table V.
Results of data stratification analysis.
| Variables | χ2 | P-value |
|---|---|---|
| pathologic_stage (I+II) | 16.8702 | 4×10−5 |
| pathologic_ stage (III+IV) | 2.2082 | 0.13728 |
| pathologic_n (N0) | 17.0432 | 4×10−5 |
| pathologic_n (N1+N2) | 2.9055 | 0.08828 |
| venous_invasion (No) | 10.8120 | 0.00101 |
| venous_invasion (Yes) | 5.6180 | 0.01778 |
The present study also reported that the prognostic value of pathologic_stage, pathologic_n and venous_invasion were independent of risk score. For this, all patients in the training set were first classified into high- and low-risk groups by the risk scoring system. Each group was then stratified into two subgroups by pathologic_stage, pathologic_n or venous_invasion, respectively (Table VI). In both low-risk and high-risk groups, RFS time was significantly different between the patients at stage I+II and the patients at stage III+IV (low-risk: P=1×10−5; high-risk: P=0.0303, respectively; Fig. 5A-C). Similar results were observed between the patients with pathologic N0 and the patients with pathologic N1+N2 (low-risk: P=0.00026; high-risk: P=0.04216, respectively; Fig. 5D-F). As presented in Fig. 5G-I, the difference in RFS time between the patients with and without venous invasion was markedly significant in the high-risk group (P=0.0068), but was not significant in the low-risk group (P-value=0.1244). These observations confirmed that risk score, pathologic_stage, pathologic_n and venous_invasion are independent prognostic factors in CRC.
Table VI.
Results of data stratification analysis.
| Risk | Clinical features | χ2 | P-value |
|---|---|---|---|
| Low risk | pathologic_stage (I+II vs. III+IV) | 19.1067 | 0.00001 |
| High risk | pathologic_stage (I+II vs. III+IV) | 4.6771 | 0.03033 |
| Low risk | pathologic_n (N0 vs. N1+N2) | 13.3250 | 0.00026 |
| High risk | pathologic_n (N0 vs. N1+N2) | 4.1289 | 0.04216 |
| Low risk | venous_invasion (No vs. Yes) | 2.3605 | 0.12444 |
| High risk | venous_invasion (No vs. Yes) | 7.3158 | 0.00684 |
Figure 5.
Kaplan-Meier survival analysis for the training set of patients first stratified by the risk scoring system and then by clinical features. All patients in the training set were first classified into H and L groups. (A) L and (B) H patients were further divided into stage I+II and stage III+IV subgroups. (C) Combined image of A and B. (D) L and (E) H patients were further divided into N0 and N1+N2 subgroups. (F) Combined image of D and E. (G) L and (H) H patients were further classified into VI0 and VI1 subgroups. (I) Combined image of G and H. H, high-risk; L, low-risk; RFS, recurrence free survival; VI0, no venous invasion; VI1, venous invasion
Analysis of co-expressed genes of prognostic lncRNAs
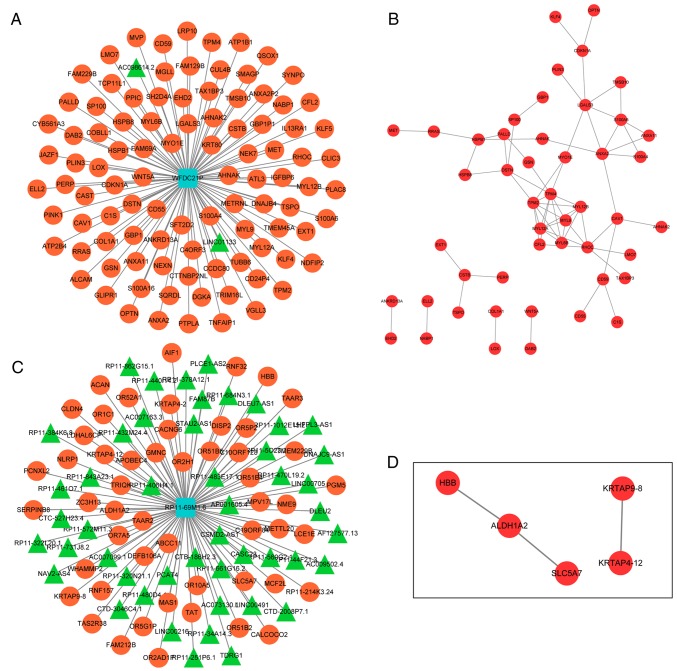
Since lncRNAs may serve their biological roles by regulating their co-expressed genes, co-expressed genes of the 5 lncRNAs were searched using the MEM tool. Only co-expressed genes of WFDC21P and RP11-69M1.6 were obtained. The top 100 co-expressed genes of WFDC21P or RP11-69M1.6 were used to construct lncRNA-gene networks. PPI networks were also generated to analyze the interactions between the genes in each lncRNA-gene network (Fig. 6).
Figure 6.
LncRNA-gene and PPI networks. (A) LncRNA-gene network for WFDC21P. (B) PPI network for WFDC21P. (C) LncRNA-gene network for RP11-69M1.6. (D) PPI network for RP11-69M1.6. In these networks, round red nodes represent genes; triangle green nodes represent lncRNAs; the association between two triangle nodes represents the interaction between two genes. LncRNA, long noncoding RNA; PPI, protein-protein interaction; WFDC21P, whey acidic protein four-disulfide core domain 2.
Function analysis
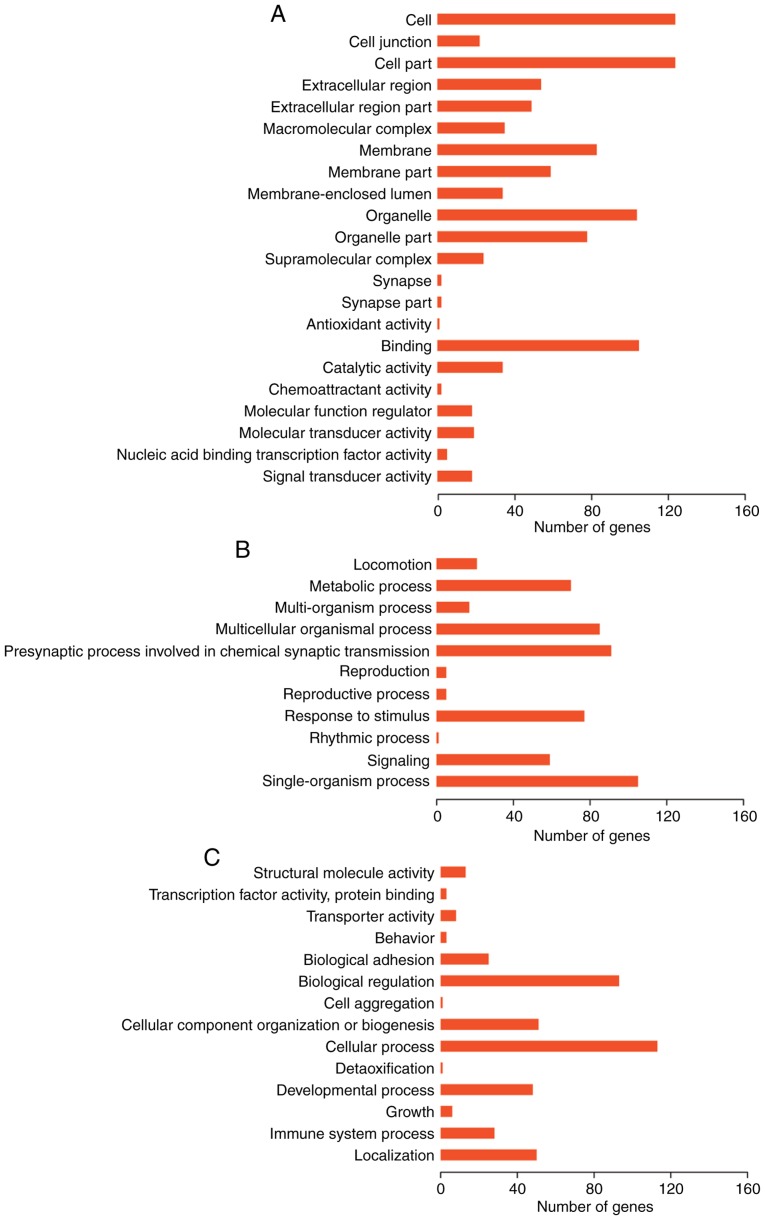
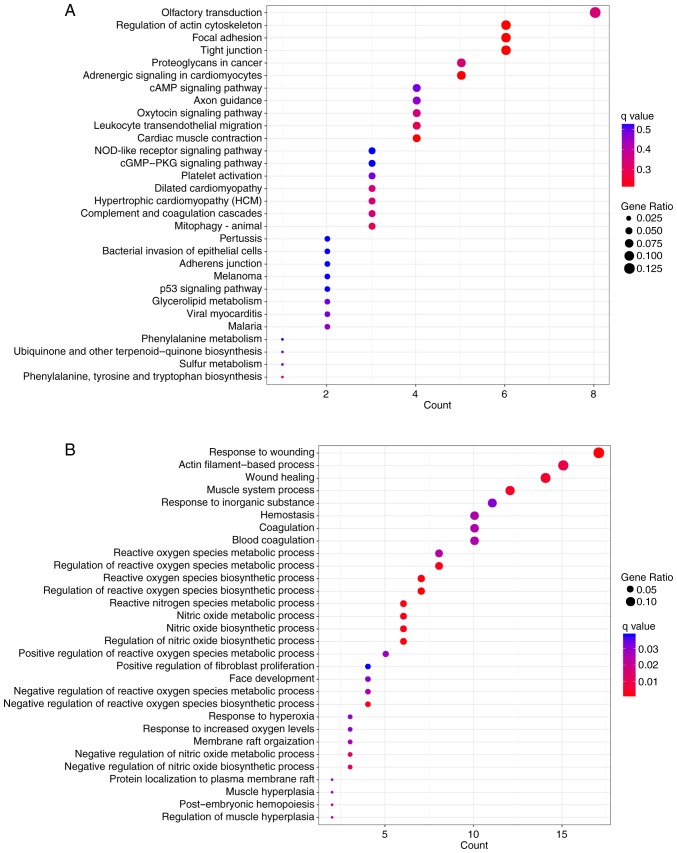
GO function and KEGG pathway enrichment analyses were performed on genes in the two lncRNA-gene networks. GO enrichment analysis using GOATOOLS suggested that these genes were possibly associated with ‘cell membrane’, ‘organelle’ and other cellular components (Fig. 7). Pathway enrichment analysis conducted by KOBAS or ClusterProfiler revealed that these genes were significantly associated with numerous pathways, including ‘tight junction’, ‘focal adhesion’, ‘regulation of actin cytoskeleton’ and ‘leukocyte transendothelial migration’ (Figs. 8 and 9A). According to the results of GO enrichment analysis by ClusterProfiler, these genes were significantly associated with numerous GO BP terms, including ‘wound healing’, ‘reactive oxygen species (ROS) metabolism’ and ‘nitric oxide metabolism’ (Fig. 9B).
Figure 7.
Results of Gene Ontolgy enrichment analysis by GOATOOLS. (A) Cellular component, (B) biological processes and (C) molecular function.
Figure 8.
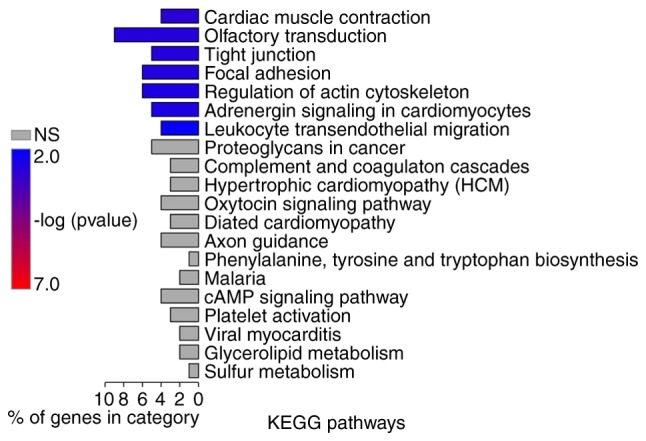
Results of KEGG pathway enrichment analysis using KOBAS. KEGG, Kyoto Encyclopedia of Genes and Genomes; NS, not significant.
Figure 9.
KEGG pathway and GO enrichment analyses using ClusterProfiler. (A) Results of KEGG pathway enrichment analysis. (B) Results of GO enrichment analysis (biological processes). Abscissa axis represents the count of genes significantly enriched in each pathway or GO term. Size of round node is in proportion to gene ratio of the enriched gene number. GO, Gene Ontology; Kyoto Encyclopedia of Genes and Genomes.
Discussion
CRC remains a major cause of mortality worldwide. Numerous lncRNAs have been reported to be involved in CRC tumorigenesis; however, there is a lack of efficient prognostic lncRNAs to refine the prediction of survival for patients with CRC (24). In order to define a prognostic lncRNA signature, the present study identified 82 DELs between CRC patients with good and poor prognoses in the training set. Among these DELs, WFDC21P, LINC02159, RP11-452L6.6, RP11-894P9.1 and RP11-69M1.6 were demonstrated to be significantly associated with prognosis via multivariate Cox regression analysis. Based on the expression of these 5 lncRNAs, a risk scoring system was generated. The results of the present study revealed that the 5-lncRNA signature-based risk scoring system may categorize patients into high- and low-risk groups with significantly varying RFS times in the training set and the testing set.
WFDC21P, namely lnc-dendritic cell (lnc-DC) has been observed to be exclusively expressed in human DCs and mediates the differentiation of monocytes to DCs via the activation of signal transducer and activator of transcription 3, which is a transcription factor that regulates numerous immune-associated genes (25,26). Wu et al (27) suggested that lnc-DC in plasma may be a possible biomarker for systemic lupus erythematosus. In addition, Zhang et al (28) reported that lnc-DC expression is increased in decidua, resulting in over-maturation of decidual DCs and increased T helper 1 cells in patients with preeclampsia. RP11-894P9.1 has been observed to be abnormally expressed in the right ventricle of the heart during heart failure (29); however, to the best of our knowledge, aberrant expression levels of WFDC21P and RP11-894P9.1 have previously not been reported in CRC. The functions of LINC02159, RP11-452L6.6, and RP11-69M1.6 remain unknown, and further investigations into lncRNA expression in cancer are required. The present study suggested that the 5-lncRNA signature may be a promising prognostic biomarker for CRC.
Previous studies have revealed the predictive value of T stage, venous invasion and lymph node metastasis in patients with CRC (30,31). Furthermore, in the present study, multivariate Cox regression analysis demonstrated that pathologic_stage, pathologic_n and venous_invasion were independently associated with prognosis. In addition, the results of data stratification analysis indicated that the prognostic value of the 5-lncRNA signature-based risk score may be independent of other clinical variables in CRC in the present study; therefore, the 5-lncRNA signature may aid in improving current prognostic approaches.
In order to uncover associated biological processes and signaling pathways of the 5-lncRNA signature, the present study attempted to identify the co-expressed genes of the 5 lncRNAs by MEM analysis. Only co-expressed genes of WFDC21P and RP11-69M1.6 were reported and lncRNA-gene networks were subsequently generated, followed by the construction of PPI networks to visualize the interactions between these genes. Furthermore, according to KEGG pathway enrichment analysis, these genes were significantly associated with ‘tight junction’, ‘focal adhesion’ and ‘regulation of actin cytoskeleton’ pathways. ‘Tight junction’ and ‘focal adhesion’ pathways are key determinants in tumor progression and metastasis (32,33). The ‘regulation of actin cytoskeleton’ pathway has been associated with cancer cell motility (34). Furthermore, the present study demonstrated that the genes in these lncRNA-gene networks were significantly associated with numerous GO BP terms that were associated with ‘ROS metabolism’ or ‘nitric oxide metabolism’. It has been established that abnormal ROS accumulation is a critical contributor to tumorigenesis (35). In addition, Colin et al (36) suggested that ROS may be involved in the development of resistance against resveratrol in colon cancer cells. Studies have also demonstrated that nitric oxide is an important regulator of tumor metabolism (37,38). These findings suggested that the 5-lncRNA signature may possibly be involved in ‘tight junction’, ‘focal adhesion’ and ‘regulation of actin cytoskeleton’ pathways, and ‘ROS metabolism’ and ‘nitric oxide metabolism’ in CRC by regulating the co-expressed genes.
Some limitations of the present study should be mentioned. Experiments were not conducted, and the size of the patient cohort may be further expanded. Therefore, future studies with larger patient cohorts are warranted to verify the prognostic significance of the 5-lncRNA signature prior to its use in clinical practice.
In conclusion, the present study identified a 5-lncRNA signature that may have great potential as a prognostic biomarker for CRC, and developed a 5-lncRNA signature-based risk scoring system as a prognostic classification system. Furthermore, the underlying signaling pathways and biological processes associated with the 5-lncRNA signature in CRC were investigated. These findings may aid in refining the stratification approach for the clinical assessment of prognosis, and provide guidance on tailored therapeutic strategies and patient management; however, further investigation is required to validate the findings of the present study.
Acknowledgements
The authors would like to thank Dr Min Yu for providing assistance in data collection and entry.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LG wrote the manuscript. LG, JY, QW, BX, LY and LJ performed the data analyses. JY and LY revised the manuscript and provided important suggestions for the analyses. HC and XZ conceptualized the study design. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 3.García Sánchez J. Colonoscopic polypectomy and long-term prevention of colorectal cancer deaths. Rev Clin Esp. 2012;212:408. (In Spanish) [PubMed] [Google Scholar]
- 4.Li X, Wu Z, Fu X, Han W. lncRNAs: Insights into their function and mechanics in underlying disorders. Mutat Res Rev Mutat Res. 2014;762:1–21. doi: 10.1016/j.mrrev.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 7.Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, Gu WL, Cai GX, Cai SJ. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7:3174–3181. [PMC free article] [PubMed] [Google Scholar]
- 8.Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H, Jia W. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med Oncol. 2013;30:588. doi: 10.1007/s12032-013-0588-6. [DOI] [PubMed] [Google Scholar]
- 9.Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C, Zhou XY, Du X. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122. doi: 10.1186/1479-5876-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozawa T, Matsuyama T, Toiyama Y, Takahashi N, Ishikawa T, Uetake H, Yamada Y, Kusunoki M, Calin G, Goel A. CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 ‘gene desert’, serve as important prognostic biomarkers in colorectal cancer. Ann Oncol. 2017;28:1882–1888. doi: 10.1093/annonc/mdx248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sillar B, Plint CW. The price of a false-negative result of mammography and an overenthusiastic lay press. Med J Aust. 1989;151:418. [PubMed] [Google Scholar]
- 13.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P, Wang Y, Bo H, Zou X, Mao JH. A novel gene expression-based prognostic scoring system to predict survival in gastric cancer. Oncotarget. 2016;7:55343–55351. doi: 10.18632/oncotarget.10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou M, Wang X, Shi H, Cheng L, Wang Z, Zhao H, Yang L, Sun J. Characterization of long non-coding RNA-associated ceRNA network to reveal potential prognostic lncRNA biomarkers in human ovarian cancer. Oncotarget. 2016;7:12598–12611. doi: 10.18632/oncotarget.7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–580. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adler P, Kolde R, Kull M, Tkachenko A, Peterson H, Reimand J, Vilo J. Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods. Genome Biol. 2009;10:R139. doi: 10.1186/gb-2009-10-12-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu MD, Qi P, Du X. Long non-coding RNAs in colorectal cancer: Implications for pathogenesis and clinical application. Mod Pathol. 2014;27:1310–1320. doi: 10.1038/modpathol.2014.33. [DOI] [PubMed] [Google Scholar]
- 25.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 26.Li Q, Liu Q. Noncoding RNAs in Cancer Immunology. Adv Exp Med Biol. 2016;927:243. doi: 10.1007/978-981-10-1498-7_9. [DOI] [PubMed] [Google Scholar]
- 27.Wu GC, Li J, Leng RX, Li XP, Li XM, Wang DG, Pan HF, Ye DQ. Identification of long non-coding RNAs GAS5, linc0597 and lnc-DC in plasma as novel biomarkers for systemic lupus erythematosus. Oncotarget. 2017;8:23650–23663. doi: 10.18632/oncotarget.15569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Zhou Y, Ding Y. Lnc-DC mediates the over-maturation of decidual dendritic cells and induces the increase in Th1 cells in preeclampsia. Am J Reprod Immunol. 2017;77 doi: 10.1111/aji.12647. [DOI] [PubMed] [Google Scholar]
- 29.Di Salvo TG, Guo Y, Su YR, Clark T, Brittain E, Absi T, Maltais S, Hemnes A. Right ventricular long noncoding RNA expression in human heart failure. Pulm Circ. 2015;5:135–161. doi: 10.1086/679721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roxburgh CS, Mcmillan DC, Richards CH, Atwan M, Anderson JH, Harvey T, Horgan PG, Foulis AK. The clinical utility of the combination of T stage and venous invasion to predict survival in patients undergoing surgery for colorectal cancer. Ann Surg. 2014;259:1156–1165. doi: 10.1097/SLA.0000000000000229. [DOI] [PubMed] [Google Scholar]
- 31.Berger AC, Sigurdson ER, Levoyer T, Hanlon A, Mayer RJ, Macdonald JS, Catalano PJ, Haller DG. Colon cancer survival is associated with decreasing ratio of metastatic to examined lymph nodes. J Clin Oncol. 2005;23:8706–8712. doi: 10.1200/JCO.2005.02.8852. [DOI] [PubMed] [Google Scholar]
- 32.Martin TA. The role of tight junctions in cancer metastasis. Semin Cell Dev Biol. 2014;36:224–231. doi: 10.1016/j.semcdb.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Eke I, Cordes N. Focal adhesion signaling and therapy resistance in cancer. Seminars in Cancer Biol. 2015;31:65–75. doi: 10.1016/j.semcancer.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Olson MF, Sahai E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis. 2009;26:273–287. doi: 10.1007/s10585-008-9174-2. [DOI] [PubMed] [Google Scholar]
- 35.Costa A, Scholerdahirel A, Mechta-Grigoriou F. The role of reactive oxygen species and metabolism on cancer cells and their microenvironment. Semin Cancer Biol. 2014;25:23–32. doi: 10.1016/j.semcancer.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 36.Colin DJ, Limagne E, Ragot K, Lizard G, Ghiringhelli F, Solary É, Chauffert B, Latruffe N, Delmas D. The role of reactive oxygen species and subsequent DNA-damage response in the emergence of resistance towards resveratrol in colon cancer models. Cell Death Dis. 2014;5:e1533. doi: 10.1038/cddis.2014.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang CF, Diers AR, Hogg N. Cancer cell metabolism and the modulating effects of nitric oxide. Free Radic Biol Med. 2015;79:324–336. doi: 10.1016/j.freeradbiomed.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wink DA, Vodovotz Y, Laval J, Laval F, Dewhirst MW, Mitchell JB. The multifaceted roles of nitric oxide in cancer. Carcinogenesis. 1998;19:711–721. doi: 10.1093/carcin/19.5.711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.