Stable isotope labeling of agricultural polyesters enables demonstration of their microbial utilization in soils.
Abstract
Plastic materials are widely used in agricultural applications to achieve food security for the growing world population. The use of biodegradable instead of nonbiodegradable polymers in single-use agricultural applications, including plastic mulching, promises to reduce plastic accumulation in the environment. We present a novel approach that allows tracking of carbon from biodegradable polymers into CO2 and microbial biomass. The approach is based on 13C-labeled polymers and on isotope-specific analytical methods, including nanoscale secondary ion mass spectrometry (NanoSIMS). Our results unequivocally demonstrate the biodegradability of poly(butylene adipate-co-terephthalate) (PBAT), an important polyester used in agriculture, in soil. Carbon from each monomer unit of PBAT was used by soil microorganisms, including filamentous fungi, to gain energy and to form biomass. This work advances both our conceptual understanding of polymer biodegradation and the methodological capabilities to assess this process in natural and engineered environments.
INTRODUCTION
Modern agriculture heavily relies on the use of plastic materials in various applications, a practice coined plasticulture. Mulching with plastic films is a major application with a global market volume of approximately 2 × 106 tons per year (1). Mulch films are placed onto agricultural soils to improve conditions for plant growth while lowering consumption of water, herbicides, and fertilizer and also minimizing soil erosion (1, 2). Because of these benefits, mulching with plastic films helps to secure food for the growing world population. However, mulch films are commonly composed of nonbiodegradable polyethylene and, therefore, accumulate in agricultural soils and surrounding receiving environments if incompletely retrieved after use. These accumulations have negative ecologic and economic impacts, including decreased soil productivity (3–5). A promising strategy to overcome these risks is to use mulch films composed of polymers that biodegrade in soils (1, 6–8).
Biodegradation of polymers requires microorganisms to metabolize all organic components of the polymer. Biodegradation in soil involves several key steps (Fig. 1): (i) colonization of the polymer surface by microorganisms, (ii) secretion of extracellular microbial enzymes that depolymerize the polymer into low–molecular weight compounds, and (iii) microbial uptake and utilization of these compounds, incorporating polymer carbon into biomass or releasing it as CO2 (9).
Fig. 1. Key steps in the biodegradation of polymers in soils.
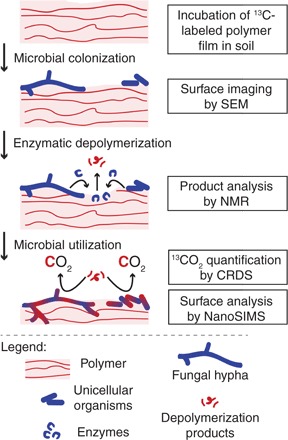
Microorganisms colonize the polymer surface and secrete extracellular enzymes that depolymerize the polymer. The formed low–molecular weight hydrolysis products are taken up by the microorganisms and used both for energy production, resulting in the formation of CO2, and for the synthesis of cellular structures and macromolecules, resulting in incorporation of polymer-derived carbon into the microbial biomass. The boxes on the right depict the analytical methods we used to study these steps. NMR, nuclear magnetic resonance.
Here, we examined each of the above steps for poly(butylene adipate-co-terephthalate) (PBAT), an aliphatic-aromatic statistical copolyester of large importance in the market of biodegradable mulch films (7). While previous studies provided indirect indications for PBAT biodegradation in soils based on determining PBAT mass loss and changes in its physicochemical properties (10–12), we here use a novel workflow using stable carbon isotope-labeled PBAT to directly and unequivocally demonstrate its biodegradation in soil (table S1). This workflow included incubation of 13C-labeled polymer films in soil with continuous quantification of polymer-derived 13CO2 by isotope-specific cavity ring-down spectroscopy (CRDS) (13). The use of 13C-labeled polymers allowed us to distinguish polymer-derived CO2 from CO2 formed by soil organic matter mineralization. After incubation, we imaged the polymer film surfaces using scanning electron microscopy (SEM) and demonstrated the incorporation of polymer-derived 13C into the biomass of film-colonizing microorganisms using element-specific, isotope-selective nanoscale secondary ion mass spectrometry (NanoSIMS) (14). We studied three PBAT variants that had similar physicochemical properties and comparable total 13C contents, but varied in the monomer that contained the 13C-label [that is, butanediol (P*BAT), adipate (PB*AT), or terephthalate (PBA*T)] (Fig. 2A and table S2). The use of these variants allowed us to follow biodegradation of all PBAT building blocks. The presented workflow is a novel approach to study the fundamental steps in polymer biodegradation in complex systems (15–17).
Fig. 2. Mineralization and surface colonization of films of three PBAT [poly(butylene adipate-co-terephthalate)] variants during a 6-week soil incubation.
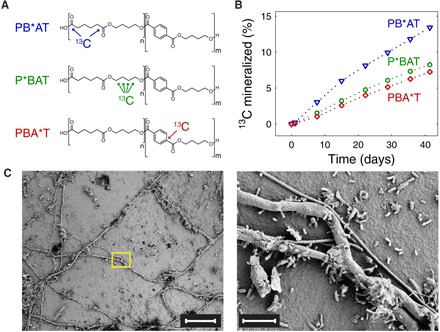
(A) Chemical structures of the tested PBAT variants, differing in the monomer unit that was 13C-labeled. PB*AT, P*BAT, and PBA*T contained 1,6-13C2-adipate, 13C4-butanediol, and 1-13C1-terephthalate, respectively. (B) Formation of 13CO2 from the three PBAT variants during their incubation in soil, monitored by 13C isotope-specific CO2 CRDS (cavity ring-down spectroscopy). PBAT variants were added to soils at time = 0 days. Results from replicate experiments performed with a slightly different setup are shown in fig. S1. (C) Representative SEM (scanning electron microscopy) images of a PBAT film after the 6-week soil incubation. The yellow rectangle in the left image defines the area shown at higher magnification in the right image. Scale bars, 50 μm (left) and 5 μm (right).
RESULTS
Soil incubation of all PBAT variants resulted in 13CO2 formation (Fig. 2B), demonstrating that soil microorganisms used carbon from all three monomer units in PBAT to gain energy. The cumulative amounts of 13CO2 formed over 6 weeks of soil incubation corresponded to approximately 13% of the 13C in PB*AT and 8% of the 13C in both P*BAT and PBA*T. We confirmed the higher extent of 13CO2 formation from PB*AT than from the other two variants in replicate soil incubations with a slightly modified setup (fig. S1). We have two complementary explanations for the faster and more extensive 13CO2 formation from PB*AT than from P*BAT and PBA*T. The first explanation builds on microscale nonuniformity in the adipate-to-terephthalate ratio within the statistical copolyester PBAT, which gives rise to microdomains with adipate-to-terephthalate ratios that deviate from the ratio of the bulk PBAT. Microdomains with higher adipate-to-terephthalate ratios are known to undergo faster enzymatic hydrolysis than those with lower adipate-to-terephthalate ratios (18–20). The preferential release of adipate and its subsequent mineralization by soil microorganisms are expected to result in faster and more extensive 13CO2 release from the variant in which the 13C-label is in the adipate (that is, PB*AT), as experimentally observed. Support for this explanation comes from incubations of unlabeled PBAT films with either Rhizopus oryzae lipase or Fusarium solani cutinase (FsC)—two fungal carboxylesterases with distinct hydrolysis mechanisms (18, 21). As expected, 1H NMR spectroscopy revealed that PBAT films that remained after partial enzymatic hydrolysis were enriched in terephthalate, while the released hydrolysis products were enriched in adipate (figs. S2 to S6). The second explanation for the higher extent of 13CO2 formation from PB*AT is that CO2 formation was more extensive for the highly oxidized carboxylate carbons in adipate than the more reduced carbons in butanediol and terephthalate. Separate soil incubations with labeled monomers demonstrated higher extents of 13CO2 formation from 1,6-13C2-adipate (~66% of added 13C) than from 13C4-butanediol and 1-13C1-terephthalate (that is, ~40% and ~55% of added 13C, respectively) (fig. S7). Furthermore, it is evident from comparing Fig. 2B with fig. S7 that the mineralization of the free monomers was much faster than that of the corresponding PBAT variants. While more than 30% of labeled carbon atoms in the monomers were mineralized within 2 days of adding the monomers to the soils, less than 1% of the labeled carbon atoms in the three PBAT specimens had been converted to CO2 over the same incubation period. This finding implies that the depolymerization step controlled the rate at which PBAT was mineralized in soils (9).
SEM images of the PBAT films retrieved from soils after the incubations showed extensive surface colonization by both filamentous fungi and unicellular organisms of diverse morphologies (selected images shown in Fig. 2C). We used NanoSIMS to image the element and isotope distribution on selected PBAT surfaces (Fig. 3 and fig. S8; the latter showing images from replicate PBAT films). Film-colonizing microorganisms were visualized on the basis of both secondary electron images (Fig. 3A) and images showing 12C14N− ion signal intensity distributions (Fig. 3B). In the latter, biomass appears in gray-white colors, while the underlying PBAT surface appears black due to the absence of nitrogen in PBAT.
Fig. 3. NanoSIMS (nanoscale secondary ion mass spectrometry) analysis of PBAT films after a 6-week soil incubation.
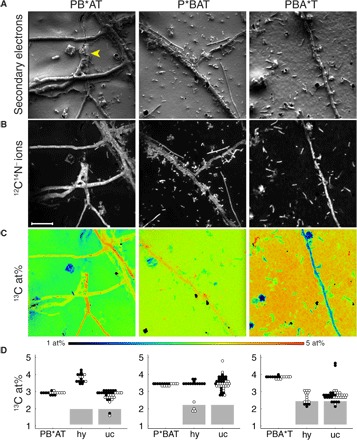
See Fig. 2 for details on PBAT variants. Images represent signal intensity distributions of secondary electrons (A), 12C14N− ions (B), and the carbon isotope composition displayed as 13C/(12C + 13C) isotope fraction, given in at%. (C) Yellow arrowhead in (A) indicates position of a burrowed hypha. Scale bar in (B) represents 10 μm and applies to all images. (D) Carbon isotope composition within ROI [categorized as background PB*AT, P*BAT, or PBA*T; fungal hyphae (hy); and unicellular organisms (uc) as specified in fig. S10]. Black (solid) dots refer to values obtained from ROI analysis of images shown in (C), and open dots refer to values obtained on replicate films (images shown in fig. S8). Gray rectangles represent the range of apparent 13C contents of microorganisms with natural 13C abundance due to carryover of 13C from the PBAT surface (see text and fig. S9 for details).
NanoSIMS-based analyses of the carbon isotope composition revealed that the 13C content of the noncolonized PBAT surfaces increased from PB*AT to P*BAT and PBA*T (Fig. 3D). This trend differs from that of the 13C content of the nonincubated bulk materials, which was 3.75 atom% (at%) for PB*AT, 3.53 at% for PBA*T, and 3.56 at% for P*BAT [determined by isotope ratio mass spectrometry (IRMS); table S2]. The difference between these trends suggests that during the initial biodegradation followed in this work, PB*AT film surfaces became depleted in 13C, while PBA*T surfaces became enriched in 13C. This finding is fully consistent with enzymatic hydrolysis of adipate-rich domains being faster than that of terephthalate-rich domains on a PBAT surface (figs. S2 to S6), and with the mineralization data showing elevated 13CO2 formation from PB*AT (Fig. 2B). A decrease in the adipate-to-terephthalate ratio during soil incubation has previously been reported also for unlabeled PBAT (10).
We demonstrated incorporation of PBAT-derived carbon into the biomass of film-colonizing microorganisms after circumventing a measurement-specific artifact: carryover of 13C from the PBAT surface onto colonizing microorganisms through atomic mixing and redeposition of sputtered atoms during NanoSIMS measurements, which would change the actual 13C content of these organisms. We assessed this carryover with a control experiment in which we determined the 13C contents of Escherichia coli cells with natural 13C abundance on the surface of a nonincubated P*BAT film (fig. S9). We used E. coli cells because we expected a larger carryover for unicellular microorganisms than for larger fungal hyphae. This control revealed a carryover, the extent of which we used to conservatively estimate an upper bound for the apparent 13C contents of soil microorganisms with natural 13C abundance on the surface of the incubated PBAT films. These bounds correspond to the upper edge of the gray rectangles in Fig. 3D (see fig. S9 for details on the control experiment). For all three PBAT variants, the majority of the film-colonizing organisms had 13C contents that were too high to have resulted from carryover, demonstrating that the organisms incorporated PBAT-derived carbon into their biomass [Fig. 3, C and D; see fig. S10 for selected regions of interest (ROIs)]. For organisms with apparent 13C contents below the threshold, it cannot be ruled out that they had natural 13C abundance and were growing, for instance, at the expense of soil organic matter. In addition, we demonstrated incorporation of PBAT carbon into microbial biomass by showing that microorganisms extracted from P*BAT films after soil incubation had 13C contents up to 6 at% when imaged by NanoSIMS on filter supports (fig. S11). NanoSIMS imaging additionally revealed the presence of fungal hyphae on PB*AT and unicellular organisms on P*BAT and PBA*T films with higher 13C contents than the underlying PBAT films (Fig. 3, C and D). For P*BAT and PBA*T, these elevated 13C contents imply that the microorganism must have preferentially incorporated the labeled carbon atoms from these two PBAT variants over other available nonlabeled carbon. For PB*AT, the high 13C contents of fungal hyphae may also have resulted from the preferential enzymatic hydrolysis of microdomains in PBAT with elevated adipate-to-terephthalate ratios. Uptake of the preferentially released 13C-labeled adipate and incorporation of its carbon into the fungal biomass would explain the elevated 13C contents of the hyphae on the PB*AT films.
While most images suggest that PBAT biodegradation primarily occurred on the film surfaces, one of the PB*AT NanoSIMS images showed a fungal hypha that burrowed into the film (yellow arrowhead in Fig. 3A). We excavated this hypha by extended surface sputtering with the primary ion beam of the NanoSIMS (Fig. 4A). Subsequent NanoSIMS analysis revealed that this burrowed hypha was highly enriched in 13C (up to 4.5 at%; Fig. 4B) relative to the surrounding PB*AT. The extended sputtering also opened some of the fungal cells on the PB*AT surface, revealing subcellular structures that were highly enriched in 13C (up to 6 at%) and depleted in nitrogen (Fig. 4, C and D, and fig. S12).
Fig. 4. NanoSIMS images acquired after extended sputter erosion of the PB*AT film shown also in Fig. 3.
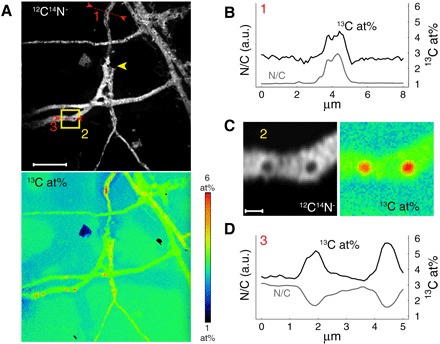
(A) Distribution of 12C14N− ion signal intensity (top) and the 13C content expressed as 13C/(12C + 13C) in at% (bottom). Scale bar, 10 μm. Yellow arrowhead indicates burrowed hypha. (B and D) Line scan analyses 1 (B) and 3 (D) of selected hyphae with respect to 13C content and relative nitrogen content (N/C ratio as inferred from C2− normalized C14N− signal intensities; see Materials and Methods for details). The positions of the line scans are shown in (A). (C) Zoom-in of the region indicated by the yellow square (2) in (A). Scale bar, 1 μm. a.u., arbitrary units.
DISCUSSION
This work presents an experimental approach to study polymer biodegradation in soils and to assess the key steps involved in this process: microbial polymer colonization, enzymatic depolymerization on the polymer surface, and microbial uptake and utilization of the released low–molecular weight compounds. Central to the approach is the use of polymer variants that are 13C-labeled in all monomer units of the polymer, thereby allowing us to assess whether all organic components of the polymer material are used by soil microorganisms. The label further allows tracing of polymer-derived carbon into both CO2 and microbial biomass. Using this approach, we demonstrate here the biodegradability of PBAT in soil. Biodegradability renders PBAT a more environmentally friendly alternative to persistent polymer materials for use in plasticulture, including single-use applications such as plastic mulching. Our results further imply that incorporation of polymer-derived carbon into microbial biomass needs to be taken into consideration in regulatory guidelines for determining biodegradability of polymers. Currently, these guidelines are solely based on extents of CO2 formation. Furthermore, the finding of subcellular structures within PBAT-colonizing fungi highly enriched in polymer-derived carbon might represent compartments in which carbon is stored (for example, in the form of neutral lipids) when fungi are limited by the availability of nutrients other than carbon (22). These limitations are plausible for microorganisms growing on PBAT and other polymers that do not contain nitrogen and phosphorous. If these limitations occur, increasing the availability of soil nutrients to microorganisms colonizing the polymer surface is expected to enhance polymer biodegradation.
This work demonstrates PBAT biodegradation in a selected agricultural soil over 6 weeks of incubation. Future studies extending on this work will need to assess variations in the rates and extents of PBAT mineralization among different agricultural soils, also over longer-time incubations. Furthermore, we propose studies that are directed toward identifying soil microorganisms that are actively involved in PBAT biodegradation. While the NanoSIMS-based approach presented here allows us to unambiguously demonstrate incorporation of polyester carbon into soil microbial biomass, it is not a high-throughput technique. Alternative approaches, including the extraction of targeted biomolecules from soils containing 13C-labeled polymers followed by quantifying the 13C contents in the extracted molecules, will allow us to analyze larger sample sets and thereby to systematically determine potential variations among soil microorganisms in the extent to which they incorporate polymer-derived carbon into their biomass.
MATERIALS AND METHODS
Experimental design
The objective of this study was to develop an experimental approach to demonstrate biodegradation of PBAT in an agricultural soil. As biodegradation includes mineralization of PBAT carbon to CO2, as well as the incorporation of PBAT-derived carbon into the biomass of soil microorganisms, we addressed both of these processes in controlled laboratory experiments. We followed PBAT mineralization during soil incubation using an isotope-specific CRDS for the quantification of formed CO2. For each of the three PBAT variants, we simultaneously incubated seven films in one incubation bottle filled with soil to allow precise quantification of PBAT mineralization to CO2. The soil incubations were terminated after 6 weeks (that is, when approximately 10% of the PBAT carbon had been mineralized) to ensure that PBAT films were still intact for the subsequent imaging analyses. We revealed incorporation of PBAT-derived carbon into biomass using NanoSIMS, which enabled identification of subcellular features and determination of the carbon isotope composition of the PBAT film surface and the colonizing microorganisms at submicrometer spatial resolution. The low throughput of this high-end topochemical analysis technique constrained the number of collected images for soil-incubated films to two images for each of the three PBAT variants including replicate films. We note that we did not exclude any data or outliers from our analysis.
Polyesters, monomers, soil, and enzymes
Polyesters were provided by BASF SE and synthesized as previously described (23, 24). The physicochemical properties of the polyesters are listed in table S2. To obtain similar 13C contents for the three PBAT variants (that is, PB*AT, P*BAT, and PBA*T), synthesis of all variants was performed with defined ratios of labeled to unlabeled monomers. The three PBAT variants were free of chemical additives.
The 13C-labeled monomers 1,6-13C2-adipate and 13C4-butanediol used for PBAT synthesis and for soil incubation studies were purchased from Sigma, with more than 99% of the indicated positions in the monomer containing 13C. We obtained 1-13C1-terephthalate from dimethyl 1-13C-terephthalate purchased from Sigma. To obtain the free diacid, we dissolved dimethyl 1-13C-terephthalate in 2:1 water/tetrahydrofuran (5 mg in 2.4 ml), added 25 μl of a sodium hydroxide solution [37% (w/w)], and stirred the solution at room temperature for 2 hours. The solvent was then carefully removed under reduced pressure to obtain the hydrolysis product 1-13C1-terephthalate (confirmed by 1H NMR).
For PBAT and monomer incubations in soils under controlled laboratory conditions, we used agricultural soils from the agricultural center Limburgerhof (Rhineland-Palatinate, Germany). Physicochemical properties of the soils are provided in table S1. The soils were air-dried to a humidity of 12% of the maximum water-holding capacity of the soil, passed through a 2-mm sieve, and stored in the dark at 4°C for 12 months before use in the incubation experiments.
R. oryzae lipase was purchased as a powder from Sigma (catalog no. 80612). FsC was obtained as a solution from ChiralVision B.V. (Novozym 51032). Stock solutions of both enzymes in water were stored at −20°C.
Preparation of PBAT films and soils for incubation experiments
We prepared two sets of solvent-cast PBAT films that differed in the way that the PBAT films were attached to the silicon wafer substrates. For the first set, we solvent-cast PBAT films by adding three times 15 μl of a PBAT solution in chloroform [concentration, 5% (w/w)] onto a square-cut antimony-doped silicon wafer platelet (7.1 mm × 7.1 mm × 0.75 mm, Active Business Company). In between the additions of the polymer solutions, we allow the chloroform to evaporate. This procedure resulted in a PBAT mass of approximately 3 mg per wafer. Before incubation in soil, the solvent-cast polyester films were stored in the dark at room temperature for 1 week to ensure complete evaporation of the solvent (chloroform). PBAT variants from this first set were used for PBAT mineralization experiments (Fig. 2B), SEM imaging (Fig. 2C), and NanoSIMS imaging (Figs. 3 and 4 and fig. S8).
For the second set of PBAT films, we pretreated the silicon wafer platelets with Vectabond (Vector Laboratories, catalog no. SP-1800) before solvent casting of the polyester films. This second set of PBAT films was included to test whether the adhesion of the PBAT to the Si surface can be improved by this modified protocol. For the pretreatment, we exposed the wafers to a 1:50 diluted solution of Vectabond in acetone, subsequently dipped them into MilliQ water (Barnstead Nanopure Diamond), and dried them in a stream of N2. PBAT variants from this set were used only to determine PBAT mineralization (fig. S1), but not for SEM and NanoSIMS imaging.
We prepared the soil for PBAT incubations by adding MilliQ water to the soil to adjust its water content to 47% of its maximum water-holding capacity. We subsequently transferred 60 g of the soil into each of the incubation vessels (100-ml glass Schott bottles). We prepared a total of nine incubation bottles in three sets of three bottles (see below). The soils were then preincubated at 25°C in the dark for 1 week.
After soil preincubation, we transferred the wafers carrying the solvent-cast polyester films into the soils in the incubation bottles. We added seven wafers to each incubation bottle. The wafers were spaced apart by at least 1 cm. All wafers were positioned upright in the soil. The three bottles of the first set each contained films of one of the three differently labeled PBAT variants obtained by direct solvent casting. The three bottles of the second set were identical to the first set except for the wafers, which were pretreated with Vectabond before solvent casting. The three bottles in the third set served as controls and contained soil but no PBAT films. All bottles were incubated for 6 weeks at 25°C in the dark. We note that our study therefore does not address potential effects of ultraviolet irradiation–induced changes in the structure of PBAT on its biodegradability. Over the course of the incubation, we gravimetrically determined the water content of the soils at defined time intervals. To sustain a constant soil water content, amounts of water that were lost from the soil through evaporation were replenished by adding corresponding amounts of MilliQ water.
Quantification of polyester mineralization
For isotope-specific quantification of the 13CO2 formed from 13C-labeled PBAT during the incubations, we used an experimental setup similar to the one described by Bai et al. (13). In brief, we attached the incubation bottles containing soils with and without the PBAT variants to a flow-through system connected to an isotope-selective CO2 CRDS (Picarro G2201-i Analyzer) for 3 days per week. The volumetric gas flow through the system was 24 ml/min, which was established by a vacuum pump connected to the system. During the remaining time, the bottles were incubated under the same conditions as specified above with closed lids. Each time that the bottles were reconnected to the CRDS, the headspace of the bottle was allowed to equilibrate for 2 days before the concentrations of 12CO2 and 13CO2 in the effluent gas of the incubation bottles were measured using the CRDS.
The amount of 13C from each PBAT variant that was mineralized to 13CO2 during the incubation was calculated as follows. First, the fraction of CO2 that originated from PBAT (fPBAT) was calculated using the following equation (25, 26)
| (1) |
where a13C,sample, a13C,control, and a13C,PBAT refer to the isotope fraction [that is, 13C/(12C + 13C) in at%] of the CO2 sampled from the incubation bottles, the CO2 sampled from the control bottles (both measured by CRDS), and PBAT (measured by IRMS; see table S2 for details), respectively.
Then, the concentration of CO2 that resulted from PBAT mineralization ([CO2]PBAT) was calculated from the total CO2 concentration measured in the effluent of the incubation bottle ([CO2]sample)
| (2) |
The concentration of PBAT-derived 13CO2 ([13CO2]PBAT) was calculated with the following equation
| (3) |
The rate of mineralization of PBAT-derived 13C [r(13C)] was calculated from [13CO2]PBAT, the volumetric gas flow rate through the CRDS (Q = 24 ml/min), the molar mass of 13C (M = 13.003 g/mol), and the molar volume of air (V = 24.465 liters/mol at 25°C)
| (4) |
Linear interpolation between data points and integration of r(13C) over time resulted in the cumulative amount of mineralized PBAT 13C. In Fig. 2 and fig. S1, this quantity was displayed as the fraction of 13C of the isotopically labeled PBAT that was added to the soils.
Preparation and SEM imaging of soil-incubated PBAT films
After 6 weeks of incubation in soil, we carefully removed the silicon wafers carrying the PBAT films from the soils. To chemically fix the cells attached to the surfaces of the PBAT films, we directly transferred the films into a freshly prepared fixation solution (pH 7.4) containing glutaraldehyde (2.5%), sodium cacodylate (0.1 M), sodium chloride (0.1 M), potassium chloride (3 mM), and sodium phosphate (0.1 M). The films were exposed to this solution for 20 min at 25°C and subsequently transferred to a solution of OsO4 in MilliQ water (1%) for 30 min of exposure on ice. Finally, we dehydrated the films in a series of water/ethanol solutions of increasing concentrations (70%, 5 min; 95%, 15 min; 100%, 2 × 20 min), followed by critical point drying of the samples with liquid CO2 (Baltec CPD 030). Critical point drying resulted in detachment of the PBAT films from the wafer. To reattach the films to the wafers for further analyses, we used a double-sided adhesive, electrically conducting carbon tape (Ted Pella, product no. 16084-1). Directly after mounting the films onto the wafers with carbon tape, thin films of platinum (thickness, 10 nm) were deposited on the samples using a sputter coater (Baltec SCD 500). SEM was conducted on a Zeiss Supra 50 VP. Imaging was performed with a secondary electron detector at a working distance of 4.0 mm and an electron high tension of 5.0 kV. These films were also used for NanoSIMS analysis (see below).
PBAT films from the second set, for which wafers were pretreated with Vectabond before solvent casting of PBAT (see above), also detached from the wafers. We decided to reject further analysis of these films (that is, SEM and NanoSIMS).
PBAT film imaging by NanoSIMS
NanoSIMS measurements were performed on a NanoSIMS NS50L (Cameca) at the Large-Instrument Facility for Advanced Isotope Research (University of Vienna). Before data acquisition, analysis areas were presputtered by scanning of a high-intensity, slightly defocused Cs+ ion beam (beam current, 400 pA; spot size, approximately 2 μm). To avoid crater edge effects, scanning during presputtering was conducted over square-sized areas with an edge length exceeding the frame size of the subsequently recorded images by at least 15 μm. Every data set acquired on the soil-incubated polymer films contains image data recorded from (at least) two distinct depth levels, accessed by sequential presputtering with Cs+ ion fluences of 5.0 × 1016 and 2.0 × 1017 ions/cm2, respectively. Application of the lower ion dose density enabled sampling of all cells within the analysis areas, irrespective of their size and/or morphology, whereas the extended presputtering allowed us to gain insight into cellular features contained within the lumen of bulky cells such as fungal hyphae (see, for example, Fig. 4).
Imaging was conducted by sequential scanning of a finely focused Cs+ primary ion beam (2-pA beam current) over areas ranging from 45 × 45 μm2 to 70 × 70 μm2 at a physical resolution of approximately 70 nm (that is, probe size) and an image resolution of 512 × 512 pixels. If not stated otherwise, images were acquired as multilayer stacks with a per-pixel dwell time of 1.5 ms per cycle. 12C−, 13C−, 12C12C−, 12C13C−, 12C14N−, 31P−, and 32S− secondary ions as well as secondary electrons were simultaneously detected, and the mass spectrometer was tuned for achieving a mass resolving power of >9.000 (according to Cameca’s definition) for detection of C2− and CN− secondary ions. Image data were analyzed with the ImageJ plugin OpenMIMS, developed by the Center for NanoImaging (27). Secondary ion signal intensities were corrected for detector dead time (44 ns) and quasi-simultaneous arrival (QSA) of secondary ions. Both corrections were performed on a per-pixel basis. QSA sensitivity factors (“beta values”) were obtained from measurements on dried yeast cells, yielding 1.1, 1.06, and 1.05 for C−, C2−, and CN− secondary ions, respectively. Before stack accumulation, images were corrected for positional variations originating from primary ion beam and/or sample stage drift. ROIs were manually defined on the basis of 12C14N− secondary ion signal intensity distribution images and cross-checked by the topographical/morphological appearance indicated in the simultaneously recorded secondary electron images (see fig. S10). While each cell from unicellular organisms was assigned to an individual ROI, image regions within the polyester surfaces and hyphae were segmented into multiple ROIs. Throughout the article, the carbon isotope composition is displayed as the 13C/(12C + 13C) isotope fraction, given in at%, calculated from the C− and C2− secondary ion signal intensities via 13C−/(12C− + 13C−) and 13C12C−/(2⋅12C12C− + 13C12C−), respectively. Owing to superior counting statistics, all carbon isotope composition data shown in the article were inferred from C2− signal intensities. We note that we did not observe any significant differences between 13C content values inferred from C2− signal intensities versus C− signal intensities. For the line scan analyses displayed in Fig. 4, C2− normalized C14N− signal intensities were used as an indicator of the relative nitrogen content {calculated via [12C14N− (1 + 13C/12C)]/[12C13C− + 12C2− (1 + (13C/12C)2)], whereby the term 13C/12C refers to the 13C-to-12C isotope ratio, calculated from the C2− signal intensities via 13C12C−/(2⋅12C12C−)}. This quantity formally refers to the relative nitrogen-to-carbon elemental ratio and was used in favor of the relative nitrogen concentration, which is inferable from C− normalized C14N− signal intensities, to minimize artifacts arising from the considerable topography within the areas of the fungal hyphae (28).
Supplementary Material
Acknowledgments
We thank S. Probst, M. Jaggi, and F. Strasser for their help with growing E. coli, performing IRMS measurements, and NanoSIMS control sample preparation and data analysis, respectively. Funding: M.T.Z., T.F.N., R.B., H.-P.E.K., K.M., and M.S. thank the Joint Research Network on Advanced Materials and Systems of BASF SE and ETH Zürich for scientific and financial support. M.W. and A.S. were supported by the European Research Council Advanced Grant project NITRICARE 294343. D.W. was supported by the European Research Council Starting Grant project DormantMicrobes 636928. SEM imaging was performed at the Center for Microscopy, University of Zurich. Author contributions: M.T.Z., A.S., D.W., H.-P.E.K., K.M., and M.S. designed the study. M.T.Z., A.S., T.F.N., and R.B. performed experiments. All authors contributed to the writing of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate our conclusions are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/7/eaas9024/DC1
Supplementary Materials and Methods
Fig. S1. Mineralization of PBAT films.
Fig. S2. NMR analysis of enzymatic hydrolysis products of PBAT films I.
Fig. S3. NMR spectra of terephthalate, adipate, and butanediol.
Fig. S4. NMR analysis of enzymatic hydrolysis products of PBAT films II.
Fig. S5. NMR analysis of enzymatic hydrolysis products of PBAT films III.
Fig. S6. NMR analysis of enzymatic hydrolysis products of PBAT films IV.
Fig. S7. Mineralization of terephthalate, adipate, and butanediol.
Fig. S8. NanoSIMS analysis of PBAT films after soil incubation I.
Fig. S9. Control experiment for NanoSIMS analysis I.
Fig. S10. Definition of ROIs.
Fig. S11. Control experiment for NanoSIMS analysis II.
Fig. S12. NanoSIMS analysis of PBAT films after soil incubation II.
Table S1. Soil characterization.
Table S2. Characterization of PBAT variants.
Supplementary Appendix. Calculations of the carryover during NanoSIMS measurements.
REFERENCES AND NOTES
- 1.Sintim H. Y., Flury M., Is biodegradable plastic mulch the solution to agriculture’s plastic problem? Environ. Sci. Technol. 51, 1068–1069 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Kasirajan S., Ngouajio M., Polyethylene and biodegradable mulches for agricultural applications: A review. Agron. Sustain. Dev. 32, 501–529 (2012). [Google Scholar]
- 3.Rillig M. C., Microplastic in terrestrial ecosystems and the soil? Environ. Sci. Technol. 46, 6453–6454 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Liu E. K., He W. Q., Yan C. R., ‘White revolution’ to ‘white pollution’—Agricultural plastic film mulch in China. Environ. Res. Lett. 9, 091001 (2014). [Google Scholar]
- 5.Briassoulis D., Dejean C., Critical review of norms and standards for biodegradable agricultural plastics part I. biodegradation in soil. J. Polym. Environ. 18, 384–400 (2010). [Google Scholar]
- 6.Gross R. A., Kalra B., Biodegradable polymers for the environment. Science 297, 803–807 (2002). [DOI] [PubMed] [Google Scholar]
- 7.A. Künkel, J. Becker, L. Börger, J. Hamprecht, S. Koltzenburg, R. Loos, M. B. Schick, K. Schlegel, C. Sinkel, G. Skupin, M. Yamamoto, Polymers, biodegradable, in Ullmann’s Encyclopedia of Industrial Chemistry (Wiley-VCH Verlag GmbH & Co. KGaA, 2016). [Google Scholar]
- 8.Albertsson A.-C., Hakkarainen M., Designed to degrade. Science 358, 872–873 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Mueller R.-J., Biological degradation of synthetic polyesters—Enzymes as potential catalysts for polyester recycling. Process Biochem. 41, 2124–2128 (2006). [Google Scholar]
- 10.Rychter P., Kawalec M., Sobota M., Kurcok P., Kowalczuk M., Study of aliphatic-aromatic copolyester degradation in sandy soil and its ecotoxicological impact. Biomacromolecules 11, 839–847 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Kijchavengkul T., Auras R., Rubino M., Alvarado E., Montero J. R. C., Rosales J. M., Atmospheric and soil degradation of aliphatic–Aromatic polyester films. Polym. Degrad. Stab. 95, 99–107 (2010). [Google Scholar]
- 12.Weng Y.-X., Jin Y.-J., Meng Q.-Y., Wang L., Zhang M., Wang Y.-Z., Biodegradation behavior of poly(butylene adipate-co-terephthalate) (PBAT), poly(lactic acid) (PLA), and their blend under soil conditions. Polym. Test. 32, 918–926 (2013). [Google Scholar]
- 13.Bai M., Köstler M., Kunstmann J., Wilske B., Gattinger A., Frede H. G., Breuer L., Biodegradability screening of soil amendments through coupling of wavelength-scanned cavity ring-down spectroscopy to multiple dynamic chambers. Rapid Commun. Mass Spectrom. 25, 3683–3689 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Herrmann A. M., Ritz K., Nunan N., Clode P. L., Pett-Ridge J., Kilburn M. R., Murphy D. V., O’Donnell A. G., Stockdale E. A., Nano-scale secondary ion mass spectrometry—A new analytical tool in biogeochemistry and soil ecology: A review article. Soil Biol. Biochem. 39, 1835–1850 (2007). [Google Scholar]
- 15.Yoshida S., Hiraga K., Takehana T., Taniguchi I., Yamaji H., Maeda Y., Toyohara K., Miyamoto K., Kimura Y., Oda K., A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351, 1196–1199 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Bombelli P., Howe C. J., Bertocchini F., Polyethylene bio-degradation by caterpillars of the wax moth Galleria mellonella. Curr. Biol. 27, R292–R293 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Weber C., Pusch S., Opatz T., Polyethylene bio-degradation by caterpillars? Curr. Biol. 27, R744–R745 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Zumstein M. T., Rechsteiner D., Roduner N., Perz V., Ribitsch D., Guebitz G. M., Kohler H.-P. E., McNeill K., Sander M., Enzymatic hydrolysis of polyester thin films at the nanoscale: Effects of polyester structure and enzyme active-site accessibility. Environ. Sci. Technol. 51, 7476–7485 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Marten E., Müller R.-J., Deckwer W.-D., Studies on the enzymatic hydrolysis of polyesters. II. aliphatic–aromatic copolyesters. Polym. Degrad. Stab. 88, 371–381 (2005). [Google Scholar]
- 20.Perz V., Bleymaier K., Sinkel C., Kueper U., Bonnekessel M., Ribitsch D., Guebitz G. M., Substrate specificities of cutinases on aliphatic–aromatic polyesters and on their model substrates. New Biotechnol. 33, 295–305 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Zumstein M. T., Kohler H.-P. E., McNeill K., Sander M., High-throughput analysis of enzymatic hydrolysis of biodegradable polyesters by monitoring cohydrolysis of a polyester-embedded fluorogenic probe. Environ. Sci. Technol. 51, 4358–4367 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Bååth E., The use of neutral lipid fatty acids to indicate the physiological conditions of soil fungi. Microb. Ecol. 45, 373–383 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Gan Z., Kuwabara K., Yamamoto M., Abe H., Doi Y., Solid-state structures and thermal properties of aliphatic–aromatic poly(butylene adipate-co-butylene terephthalate) copolyesters. Polym. Degrad. Stab. 83, 289–300 (2004). [Google Scholar]
- 24.Lindström A., Albertsson A.-C., Hakkarainen M., Quantitative determination of degradation products an effective means to study early stages of degradation in linear and branched poly(butylene adipate) and poly(butylene succinate). Polym. Degrad. Stab. 83, 487–493 (2004). [Google Scholar]
- 25.Staddon P. L., Carbon isotopes in functional soil ecology. Trends Ecol. Evol. 19, 148–154 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Fernandez I., Cabaneiro A., González-Prieto S. J., Partitioning CO2 effluxes from an Atlantic pine forest soil between endogenous soil organic matter and recently incorporated 13C-enriched plant material. Environ. Sci. Technol. 40, 2552–2558 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Gormanns P., Reckow S., Poczatek J. C., Turck C. W., Lechene C., Segmentation of multi-isotope imaging mass spectrometry data for semi-automatic detection of regions of interest. PLOS ONE 7, e30576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomen A., Robert F., Remusat L., Determination of the nitrogen abundance in organic materials by NanoSIMS quantitative imaging. J. Anal. At. Spectrom 29, 512–519 (2014). [Google Scholar]
- 29.R. Baumgartner, Transformations of Novel and Unconventional Organic Compounds in Engineered and Natural Systems: Fluoroaromatics and Biodegradable Polyesters (ETH Zurich, 2015). [Google Scholar]
- 30.Ugolini F., Henneberger R., Bürgmann H., Zeyer J., Schroth M. H., In-situ sonication for enhanced recovery of aquifer microbial communities. Groundwater 52, 737–747 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Banfi D., Patiny L., www.nmrdb.org: Resurrecting and processing NMR spectra on-line. CHIMIA 62, 280–281 (2008). [Google Scholar]
- 32.Aires-de-Sousa J., Hemmer M. C., Gasteiger J., Prediction of 1H NMR chemical shifts using neural networks. Anal. Chem. 74, 80–90 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Briones M. J. I., Ineson P., Sleep D., Use of δ13C to determine food selection in collembolan species. Soil Biol. Biochem. 31, 937–940 (1999). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/7/eaas9024/DC1
Supplementary Materials and Methods
Fig. S1. Mineralization of PBAT films.
Fig. S2. NMR analysis of enzymatic hydrolysis products of PBAT films I.
Fig. S3. NMR spectra of terephthalate, adipate, and butanediol.
Fig. S4. NMR analysis of enzymatic hydrolysis products of PBAT films II.
Fig. S5. NMR analysis of enzymatic hydrolysis products of PBAT films III.
Fig. S6. NMR analysis of enzymatic hydrolysis products of PBAT films IV.
Fig. S7. Mineralization of terephthalate, adipate, and butanediol.
Fig. S8. NanoSIMS analysis of PBAT films after soil incubation I.
Fig. S9. Control experiment for NanoSIMS analysis I.
Fig. S10. Definition of ROIs.
Fig. S11. Control experiment for NanoSIMS analysis II.
Fig. S12. NanoSIMS analysis of PBAT films after soil incubation II.
Table S1. Soil characterization.
Table S2. Characterization of PBAT variants.
Supplementary Appendix. Calculations of the carryover during NanoSIMS measurements.


