Abstract
The prognosis of the majority of patients with papillary thyroid cancer (PTC) is excellent, although there are patients who experience disease recurrence and progression. The aim of the present study was to identify potential prognostic risk markers in PTC. Differentially expressed genes (DEGs), identified from four Genome Expression Omnibus cohorts were subjected to functional enrichment analyses with Gene Ontology terms and the Kyoto Encyclopedia of Genes and Genome pathways. Hub genes, filtered from cytoHubba, were validated using the The Cancer Genome Atlas (TCGA) cohort, and their associations with clinicopathological features and prognosis were analyzed. A total of 277 DEGs were identified following data preprocessing. DEGs were primarily enriched in ‘small cell lung cancer’, ‘ECM-receptor interaction’, ‘pathways in cancer’ and ‘tyrosine metabolism’. Hub genes [APOE, cathepsin S (CTSS), insulin receptor substrate 1 (IRS1), KIT, LGALS3, RUNX2 and TGFBR1] were extracted from cytoHubba. Their expression in the TCGA cohort was consistent with that in the GEO cohorts. CTSS (P=0.006) and IRS1 (P=0.005) were associated with disease-free survival, as determined using the Kaplan-Meier analysis. CTSS was an independent risk factor for poor disease-free survival (HR, 2.649; 95% CI, 1.095–6.409; P=0.031). Patients with high expression of CTSS exhibited different histological types (increased tall-cell subtype and reduced follicular subtype; P<0.001), more frequent lymph node metastasis (P<0.001) and advanced tumor-node-metastasis stages (P=0.049) compared with the low-expression group. High expression of CTSS was independently associated with lymph node metastasis (OR, 2.015; 95% CI, 1.225–3.315; P=0.006). Therefore, CTSS may serve as a predictive risk marker for the progression and prognosis of PTC.
Keywords: CTSS, papillary thyroid cancer, lymph node metastasis, prognosis, bioinformatics analysis, differentially-expressed genes
Introduction
Thyroid cancer is the most common endocrine malignancy worldwide, with the most rapidly increasing incidence (>5% per year) (1). It is the fifth most common type of cancer in women in the United States, with ~56,870 cases being newly diagnosed in 2017 and ~2,010 mortalities resulting from thyroid cancer (2). Papillary thyroid cancer (PTC) accounts for ~80% of all thyroid cancer cases (3). The majority of patients with this indolent tumor have an excellent prognosis, however >25% of patients experience disease recurrence during long-term follow-up (4). Therefore, it is important to identify the biomarkers associated with the progression and prognosis of PTC.
BRAF and RAS mutations and fusions involving the RET and NTRK1 tyrosine kinases accelerate PTC tumorigenesis and progression by aberrantly activating the mitogen-activated protein kinase (MAPK) signaling pathway (5–8). Mutations in members of the phosphoinositide 3-kinase (PI3K) pathway, including PTEN, PIK3CA and AKT1, play a fundamental role in thyroid tumorigenesis (9,10), and the NF-κB pathway controls proliferative and anti-apoptotic signaling pathways in thyroid cancer cells (11,12). Furthermore, the WNT-β-catenin pathway may play a particularly important role in the aggressiveness of thyroid cancer (10,13). In addition, the overexpression of microRNA-21 is associated with aggressive tall-cell histology and may play a crucial role in the pathogenesis of PTC. In addition, TERT promoter mutations identify a subset of aggressive and poorly differentiated PTCs (14). Despite significant research progress, the mechanisms underlying PTC tumorigenesis remain elusive. Therefore, a systemic approach is required to identify predictive biomarkers for disease progression and prognosis.
In the present study, a comprehensive bioinformatics analysis was used to identify key genes associated with PTC. A comparison of the expression profiles of PTC and normal tissues uncovered differentially expressed genes (DEGs). The key genes were extracted using cytoHubba (15) and their expression in PTC and normal tissues was compared using The Cancer Genome Atlas (TCGA) database. To determine whether their expression was correlated with a poor prognosis, a series of survival analyses were conducted. The relationships between key genes that were aberrantly expressed and clinicopathological factors, as well as tumor progression in patients with PTC, were also analyzed.
Materials and methods
Microarray data
Four gene expression profiles, GSE3467, GSE29265, GSE33630 and GSE50901, were acquired from the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). GSE3467 included 9 PTC tissue samples and 9 normal samples. GSE29265 consisted of 20 PTC tissue samples and 20 normal samples. GSE33630 comprised 49 PTC tissue samples and 45 normal samples. GSE50901 included 61 PTC tissue samples and 4 normal samples.
Screening DEGs
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) is available to analyze almost any GEO series between two sample groups under the same experimental conditions (16). This tool uses established Bioconductor R packages to analyze GEO data. In the present study, GEO2R was applied to screen for DEGs between PTC and normal tissue samples. An adjusted P-value (adj. P) <0.01 and |logFC| >1 were set as the cut-off criteria. Common DEGs from the four datasets were selected for further analyses.
Identification of key genes and predictions of function
Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) online tool, with an enrichment threshold of P<0.05. GO analysis comprises biological process (BP), cellular component (CC) and molecular function (MF) terms. A DEG-associated protein-protein interaction (PPI) network was constructed using the STRING database (https://string-db.org/) (17) and was subsequently visualized using Cytoscape (18). Hub genes were filtered from the intersection of the top 55 genes of 12 topological analysis methods using the Cytoscape plugin cytoHubba.
Patients and clinicopathological data
Normalized RNA sequencing data (Illumina HiSeq) and the corresponding clinicopathological data for PTCs from the TCGA dataset were downloaded from Firebrowse (http://firebrowse.org/) and cBioPortal for Cancer Genomics (http://www.cbioportal.org/). Normalized expression levels of mRNAs were log2-transformed and used for further analyses. The expression of key genes was validated using RNA sequencing data from 501 PTC and 59 normal thyroid samples. The clinicopathological data included the following variables: sex, age at diagnosis, maximum tumor size, multifocality, extrathyroidal extension (ETE), histological subtypes, lymph node metastasis (LNM), and tumor-node-metastasis (TNM) stage.
Statistical analysis
All statistical analyses were performed using SPSS 23.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 6.0 software (GraphPad, Inc., San Diego, CA, USA). An independent-sample Student's t-test was applied to compare the differential expression of DEGs between two groups. Disease-free survival (DFS), which was defined as the time from registration to detection of tumor recurrence or progression or until the last follow-up date, was calculated using Kaplan-Meier analysis followed by the log-rank test. Non-parametric receiver operating characteristic (ROC) analyses were performed to calculate the best cut-off value for hub gene expression levels predictive of LNM and DFS. The multivariable Cox proportional hazards regression model was used to determine independent predictive factors for DFS. Initial candidate variables with P<0.5 in the univariate analyses were included in further multiple logistic regression. To analyze the association between key genes and clinicopathological parameters, patients were divided into low-expression and high-expression groups according to the median expression levels of the key genes. The associations between the expression of key genes and clinicopathological characteristics were evaluated using the χ2 test. Multivariable logistic regression was performed to identify independent predictors of LNM. Multivariate survival analysis was performed on all parameters that were revealed to be significant on univariate analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of DEGs
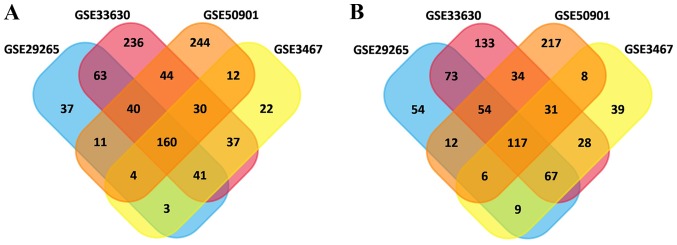
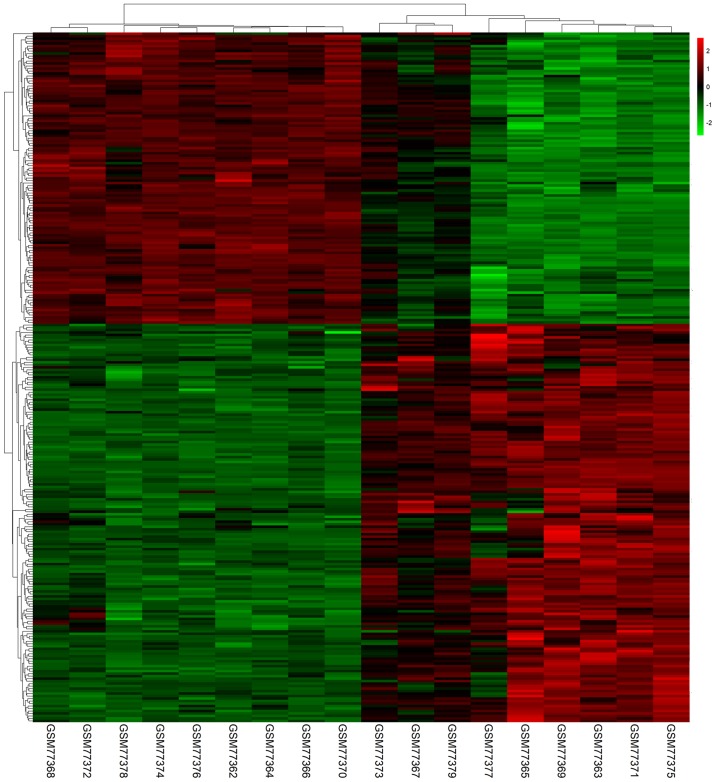
In total, 615, 752, 1,189 and 1,025 DEGs were identified in the GSE3467, GSE29265, GSE33630 and GSE50901 gene expression profile datasets, respectively. Among them, 277 genes showed identical expression trends in the four datasets, including 160 upregulated genes (Fig. 1A) and 117 downregulated genes (Fig. 1B) in PTC tissues compared with normal tissues. A heat map demonstrated the significant differential distribution of the DEGs using data profile GSE3467 as a reference (Fig. 2).
Figure 1.
Identification of DEGs in mRNA expression profiling datasets GSE3467, GSE29265, GSE33630 and GSE50901. (A) The crossed areas indicated the commonly upregulated DEGs. (B) The crossed areas indicated the commonly downregulated DEGs. DEGs, differentially expressed genes.
Figure 2.
Heatmap plot of the 277 overlapped genes between PTC and normal samples in GSE3467 dataset. Red represents higher expression and green represents lower expression. PTC, papillary thyroid cancer; DEGs, differentially expressed genes.
Identification of key genes and prediction of function
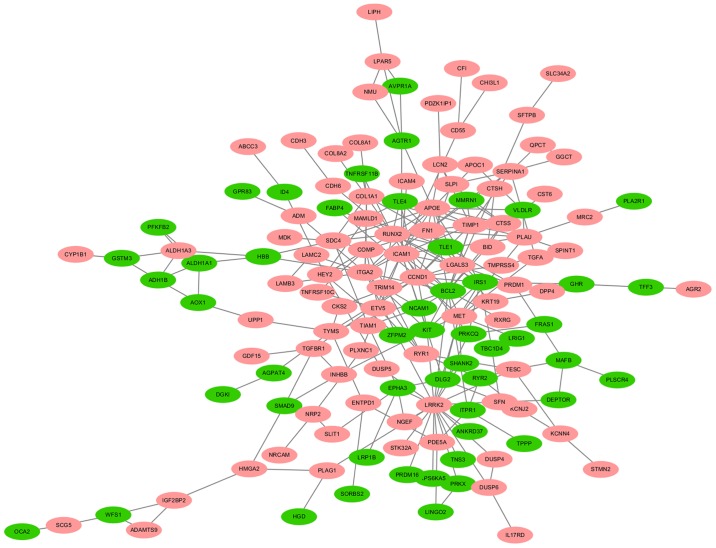
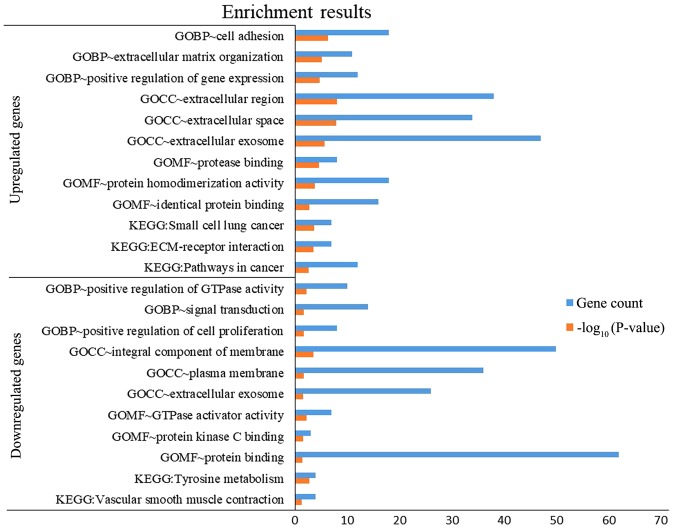
A total of 140 DEGs (89 upregulated and 51 downregulated genes) were filtered into the PPI network, which comprised 140 nodes and 255 edges, using the STRING online database and were visualized using Cytoscape software (Fig. 3). Seven key genes were selected from the intersection of the top 55 genes from 12 algorithms by cytoHubba, including apolipoprotein E (APOE), CTSS, IRS1, KIT proto-oncogene receptor tyrosine kinase (KIT), LGALS3, RUNX2 and transforming growth factor-β receptor type 1 (TGFBR1). DEGs were subjected to functional enrichment analyses, which indicated that the upregulated genes were mainly enriched in the terms ‘cell adhesion’, ‘protease binding’, ‘small cell lung cancer’, ‘ECM-receptor interaction’ and ‘pathways in cancer’, while the downregulated genes were mainly enriched in the terms ‘signal transduction’, ‘protein binding’ and ‘tyrosine metabolism’ (Fig. 4). Therefore, these DEGs may be associated with thyroid tumorigenesis.
Figure 3.
A protein-protein interaction (PPI) network of DEGs. Red nodes stand for upregulated genes, while green nodes stand for downregulated genes. DEGs, differentially expressed genes.
Figure 4.
The cellular component (CC), biological process (BP) and molecular function (MF) terms, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched by DEGs. DEGs, differentially expressed genes.
Expression of key genes in the TCGA cohort
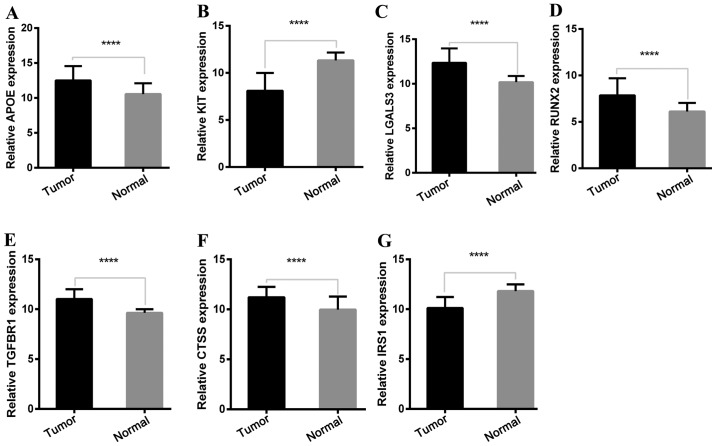
To determine the reliability of the identified DEGs from the four cohorts, their expression levels in 501 PTC samples and 59 normal thyroid tissues included in the TCGA database were evaluated. Consistent with the GEO analysis, 5 upregulated and 2 downregulated genes were significantly overexpressed and underexpressed respectively, in the PTC samples compared with the normal tissue samples (P<0.0001; Fig. 5).
Figure 5.
mRNA expression level of key genes in TCGA cohort. (A) APOE was significantly upregulated in 501 PTC samples compared with 59 normal thyroid samples. (B) KIT was significantly downregulated in 501 PTC samples compared with 59 normal thyroid samples. (C) LGALS3 was significantly upregulated in 501 PTC samples compared with 59 normal thyroid samples. (D) RUNX2 was significantly upregulated in 501 PTC samples compared with 59 normal thyroid samples. (E) TGFBR1 was significantly upregulated in 501 PTC samples compared with 59 normal thyroid samples. (F) CTSS was significantly upregulated in 501 PTC samples compared with 59 normal thyroid samples. (G) IRS1 was significantly downregulated in 501 PTC samples compared with 59 normal thyroid samples. Statistical differences were analyzed using independent t-test. ****P<0.0001. APOE, apolipoprotein E; KIT, KIT proto-oncogene receptor tyrosine kinase; LGALS3, galectin-3; RUNX2, runt-related transcription factor 2; TGFBR1, transforming growth factor-β receptor type 1; CTSS, cathepsin S; IRS1, insulin receptor substrate 1.
Key genes associated with DFS
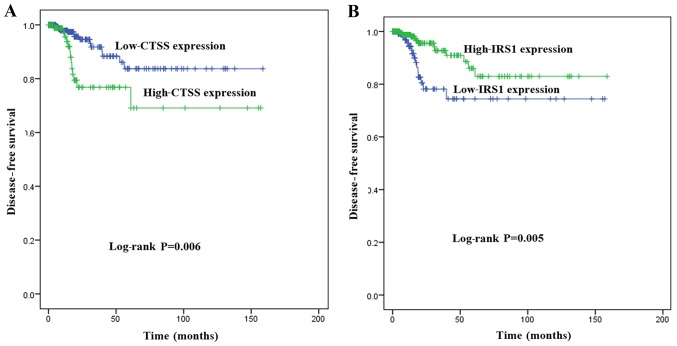
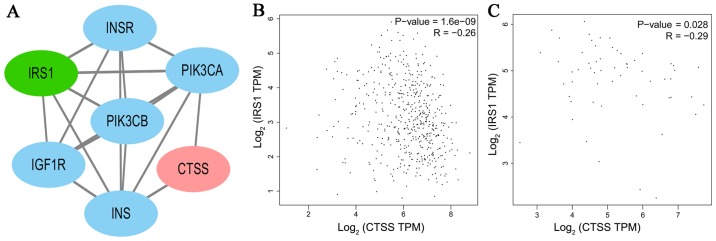
To investigate the association between the expression of key genes and DFS in patients with PTC, Kaplan-Meier survival analyses comparing the expression of the seven key genes and the DFS in the TCGA cohort were performed. From the Kaplan-Meier survival curves, CTSS (Fig. 6A; P=0.006) and IRS1 (Fig. 6B; P=0.005) were observed to exhibit significant correlations with DFS. A PPI network analysis revealed that there was no direct association between IRS1 and CTSS (Fig. 7A). A significant positive correlation was observed between CTSS and IRS1 mRNAs, both in PTC tissues and normal thyroid tissues (Pearson's correlation, in PTC tissues, r=−0.26, P<0.001, Fig. 7B; in normal thyroid tissues, r=−0.29, P<0.05, Fig. 7C).
Figure 6.
Kaplan-Meier survival analysis of the association between key gene expression level and DFS. (A) Patients with high expression of CTSS demonstrated significantly shorter DFS than those with low level of CTSS expression (P=0.006). (B) Patients with low expression of IRS1 demonstrated significantly shorter DFS than those with high level of IRS1 expression (P=0.005). Survival curves were compared using log-rank test. DFS, disease-free survival.
Figure 7.
Correlation between the expression of CTSS and IRS1 in the protein-protein interaction network (PPI) and the Gene Expression Profiling Interactive Analysis (GEPIA) database(http://gepia.cancer-pku.cn/). (A) A PPI of IRS1 and CTSS. (B) The expression of CTSS was negatively correlated with IRS1 mRNA expression in PTC tissues (r=−0.26, P<0.001). (C) The expression of CTSS was negatively correlated with IRS1 mRNA expression in normal thyroid tissues (r=−0.29, P<0.05).
Key genes independently predicting DFS
The Cox proportional hazards regression model was used to determine whether aberrant expression levels of CTSS and IRS1 may be independent risk factors for DFS in PTC. Univariate analysis revealed that the significant variables associated with DFS were increased LNM (HR, 3.242; 95% CI, 1.300–8.083; P=0.012), advanced TNM stage (HR, 2.323; 95% CI, 1.102–4.895; P=0.027), higher expression of CTSS (HR, 2.705; 95% CI, 1.287–5.688; P=0.009), and lower expression of IRS1 (HR, 0.360, 95% CI, 0.171–0.756; P=0.007). Multivariate analysis adjusted for sex, histological type, T stage, LNM, TNM stage, as well as the expression of CTSS and IRS1 indicated that only the increased expression of CTSS was an independent predictive factor for DFS (HR, 2.649; 95% CI, 1.095–6.409; P=0.031). Therefore, the increased expression of CTSS was considered to have a significant negative impact on the DFS of patients with PTC (Table I).
Table I.
Univariate and multivariate Cox regression analysis of factors associated with disease-free survival (DFS).
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| All variables | P-value | HR | 95% CI | P-value | HR | 95% CI |
| Age (years) | ||||||
| <45 | 1 | |||||
| ≥45 | 0.939 | 1.030 | 0.488–2.172 | nd | nd | nd |
| Sex | ||||||
| Male | 1 | 1 | ||||
| Female | 0.273b | 0.641 | 0.290–1.418 | 0.152 | 0.530 | 0.222–1.265 |
| Maximum tumor size (cm) | ||||||
| <4 | 1 | |||||
| ≥4 | 0.619 | 1.229 | 0.545–2.771 | nd | nd | nd |
| Multifocality | ||||||
| Unifocal | 1 | |||||
| Multifocal | 0.794 | 1.108 | 0.515–2.383 | nd | nd | nd |
| ETE | ||||||
| No | 1 | |||||
| Yes | 0.952 | 1.024 | 0.470–2.230 | nd | nd | nd |
| LNM | ||||||
| N0 | 1 | 1 | ||||
| N1 | 0.012c | 3.242 | 1.300–8.083 | 0.112 | 2.400 | 0.816–7.056 |
| T stage | ||||||
| T1-T2 | 1 | 1 | ||||
| T3-T4 | 0.326b | 1.451 | 0.690–3.052 | 0.611 | 1.239 | 0.542–2.835 |
| TNM stage | ||||||
| I–II | 1 | 1 | ||||
| III–IV | 0.027c | 2.323 | 1.102–4.895 | 0.100 | 2.117 | 0.867–5.173 |
| Histological type | ||||||
| Classical PTC | 1 | 1 | ||||
| Follicular PTC | 0.369b | 0.575 | 0.172–1.925 | 0.672 | 1.312 | 0.373–4.610 |
| Tall-cell PTC | 0.557 | 0.548 | 0.073–4.086 | 0.190 | 0.251 | 0.032–1.980 |
| CTSS | ||||||
| ≥ cut-offa | 0.009d | 2.705 | 1.287–5.688 | 0.031c | 2.649 | 1.095–6.409 |
| < cut-offa | 1 | 1 | ||||
| IRS1 | ||||||
| ≥ cut-offa | 0.007d | 0.360 | 0.171–0.756 | 0.208 | 0.551 | 0.218–1.392 |
| < cut-offa | 1 | 1 | ||||
Cut-off was calculated by non-parametric receiver operating characteristic analyses to predict disease-free survival; PTC, papillary thyroid cancer; ETE, extrathyroidal extension; LNM, lymph node metastasis; TNM, tumor-node-metastases; HR, hazard ratio; CI, confidence interval; nd, not done
P<0.5
P<0.05
P<0.01.
Expression of CTSS and clinicopathological factors
To further analyze the association between the expression of CTSS and clinicopathological characteristics in patients with PTC, patients were divided into high-expression and low-expression groups based on CTSS levels. It was revealed that high expression of CTSS was significantly associated with different histological subtypes (increased tall-cell subtype and reduced follicular subtype; P<0.001), more frequent LNM (P<0.001) and advanced TNM stage (P=0.049). However, there was no significant association between the expression of CTSS and any other clinicopathological characteristic, including age, sex, maximum size of tumor, multifocality and T stage (P>0.05; Table II).
Table II.
Correlation between the expression of CTSS and patient clinicopathological characteristics.
| CTSS subgroupa | |||
|---|---|---|---|
| Variables | Low (%) | High (%) | P-valueb |
| No. of cases | 251 | 250 | |
| Age (years) | 0.140 | ||
| <45 | 106 (46.5) | 122 (53.5) | |
| ≥45 | 145 (53.1) | 128 (46.9) | |
| Sex | 0.244 | ||
| Male | 62 (45.9) | 73 (54.1) | |
| Female | 187 (51.8) | 174 (48.2) | |
| Maximum tumor size (cm) | 0.529 | ||
| <4 | 177 (49.6) | 180 (50.4) | |
| ≥4 | 64 (52.9) | 57 (47.1) | |
| Multifocality | 0.495 | ||
| Unifocal | 129 (48.7) | 136 (51.3) | |
| Multifocal | 117 (51.8) | 109 (48.2) | |
| ETE | 0.230 | ||
| No | 183 (55.1) | 149 (44.9) | |
| Yes | 59 (48.8) | 62 (51.2) | |
| T stage | 0.133 | ||
| T1-T2 | 162 (52.8) | 145 (47.2) | |
| T3-T4 | 89 (45.9) | 105 (54.1) | |
| LNM | <0.001d | ||
| N0 | 129 (57.1) | 97 (42.9) | |
| N1 | 87 (38.7) | 138 (61.3) | |
| TNM stage | 0.049c | ||
| I–II | 177 (53.0) | 157 (47.0) | |
| III–IV | 72 (43.6) | 93 (56.4) | |
| Histological type | <0.001d | ||
| Classical PTC | 144 (44.6) | 179 (55.4) | |
| Follicular PTC | 77 (77.8) | 22 (22.2) | |
| Tall-cell PTC | 8 (23.5) | 26 (76.5) | |
Median expression level was used as a cut-off to divide the 501 patients into low-CTSS group (n=251) and high-CTSS group (n=250)
Chi-square test; ETE, extrathyroidal extension; LNM, lymph node metastasis; TNM, tumor-node-metastasis
P<0.05.
P<0.001.
Correlation between the expression of CTSS and LNM
Binary logistic regression analysis was performed to determine whether the overexpression of CTSS may be an independent risk factor for LNM in PTC. As listed in Table III, univariate analysis revealed that LNM was associated with older age (OR, 0.610; 95% CI, 0.420–0.887; P=0.010), larger tumor size (OR, 1.884; 95% CI, 1.203–2.951; P=0.006), ETE (OR, 2.751; 95% CI, 1.809–4.183; P<0.001), advanced T stage (OR, 2.646; 95% CI, 1.792–3.908; P<0.001), histological subtype (OR, 0.172; 95% CI, 0.091–0.326; P<0.001), overexpression of IRS1 (OR, 0.335; 95% CI, 0.228–0.492; P<0.001) and overexpression of CTSS (OR, 2.688; 95% CI, 1.780–4.059; P<0.001). After adjusting for these variables, multivariate analysis indicated that overexpression of CTSS (OR, 2.015; 95% CI, 1.225–3.315; P=0.006) was an independent risk factor for LNM, whereas age ≥45 years was a significant protective predictive factor (OR, 0.527; 95% CI, 0.331–0.840; P=0.007), as was the overexpression of IRS1 (OR, 0.579; 95% CI, 0.362–0.926; P=0.022) and follicular-variant histology was significantly associated with a lower incidence of LNM compared with classical PTC (OR, 0.205; 95% CI, 0.102–0.414; P<0.001). No significant associations were observed between LNM and other factors, including tumor size, ETE and T stage (P>0.05; Table III).
Table III.
Univariate and multivariate logistic regression analysis of the factors associated with lymph node metastasis (LNM).
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variables | P-value | OR | 95% CI | P-value | OR | 95% CI |
| Age (years) | ||||||
| <45 | 1 | 1 | ||||
| ≥45 | 0.010b | 0.610 | 0.420–0.887 | 0.007c | 0.527 | 0.331–0.840 |
| Sex | ||||||
| Male | 1 | |||||
| Female | 0.051 | 0.659 | 0.434–1.001 | nd | nd | nd |
| Maximum tumor size (cm) | ||||||
| <4 | 1 | 1 | ||||
| ≥4 | 0.006c | 1.884 | 1.203–2.951 | 0.246 | 1.443 | 0.776–2.682 |
| Multifocality | ||||||
| Unifocal | 1 | |||||
| Multifocal | 0.088 | 1.386 | 0.953–2.017 | nd | nd | nd |
| ETE | ||||||
| No | 1 | 1 | ||||
| Yes | <0.001d | 2.751 | 1.809–4.183 | 0.392 | 1.430 | 0.630–3.246 |
| T stage | ||||||
| T1-T2 | 1 | 1 | ||||
| T3-T4 | <0.001d | 2.646 | 1.792–3.908 | 0.346 | 1.495 | 0.648–3.448 |
| Histological type | ||||||
| Classical PTC | 1 | 1 | ||||
| Follicular PTC | <0.001d | 0.172 | 0.091–0.326 | <0.001d | 0.205 | 0.102–0.414 |
| Tall-cell PTC | 0.185 | 1.671 | 0.783–3.567 | 0.932 | 1.038 | 0.439–2.458 |
| CTSS | ||||||
| ≤ cut-offa | 1 | 1 | ||||
| > cut-offa | <0.001d | 2.688 | 1.780–4.059 | 0.006c | 2.015 | 1.225–3.315 |
| IRS1 | ||||||
| ≤ cut-offa | 1 | 1 | ||||
| > cut-offa | <0.001d | 0.335 | 0.228–0.492 | 0.022b | 0.579 | 0.362–0.926 |
Cut-off was calculated by non-parametric receiver operating characteristic analyses to predict lymph node metastasis. ETE, extrathyroidal extension; PTC, papillary thyroid cancer; OR, odds ratio; CI, confidence interval; nd, not done
P<0.05
P<0.01
P<0.001.
Discussion
The present study identified 277 DEGs in tumor samples from four GEO cohorts. Enrichment analyses indicated that the majority of the DEGs could be associated with thyroid tumorigenesis. The top DEGs [APOE, CTSS, IRS1, KIT, galectin-3 (LGALS3), runt-related transcription factor 2 (RUNX2) and TGFBR1] were extracted as hub genes, and their aberrant expression in the TCGA cohort was consistent with the GEO analysis, supporting the validity of the results.
Apolipoprotein E (APOE) is a 299-amino acid glycoprotein that is known to play a role in cholesterol transport and metabolism. APOE can play diverse roles in a number of biological processes, including cell growth, differentiation, immune stress and survival (19). APOE has been identified to be overexpressed in lung (20) and ovarian cancer (21), however, the data available regarding APOE in PTC are limited. The present study revealed that APOE was significantly upregulated in PTC tissues. Therefore, APOE may be a typical biological characteristic of PTC.
KIT proto-oncogene receptor tyrosine kinase (KIT), a type-III receptor tyrosine kinase (RTK), plays a crucial role in the occurrence of cancer. Dysregulation of KIT (including overexpression and gain-of-function, loss-of-function and point mutations) has been detected in several types of human cancer (22–25). In thyroid tumors, the expression levels of KIT were significantly lower in malignant than in benign tumors, which was considered a very strong predictive indicator for discriminating malignant from benign tumors (26,27). Consistent with the results of previous studies, the present study revealed that KIT was significantly downregulated in PTC tissues. Therefore, KIT may be a useful diagnostic marker in PTC.
LGALS3, a member of a β-galactoside-binding protein family, is involved in normal growth development, and cancer progression and metastasis, as well as implicated in cell growth, differentiation, adhesion and malignant transformation. Previous studies have demonstrated that LGALS3 may serve as a diagnostic marker for thyroid cancer (28,29). The present study demonstrated that the expression levels of LGALS3 were significantly higher in PTC tissues than in normal tissues, which supported the hypothesis that LGALS3 is a valuable diagnostic marker for thyroid cancer.
Runt-related transcription factor 2 (RUNX2) is a transcription factor required for bone development. The involvement of RUNX2 in tumor progression and metastasis has been increasingly recognized in different types of tumor cells (30–35). The present study revealed that RUNX2 was upregulated in PTC tissues. Additionally, RUNX2 was enriched in the GO BP terms ‘positive regulation of cell proliferation’ and in the KEGG pathway ‘transcriptional misregulation in cancer’. In agreement with this, previous studies have identified an association between the expression of RUNX2 and thyroid cancer progression (36–39). Therefore, RUNX2 may play an important role in thyroid tumorigenesis.
Transforming growth factor-β (TGF-β) is a member of the disulfide-bonded cytokine family and plays an important role in cell growth, differentiation, apoptosis and survival. TGFBR1 is a receptor for TGF-β ligands. Following stimulation with TGF-β ligands, TGFBR1 activates several different signaling pathways, including the Smad, MAPK, PI3K and serine/threonine protein kinase (Akt) pathways, and contributes to the invasion, metastasis and progression of cancer (40–42). However, the oncogenic role of TGFBR1 in thyroid cancer has not been adequately investigated. The present study revealed that TGFBR1 was associated with the GO BP terms ‘positive regulation of apoptotic signaling pathway’, ‘positive regulation of cell proliferation’, ‘positive regulation of cell migration’ and the KEGG term ‘pathways in cancer’. Furthermore, the present study demonstrated that TGFBR1 was highly expressed in PTC tissues, indicating a potential role in thyroid tumorigenesis.
In the present study, aberrant expression of CTSS and IRS1 was revealed to be associated with DFS. To the best of our knowledge, there have been no previous studies on the relationship between IRS1 and CTSS. Although there was no direct association between IRS1 and CTSS, a significant positive correlation was observed between CTSS and IRS1 mRNAs, both in PTC and normal thyroid tissues. The mechanisms underlying the relationship between CTSS and IRS1 must be explored in future studies. Insulin receptor substrate 1 (IRS1) is an adaptor protein that functions as a downstream messenger from activated cell surface receptors to numerous signaling pathway cascades. Early studies on IRS1 introduced its role in metabolism, but a great deal of recent research has focused on its role in cancer progression and metastasis. The PI3K/Akt pathway is one of the main downstream signaling pathways activated by phosphorylated IRS1 (43). However, the regulatory roles of IRS1 in tumorigenesis are controversial. IRS1 has been shown to promote the proliferation and metastasis of pancreatic (44) and colon cancer (45). However, in other studies, suppression of IRS1 promoted the metastasis of head and neck squamous cell carcinoma (46), neutrophil elastase-mediated degradation of IRS1 induced lung tumor cell proliferation (47). Previous studies have also demonstrated that IRS1 may be both a tumor promoter and suppressor in breast cancer (48,49). The present study revealed that IRS1 was a protective factor for LNM, and the low-IRS1 expression group exhibited a shorter DFS time compared with the high-IRS1 expression group, indicating it as a negative regulator in PTC. This finding was consistent with studies that have indicated that IRS1 was a metastatic suppressor (50). Therefore, the role of IRS1 may depend on the type of cancer.
In the present study, the overexpression of CTSS was the only independent predictive risk factor for DFS. The high-CTSS expression group exhibited different histological types, advanced TNM stage and increased LNM compared with the low-CTSS expression group. Furthermore, CTSS was identified as an independent risk factor for LNM in PTC. The results of the present study indicated that CTSS may play an important role in tumorigenesis, and that high CTSS expression may be associated with disease recurrence and progression in PTC.
Cysteine cathepsin proteases, which are endo-lysosomal proteases, contribute to diseases such as cancer, osteoporosis and arthritis (51). The initial attention of researchers in cathepsins in cancer is attributed to the overexpression of cathepsin in tumors in comparison with normal tissues. Previous studies spanning decades have suggested that cathepsins make crucial contributions to tumor progression of numerous cancers by diverse mechanisms. For example, cysteine cathepsins are associated with protein catabolism and autophagy, degradation of the extracellular matrix (ECM) and activation and degradation of growth factors, cytokines and chemokines (52–55).
The human family of cysteine cathepsins comprises 11 members that are involved in diverse physiological processes, including bone protein degradation, autophagy, antigen presentation, growth factor receptor recycling, cellular stress signaling and lysosome-mediated cell death (56). CTSS is a lysosomal cysteine protease of the papain superfamily of cysteine proteases, which plays an important role in cancer by contributing to apoptosis, autophagy, angiogenesis, invasion, migration and clinical prognosis (57–60). CTSS has characteristics distinct from numerous other family members. Firstly, it remains active at neutral and acidic pH (61), whereas many other cysteine cathepsins are active at an acidic pH. Secondly, the expression of CTSS is unique among the cysteine cathepsin family, as it is specifically expressed by antigen-presenting cells (APCs) (62). Thirdly, CTSS exerts more physiological effects than other cathepsins in the processing of proteins in the extracellular microenvironment (63).
Previous studies have indicated that CTSS was frequently overexpressed in colorectal (58,64), lung, gastric (65) and prostate cancer (66), as well as in hepatocellular carcinoma (67), glioma (68) and breast cancer (69). Although the results of these recent in vitro and in vivo studies demonstrated that CTSS played an important role in tumorigenesis, no data are currently available regarding the abnormal expression of CTSS or its association with the clinical outcomes of patients with PTC. The present study revealed that CTSS was frequently upregulated in PTC samples compared with normal tissues. Furthermore, it was revealed that high CTSS expression was significantly associated with more frequent LNM and a poorer DFS. However, the exact mechanisms underlying how the overexpression of CTSS influenced LNM remained unclear. It has been reported that the expression of CTSS was regulated by the PI3K/Akt and Ras/Raf/MAPK signaling pathways through the activation of NF-κB (69), which has been well established to contribute to thyroid tumorigenesis (70). Therefore, all these results indicated that the expression of CTSS plays a key role in thyroid tumorigenesis and its overexpression may be associated with aggressiveness and progression in thyroid cancer.
Accumulating evidence has demonstrated that the expression of CTSS is an independent predictor of a poor prognosis in some types of human cancer, including gastric (65) and colorectal cancer (58), as well as glioma (71) and lung cancer (72). The present study extended this observation to PTC. A shorter DFS time was observed in patients with high expression of CTSS compared with those with low expression of CTSS. These results indicated that the overexpression of CTSS may provide guidance for identifying postoperative patients at high risk of disease recurrence or progression who may benefit from effective adjuvant therapy.
To the best of our knowledge, the present study was the first to demonstrate that CTSS was significantly upregulated in PTC samples compared with normal tissues. Furthermore, a high expression of CTSS was significantly associated with LNM and disease recurrence or progression in patients with PTC. Further studies are required to confirm the results of the present study and to identify the mechanism underlying the association between CTSS expression and the pathogenesis of PTC. Confirmation of the results of the present study may establish the expression of CTSS as a diagnostic and prognostic marker of PTC.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CTSS
cathepsin S
- PTC
papillary thyroid cancer
- DEGs
differentially expressed genes
- DFS
disease-free survival
- LNM
lymph node metastasis
- ETE
extrathyroidal extension
- TNM
tumor-node-metastasis
- CI
confidence interval
- HR
hazard ratio
- OR
odds ratio
Funding
The present study was supported in part by the Key Research and Development Project of Jiangsu Province (BE2015723) and Six Talent Peak Funding Plan (WSN-061).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
JT and YL conceived and designed the study. XA, TC, BS and HL analyzed the data. JT and HL wrote the paper. XQ, DD and ZZ were also involved in the conception of the study. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors state that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Haugen BR. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: What is new and what has changed? Cancer. 2017;123:372–381. doi: 10.1002/cncr.30360. [DOI] [PubMed] [Google Scholar]
- 4.Grogan RH, Kaplan SP, Cao H, Weiss RE, Degroot LJ, Simon CA, Embia OM, Angelos P, Kaplan EL, Schechter RB. A study of recurrence and death from papillary thyroid cancer with 27 years of median follow-up. Surgery. 2013;154:1436–1446. doi: 10.1016/j.surg.2013.07.008. discussion 1446-7. [DOI] [PubMed] [Google Scholar]
- 5.Giordano TJ, Kuick R, Thomas DG, Misek DE, Vinco M, Sanders D, Zhu Z, Ciampi R, Roh M, Shedden K, et al. Molecular classification of papillary thyroid carcinoma: Distinct BRAF, RAS, and RET/PTC mutation-specific gene expression profiles discovered by DNA microarray analysis. Oncogene. 2005;24:6646–6656. doi: 10.1038/sj.onc.1208822. [DOI] [PubMed] [Google Scholar]
- 6.Adeniran AJ, Zhu Z, Gandhi M, Steward DL, Fidler JP, Giordano TJ, Biddinger PW, Nikiforov YE. Correlation between genetic alterations and microscopic features, clinical manifestations, and prognostic characteristics of thyroid papillary carcinomas. Am J Surg Pathol. 2006;30:216–222. doi: 10.1097/01.pas.0000176432.73455.1b. [DOI] [PubMed] [Google Scholar]
- 7.Greco A, Miranda C, Pierotti MA. Rearrangements of NTRK1 gene in papillary thyroid carcinoma. Mol Cell Endocrinol. 2010;321:44–49. doi: 10.1016/j.mce.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Ellis RJ, Wang Y, Stevenson HS, Boufraqech M, Patel D, Nilubol N, Davis S, Edelman DC, Merino MJ, He M, et al. Genome-wide methylation patterns in papillary thyroid cancer are distinct based on histological subtype and tumor genotype. J Clin Endocrinol Metab. 2014;99:E329–E337. doi: 10.1210/jc.2013-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saji M, Ringel MD. The PI3K-Akt-mTOR pathway in initiation and progression of thyroid tumors. Mol Cell Endocrinol. 2010;321:20–28. doi: 10.1016/j.mce.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13:184–199. doi: 10.1038/nrc3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Abdel-Mageed AB, Mondal D, Kandil E. The nuclear factor kappa-B signaling pathway as a therapeutic target against thyroid cancers. Thyroid. 2013;23:209–218. doi: 10.1089/thy.2012.0237. [DOI] [PubMed] [Google Scholar]
- 12.Bauerle KT, Schweppe RE, Lund G, Kotnis G, Deep G, Agarwal R, Pozdeyev N, Wood WM, Haugen BR. Nuclear factor κB-dependent regulation of angiogenesis, and metastasis in an in vivo model of thyroid cancer is associated with secreted interleukin-8. J Clin Endocrinol Metab. 2014;99:E1436–E1444. doi: 10.1210/jc.2013-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Agrawal N, Akbani R, Aksoy BA, Ally A, Arachchi H, Asa SL, Auman JT, Balasundaram M, Balu S, Baylin SB, et al. Cancer Genome Atlas Research Network: Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets - update. Nucleic Acids Res. 2013;41D:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41D:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Gao Y, Hao F, Lou X, Zhang X, Li Y, Wu D, Xiao T, Yang L, Li Q, et al. Secretomes are a potential source of molecular targets for cancer therapies and indicate that APOE is a candidate biomarker for lung adenocarcinoma metastasis. Mol Biol Rep. 2014;41:7507–7523. doi: 10.1007/s11033-014-3641-4. [DOI] [PubMed] [Google Scholar]
- 20.Su WP, Chen YT, Lai WW, Lin CC, Yan JJ, Su WC. Apolipoprotein E expression promotes lung adenocarcinoma proliferation and migration and as a potential survival marker in lung cancer. Lung Cancer. 2011;71:28–33. doi: 10.1016/j.lungcan.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Boylan KL, Andersen JD, Anderson LB, Higgins L, Skubitz AP. Quantitative proteomic analysis by iTRAQ(R) for the identification of candidate biomarkers in ovarian cancer serum. Proteome Sci. 2010;8:31. doi: 10.1186/1477-5956-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen EC, Karl TA, Kalisky T, Gupta SK, O'Brien CA, Longacre TA, van de Rijn M, Quake SR, Clarke MF, Rothenberg ME. KIT signaling promotes growth of colon xenograft tumors in mice and is up-regulated in a subset of human colon cancers. Gastroenterology. 2015;149:705–17.e2. doi: 10.1053/j.gastro.2015.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joensuu H, Rutkowski P, Nishida T, Steigen SE, Brabec P, Plank L, Nilsson B, Braconi C, Bordoni A, Magnusson MK, et al. KIT and PDGFRA mutations and the risk of GI stromal tumor recurrence. J Clin Oncol. 2015;33:634–642. doi: 10.1200/JCO.2014.57.4970. [DOI] [PubMed] [Google Scholar]
- 24.Gavert N, Shvab A, Sheffer M, Ben-Shmuel A, Haase G, Bakos E, Domany E, Ben-Ze'ev A. c-Kit is suppressed in human colon cancer tissue and contributes to L1-mediated metastasis. Cancer Res. 2013;73:5754–5763. doi: 10.1158/0008-5472.CAN-13-0576. [DOI] [PubMed] [Google Scholar]
- 25.Mainetti LE, Zhe X, Diedrich J, Saliganan AD, Cho WJ, Cher ML, Heath E, Fridman R, Kim HR, Bonfil RD. Bone-induced c-kit expression in prostate cancer: A driver of intraosseous tumor growth. Int J Cancer. 2015;136:11–20. doi: 10.1002/ijc.28948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panebianco F, Mazzanti C, Tomei S, Aretini P, Franceschi S, Lessi F, Di Coscio G, Bevilacqua G, Marchetti I. The combination of four molecular markers improves thyroid cancer cytologic diagnosis and patient management. BMC Cancer. 2015;15:918. doi: 10.1186/s12885-015-1917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pusztaszeri MP, Sadow PM, Faquin WC. CD117: A novel ancillary marker for papillary thyroid carcinoma in fine-needle aspiration biopsies. Cancer Cytopathol. 2014;122:596–603. doi: 10.1002/cncy.21437. [DOI] [PubMed] [Google Scholar]
- 28.Trimboli P, Virili C, Romanelli F, Crescenzi A, Giovanella L. Galectin-3 performance in histologic a cytologic assessment of thyroid nodules: A systematic review and meta-analysis. Int J Mol Sci. 2017;18:18. doi: 10.3390/ijms18081756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xing M, Haugen BR, Schlumberger M. Progress in molecular-based management of differentiated thyroid cancer. Lancet. 2013;381:1058–1069. doi: 10.1016/S0140-6736(13)60109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Owens TW, Rogers RL, Best S, Ledger A, Mooney AM, Ferguson A, Shore P, Swarbrick A, Ormandy CJ, Simpson PT, et al. Runx2 is a novel regulator of mammary epithelial cell fate in development and breast cancer. Cancer Res. 2014;74:5277–5286. doi: 10.1158/0008-5472.CAN-14-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sase T, Suzuki T, Miura K, Shiiba K, Sato I, Nakamura Y, Takagi K, Onodera Y, Miki Y, Watanabe M, et al. Runt-related transcription factor 2 in human colon carcinoma: A potent prognostic factor associated with estrogen receptor. Int J Cancer. 2012;131:2284–2293. doi: 10.1002/ijc.27525. [DOI] [PubMed] [Google Scholar]
- 32.Boregowda RK, Olabisi OO, Abushahba W, Jeong BS, Haenssen KK, Chen W, Chekmareva M, Lasfar A, Foran DJ, Goydos JS, et al. RUNX2 is overexpressed in melanoma cells and mediates their migration and invasion. Cancer Lett. 2014;348:61–70. doi: 10.1016/j.canlet.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge C, Zhao G, Li Y, Li H, Zhao X, Pannone G, Bufo P, Santoro A, Sanguedolce F, Tortorella S, et al. Role of Runx2 phosphorylation in prostate cancer and association with metastatic disease. Oncogene. 2016;35:366–376. doi: 10.1038/onc.2015.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pratap J, Lian JB, Stein GS. Metastatic bone disease: Role of transcription factors and future targets. Bone. 2011;48:30–36. doi: 10.1016/j.bone.2010.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sancisi V, Gandolfi G, Ambrosetti DC, Ciarrocchi A. Histone deacetylase inhibitors repress tumoral expression of the proinvasive factor RUNX2. Cancer Res. 2015;75:1868–1882. doi: 10.1158/0008-5472.CAN-14-2087. [DOI] [PubMed] [Google Scholar]
- 36.Sancisi V, Borettini G, Maramotti S, Ragazzi M, Tamagnini I, Nicoli D, Piana S, Ciarrocchi A. Runx2 isoform I controls a panel of proinvasive genes driving aggressiveness of papillary thyroid carcinomas. J Clin Endocrinol Metab. 2012;97:E2006–E2015. doi: 10.1210/jc.2012-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalle Carbonare L, Frigo A, Francia G, Davì MV, Donatelli L, Stranieri C, Brazzarola P, Zatelli MC, Menestrina F, Valenti MT. Runx2 mRNA expression in the tissue, serum, and circulating non-hematopoietic cells of patients with thyroid cancer. J Clin Endocrinol Metab. 2012;97:E1249–E1256. doi: 10.1210/jc.2011-2624. [DOI] [PubMed] [Google Scholar]
- 38.Kaptan E, Bas Sancar S, Sancakli A, Aktas HG, Bayrak BB, Yanardag R, Bolkent S. Runt-related transcription factor 2 (Runx2) Is responsible for galectin-3 overexpression in human thyroid carcinoma. J Cell Biochem. 2017;118:3911–3919. doi: 10.1002/jcb.26043. [DOI] [PubMed] [Google Scholar]
- 39.Carr FE, Tai PW, Barnum MS, Gillis NE, Evans KG, Taber TH, White JH, Tomczak JA, Jaworski DM, Zaidi SK, et al. Thyroid hormone receptor-β (TRβ) mediates runt-related transcription factor 2 (Runx2) expression in thyroid cancer cells: A novel signaling pathway in thyroid cancer. Endocrinology. 2016;157:3278–3292. doi: 10.1210/en.2015-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bian Y, Terse A, Du J, Hall B, Molinolo A, Zhang P, Chen W, Flanders KC, Gutkind JS, Wakefield LM, et al. Progressive tumor formation in mice with conditional deletion of TGF-beta signaling in head and neck epithelia is associated with activation of the PI3K/Akt pathway. Cancer Res. 2009;69:5918–5926. doi: 10.1158/0008-5472.CAN-08-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu T, Chen X, Peng R, Liu H, Yin P, Peng H, Zhou Y, Sun Y, Wen L, Yi H, et al. Let-7a suppresses cell proliferation via the TGF-β/SMAD signaling pathway in cervical cancer. Oncol Rep. 2016;36:3275–3282. doi: 10.3892/or.2016.5160. [DOI] [PubMed] [Google Scholar]
- 42.He J, Jin Y, Zhou M, Li X, Chen W, Wang Y, Gu S, Cao Y, Chu C, Liu X, Zou Q. Solute carrier family 35 member F2 is indispensable for papillary thyroid carcinoma progression through activation of transforming growth factor-β type I receptor/apoptosis signal-regulating kinase 1/mitogen-activated protein kinase signaling axis. Cancer Sci. 2018;109:642–655. doi: 10.1111/cas.13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zha J, Lackner MR. Targeting the insulin-like growth factor receptor-1R pathway for cancer therapy. Clin Cancer Res. 2010;16:2512–2517. doi: 10.1158/1078-0432.CCR-09-2232. [DOI] [PubMed] [Google Scholar]
- 44.Wang G, Pan J, Zhang L, Wei Y, Wang C. Long non-coding RNA CRNDE sponges miR-384 to promote proliferation and metastasis of pancreatic cancer cells through upregulating IRS1. Cell Prolif. 2017;50:50. doi: 10.1111/cpr.12389. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Bailey KL, Agarwal E, Chowdhury S, Luo J, Brattain MG, Black JD, Wang J. TGFβ/Smad3 regulates proliferation and apoptosis through IRS-1 inhibition in colon cancer cells. PLoS One. 2017;12:e0176096. doi: 10.1371/journal.pone.0176096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo X, Fan S, Huang W, Zhai S, Ma Z, Li P, Sun SY, Wang X. Downregulation of IRS-1 promotes metastasis of head and neck squamous cell carcinoma. Oncol Rep. 2012;28:659–667. doi: 10.3892/or.2012.1846. [DOI] [PubMed] [Google Scholar]
- 47.Houghton AM, Rzymkiewicz DM, Ji H, Gregory AD, Egea EE, Metz HE, Stolz DB, Land SR, Marconcini LA, Kliment CR, et al. Neutrophil elastase-mediated degradation of IRS-1 accelerates lung tumor growth. Nat Med. 2010;16:219–223. doi: 10.1038/nm.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Zhang X, Zou C, Kung HF, Lin MC, Dress A, Wardle F, Jiang BH, Lai L. miR-195 inhibits tumor growth and angiogenesis through modulating IRS1 in breast cancer. Biomed Pharmacother. 2016;80:95–101. doi: 10.1016/j.biopha.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 49.Ma Z, Gibson SL, Byrne MA, Zhang J, White MF, Shaw LM. Suppression of insulin receptor substrate 1 (IRS-1) promotes mammary tumor metastasis. Mol Cell Biol. 2006;26:9338–9351. doi: 10.1128/MCB.01032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi J, Wang DM, Wang CM, Hu Y, Liu AH, Zhang YL, Sun B, Song JG. Insulin receptor substrate-1 suppresses transforming growth factor-beta1-mediated epithelial-mesenchymal transition. Cancer Res. 2009;69:7180–7187. doi: 10.1158/0008-5472.CAN-08-4470. [DOI] [PubMed] [Google Scholar]
- 51.Vasiljeva O, Reinheckel T, Peters C, Turk D, Turk V, Turk B. Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Curr Pharm Des. 2007;13:387–403. doi: 10.2174/138161207780162962. [DOI] [PubMed] [Google Scholar]
- 52.Clark AK, Yip PK, Grist J, Gentry C, Staniland AA, Marchand F, Dehvari M, Wotherspoon G, Winter J, Ullah J, et al. Inhibition of spinal microglial cathepsin S for the reversal of neuropathic pain. Proc Natl Acad Sci USA. 2007;104:10655–10660. doi: 10.1073/pnas.0610811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dennemärker J, Lohmüller T, Müller S, Aguilar SV, Tobin DJ, Peters C, Reinheckel T. Impaired turnover of autophagolysosomes in cathepsin L deficiency. Biol Chem. 2010;391:913–922. doi: 10.1515/bc.2010.097. [DOI] [PubMed] [Google Scholar]
- 54.Pan L, Li Y, Jia L, Qin Y, Qi G, Cheng J, Qi Y, Li H, Du J. Cathepsin S deficiency results in abnormal accumulation of autophagosomes in macrophages and enhances Ang II-induced cardiac inflammation. PLoS One. 2012;7:e35315. doi: 10.1371/journal.pone.0035315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin M, Soikkeli J, Jahkola T, Virolainen S, Saksela O, Hölttä E. TGF-β signaling, activated stromal fibroblasts, and cysteine cathepsins B and L drive the invasive growth of human melanoma cells. Am J Pathol. 2012;181:2202–2216. doi: 10.1016/j.ajpath.2012.08.027. [DOI] [PubMed] [Google Scholar]
- 56.Olson OC, Joyce JA. Cysteine cathepsin proteases: Regulators of cancer progression and therapeutic response. Nat Rev Cancer. 2015;15:712–729. doi: 10.1038/nrc4027. [DOI] [PubMed] [Google Scholar]
- 57.Chen KL, Chang WS, Cheung CH, Lin CC, Huang CC, Yang YN, Kuo CP, Kuo CC, Chang YH, Liu KJ, et al. Targeting cathepsin S induces tumor cell autophagy via the EGFR-ERK signaling pathway. Cancer Lett. 2012;317:89–98. doi: 10.1016/j.canlet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 58.Gormley JA, Hegarty SM, O'Grady A, Stevenson MR, Burden RE, Barrett HL, Scott CJ, Johnston JA, Wilson RH, Kay EW, et al. The role of Cathepsin S as a marker of prognosis and predictor of chemotherapy benefit in adjuvant CRC: A pilot study. Br J Cancer. 2011;105:1487–1494. doi: 10.1038/bjc.2011.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Small DM, Burden RE, Jaworski J, Hegarty SM, Spence S, Burrows JF, McFarlane C, Kissenpfennig A, McCarthy HO, Johnston JA, et al. Cathepsin S from both tumor and tumor-associated cells promote cancer growth and neovascularization. Int J Cancer. 2013;133:2102–2112. doi: 10.1002/ijc.28238. [DOI] [PubMed] [Google Scholar]
- 60.Yang M, Liu J, Shao J, Qin Y, Ji Q, Zhang X, Du J. Cathepsin S-mediated autophagic flux in tumor-associated macrophages accelerate tumor development by promoting M2 polarization. Mol Cancer. 2014;13:43. doi: 10.1186/1476-4598-13-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sage J, Mallèvre F, Barbarin-Costes F, Samsonov SA, Gehrcke JP, Pisabarro MT, Perrier E, Schnebert S, Roget A, Livache T, et al. Binding of chondroitin 4-sulfate to cathepsin S regulates its enzymatic activity. Biochemistry. 2013;52:6487–6498. doi: 10.1021/bi400925g. [DOI] [PubMed] [Google Scholar]
- 62.Shi GP, Webb AC, Foster KE, Knoll JH, Lemere CA, Munger JS, Chapman HA. Human cathepsin S: Chromosomal localization, gene structure, and tissue distribution. J Biol Chem. 1994;269:11530–11536. [PubMed] [Google Scholar]
- 63.Jordans S, Jenko-Kokalj S, Kühl NM, Tedelind S, Sendt W, Brömme D, Turk D, Brix K. Monitoring compartment-specific substrate cleavage by cathepsins B, K, L, and S at physiological pH and redox conditions. BMC Biochem. 2009;10:23. doi: 10.1186/1471-2091-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burden RE, Gormley JA, Kuehn D, Ward C, Kwok HF, Gazdoiu M, McClurg A, Jaquin TJ, Johnston JA, Scott CJ, et al. Inhibition of Cathepsin S by Fsn0503 enhances the efficacy of chemotherapy in colorectal carcinomas. Biochimie. 2012;94:487–493. doi: 10.1016/j.biochi.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 65.Liu WL, Liu D, Cheng K, Liu YJ, Xing S, Chi PD, Liu XH, Xue N, Lai YZ, Guo L, et al. Evaluating the diagnostic and prognostic value of circulating cathepsin S in gastric cancer. Oncotarget. 2016;7:28124–28138. doi: 10.18632/oncotarget.8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lindahl C, Simonsson M, Bergh A, Thysell E, Antti H, Sund M, Wikström P. Increased levels of macrophage-secreted cathepsin S during prostate cancer progression in TRAMP mice and patients. Cancer Genomics Proteomics. 2009;6:149–159. [PubMed] [Google Scholar]
- 67.Wang X, Xiong L, Yu G, Li D, Peng T, Luo D, Xu J. Cathepsin S silencing induces apoptosis of human hepatocellular carcinoma cells. Am J Transl Res. 2015;7:100–110. [PMC free article] [PubMed] [Google Scholar]
- 68.Gole B, Huszthy PC, Popović M, Jeruc J, Ardebili YS, Bjerkvig R, Lah TT. The regulation of cysteine cathepsins and cystatins in human gliomas. Int J Cancer. 2012;131:1779–1789. doi: 10.1002/ijc.27453. [DOI] [PubMed] [Google Scholar]
- 69.Gautam J, Bae YK, Kim JA. Up-regulation of cathepsin S expression by HSP90 and 5-HT7 receptor-dependent serotonin signaling correlates with triple negativity of human breast cancer. Breast Cancer Res Treat. 2017;161:29–40. doi: 10.1007/s10549-016-4027-1. [DOI] [PubMed] [Google Scholar]
- 70.Kondo T, Ezzat S, Asa SL. Pathogenetic mechanisms in thyroid follicular-cell neoplasia. Nat Rev Cancer. 2006;6:292–306. doi: 10.1038/nrc1836. [DOI] [PubMed] [Google Scholar]
- 71.Flannery T, McQuaid S, McGoohan C, McConnell RS, McGregor G, Mirakhur M, Hamilton P, Diamond J, Cran G, Walker B, et al. Cathepsin S expression: An independent prognostic factor in glioblastoma tumours - A pilot study. Int J Cancer. 2006;119:854–860. doi: 10.1002/ijc.21911. [DOI] [PubMed] [Google Scholar]
- 72.Kos J, Sekirnik A, Kopitar G, Cimerman N, Kayser K, Stremmer A, Fiehn W, Werle B. Cathepsin S in tumours, regional lymph nodes and sera of patients with lung cancer: Relation to prognosis. Br J Cancer. 2001;85:1193–1200. doi: 10.1054/bjoc.2001.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.