Abstract
Glioblastoma multiforme (GBM) is one of the most lethal types of tumour, despite severe treatment methods. The Cancer Genome Atlas has categorised GBMs into proneural, neural, classical and mesenchymal subtypes; the mesenchymal subgroup has the worst prognosis. CXCR4 has been reported as selectively overexpressed in the mesenchymal subtype and positively associated with MES markers. However, to the best of our knowledge the underlying mechanisms regarding how CXCR4 may regulate mesenchymal GBM are still unknown. The present study aimed to investigate the critical pathways mediated by CXCR4 in mesenchymal GBM using bioinformatic analyses. The results suggested that CXCR4 is a predictor of poor prognosis and may serve as a biomarker of the mesenchymal subtype in patients with GBM. In addition, CXCR4 mediated the mitogen-activated protein kinase signaling pathway, which was identified specifically in patients with mesenchymal GBM. CXCR4 associated genes or pathways may be a ‘basket trial’ option for the management of melanoma, prostate cancer and mesenchymal GBM.
Keywords: bioinformatic analyses, CXCR4, prognosis, differentially expressed gene, mesenchymal glioblastoma
Introduction
Glioblastoma multiforme (GBM) is the most common malignant tumour of central nervous system (CNS), and it currently remains incurable. Despite drastic treatment methods, including maximal safe resection followed by radiotherapy in combination with temozolomide, the patient survival rate has not effectively improved, with a 5-year survival rate of only 5.5% being observed (1,2). Therefore, more refined studies are vital to elucidate the potential mechanism of therapeutic tumourigenesis and resistance of this malignancy.
In the previous classification scheme of gliomas, GBM were commonly diagnosed as astrocytomas or oligodendrogliomas based on their morphological resemblance and further distinguished by malignant grades (I to IV) according to cellular features (proliferation, angiogenesis and necrosis) (3). However, this partition method is highly subjective and inconsistent in its designation in glioma subtypes and grades (4). In addition, the pathological criteria diagnosis of gliomas especially GBMs, which has caused artificial heterogeneity and complexities in investigations, did not bring valid effects to our clinical management (5). Recent studies focused on the molecule-based GBM classification which benefited from the availability of the datasets generated by The Cancer Genome Atlas (TCGA) workgroup have established the groundwork for a better understanding of the etiology and improved personalized therapy for subgroup patients (6–9). In 2016, the new WHO classification of diffuse gliomas was refined, and formally brought the histomolecular conception into our sight for the first time, which incorporated 1p/19q codeletion, IDH1/2 mutation, and histone H3-K27M mutation into previous diagnostic criteria (10–12).
Because of robust gene expression, TCGA classified GBMs into four subtypes: proneural, neural, classical and mesenchymal. Each subtype differs greatly in terms of its cellular features, genetic contexts and signalling pathways involved (7). Due to a more aggressive biological nature and higher overall fraction of necrosis evident in these tumours, the mesenchymal group is usually placed in a more malignant base (7,13). It is proposed that not a single molecule but the alterations of a small regulatory module can induce and maintain a specific phenotypic state in glioma cells (14). For instance, the activation of a small group of hub genes may facilitate mesenchymal transformation, which is characterized by extensive necrosis, angiogenesis, and an enhanced inflammatory/immune response (15,16).
CXCR4, a cell surface chemokine receptor, is involved in many cell fate decisions, such as growth, invasion, angiogenesis and metastasis in a wide range of malignant cancers, including leukaemia, breast cancer and, recently, in glioma (17–19). In this study, we clarified the prognosis and clinical significance of CXCR4 in glioma and observed that CXCR4 has shown to be selectively overexpressed in the mesenchymal subtype compared with other phenotypes of GBMs, which indicates that CXCR4 may participate in phenotype transformation of the mesenchymal GBMs.
Methods to classify tumours according to key molecular events that manage growth of their most aggressive cellular component and to search for the genetic alterations that accompany disease recurrence might greatly facilitate development of targeted therapies (13,20). In the present study, we aimed to investigate the associations and uncover the critical pathways that CXCR4 mediated in mesenchymal glioblastoma using computational methods to analyse the relationship between a wide range of expression patterns and the activation of specific hub genes or pathways.
Materials and methods
Glioma specimen and brain tissue collection
Glioma surgical specimens were collected in Tianjin Medical University General Hospital between October 2011 and November 2017 in accordance with institution-approved protocols. All patients signed and approved consent forms prior to the surgery and the study was approved by the Ethics Committee of Tianjin Medical University Hospital (Tianjin, China). These tissue samples were analyzed retrospectively in the present study. Collected specimens were split into two parts for 4% paraformaldehyde (PFA) fixation/cryo-sectioning and primary tissues culture establishment respectively. Specimens were examined by pathologists to verify tumour types and grades.
Analysis of glioma patients' survival and expression data
Tumour gene expression and clinical data of glioma patients were retrieved from The Cancer Genome Atlas (TCGA) and French dataset which were available on R2 analysis and visualization platform (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi). Patients were classified into CXCR4low and CXCR4high expression groups by the mean expression levels.
Differential expression genes analysis of TCGA microarray data
The gene expression profiles of TCGA were downloaded from UCSC Xena Browser (https://xenabrowser.net/heatmap/), which contains 539 GBM samples including 145 classical subtype, 158 mesenchymal subtype, 87 neural subtype and 139 proneural subtype samples. Morpheus (https://software.broadinstitute.org/morpheus/) online software was used to perform heat maps, and differential expression genes (DEGs) were determined using a threshold P-value of 0.05.
Immunohistochemistry staining
Paraffin embedded tumour tissues were sectioned at 6 µm, for immunohistochemistry After quenching the endogenous peroxidase activity and blocking with normal goat serum, sections were incubated sequentially with the primary antibodies at 4°C overnight, the next day, after rewarming for 1 h, sections were incubated with secondary antibodies (ZSGB-Bio, Beijing, China) for 1 h at 37°C. Immunostaining was performed using DAB kit (ZSGB-Bio), which resulted in a brown precipitate at the antigen site. Subsequently, sections were counterstained with Mayer Haematoxylin solution (ZSGB-Bio) and mounted in mounting medium. After dehydration, sections were examined using a light microscope. Protein expression levels were quantified on the basis of a multiplicative index of staining extent (0–3) and the average staining intensity (0–3). The staining score is the product of staining extent and the staining intensity.
Gene ontology and pathway enrichment analysis
DAVID database (https://david.ncifcrf.gov/) is an essential foundation for the success of any high-throughput gene functional analysis. To understand the significance of genes, we performed the Gene Ontology (GO) classification, making use of the following categories: BP_Fat (biological process), CC_Fat (cellular component), and MF_Fat (molecular function). We also performed the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis to detect the potential pathway of target genes and had the hypergeometric test with P-value <0.05. GO enrichment and KEGG pathway analysis were performed using the DAVID online tool. P<0.05 was considered to indicate a statistically significant difference.
Statistical analysis
The Kaplan-Meier survival analysis was used to estimate the survival distributions, and the log-rank test was used to assess the statistical significance between stratified survival groups using the mean value as the cut-off. The Pearson correlation array was performed to determine significant differences. One-way ANOVA was used to test for differences among at least 3 groups. The Newman-Keuls multiple comparisons test was performed after ANOVA. The t-test was used to determine differences in each 2-group comparison. All data are presented as mean ± standard error.
Results
CXCR4 is a strong predictor of poor prognosis in GBM patients
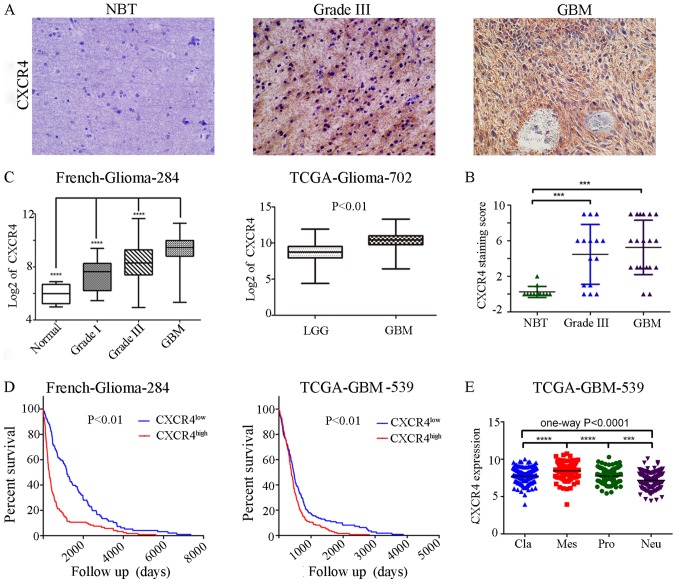
To testify whether alterations at the genetic locus of CXCR4 could be implicated as a predictor in GBM patients prognosis. We checked CXCR4 expressions in various glioma specimens (12 normal brain tissues (NBT), 15 grade III and 20 GBMs) on a tissue array using immunohistochemistry followed by quantitation. As a result, in contrast to negligible CXCR4 expressions in NBT, profound CXCR4 expression can be seen in high grade glioma (HGG, WHO Grade III–IV) samples, especially in GBMs (WHO Grade IV) (Fig. 1A). At the same time, CXCR4 owns a distinctly 85.7% (30/35) to 16.7% (2/12) positive expression samples between HGG and NBT (Fig. 1B). TCGA and French datasets retrieved from the R2 genomics analysis and visualization platform also showed that compared to lower grade glioma or normal brain tissue, the highest expression levels were detected in GBMs (Fig. 1C). TCGA and French datasets was employed to evaluate the effects of CXCR4 on overall patient survival using the KM curve. The mRNA expression levels of CXCR4 from the two databases were used to classify patients into upregulation and downregulation groups according to their mean expression values. Compared to the downregulation CXCR4 cases, patients with high levels of CXCR4 were associated with a significantly shorter overall survival time (Fig. 1D). These data indicated that CXCR4 expression correlates with glioma grade at the protein and transcriptional level and is a strong predictor of poor prognosis in GBM patients.
Figure 1.
CXCR4 is a prognostic factor of patients with glioma patients and a marker for the glioma mesenchymal subtype. (A) Immunohistochemical staining of CXCR4 in human NBT, anaplastic glioma and GBM tissues (magnification ×200). (B) The levels of CXCR4 were analyzed in the glioma tissues of the French and TCGA glioma dataset. (C) Kaplan-Meier survival curves for CXCR4 expression in of the French glioma and TCGA GBM dataset. (D) Scattered dot plots of CXCR4 staining score in NBT and high grade gliomas (grade III–IV). (E) The CXCR4 mRNA expression in four subtypes of GBM in TCGA database. ***P<0.001 and ****P<0.0001. NBT, normal brain tissue; GBM, glioblastoma multiforme; LGG, lower grade glioma; TCGA, The Cancer Genome Atlas; Pro, proneural; Neu, neural; Cla, classical; Mes, mesenchymal.
CXCR4 is a clinical prognostic factor in glioma patients
In the French glioma dataset (n=284), a high expression of CXCR4 was remarkably associated with an older age at diagnosis, shorter overall survival years, non IDH1 mutation, lower expression of PTEN, and a high expression of KI67 and EGFR amplification (Table I). While in the TCGAs 539 GBM samples, a high expression of CXCR4 was selectively associated with a shorter overall survival and days to recurrence (Table II). Consistent with the theory that recurrence of glioblastoma after radio-chemotherapy is associated with an angiogenic switch to the CXCL12-CXCR4 pathway (21), CXCR4high patients experience a shorter days to recurrence. These results demonstrate that CXCR4 may be involved in the recurrence of the glioma and could be conferred as a new clinical prognostic factor of GBM.
Table I.
Clinical and molecular pathology features of French glioma samples in association with CXCR4 expression.
| Variables | Low | High | P-value |
|---|---|---|---|
| Age | 46.1±13.4 | 53.4±14.9 | <0.0001a |
| Gender, female/male | 43/72 | 47/106 | 0.2283b |
| KPS ≥80/<80 | 82/28 | 100/54 | 0.1229b |
| OS, years ± SD | 4.1±4.0 | 1.7±2.7 | <0.0001c |
| Resection complete/partial | 40/57 | 46/89 | 0.3643b |
| IDH1 mutation/no mutation | 50/49 | 33/94 | 0.0001b |
| 1p mutation/no mutation | 39/39 | 11/59 | 0.5433b |
| 19q mutation/no mutation | 39/38 | 13/57 | 0.6312b |
| KI67 low/high | 66/57 | 52/107 | 0.0009b |
| PTEN low/high | 42/81 | 93/68 | <0.0001b |
| EGFR amplification/wild | 12/63 | 31/45 | 0.0004b |
| EGFR low/high | 55/68 | 83/78 | 0.8005b |
Student's t-test
χ2 test or Fisher's exact test
Log-rank test. IDH1, isocitrate dehydrogenase 1; KPS, Karnofsky Performance Scale; OS, overall survival; EGFR, epidermal growth factor receptor; PTEN, phosphatase and tensin homologue; SD, standard deviation. The level of CXCR4 was classified by the mean value.
Table II.
Clinical and molecular pathology features of TCGA GBM samples in association with CXCR4 expression.
| Variables | Low | High | P-value |
|---|---|---|---|
| Age (years ± SD) | 56.4±13.1 | 58.6±13.1 | 0.1496a |
| Gender, female/male | 75/136 | 122/178 | 0.2424b |
| OS (days ± SD) | 521.8±391.9 | 413.5±610.2 | 0.0129c |
| KPS ≥80/<80 | 117/80 | 146/124 | 0.2536b |
| Treated/untreated | 10/201 | 10/289 | 0.4251b |
| Radiation therapy, yes/no | 20/170 | 33/247 | 0.6726b |
| Chemo therapy, yes/no | 58/112 | 105/157 | 0.4703b |
| Days to progression ± SD | 311.8 to pro | 242.3 to pro | 0.1140a |
| Days to recurrence ± SD | 515.6 to 1.0 | 257.3 to 1.0 | 0.0036a |
| MGMT promoter methylation (unmethylation/methylation) | 9/17 | 13/46 | 0.0573b |
Student's t-test
χ2 test or Fisher's exact test
Log-rank test. IDH1, isocitrate dehydrogenase 1; KPS, Karnofsky Performance Scale; MGMT, O(6)-methylguanine DNA methyltransferase; OS, overall survival; GBM, glioblastoma multiforme; TCGA, The Cancer Genome Atlas; SD, standard deviation. The level of CXCR4 was classified by the mean value.
CXCR4 is preferentially expressed in mesenchymal subtype of glioma
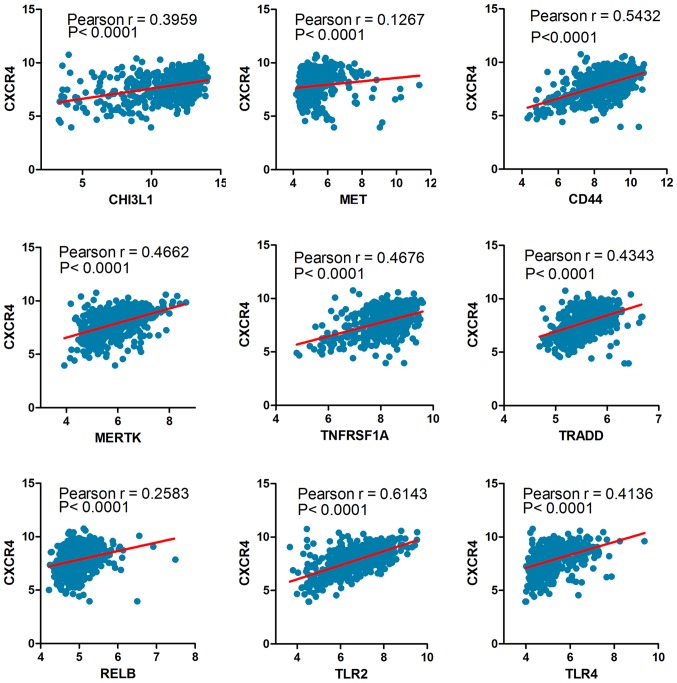
The Cancer Genome Atlas (TCGA) network described a robust gene expression based molecular classification. We retrieved 539 GBM samples from TCGA, including 145 classical subtype, 158 mesenchymal subtype, 87 neural subtype and 139 proneural subtype samples. One-way ANOVA indicated a markedly significant difference in CXCR4 expression between the four glioma subtypes in the TCGA datasets. In particular, CXCR4 is preferentially expressed in the mesenchymal subtype of glioma with a notable statistical significance (P<0.0001; Fig. 1E). Similarly, to confirm the CXCR4 as a biomarker of mesenchymal subtype of glioma, we further analysed the association between CXCR4 and the mesenchymal markers such as CHI3L1 (also known as YKL40), MET, CD44, MERTK and NF-κB pathway genes (TRADD, RELB, TNFRSF1A) using a Pearson correlation array (7). Likewise, CXCR4 was positively correlated with MES markers (CHI3L1, R=0.3959; MET, R=0.1267; CD44, R=0.5432; MERTK, R=0.4662; TLR2, R=0.6143; TLR4, R=0.4136; TRADD, R=0.4343; RELB, R=0.2583; TNFRSF1A, R=0.4676) (Fig. 2). These results foreshadowed that CXCR4 acts as a marker for the glioma molecular subtype and may regulate gene expression patterns in glioma mesenchymal transition.
Figure 2.
The Pearson correlation coefficient between CXCR4 and mesenchymal markers. TLR, toll-like receptor; CHI3L1, Chitinase 3 Like 1; MET, MET proto-oncogene, receptor tyrosine kinase; CD, cluster of differentiation; MERTK, MER proto-oncogene, tyrosine kinase; TRADD, TNFRSF1A associated via death domain; RELB, RELB proto-oncogene; TNFRSF1A, TNF receptor superfamily member 1A.
Selection of genes associated with CXCR4 signalling in mesenchymal subtype of GBM
To study the underlying gene and pathway pattern that CXCR4 regulated in mesenchymal glioma. First, the students t-test was performed on the DEGs to test the difference between the mesenchymal subtype and other subtypes of glioblastoma. The differential expression was determined using a threshold P-value of 0.05, and the top 100 genes were shown in the heatmaps (Fig. 3A-C). Next, we selected 3198 genes (Fig. 3D) that were highly related in CXCR4high patients (classified by the mean value of CXCR4 mRNA expression) of mesenchymal GBM, namely, CXCR4 correlated genes in mesenchymal glioblastoma (CCGIM). Therefore, we found that 4004 genes that were differentially expressed between the classical subtype and the mesenchymal subtype, 3455 genes that were differentially expressed between the neural subtype and the mesenchymal subtype and 3289 genes were differentially expressed between the proneural subtype and the mesenchymal subtype. Then, we compared the genes that were differentially expressed in mesenchymal CXCR4high patients with the genes that were differentially expressed between mesenchymal and three other subtypes (classical, neural, proneural), which comprised 838, 1397, 1187 overlapping genes respectively (Fig. 3E-G). Finally, we compared the abovementioned 838, 1397 and 1187 genes that overlapped between groups to identify DEGs that were specific to the mesenchymal CXCR4high subgroup, yielding a total of 34 genes (Fig. 3H).
Figure 3.
The heat maps displaying the DEGs between MES and (A) CLA, (B) NEU and (C) PRO subtypes of GBM. (D) Heat map displaying the DEGs between the CXCR4high and CXCR4low group in MES GBM. (E) Total of 3198 CCGIM subtype were compared with 4004 genes DEGs between the mesenchymal and classical groups, yielding a set of 838 overlapping genes. (F) 3455 DEGs expressed between the mesenchymal and neural groups, yielding a set of 1397 overlapping genes. (G) 3289 DEGs between the mesenchymal and proneural groups, yielding a set of 1187 overlapping genes. (H) A comparison of the 838, 1397 and 1187 genes revealed 34 common genes specific to the CXCR4 correlated mesenchymal subgroup. M&C, the overlap between mesenchymal and classical subtype; M&N, the overlap between mesenchymal and neural subtype; M&P, the overlap between mesenchymal and proneural subtype; CCGIM, CXCR4 correlated genes in the mesenchymal GBM; GBM, glioblastoma; DEG, different expressed genes.
Gene ontology and KEGG pathway analysis of CXCR4 associated genes in mesenchymal GBM
We uploaded all 34 DEGs to the online software DAVID to identify overrepresented Gene Ontology (GO) categories and KEGG pathways. The GO categories and KEGG pathways were ranked by P-value, and the top 5 items were displayed in the table. GO cell component analysis displayed that CXCR4 associated genes were significantly enriched in the golgi apparatus, cytoplasmtic membrane-bounded vesicle, vacuole, golgi apparatus part and endoplasmic reticulum with a notable statistical significance of P<0.05. For biological processes (BP) CXCR4 associated genes were significantly enriched in regulation of MAP kinase activity, regulation of protein serine/threonine kinase activity, regulation of protein kinase activity, regulation of kinase activity and mitogen-activated protein kinase (MAPK) cascade, while in the GO molecular function (MF) analysis no item was involved with a threshold of P<0.05 (Table III). Table III also contains the most significantly enriched pathways of the CXCR4 associated genes in mesenchymal GBM analyzed by KEGG analysis. The 34 DEGs were enriched in melanoma, prostate cancer, pathways in cancer, protein processing in endoplasmic reticulum and regulation of actin cytoskeleton. In collection, Gene ontology and KEGG pathway analyses demonstrated that MAPK signalling pathway is associated with CXCR4 activation in the mesenchymal subtype. Meanwhile, the activation of CXCR4 is also enriched in melanoma and prostate cancer, which demonstrated there might be a co-therapeutic target or strategies in the management of melanoma, prostate cancer and mesenchymal GBM.
Table III.
Gene ontology and KEGG pathway analysis of CXCR4 correlated genes in mesenchymal glioblastoma.
| Category | Term, function | Count | Percentage | P-value | Genes |
|---|---|---|---|---|---|
| GOTERM_CC_FAT | GO:0005794, golgi apparatus | 13 | 23.6 | 7.78×10−6 | NCSTN, NUCB1, SPRY1, GANAB, ARFRP1, PDGFC, ADAM19, TM9SF4, TRIP10, KDELR1, SPRY4, HS2ST1, AP3B1 |
| GOTERM_CC_FAT | GO:0016023, cytoplasmic, membrane-bounded vesicle | 7 | 12.7 | 2.30×10−2 | NCSTN, NUCB1, FGFR1, GANAB, SPRED2, KDELR1, AP3B1 |
| GOTERM_CC_FAT | GO:0005773, vacuole | 7 | 12.7 | 2.55×10−2 | NCSTN, NUCB1, TM9SF1, TRAF6, TM9SF4, TRIP10, AP3B1 |
| GOTERM_CC_FAT | GO:0044431, golgi apparatus part | 6 | 10.9 | 2.81×10−2 | NUCB1, ARFRP1, PDGFC, TRIP10, KDELR1, HS2ST1 |
| GOTERM_CC_FAT | GO:0005783, endoplasmic reticulum | 8 | 14.5 | 3.44×10−2 | NCSTN, NUCB1, RAB3GAP2, GANAB, BAX, PDGFC, EDEM3, KDELR1 |
| GOTERM_BP_FAT | GO:0043405, regulation of MAP kinase activity | 6 | 10.9 | 2.70×10−4 | FGFR1, SPRY1, SPRED2, PDGFC, TRAF6, SPRY4 |
| GOTERM_BP_FAT | GO:0071900, regulation of protein serine/threonine kinase activity | 6 | 10.9 | 1.48×10−3 | FGFR1, SPRY1, SPRED2, PDGFC, TRAF6, SPRY4 |
| GOTERM_BP_FAT | GO:0045859, regulation of protein kinase activity | 7 | 12.7 | 1.68×10−3 | FGFR1, SPRY1, BAX, SPRED2, PDGFC, TRAF6, SPRY4 |
| GOTERM_BP_FAT | GO:0043549, regulation of kinase activity | 7 | 12.7 | 2.47×10−3 | FGFR1, SPRY1, BAX, SPRED2, PDGFC, TRAF6, SPRY4 |
| GOTERM_BP_FAT | GO:0000165, MAPK cascade | 7 | 12.7 | 3.42×10−3 | FGFR1, SPRY1, ARAF, SPRED2, PDGFC, TRAF6, SPRY4 |
| GOTERM_MF_FAT | GO:0042802, identical protein binding | 6 | 10.9 | 8.25×10−2 | BCAT1, FGFR1, BAX, PDGFC, TRAF6, TRIP10 |
| KEGG_PATHWAY | hsa05218, melanoma | 3 | 5.4 | 8.75×10−3 | FGFR1, ARAF, PDGFC |
| KEGG_PATHWAY | hsa05215, prostate cancer | 3 | 5.4 | 1.32×10−2 | FGFR1, ARAF, PDGFC |
| KEGG_PATHWAY | hsa05200, pathways in cancer | 4 | 7.3 | 4.16×10−2 | FGFR1, BAX, ARAF, TRAF6 |
| KEGG_PATHWAY | hsa04141, protein processing in endoplasmic reticulum | 3 | 5.4 | 4.46×10−2 | GANAB, BAX, EDEM3 |
| KEGG_PATHWAY | hsa04810, regulation of actin cytoskeleton | 3 | 5 | 6.64×10−2 | FGFR1, ARAF, PDGFC |
The GO categories and KEGG pathways were ranked by P-value, the top 5 items were displayed in the table. Only one item of MF go analysis was involved in the results. GO, gene ontology; BP, biological processes; MF, molecular function; CC, cell component.
Discussion
Glioblastoma is the most malignant primary tumour of the CNS with a devastating outcome. This tumour often invades healthy brain tissue leading to tumour progression or recurrence despite drastic treatments such as chemotherapy and radiotherapy (22,23). The classification of GBMs into classical, mesenchymal, neural and proneural subtypes based on gene expression profiles bring a new insight into understanding of the molecular mechanisms of GBM. This phenomenon is reminiscent of a newly emerging concept that differential activation of critical signalling pathways induces and maintains each subtype in cancer biology. Extensive reports suggested that CXCR4 is required for tumour proliferation, invasion, angiogenesis, modulation of the immune response and recently enriched in neural stem cells (24,25). In this study, we demonstrated that CXCR4 could serve as a prognostic factor in characterizing subsets of GBM, as patients with high expression of CXCR4 gliomas seem to have poorer prognosis. Additionally, CXCR4 is preferentially expressed in the MES subtype of GBM and highly consistent with MES makers such as CHI3L1 (also known as YKL40) and MET. However, little has been reported regarding about the role of activated CXCR4 in mesenchymal GBM.
In the present study, gene expression data of 539 GBM patients were retrieved from the TCGA dataset. We identified 3198 DEGs associated with CXCR4 in mesenchymal GBM, 4004 DEGs between mesenchymal and classical GBM, 3455 DEGs between mesenchymal and neural GBM, and 3289 DEGs between mesenchymal and proneural GBM. Subsequently, we intersected of the gene sets mentioned above to determine key pathways and genes in the CXCR4 mediated mesenchymal subtype of glioblastoma. As a result, 34 overlapped genes were found to be distinctively specific to the CXCR4high group of mesenchymal patients.
In addition, Gene ontology and KEGG pathway analyses were performed to compare functional annotations associated with CXCR4 signalling across the mesenchymal GBM subtype. In accordance with expectations, CXCR4 was found to be enriched in the cell component of golgi apparatus, cytoplasmic membrane, vacuole and endoplasmic reticulum. Interestingly, CXCR4 was demonstrated to be involved in the regulation of the MAPK pathway, which is reported to be associated with the many cell fate decision including proliferation, differentiation, migration, senescence and apoptosis (26,27). Numerous studies investigated the linkage between CXCR4 and MAPK in a wide range of malignant tumours, covering small-cell lung cancer, breast cancer and, recently, osteosarcoma (28–30). These studies indicated that the CXCL12/CXCR4 axis could act as a mediator to promote tumour biology through the activation of the MAPK pathway. Sun et al (31), reported that the expression of β-arrestin2 strengthened the CXCR4-mediated activation of both p38 MAPK and ERK in HeLa cells. Rhodes et al (28), showed that enhanced CXCR4 signalling is sufficient to drive ER-positive breast cancers to a metastatic and endocrine therapy-resistant phenotype via increased MAPK signalling. However, fewer literatures are written about the potential association between CXCR4 and MAPK pathway in the mesenchymal subtype of GBM. Our study demonstrated that the MAPK pathway may play a pivotal role in the progression of CXCR4-mediated mesenchymal GBM or the mesenchymal phenotype transition. Given the poor prognosis and aggressiveness of mesenchymal GBM, it is anticipated that these patients would benefit most from gene based chemotherapeutic strategies through targeting CXCR4/MAPK related factors.
In conclusion, our data provide a comprehensive bioinformatics analysis of CXCR4 and its DEGs, which may play a functional role in the development of GBM and the maintenance of the mesenchymal phenotype. The study also provides preliminary evidence that the CXCR4 mediated MAPK pathway was identified specifically in patients with mesenchymal GBM and CXCR4 be a co-therapeutic target in the management of melanoma, prostate cancer and mesenchymal GBM. Therefore, targeting CXCR4 and its related cooperative pathways and genes could be a promising approach for the efficient suppression or elimination of this devastating cancer.
Acknowledgements
Not applicable.
Funding
The research was funded by the National Natural Science Foundation of China (grant no. 81472352) and the Natural Science Foundation of Tianjin City (grant no. 15JCZDJC36200).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the R2 analysis and visualization platform (hgserver1.amc.nl/cgi-bin/r2/main.cgi) and UCSC Xena (xenabrowser.net/datapages/).
Authors' contributions
LY drafted the manuscript. LY and LT performed the experiments. PL and IRA participated in the design of the study. TL and LH downloaded and interpreted the raw data. ZT and HM analyzed the data. YX, SY, JL and FY contributed to the data collection and processing. XY helped design the study and revised and approved the final version of manuscript.
Ethics approval and consent to participate
All participants provided written informed consent prior to their inclusion within the study and the study was approved by the Ethics Committee of Tianjin Medical University Hospital.
Consent for publication
All participants provided written informed consent for the publication of their data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro Oncol. 2016;18(Suppl_5):v1–v75. doi: 10.1093/neuonc/now207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, Carpentier AF, Hoang-Xuan K, Kavan P, Cernea D, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370:709–722. doi: 10.1056/NEJMoa1308345. [DOI] [PubMed] [Google Scholar]
- 3.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coons SW, Johnson PC, Scheithauer BW, Yates AJ, Pearl DK. Improving diagnostic accuracy and interobserver concordance in the classification and grading of primary gliomas. Cancer. 1997;79:1381–1393. doi: 10.1002/(SICI)1097-0142(19970401)79:7<1381::AID-CNCR16>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 5.Sun Y, Zhang W, Chen D, Lv Y, Zheng J, Lilljebjörn H, Ran L, Bao Z, Soneson C, Sjögren HO, et al. A glioma classification scheme based on coexpression modules of EGFR and PDGFRA. Proc Natl Acad Sci USA. 2014;111:3538–3543. doi: 10.1073/pnas.1313814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML, Smirnov IV, et al. glioma groups based on 1p/19q, IDH and TERT promoter mutations in tumors. N Engl J Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 11.Tabouret E, Nguyen AT, Dehais C, Carpentier C, Ducray F, Idbaih A, Mokhtari K, Jouvet A, Uro-Coste E, Colin C, et al. Prognostic impact of the 2016 WHO classification of diffuse gliomas in the French POLA cohort. Acta Neuropathol. 2016;132:625–634. doi: 10.1007/s00401-016-1611-8. [DOI] [PubMed] [Google Scholar]
- 12.Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, Agarwalla PK, Chheda MG, Campos B, Wang A, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Cheng W, Zhang C, Ren X, Jiang Y, Han S, Liu Y, Cai J, Li M, Wang K, Liu Y, et al. Bioinformatic analyses reveal a distinct Notch activation induced by STAT3 phosphorylation in the mesenchymal subtype of glioblastoma. J Neurosurg. 2017;126:249–259. doi: 10.3171/2015.11.JNS15432. [DOI] [PubMed] [Google Scholar]
- 15.Cooper LA, Gutman DA, Chisolm C, Appin C, Kong J, Rong Y, Kurc T, Van Meir EG, Saltz JH, Moreno CS, Brat DJ. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am J Pathol. 2012;180:2108–2119. doi: 10.1016/j.ajpath.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krock BL, Skuli N, Simon MC. Hypoxia-induced angiogenesis: Good and evil. Genes Cancer. 2011;2:1117–1133. doi: 10.1177/1947601911423654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gagliardi F, Narayanan A, Reni M, Franzin A, Mazza E, Boari N, Bailo M, Zordan P, Mortini P. The role of CXCR4 in highly malignant human gliomas biology: Current knowledge and future directions. Glia. 2014;62:1015–1023. doi: 10.1002/glia.22669. [DOI] [PubMed] [Google Scholar]
- 18.Passaro D, Irigoyen M, Catherinet C, Gachet S, Da Costa De Jesus C, Lasgi C, Quang Tran C, Ghysdael J. CXCR4 Is required for leukemia-initiating cell activity in t cell acute lymphoblastic leukemia. Cancer Cell. 2015;27:769–779. doi: 10.1016/j.ccell.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Luker KE, Lewin SA, Mihalko LA, Schmidt BT, Winkler JS, Coggins NL, Thomas DG, Luker GD. Scavenging of CXCL12 by CXCR7 promotes tumor growth and metastasis of CXCR4-positive breast cancer cells. Oncogene. 2012;31:4750–4758. doi: 10.1038/onc.2011.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seo CH, Kim JR, Kim MS, Cho KH. Hub genes with positive feedbacks function as master switches in developmental gene regulatory networks. Bioinformatics. 2009;25:1898–1904. doi: 10.1093/bioinformatics/btp316. [DOI] [PubMed] [Google Scholar]
- 21.Tabouret E, Tchoghandjian A, Denicolai E, Delfino C, Metellus P, Graillon T, Boucard C, Nanni I, Padovani L, Ouafik L, et al. Recurrence of glioblastoma after radio-chemotherapy is associated with an angiogenic switch to the CXCL12-CXCR4 pathway. Oncotarget. 2015;6:11664–11675. doi: 10.18632/oncotarget.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huse JT, Holland EC. Targeting brain cancer: Advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10:319–331. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, McKay RM, Parada LF. Malignant glioma: Lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Chang CJ, Lathia JD, Wang L, Pacenta HL, Cotleur A, Ransohoff RM. Chemokine receptor CXCR4 signaling modulates the growth factor-induced cell cycle of self-renewing and multipotent neural progenitor cells. Glia. 2011;59:108–118. doi: 10.1002/glia.21080. [DOI] [PubMed] [Google Scholar]
- 25.Ho SY, Ling TY, Lin HY, Liou JT, Liu FC, Chen IC, Lee SW, Hsu Y, Lai DM, Liou HH. SDF-1/CXCR4 signaling maintains stemness signature in mouse neural stem/progenitor cells. Stem Cells Int. 2017;2017:2493752. doi: 10.1155/2017/2493752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 27.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 28.Rhodes LV, Short SP, Neel NF, Salvo VA, Zhu Y, Elliott S, Wei Y, Yu D, Sun M, Muir SE, et al. Cytokine receptor CXCR4 mediates estrogen-independent tumorigenesis, metastasis, and resistance to endocrine therapy in human breast cancer. Cancer Res. 2011;71:603–613. doi: 10.1158/0008-5472.CAN-10-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burger M, Glodek A, Hartmann T, Schmitt-Gräff A, Silberstein LE, Fujii N, Kipps TJ, Burger JA. Functional expression of CXCR4 (CD184) on small-cell lung cancer cells mediates migration, integrin activation, and adhesion to stromal cells. Oncogene. 2003;22:8093–8101. doi: 10.1038/sj.onc.1207097. [DOI] [PubMed] [Google Scholar]
- 30.Liao YX, Fu ZZ, Zhou CH, Shan LC, Wang ZY, Yin F, Zheng LP, Hua YQ, Cai ZD. AMD3100 reduces CXCR4-mediated survival and metastasis of osteosarcoma by inhibiting JNK and Akt, but not p38 or Erk1/2, pathways in in vitro and mouse experiments. Oncol Rep. 2015;34:33–42. doi: 10.3892/or.2015.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y, Cheng Z, Ma L, Pei G. Beta-arrestin2 is critically involved in CXCR4-mediated chemotaxis, and this is mediated by its enhancement of p38 MAPK activation. J Biol Chem. 2002;277:49212–49219. doi: 10.1074/jbc.M207294200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the R2 analysis and visualization platform (hgserver1.amc.nl/cgi-bin/r2/main.cgi) and UCSC Xena (xenabrowser.net/datapages/).