Abstract
Oral squamous cell carcinoma (OSCC), one of the 10 most common types of neoplasms in the US, constitutes ~90% of all cases of oral malignancies. Chemoresistance and metastasis are difficult to avoid during the course of treatment, leading to a poor prognosis and a high mortality rate for patients with OSCC. Autophagy, a critical conserved cellular process, has been reported to be highly associated with the regulation of chemoresistance and metastasis of cancer cells. The present study investigated the role of karyopherin α2 (KPNA2), a member of the importin α family, which may serve an important role in p53 nucleocytoplasmic transport in the process of OSCC autophagy. In the CAL-27, SCC-15 and Tca8113 OSCC cell lines, we observed that the downregulation of KPNA2 suppressed cell migration and cisplatin resistance, using wound-healing, Transwell and CCK-8 assays. Additionally, the results of western blot analysis and transmission electron microscopy (TEM) analysis indicated that the knockdown of KPNA2 inhibited autophagy. We confirmed that the inhibition of autophagy with anti-autophagy agents decreased the migration and cisplatin resistance of OSCC cells. We hypothesized that the suppression of cell migration and cisplatin resistance induced by KPNA2 knockdown may be associated with the inhibition of autophagy. To identify the underlying mechanism, further experiments determined that KPNA2 affects the level of autophagy via regulating the p53 nuclear import. Thus, the present study demonstrated that the function of KPNA2 in the process of autophagy may be p53-dependent, and by regulating the translocation of p53, KPNA2 can support autophagy to promote the chemoresistance and metastasis of OSCC cells.
Keywords: karyopherin α2, autophagy, oral squamous cell carcinoma, metastasis, chemoresistance
Introduction
Oral squamous cell carcinoma (OSCC) is one of the 10 most common types of neoplasms in the USA (1). OSCC, a major factor of morbidity and mortality among head and neck cancers, constitutes ~90% of all cases of oral malignancies (2). At present, the treatment methods for OSCC, primarily chemotherapy, radiotherapy and surgery, are insufficient to overcome the issues of drug resistance, recurrence and metastasis (3), leading to a poor prognosis and a high mortality rate. Therefore, the investigation of the molecular pathogenesis, including the survival mechanisms of cells under stress, may provide prospective targets for reducing metastasis and resistance to therapy, thereby improving the survival and prognosis of patients with OSCC.
Autophagy, cellular ‘self-eating’, is the process of intracellular lysosomal degradation to recycle proteins and organelles, which is regulated by autophagy-related genes (4). Autophagy is critical to prevent the toxic accumulation of damaged proteins and organelles, and stabilizes the metabolism to maintain energy homeostasis and ensure cell survival (5). Therefore, autophagy is predominantly a pro-survival adaptive response that enables cancer cells to withstand the unfavorable conditions to which they are exposed, such as starvation, ischemia, hypoxia and chemotherapy (6–8). Consequently, autophagy can promote malignant processes after tumorigenesis (7), and facilitate chemotherapy and radiotherapy resistance (8–11). It has been reported that resistant cells can be re-sensitized to chemotherapy drugs by using autophagy inhibitors or affecting the molecular regulators of autophagy (9). The role of autophagy in cancer metastasis is a double-edged sword, as it can promote both pro-metastasis and anti-metastasis processes. The cellular response to autophagy during cancer metastasis is stage-specific (12–14). Autophagy is regarded as a potential target in cancer treatment and may provide a promising therapeutic strategy for overcoming resistance and enhancing the effect of chemotherapy. However, as autophagy is a complex process involving many molecules and pathways, the specific mechanisms and molecules involved remain under continuous research and expansion.
Karyopherin α2 (KPNA2), which is a member of the importin α family, plays an important role in nucleocytoplasmic transport, as previously reported (15–18). KPNA2 may mediate the translocation of cancer-associated functional proteins to affect tumorigenesis (19). Additionally, KPNA2 has been demonstrated to be involved in the translocation of various proteins, including transcription factors or cargo proteins associated with DNA repair and cell-cycle regulation (16). These proteins are involved in a multitude of cellular processes, such as proliferation, apoptosis and metastasis. Recently, the biological function of KPNA2 has been confirmed in oncological clinical studies and cell experiments (20–24). For example, KPNA2 can enhance the migratory ability and viability of breast cancer cells (20,23). In addition, the knockdown of KPNA2 can inhibit the proliferation of cells derived from prostate and ovarian cancer (22,24). Thus, KPNA2 is regarded as a candidate oncogene. However, the role of KPNA2 in the progression of OSCC remains unclear and limited information is available regarding the role of KPNA2 in the process of autophagy. Thus, there are additional molecular mechanisms of KPNA2 that need to be further investigated.
In the present study, we reported that KPNA2 knockdown inhibited the migration, cisplatin resistance and autophagy of OSCC cells, and that the mechanisms involved were associated with the blockade of p53 translocation. Collectively, our findings may provide an original perspective for improving the therapeutic efficacy of OSCC.
Materials and methods
Reagents
Antibodies for LC3B and KPNA2 were purchased from Abcam (Cambridge, UK), autophagy-related gene (Atg)3, Atg5, Atg7, SQSTM1/p62, p53 and β-actin were obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA). Rapamycin, chloroquine phosphate, 3-methyladenine (3-MA) and cisplatin were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). Cell Counting Kit-8 (CCK-8) was acquired from Dojindo Molecular Technologies, Inc. (Kumamoto, Japan). The catalogue numbers of the agents are listed in Table I.
Table I.
The catalog numbers of the agents used in the present study.
| Agent | Source | Catalogue number |
|---|---|---|
| Anti-KPNA2 | Abcam | ab173295 |
| Anti-LC3B | Abcam | ab51520 |
| Anti-Atg3 | CST | 3415T |
| Anti-Atg5 | CST | 12994T |
| Anti-Atg7 | CST | 2631T |
| Anti-SQSTM1/p62 | CST | 5114T |
| Anti-p53 | CST | 2527T |
| Anti-β-actin | CST | 4970T |
| HRP-conjugated goat anti-rabbit IgG | ZSGB-Bio | ZB-2301 |
| TRITC-conjugated goat anti-rabbit IgG | ZSGB-Bio | ZF-0316 |
| Rapamycin | Sigma-Aldrich | V900930-1MG |
| Chloroquine phosphate | Sigma-Aldrich | C6628-25G |
| 3-Methyladenine | Sigma-Aldrich | M9281-100MG |
| Cisplatin | Sigma-Aldrich | P4394-25MG |
| CCK-8 | Dojindo | CK04-500T |
CST, Cell Signaling Technology; HRP, horseradish peroxidase; TRITC, Rhodamine; ZSGB-Bio, Zhongshan Jinqiao Biological Technology, Co., Ltd.
Cell culture
The three OSCC cell lines, CAL-27, SCC-15 and Tca8113, were donated by the Institute of Hard Tissue Development and Regeneration, The Second Affiliated Hospital of Harbin Medical University (Harbin, China). Cells were cultured in RPMI-1640 medium (HyClone Laboratories; GE Healthcare Life Science, Logan, UT, USA) with 10% (v/v) fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 1% (v/v) penicillin-streptomycin solution (Gibco; Thermo Fisher Scientific). Cells were incubated at 37°C with 5% (v/v) CO2 in humidified air.
Lentivirus transfection
KPNA2 shRNA, p53 shRNA and scramble control shRNAs packaged in a lentiviral vector were purchased from Shanghai GeneChem Co., Ltd. (Shanghai, China). The sequences were synthesized and verified by Shanghai GeneChem Co., Ltd. According to the manufacturer's protocol, cells were seeded in a 6-well plate and incubated in infection medium with the lentiviral vectors for 12 h. The medium was then replaced with fresh complete medium. The cells were screened with puromycin (Shanghai GeneChem Co., Ltd.) at a dose of 2 µg/ml after 72 h transfection. The transfection efficiency was confirmed by western blotting. The sequences of the three shRNAs are listed in Table II.
Table II.
The sequences of the three shRNA insert elements.
| shRNA | Sequence |
|---|---|
| KPNA2-shRNA | 5′-CCGGCCGTTGATGAACCTCTTAACTCGAGTTAAGAGGTTCATCAACGGTTTTTG-3′ |
| p53-shRNA | 5′-CCGGCGGCGCACAGAGGAAGAGAATCTCGAGATTCTCTTCCTCTGTGCGCCGTTTTT-3′ |
| Scramble control shRNA | 5′-CCGGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTG-3′ |
CCK-8 detection of cell viability
The three treated OSCC cell lines were seeded in 96-well plates (104 cells/well) and cultured in 100 µl serum-free medium for 24 h. Then, the medium was replaced with 0, 0.01, 0.1, 1, 10 and 100 µg/ml cisplatin diluted in RPMI-1640 medium with 10% FBS (v/v) and cultured for a further 24 h. Subsequently, the cells were incubated with 10% CCK-8 solution for another 2 h, and the absorbance of each well was assessed at 450 nm using a microplate reader (Bio-Rad iMark; Bio-Rad Laboratories, Hercules, CA, USA). Each experiment was repeated three times independently.
Wound-healing assay
The three treated OSCC cell lines were reseeded onto a 6-well plate and cultured to 100% confluence. The cell monolayer was scratched using a pipette tip, and the medium was replaced with serum-free medium. The cells were incubated at 37°C in 5% CO2 for 24 h and representative images of the wound were captured by a digital camera installed on an inverted microscope (Nikon Eclipse Ti-E; Nikon, Kobe, Japan). The scratch area was quantified using ImageJ 1.46r (National Institutes of Health, Bethesda, MA, USA). Each experiment was repeated three times independently.
Transwell migration assay
Migration assays were performed in 6.5 mm Transwell inserts with 8.0-µm pore polycarbonate membranes purchased from Corning Inc. (Corning, NY, USA). A total of 1×105 treated cells/well from each of the three OSCC cell lines were seeded in the upper chambers with 200 µl serum-free medium, while the lower chambers were filled with 500 µl medium supplemented with FBS. Following incubation at 37°C in 5% CO2 for 24 h, the cells were fixed with methanol for 15 min. Cells were removed from the upper chambers gently using cotton swabs, and the membranes were stained with crystal violet (Beyotime Institute of Biotechnology, Shanghai, China). The membranes were photographed and the images were analyzed with ImageJ 1.46r (National Institutes of Health). Each experiment was repeated three times independently.
Western blot analysis
Cells were harvested and lysed in ice-cold buffer, including RIPA (Beyotime Institute of Biotechnology), 1% PMSF (Beyotime Institute of Biotechnology) and a protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany) for 30 min to extract the total protein. The concentration of protein was quantified using a BCA protein assay kit (Beyotime Institute of Biotechnology) and the protein samples were denatured. Nuclear proteins were extracted using a nuclear protein extraction kit (Beyotime Institute of Biotechnology). Protein samples (96 µg/lane) were separated by 10, 12 or 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Beyotime Institute of Biotechnology) and blotted onto polyvinylidene difluoride membranes (Roche Diagnostics GmbH). After blocking with 5% skimmed milk (Beyotime Institute of Biotechnology), the membranes were incubated with the primary antibodies overnight at 4°C. The membranes were incubated with a horseradish peroxidase-conjugated secondary antibody (Zhongshan Jinqiao Biological Technology, Co., Ltd., Beijing, China) for 1.5 h, followed by an electrochemiluminescence reagent (Beyotime Institute of Biotechnology) for 3 min. Images of the bands were captured with SmartChemiI (Beijing Sage Creation Science, Co., Beijing, China) and the gray value was assessed with ImageJ 1.46r (National Institutes of Health). Each experiment was repeated three times independently. The antibodies used for western blot analysis are listed in Table III.
Table III.
The antibodies used in the present study.
| Antibody | Type | Dilution | Source | Application |
|---|---|---|---|---|
| KPNA2 | Rabbit-mAb | 1:1,000 | CST | WB |
| LC3 | Rabbit-mAb | 1:1,000 | Abcam | WB |
| Atg3 | Rabbit-mAb | 1:1,000 | CST | WB |
| Atg5 | Rabbit-mAb | 1:1,000 | CST | WB |
| Atg7 | Rabbit-mAb | 1:1,000 | CST | WB |
| SQSTM1/P62 | Rabbit-mAb | 1:1,000 | CST | WB |
| β-actin | Rabbit-mAb | 1:1,000 | CST | WB |
| P53 | Rabbit-mAb | 1:160 | Abcam | ICC/IF |
| HRP-conjugated goat anti-rabbit IgG | Goat-mAb | 1:20,000 | ZSGB-Bio | WB |
| TRITC-conjugated goat anti-rabbit IgG | Goat-mAb | 1:50 | ZSGB-Bio | ICC/IF |
CST, Cell Signaling Technology; WB, western blot analysis; ICC/IF, immunocytochemistry/immunofluorescence; HRP, horseradish peroxidase; TRITC, Rhodamine; ZSGB-Bio, Zhongshan Jinqiao Biological Technology, Co., Ltd.
Transmission electron microscopy (TEM)
Cells were collected, fixed with 2.5% glutaraldehyde overnight at 4°C and post-fixed with 1% osmium acid for 3 h. The cells were dehydrated in a graded ethanol series (30, 50, 70, 80, 90, 95 and 100%) followed by a treatment with absolute acetone for 20 min after each incubation. The samples were incubated in a 3:1 mixture of absolute acetone and Spurr resin (Shanghai GenMed, Co., Ltd., Shanghai, China) (1 h), a 1:1 mixture of absolute acetone and Spurr resin (1 h), a 1:3 mixture of absolute acetone and Spurr resin (1 h) once again, and a final Spurr resin incubation (4 h). The samples were embedded in Spurr resin at room temperature overnight, then heated to 70°C for 9 h in capsules. Finally, the samples were cut into 70-nm-thick sections using an ultramicrotome (Leica Ultracut R; Leica Microsystems GmbH, Wetzlar, Germany), and the specimens were double-stained with uranyl acetate and lead citrate for 10 min each. The samples were observed and images were captured using a transmission electron microscope (JEM-1230; JEOL, Ltd., Tokyo, Japan).
Immunofluorescence assay
Treated cells were seeded onto glass slides and cultured for 24 h. Then, cells were fixed with 4% paraformaldehyde for 10 min and permeabilized with 0.2% Triton X-100 (Beyotime Institute of Biotechnology) for 5 min on ice. After blocking with 5% normal goat serum (NGS; Wuhan Boster Biological Technology, Co., Ltd., Wuhan, China) for 1 h, slides were incubated with primary antibodies overnight at 4°C, followed by incubation with TRITC-conjugated secondary antibodies (Zhongshan Jinqiao Biological Technology, Co., Ltd., Beijing, China) for 1.5 h in the dark. Subsequent to mounting with DAPI (Beyotime Institute of Biotechnology) for nuclear staining, the slides were covered on microscope slides with anti-quench mount reagents (Beyotime Institute of Biotechnology). The slides were examined and photographed using a fluorescence microscope (Nikon Eclipse Ci-S; Nikon). The antibodies used in these experiments are listed in Table III.
Statistical analysis
The data are presented as the means ± standard deviation (SD). Differences between the groups were examined using a one-way ANOVA in GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Lentiviral transfection of shRNA can effectively knock down the expression of KPNA2 and p53
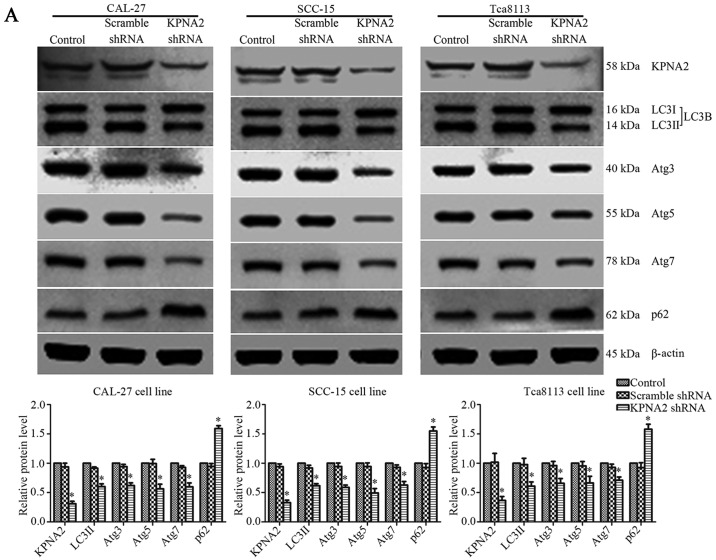
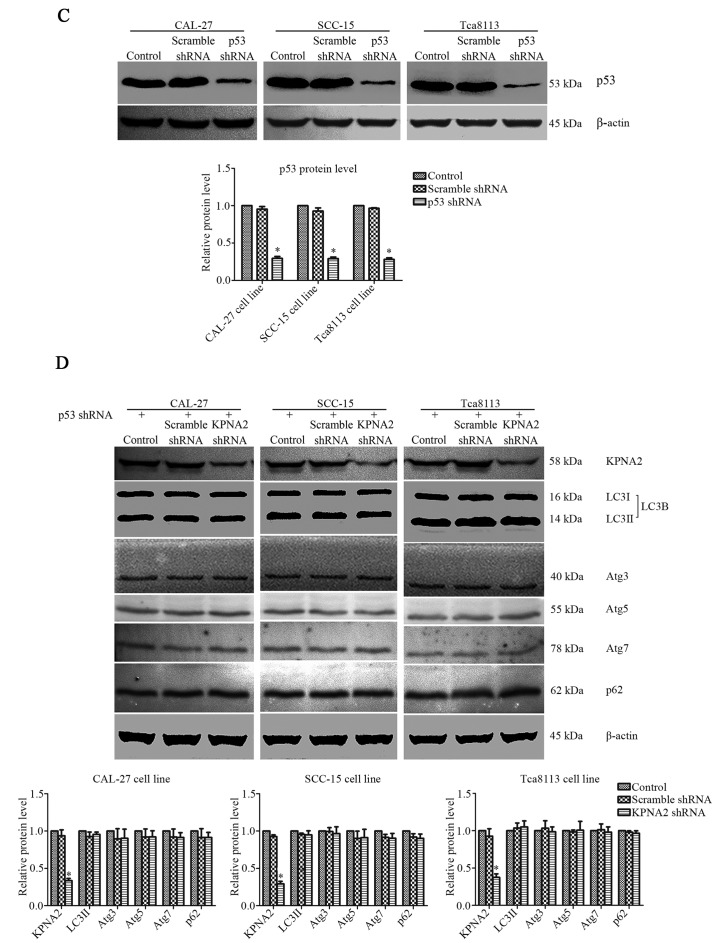
The efficiency of shRNA lentivirus transfection was evaluated at the protein level by western blotting. KPNA2-targeted shRNA significantly downregulated the level of KPNA2 protein expression in CAL-27, SCC-15 and Tca8113 cells by 68.9±4.1, 67.1±4.1 and 63.4±6.7%, respectively (P<0.05, n=3). Additionally, p53-targeted shRNA reduced the level of p53 protein expression in CAL-27, SCC-15 and Tca8113 cells by 70.5±2.6, 70.8±2.0 and 71.8±1.9%, respectively (P<0.05, n=3). Representative bands, and histograms of the relative protein expression level are displayed in Figs. 3A and 6C.
Figure 3.
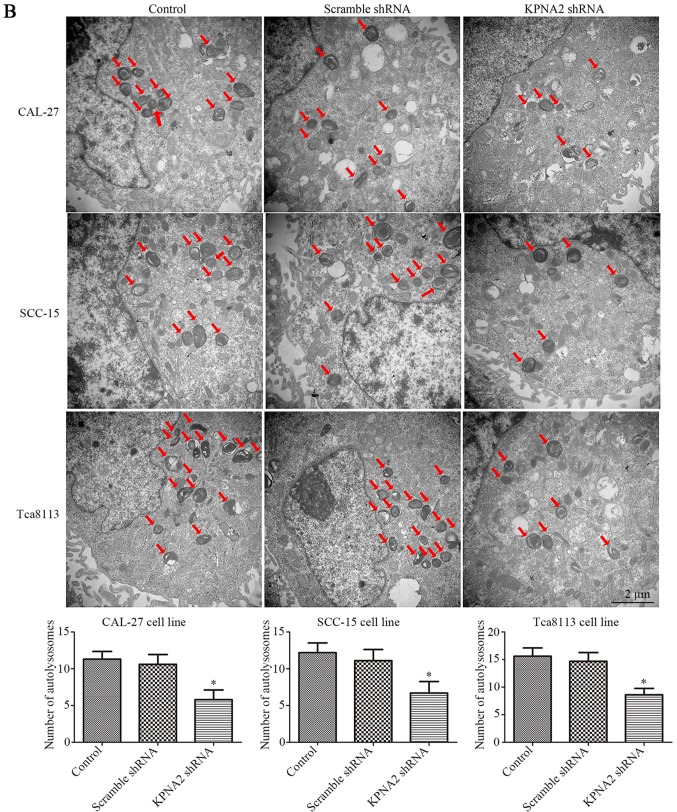
Knockdown of KPNA2 inhibits the autophagy of OSCC cells. (A) Following the establishment of CAL-27, SCC-15 and Tca8113 cells exhibiting KPNA2 knockdown, autophagy-related protein expression levels of each group were assessed by western blotting. Densitometry analysis of the western blots is also shown. *P<0.05 compared with the control. Data are presented as the mean ± standard deviation (n=3). (B) TEM images of CAL-27, SCC-15 and Tca8113 cells following KPNA2 knockdown. Red arrows indicate formation of autolysosomes. The number of autolysosomes counted in each view are presented in histograms. *P<0.05 compared with the control group. Data are presented as the mean ± standard deviation (n=10 views of each group).
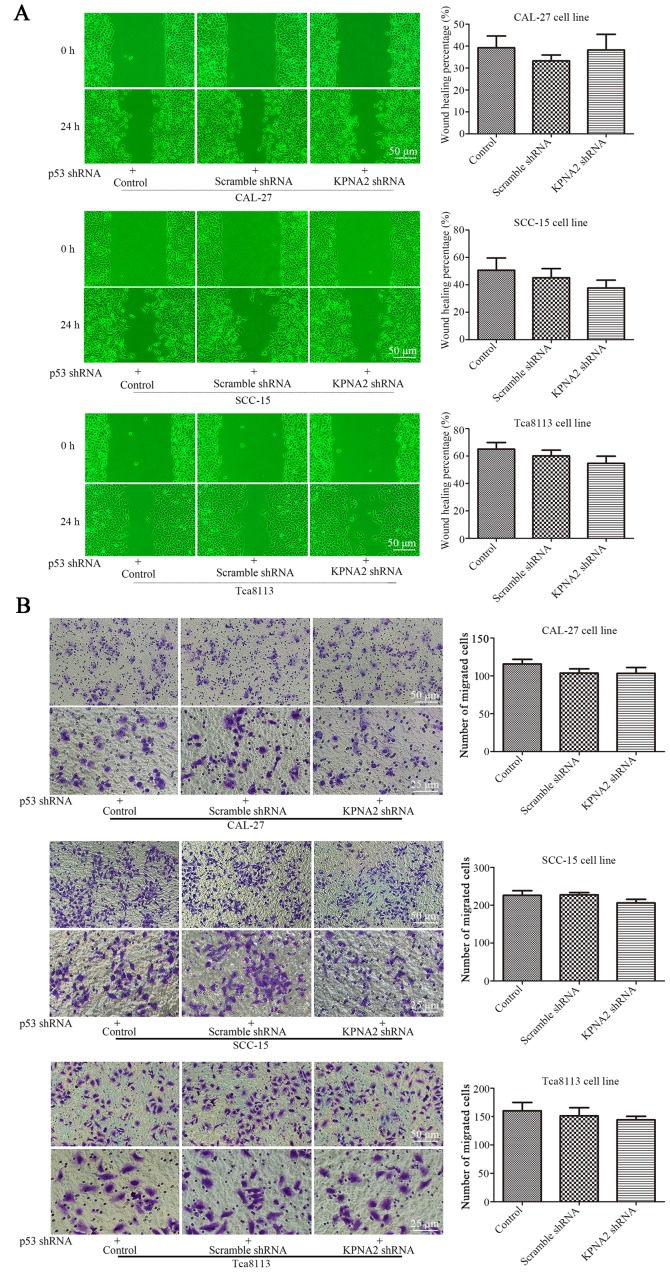
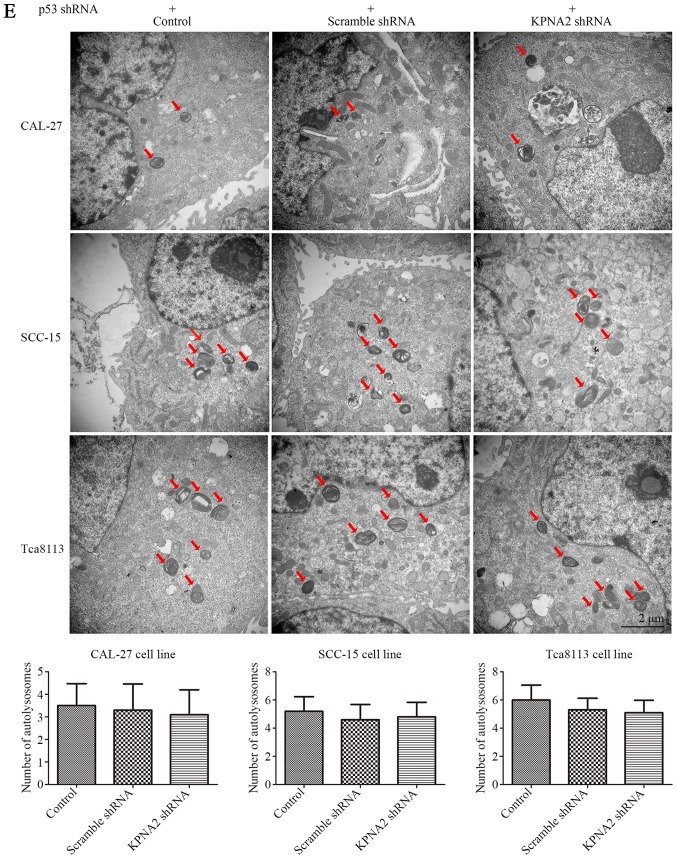
Figure 6.
KPNA2 knockdown does not inhibit autophagy in p53-knockdown cells. The expression of p53 in all groups of cells was downregulated by p53 shRNA. (A) Wound-healing assay of CAL-27, SCC-15 and Tca8113 cells in the presence or absence of KPNA2 shRNA, and quantification of the wound closure percentage for each group (n=3). (B) Confirmation of the migration ability of cells in each group via a Transwell assay; quantification of the assay is shown in histograms (n=3). (C) The level of p53 protein levels of each group were assessed by western blotting to confirm the effectiveness of the knockdown. (D) KPNA2 shRNA and scramble control shRNA were transfected into p53-knockdown CAL-27, SCC-15 and Tca8113 cells, and the levels of KPNA2 and autophagy-related protein expression were detected by western blot analysis. Densitometry analysis of the western blots was displayed in histograms (n=3). (E) TEM images of p53-knockdown CAL-27, SCC-15 and Tca8113 cells treated with or without KPNA2 shRNA, and the formation of autolysosomes indicated by red arrows. The histogram represents the number of autolysosomes counted in 10 views of each group. *P<0.05 compared with the control, and error bars represent the standard deviation.
Knockdown of KPNA2 inhibits the migration of OSCC cells
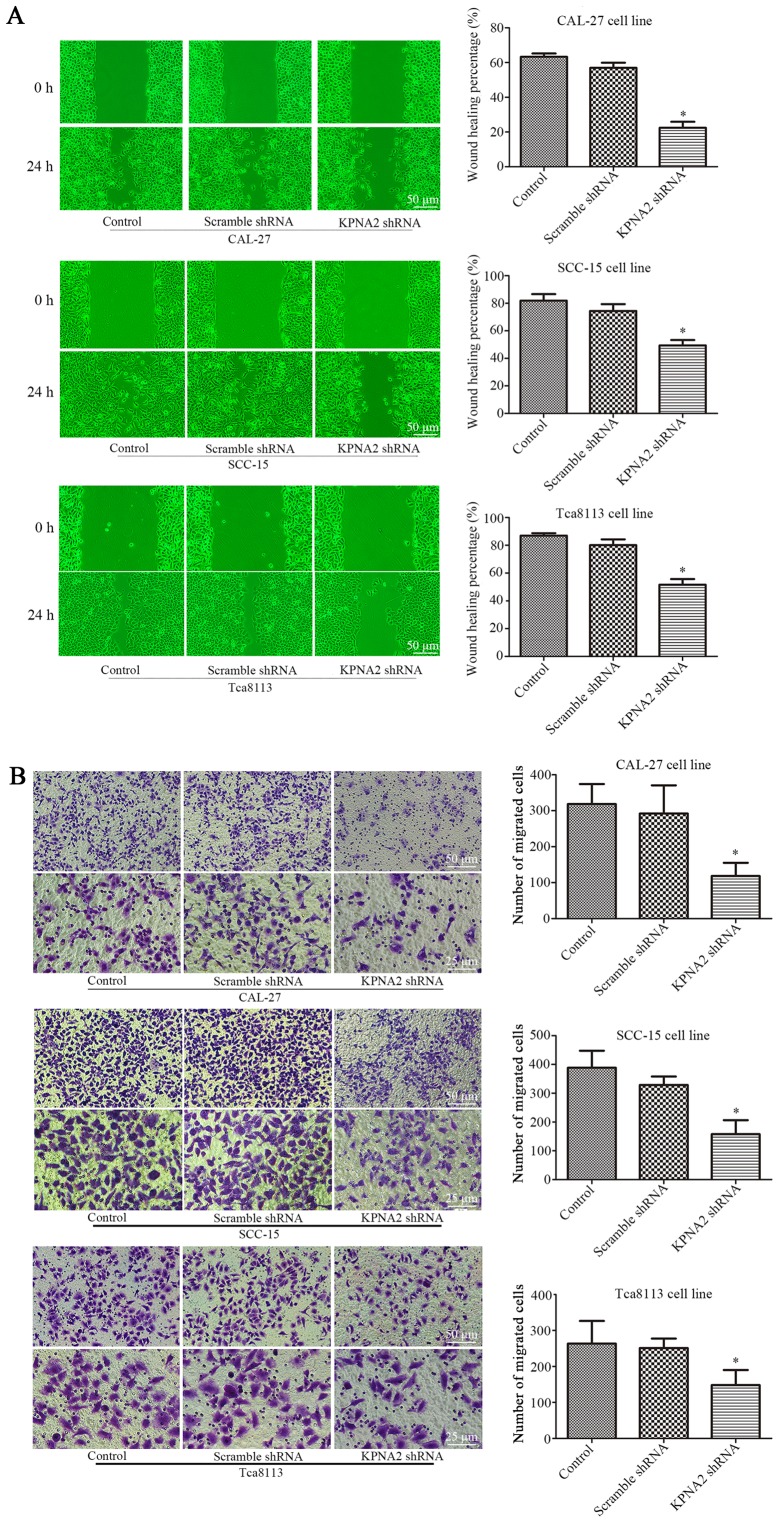
To determine the role of KPNA2 in the migration of autophagic cells, KPNA2 was downregulated in the three OSCC cell lines. Wound healing and Transwell assays were performed for each group of cells treated with 100 nM rapamycin for 24 h (25,26). As displayed in Fig. 1A the wound healing percentage was reduced from 63.3±1.9 to 22.4±3.5%, from 81.9±4.8 to 49.4±4.0% and from 87.0±1.7 to 51.7±4.1% in the KPNA2-knockdown cells at 24 h, in CAL-27, SCC-15 and Tca8113 cells, respectively, compared with the corresponding control groups (P<0.05, n=3). Furthermore, as demonstrated in Fig. 1B, the knockdown of KPNA2 reduced the number of migrating cells by 63.8, 59.4 and 43.8% in CAL-27, SCC-15 and Tca8113 cells, respectively, consistent with the results of the wound healing assay (P<0.05, n=3). These data demonstrated that the knockdown of KPNA2 significantly inhibited cell migration in the OSCC cell lines.
Figure 1.
KPNA2 knockdown inhibits the migration of OSCC cells. CAL-27, SCC-15 and Tca8113 cells were transfected with KPNA2 shRNA. (A) Wound-healing assay of CAL-27, SCC-15 and Tca8113 cells, and quantification of the wound closure percentage. (B) The number of migrating cells in Transwell assays. *P<0.05 compared with the control group. Data are expressed as the mean ± standard deviation of three independent experiments for each group.
Knockdown of KPNA2 reduces the cisplatin resistance of OSCC cells
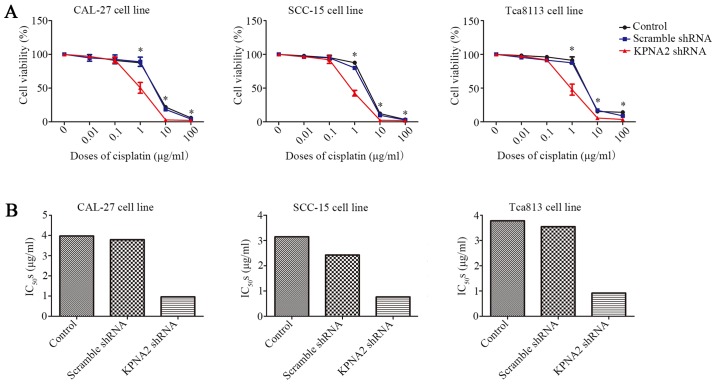
In addition to cell migration, we also examined the effect of KPNA2 on the resistance to cisplatin using a CCK-8 assay. As displayed in Fig. 2A KPNA2 knockdown significantly decreased cell viability in a series of cisplatin concentrations, particularly 1 µg/ml, whereas transfection with the scramble control shRNA had little effect on cell vitality compared with untransfected cells. Subsequently, IC50 for each group was calculated, as presented in Fig. 2B and Table IV. KPNA2 knockdown reduced the IC50 from 3.97 to 0.96 µg/ml in CAL-27 cells, from 3.14 to 0.76 µg/ml in SCC-15 cells and from 3.78 to 0.92 µg/ml in Tca8113 cells. These data revealed that KPNA2 knockdown significantly reduced cisplatin resistance in the OSCC cell lines.
Figure 2.
Knockdown of KPNA2 reduces the cisplatin resistance of OSCC cells. (A) Relative cell viability of each group as determined by a CCK-8 assay in a graded concentration of cisplatin. (B) IC50s for each group of cells. *P<0.05 vs. the control group. Data are expressed as the mean ± standard deviation of three independent experiments for each group.
Table IV.
IC50s of KPNA2-knockdown cells.
| Control | Scramble shRNA | KPNA2 shRNA | |
|---|---|---|---|
| CAL-27 | 3.971 | 3.791 | 0.9596 |
| SCC-15 | 3.144 | 2.426 | 0.7639 |
| Tca8113 | 3.781 | 3.549 | 0.9181 |
IC50, the half maximal inhibitory concentration.
KPNA2 knockdown inhibits autophagy in OSCC cells
The effect of KPNA2 on autophagy remains unclear. Following KPNA2 knockdown in three OSCC cell lines, western blotting and TEM were used to detect the extent of autophagy induced by treatment with rapamycin in each group. Representative western blotting bands and gray values for autophagy-related proteins are displayed in Fig. 3A. KPNA2 knockdown induced a reduction in the relative level of LC3II by 39.7±4.7, 38.3±3.4, and 39.0±7.1%; in Atg3 by 37.9±4.4, 41.1±3.8 and 34.0±8.0%; in Atg5 by 43.5±7.8, 50.5±7.4 and 33.6±11.4%; and in Atg7 by 40.3±6.7, 37.1±6.0 and 29.0±5.5%, and reduced the extent of SQSTM1/p62 degradation in CAL-27, SCC-15 and Tca8113 cells (n=3). The relative level of SQSTM1/p62 protein in the control group cells was 159.3±4.8, 155.1±6.8 and 158.1±8.5% of that in the KPNA2-knockdown cells in the three OSCC cell lines (P<0.05, n=3). The results of western blot analysis revealed that KPNA2 knockdown induced the suppression of autophagy in the three OSCC cell lines at the protein level.
In order to further investigate autophagy at the cellular microstructure level, TEM was performed for each group of cells. The results from TEM are displayed in Fig. 3B. As identified with TEM, the cells with KPNA2 knockdown displayed a decreased number of autolysosomes in the cytoplasm, the characteristic vacuole-like structures indicated by red arrows in Fig. 3B. The results of TEM demonstrated that KPNA2 knockdown suppressed autophagy in the three OSCC cell lines at a microstructure level, consistent with the results of western blot analysis. These data demonstrated that KPNA2 knockdown inhibited the process of autophagy in OSCC cells.
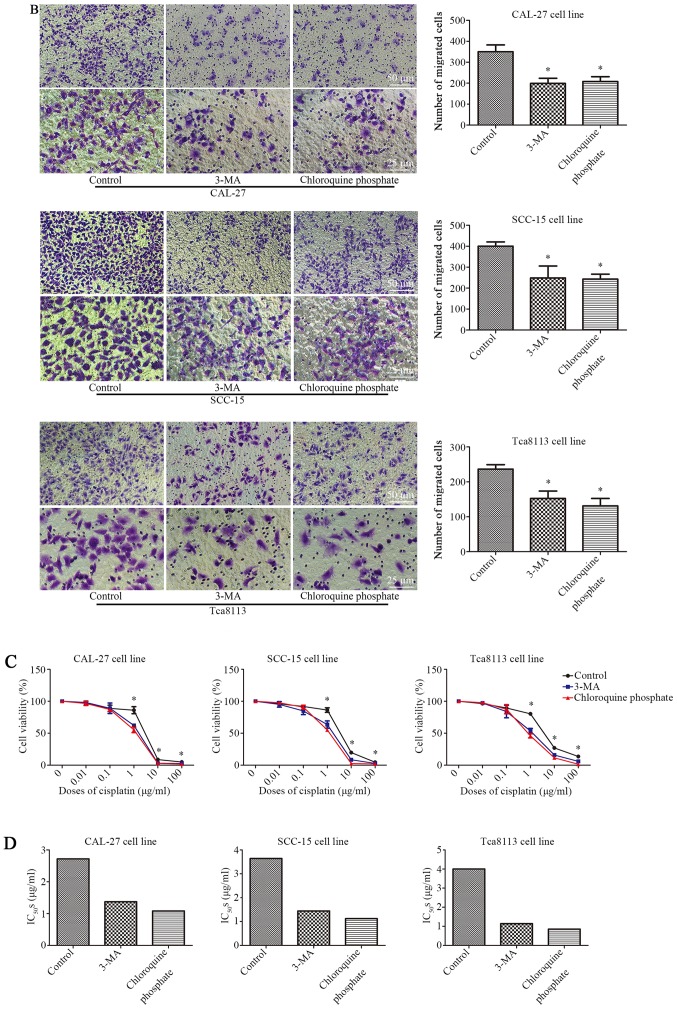
Inhibition of autophagy decreases the migration and cisplatin resistance of OSCC cells
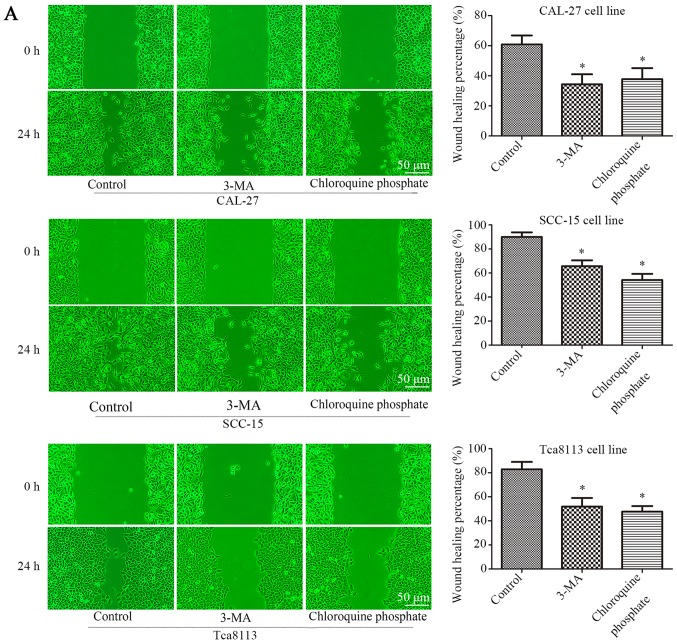
The three cell lines were treated with 100 nM rapamycin for 24 h. When 3-MA (2 mM) (27) or chloroquine phosphate (10 µM) (28) were used in combination with rapamycin, the rate of cell migration was significantly reduced. As displayed in Fig. 4A, 3-MA or chloroquine phosphate treatment reduced the rate of wound closure. Wound-healing was reduced from 60.8±6.0 to 34.36±6.7% (3-MA) or 37.8±7.4 (chloroquine phosphate) in CAL-27 cells, 89.9±3.9 to 65.6±5.0% (3-MA) or 54.2±5.0% (chloroquine phosphate) in SCC-15 cells, and 82.8±6.2 to 51.9±7.2% (3-MA) or 47.7±4.6% (chloroquine phosphate) in Tca8113 cells (P<0.05, n=3). Fig. 4B presents the results of a Transwell assay, in which 3-MA or chloroquine phosphate treatment reduced the number of migrating cells by ~43.1% (3-MA) and 40.5% (chloroquine phosphate), 37.8% (3-MA) and 39.2% (chloroquine phosphate), 35.5% (3-MA) and 44.6% (chloroquine phosphate) in the three OSCC cell lines (P<0.05, n=3). These data demonstrated that the inhibition of autophagy can decrease the rate of cell migration in OSCC cells. Subsequently, whether the inhibition of autophagy affected the cisplatin resistance of OSCC cells was further evaluated with a CCK-8 assay. As displayed in Fig. 4C 3-MA or chloroquine phosphate treatment decreased the cell viability of the three cell lines in combination with a range of cisplatin concentrations, particularly 1 µg/ml, according to the results of the CCK-8 assay. Additionally, the IC50 was reduced from 2.72 to 1.37 µg/ml (3-MA) and 1.08 µg/ml (chloroquine phosphate) in CAL-27 cells, 3.64 to 1.44 µg/ml (3-MA) and 1.12 µg/ml (chloroquine phosphate) in SCC-15 cells, and 4.00 to 1.14 µg/ml (3-MA) and 0.85 µg/ml (chloroquine phosphate) in Tca8113 cells, as displayed in Fig. 4D and Table V. These data demonstrated that the inhibition of autophagy decreased cisplatin resistance of OSCC cells.
Figure 4.
Inhibition of autophagy decreases the migration and cisplatin resistance of OSCC cells. (A) Wound-healing assay of CAL-27, SCC-15 and Tca8113 cells in the presence or absence of autophagy inhibitors, and quantification of the wound closure percentage in each group of cells. (B) Transwell assays were conducted to evaluate the migration capacity in CAL-27, SCC-15 and Tca8113 cells treated with or without autophagy inhibitors. Histograms represent the number of migrating cells from 3 independent experiments. (C) Relative cell viability detected via a CCK-8 assay for cells treated with or without anti-autophagy agents in a graded concentration of cisplatin. (D) The IC50s of each group from (C) were calculated and are displayed in histograms. *P<0.05 compared with the control. Data are presented as the mean and error bars represent the standard deviation (n=3 for each experiment).
Table V.
IC50s of cells treated with 3-MA or chloroquine phosphate.
| Control | 3-MA | Chloroquine phosphate | |
|---|---|---|---|
| CAL-27 | 2.718 | 1.371 | 1.081 |
| SCC-15 | 3.642 | 1.443 | 1.121 |
| Tca8113 | 4.001 | 1.14 | 0.8534 |
IC50, the half maximal inhibitory concentration.
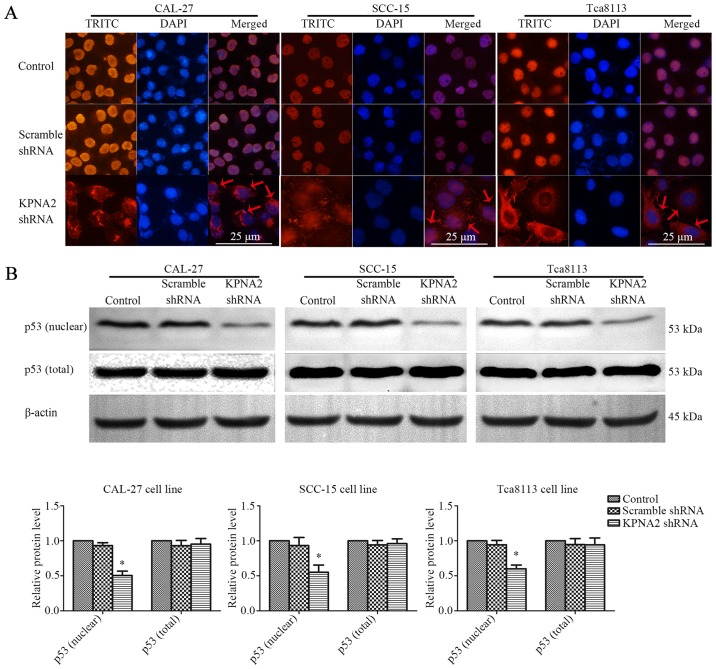
KPNA2 knockdown inhibits p53 nuclear translocation
KPNA2 plays an important role in the nuclear import of transcription factors, including p53 (15). The interaction between KPNA2 and p53 was assessed by an immunofluorescence assay and western blot analysis of nuclear or total p53 protein in the three OSCC cell lines. The results of immunofluorescence are displayed in Fig. 5A. p53 protein localized to the nucleus of the control cells, whereas KPNA2 knockdown caused the accumulation of p53 protein in the cytoplasm. As displayed in Fig. 5B, KPNA2 knockdown only marginally affected the total p53 protein level. However, KPNA2 knockdown reduced the level of nuclear p53 by 49.6±6.3, 44.9±10.3 and 40.1±5.5% in the three OSCC cell lines (P<0.05, n=3). These data demonstrated that KPNA2 knockdown interrupted p53 nuclear import in OSCC cells.
Figure 5.
Knockdown of KPNA2 inhibits p53 nuclear translocation. (A) Representative immunofluorescence images of p53 in CAL-27, SCC-15 and Tca8113 cells following the knockdown of KPNA2. Immunofluorescence analysis was conducted to determine the localization of p53. (B) The level of p53 protein in the nucleus and in total were analyzed by western blotting. *P<0.05 compared with the control, error bars represent the standard deviation (n=3).
KPNA2 knockdown does not inhibit autophagy in p53-knockdown cells
To further validate the roles of KPNA2 and p53 in the process of autophagy, p53 was knocked down in the three OSCC cell lines. Cell migration, cisplatin resistance and the level of autophagy were detected in the p53− OSCC cells with or without KPNA2 knockdown. As displayed in Fig. 6A, despite the downregulation of KPNA2, there was no significant difference in the wound closure among the groups of cells in the three p53− OSCC cell lines (P>0.05, n=3). The results were confirmed by a Transwell assay as displayed in Fig. 6B, indicating that the number of migrating cells in the KPNA2-knockdown groups did not differ from that in the control groups for the three p53− OSCC cell lines. These data demonstrated that KPNA2 knockdown failed to inhibit cell migration in the p53-knockdown cells of the three OSCC cell lines (P>0.05, n=3). Furthermore, in Fig. 6D, a western blot analysis for autophagy-related proteins including LC3I/II, Atg3, Atg5, Atg7 and SQSTM1/p62 identified no difference among the groups for the three p53− OSCC cell lines (P>0.05, n=3). The results were confirmed by TEM, as displayed in Fig. 6E, indicating that the number of autolysosomes was similar among the groups in the three p53− OSCC cell lines (P>0.05, n=3). These data confirmed that KPNA2 knockdown failed to inhibit autophagy in p53-knockdown cells of the three OSCC cell lines. Collectively, it was demonstrated that the KPNA2 knockdown-induced inhibition of autophagy and suppression of cell migration are both p53-dependent.
In conclusion, these data demonstrated that KPNA2 plays an important role in regulating cell migration and cisplatin resistance by altering the level of autophagy in OSCC cells by mediating p53 nucleocytoplasmic translocation.
Discussion
In the present study, the role of KPNA2 knockdown in the migration, cisplatin resistance and autophagy of OSCC cells was examined in vitro. It was identified that the knockdown of KPNA2 inhibited cell migration and autophagy, and decreased cisplatin resistance through the inhibition of p53 nucleocytoplasmic translocation. Therefore, this study may present a novel insight into the clinical role of KPNA2 in the treatment of OSCC.
According to recent research, KPNA2 has been regarded as a potential diagnostic biomarker in multiple cancers, and the aberrant overexpression of KPNA2 in tumor tissue is frequently associated with a poor prognosis or adverse outcome, as reviewed by Christiansen and Dyrskjøt (16). This has been confirmed in clinical trials with various types of malignant tumors, including lung, breast, hepatocellular, gastric, colorectal, ovarian and prostate carcinoma (19,29–32). However, the effect of KPNA2 on the oncogenesis and progression of OSCC remains unclear. Previous studies focused more on the pro-survival and pro-proliferation functions of KPNA2, whereas the present study investigated the impact of KPNA2 knockdown on the metastasis and cisplatin resistance of OSCC cells. The knockdown of KPNA2, a nuclear transport protein regulating the translocation of target proteins, was expected to alter the nuclear levels of several proteins implicated in the processes of DNA repair, cell cycle and transcriptional regulation of a range of target genes in cancer cells (16). Additionally, it has been confirmed in several types of cancer cell lines that KPNA2 knockdown decreased proliferation and increased apoptosis and cell mobility, and that increasing the level of KPNA2 may enhance proliferation (18,22,23,33,34). However, the role of KPNA2 in the process of autophagy is unclear and remains to be further studied.
In the present study, western blot analysis of autophagy-related proteins and TEM of cellular microstructure revealed that KPNA2 knockdown significantly reduced the level of autophagy in OSCC cells. It is established that autophagy is a protective cellular process that occurs frequently in oncogenesis and chemotherapy. Generally, autophagy supports the survival of cancer cells during the stress of anticancer therapies, such as chemotherapy, radiotherapy or targeted agents, thus promoting resistance (35–37). A range of evidence indicates that chemotherapy resistance develops following autophagy. Increased levels of autophagy after chemotherapy have been detected in patients with a poor prognosis, indicating that autophagy may enhance chemoresistance (38). Previous research has indicated that autophagy induction by chemotherapy may promote the resistance of cancer cells to a range of anti-neoplastic drugs, including paclitaxel, tamoxifen and epirubicin (39–41). As autophagy degrades various types of substrates into small molecules for reuse by the cell, autophagy has the potential to provide fuel for almost all aspects of central carbon metabolism (42–44). In brief, the mechanism for autophagy participation in chemotherapy resistance may be that autophagy supplies a multitude of metabolic and biosynthetic pathways, providing tremendous metabolic plasticity to tumor cells and thus ensuring their survival under chemotherapeutic stress. In addition, there is a wealth of preclinical evidence supporting the notion that the inhibition of autophagy can improve clinical outcomes in cancer treatment, especially in solid tumors (45,46). Thus, there is sufficient evidence to suggest that the inhibition of autophagy can reduce the resistance to chemotherapy. The results of the present study confirmed that inhibiting autophagy with anti-autophagy agents, such as 3-MA and chloroquine phosphate, significantly decreased the cisplatin resistance of OSCC cells. Furthermore, the results of the present study demonstrated that KPNA2 knockdown reduced the cisplatin resistance of OSCC cells. In the cells transfected with KPNA2 shRNA, the viability of the cisplatin-treated group was reduced at all concentrations, and the cisplatin IC50 was significantly decreased. Given that KPNA2 knockdown inhibits autophagy, as determined in the present study, it is reasonable to conclude that the reduction of cisplatin resistance by KPNA2 knockdown may be associated with the inhibition of autophagy.
As aforementioned, the role of autophagy in metastasis is controversial. In previous studies, evidence has suggested that the role of autophagy in metastasis can be both facilitative and inhibitive, and that the role of autophagy in metastasis depends on the context and stage (12–14). The anti-metastatic function of autophagy may be related to the inhibition of necrosis and the mediation of autophagic cell death. In addition, autophagy promotes metastasis by enhancing the adaptability of cancer cells in response to stress (47). According to previous research, autophagy protects cancer cells from anoikis or apoptosis, promotes the dormancy of cancer cell to avoid detection or destruction by the immune system and promotes the survival of cancer stem cells (47–51). As previously described, autophagy can also promote cancer metastasis depending on the context, therefore an inhibitor of autophagy can simultaneously inhibit metastasis and increase the cytotoxicity of the anti-metastatic agents, and efficiently block tumor invasion and metastasis (52–54). Additionally, according to the present study, cell migration was reduced in OSCC cells treated with anti-autophagy agents, including 3-MA or chloroquine phosphate, which was consistent with previous research that was referred above. Furthermorre, the present study revealed anti-metastatic effects of KPNA2 knockdown on OSCC cells. We identified that OSCC cells transfected with KPNA2 shRNA exhibited reduced cell migration, using wound healing and Transwell assays. Since KPNA2 knockdown has been demonstrated to suppress autophagy in the present study, it can be initially suggested that the anti-metastatic function of KPNA2 knockdown may be mediated by autophagy inhibition. Furthermore, substantial evidence has indicated that the level of KPNA2 itself affects the rate of cell migration, which has been confirmed in several cancer cell lines (19,31,34,55,56). This is dependent on the nuclear transport function of KPNA2, as several KPNA2-regulating proteins are associated with the movements of cellular components and cell migration. However, the relationship between KPNA2 knockdown-induced decrease in cell migration and the inhibition of autophagy is still unverified and remains to be further investigated. Although the results of the present study indicated that KPNA2 plays a role in cancer metastasis, cisplatin resistance and autophagy, the underlying mechanisms for the exact function of KPNA2 still require further research.
In the present study, in order to understand the molecular mechanisms underlying the role of KPNA2 knockdown in autophagy and metastasis, further experiments considered the effect on p53 which plays a critical role in autophagy. Not only the regulatory functions of p53 itself, but also the subcellular localization of p53 are particularly important in the process of autophagy. In response to the stressors that cells are faced with, such as metabolic or oxidative stress, and DNA damage, p53 is activated to regulate the transcription of genes, or act through a non-transcriptional mechanism, in order to either assist in stress adaptation or to remove irreparable cells through mediating apoptosis or senescence. One of the components of the transcriptional responses mediated by the p53 gene is the activation of autophagy (57). It has been reported that p53 regulates autophagy depending on the context. The transcriptional mechanism of p53 in regulating autophagy is widely recognized. According to previous research, the expression of several target genes induced by p53 were involved in autophagy, including DRAM, ISG20L1, Ei24, Bax and PUMA. Negatively regulating the mTOR signaling pathway is another mechanism through which p53 promotes autophagy (58). Whether p53 is localized in the nucleus or cytoplasm plays a particular role in the regulation of autophagy. There is evidence that p53 in the nucleus can facilitate autophagy in a transcription-dependent or -independent manner, whereas p53 in the cytoplasm inhibits autophagy induction; this has been confirmed in glioblastoma and colon cancer cell lines (59–61). As the effect of p53 on autophagy depends on its localization, the nuclear import and export of p53 are tightly regulated. A component of the nuclear transporter mediating the import of p53 is the importin-α family (62). KPNA2, a member of the importin-α family, is involved in the nucleocytoplasmic transport of a number of critical transcription factors, such as p53, E2F1 and PLAG1 (16). In the present study, an immunofluorescence analysis of OSCC cells with KPNA2 knockdown was conducted. The results confirmed the p53-importing function of KPNA2. Furthermore, there is evidence that the truncated form of importin-α can impair the nuclear import of p53 to result in the cytoplasmic accumulation of p53 (20,63–66). Another previous study demonstrated that the level of KPNA2 affected not only p53, but also the downstream targets of p53 (19). The present study has revealed that KPNA2 knockdown may suppress autophagy via the disruption of p53 nuclear import in OSCC cells, and these results are consistent with those of previous studies. Therefore, we hypothesized that the reduction of cell migration induced by KPNA2 knockdown may be partly associated with the autophagy inhibition caused by the interruption of p53 nuclear translocation. Furthermore, it has been confirmed in recent research that blocking the intra-nuclear transport of p53 suppresses autophagy, resulting in a reduction in epithelial-mesenchymal transition-related migration and invasion (59). Furthermore, the present study also illustrated that KNPA2 knockdown cannot affect the level of autophagy and the cell migration ability following p53 knockdown. The results of the present study confirmed that the autophagy inhibition and cell migration reduction induced by KPNA2 knockdown was p53-dependent, and that occured through the mechanism of p53 translocation blockade.
In conclusion, the present study demonstrated that the migration and cisplatin resistance of OSCC cells can be influenced by the level of KPNA2. By hindering the nuclear import of p53, the downregulation of KPNA2, a component of the key import proteins, can interfere with the process of autophagy, resulting in a reduction in cisplatin resistance and cell migration, inducing a significant decrease in the malignant characteristics of OSCC cells. The results of the present study indicated that KPNA2, a widely recognized potential marker of prognosis and therapeutic sensitivity, can be regarded as a therapeutic target aimed at suppressing metastasis and chemoresistance in OSCC that is worth studying for therapeutic interventions. However, the potential participation of unknown factors or pathways as underlying mechanisms of anti-autophagy, anti-metastasis and chemotherapy resensitization effects of KPNA2 knockdown in OSCC cells requires further investigation.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- OSCC
oral squamous cell carcinoma
- KPNA2
karyopherin alpha 2
- 3-MA
3-methyladenine
- CCK-8
Cell Counting Kit-8
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- PVDF
polyvinylidene difluoride
- ECL
electrochemiluminescence
- TEM
transmission electron microscopy
- IC50
the half maximal inhibitory concentration
Funding
The present study was partly funded by the National Natural Science Foundation of China (grant nos. 81570951 and 81500816), the Postgraduate Research Innovation Fund of Harbin Medical University (grant no. YJSCX2016-49HYD), the Special Foundation for Sino-Russian Translational Medicine Research Center of Harbin Medical University (grant nos. CR201412 and CR201504), the Natural Science Foundation of Heilongjiang Province of China (grant no. H2015103) and the Science Foundation of the Second Affiliated Hospital of Harbin Medical University (grant no. CX2016-20).
Availability of data and materials
All data generated or analyzed during this study are included in this published article. Still, more details about the datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
FL and LG conceived and designed the study. FL, ZS and YZ performed the experiments. XC, YL and BZ were also involved in the conception of the study. FL wrote the paper. FL, XC, YL and BZ reviewed and edited the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors state that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Sarode GS, Sarode SC, Maniyar N, Anand R, Patil S. Oral cancer databases: A comprehensive review. J Oral Pathol Med. 2017 doi: 10.1111/jop.12667. [DOI] [PubMed] [Google Scholar]
- 3.Gharat SA, Momin M, Bhavsar C. Oral squamous cell carcinoma: Current treatment strategies and nanotechnology-based approaches for prevention and therapy. Crit Rev Ther Drug Carrier Syst. 2016;33:363–400. doi: 10.1615/CritRevTherDrugCarrierSyst.2016016272. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 5.White E, Mehnert JM, Chan CS. Autophagy, metabolism, and cancer. Clin Cancer Res. 2015;21:5037–5046. doi: 10.1158/1078-0432.CCR-15-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorin S, Hamai A, Mehrpour M, Codogno P. Autophagy regulation and its role in cancer. Semin Cancer Biol. 2013;23:361–379. doi: 10.1016/j.semcancer.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Jiang X, Overholtzer M, Thompson CB. Autophagy in cellular metabolism and cancer. J Clin Invest. 2015;125:47–54. doi: 10.1172/JCI73942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villar VH, Merhi F, Djavaheri-Mergny M, Duran RV. Glutaminolysis and autophagy in cancer. Autophagy. 2015;11:1198–1208. doi: 10.1080/15548627.2015.1053680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar A, Singh UK, Chaudhary A. Targeting autophagy to overcome drug resistance in cancer therapy. Future Med Chem. 2015;7:1535–1542. doi: 10.4155/fmc.15.88. [DOI] [PubMed] [Google Scholar]
- 10.Huang Z, Zhou L, Chen Z, Nice EC, Huang C. Stress management by autophagy: Implications for chemoresistance. Int J Cancer. 2016;139:23–32. doi: 10.1002/ijc.29990. [DOI] [PubMed] [Google Scholar]
- 11.Gomes LR, Vessoni AT, Menck CFM. Microenvironment and autophagy cross-talk: Implications in cancer therapy. Pharmacol Res. 2016;107:300–307. doi: 10.1016/j.phrs.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 12.Su Z, Yang Z, Xu Y, Chen Y, Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol Cancer. 2015;14:48. doi: 10.1186/s12943-015-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mowers EE, Sharifi MN, Macleod KF. Autophagy in cancer metastasis. Oncogene. 2017;36:1619–1630. doi: 10.1038/onc.2016.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcucci F, Ghezzi P, Rumio C. The role of autophagy in the cross-talk between epithelial-mesenchymal transitioned tumor cells and cancer stem-like cells. Mol Cancer. 2017;16:3. doi: 10.1186/s12943-016-0573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yasuhara N, Kumar PK. Aptamers that bind specifically to human KPNA2 (importin-α1) and efficiently interfere with nuclear transport. J Biochem. 2016;160:259–268. doi: 10.1093/jb/mvw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christiansen A, Dyrskjøt L. The functional role of the novel biomarker karyopherin α 2 (KPNA2) in cancer. Cancer Lett. 2013;331:18–23. doi: 10.1016/j.canlet.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teng SC, Wu KJ, Tseng SF, Wong CW, Kao L. Importin KPNA2, NBS1, DNA repair and tumorigenesis. J Mol Histol. 2006;37:293–299. doi: 10.1007/s10735-006-9032-y. [DOI] [PubMed] [Google Scholar]
- 18.Gao Li, Yu L, Li CM, Li Y, Jia BL, Zhang B. Karyopherin α2 induces apoptosis in tongue squamous cell carcinoma CAL-27 cells through the p53 pathway. Oncol Rep. 2016;35:3357–3362. doi: 10.3892/or.2016.4750. [DOI] [PubMed] [Google Scholar]
- 19.Wang CI, Chien KY, Wang CL, Liu HP, Cheng CC, Chang YS, Yu JS, Yu CJ. Quantitative proteomics reveals regulation of karyopherin subunit alpha-2 (KPNA2) and its potential novel cargo proteins in nonsmall cell lung cancer. Mol Cell Proteomics. 2012;11:1105–1122. doi: 10.1074/mcp.M111.016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gluz O, Wild P, Meiler R, Frick M, Ting E, Mohrmann S, Schuett G, Dahl E, Fuchs T, Herr A, et al. Nuclear karyopherin alpha2 expression predicts poor survival in patients with advanced breast cancer irrespective of treatment intensity. Int J Cancer. 2008;123:1433–1438. doi: 10.1002/ijc.23628. [DOI] [PubMed] [Google Scholar]
- 21.Sakai M, Sohda M, Miyazaki T, Suzuki S, Sano A, Tanaka N, Inose T, Nakajima M, Kato H, Kuwano H. Significance of karyopherin-{alpha} 2 (KPNA2) expression in esophageal squamous cell carcinoma. Anticancer Res. 2010;30:851–856. [PubMed] [Google Scholar]
- 22.Mortezavi A, Hermanns T, Seifert HH, Baumgartner MK, Provenzano M, Sulser T, Burger M, Montani M, Ikenberg K, Hofstädter F, et al. KPNA2 expression is an independent adverse predictor of biochemical recurrence after radical prostatectomy. Clin Cancer Res. 2011;17:1111–1121. doi: 10.1158/1078-0432.CCR-10-0081. [DOI] [PubMed] [Google Scholar]
- 23.Noetzel E, Rose M, Bornemann J, Gajewski M, Knüchel R, Dahl E. Nuclear transport receptor karyopherin-alpha2 promotes malignant breast cancer phenotypes in vitro. Oncogene. 2012;31:2101–2114. doi: 10.1038/onc.2011.403. [DOI] [PubMed] [Google Scholar]
- 24.Huang L, Wang HY, Li JD, Wang JH, Zhou Y, Luo RZ, Yun JP, Zhang Y, Jia WH, Zheng M. KPNA2 promotes cell proliferation and tumorigenicity in epithelial ovarian carcinoma through upregulation of c-Myc and downregulation of FOXO3a. Cell Death Dis. 2013;4:e745. doi: 10.1038/cddis.2013.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rezabakhsh A, Ahmadi M, Khaksar M, Montaseri A, Malekinejad H, Rahbarghazi R, Garjani A. Rapamycin inhibits oxidative/nitrosative stress and enhances angiogenesis in high glucose-treated human umbilical vein endothelial cells: Role of autophagy. Biomed Pharmacother. 2017;93:885–894. doi: 10.1016/j.biopha.2017.07.044. [DOI] [PubMed] [Google Scholar]
- 26.Ko JH, Yoon SO, Lee HJ, Oh JY. Rapamycin regulates macrophage activation by inhibiting NLRP3 inflammasome-p38 MAPK-NFkappaB pathways in autophagy- and p62-dependent manners. Oncotarget. 2017;8:40817–40831. doi: 10.18632/oncotarget.17256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Z, Huang CM, Shao Z, Zhao XP, Wang M, Yan TL, Zhou XC, Jiang EH, Ke Liu, Shang ZJ. Autophagy induced by areca nut extract contributes to decreasing cisplatin toxicity in oral squamous cell carcinoma cells: Roles of reactive oxygen species/AMPK signaling. Int J Mol Sci. 2017;18:E524. doi: 10.3390/ijms18030524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia L, Wang J, Wu T, Wu J, Ling J, Cheng B. In vitro and in vivo antitumor effects of chloroquine on oral squamous cell carcinoma. Mol Med Rep. 2017;16:5779–5786. doi: 10.3892/mmr.2017.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alshareeda AT, Negm OH, Green AR, Nolan CC, Tighe P, Albarakati N, Sultana R, Madhusudan S, Ellis IO, Rakha EA. KPNA2 is a nuclear export protein that contributes to aberrant localisation of key proteins and poor prognosis of breast cancer. Br J Cancer. 2015;112:1929–1937. doi: 10.1038/bjc.2015.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altan B, Yokobori T, Mochiki E, Ohno T, Ogata K, Ogawa A, Yanai M, Kobayashi T, Luvsandagva B, Asao T, Kuwano H. Nuclear karyopherin-alpha2 expression in primary lesions and metastatic lymph nodes was associated with poor prognosis and progression in gastric cancer. Carcinogenesis. 2013;34:2314–2321. doi: 10.1093/carcin/bgt214. [DOI] [PubMed] [Google Scholar]
- 31.Huang L, Zhou Y, Cao XP, Lin JX, Zhang L, Huang ST, Zheng M. KPNA2 is a potential diagnostic serum biomarker for epithelial ovarian cancer and correlates with poor prognosis. Tumour Biol. 2017;39:1010428317706289. doi: 10.1177/1010428317706289. [DOI] [PubMed] [Google Scholar]
- 32.Zhou LN, Tan Y, Li P, Zeng P, Chen MB, Ye Tian, Zhu YQ. Prognostic value of increased KPNA2 expression in some solid tumors: A systematic review and meta-analysis. Oncotarget. 2017;8:303–314. doi: 10.18632/oncotarget.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang CI, Wang CL, Wang CW, Chen CD, Wu CC, Liang Y, Tsai YH, Chang YS, Yu JS, Yu CJ. Importin subunit alpha-2 is identified as a potential biomarker for non-small cell lung cancer by integration of the cancer cell secretome and tissue transcriptome. Int J Cancer. 2011;128:2364–2372. doi: 10.1002/ijc.25568. [DOI] [PubMed] [Google Scholar]
- 34.Tsai MM, Huang HW, Wang CS, Lee KF, Tsai CY, Lu PH, Chi HC, Lin YH, Kuo LM, Lin KH. MicroRNA-26b inhibits tumor metastasis by targeting the KPNA2/c-jun pathway in human gastric cancer. Oncotarget. 2016;7:39511–39526. doi: 10.18632/oncotarget.8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buchser WJ, Laskow TC, Pavlik PJ, Lin HM, Lotze MT. Cell-mediated autophagy promotes cancer cell survival. Cancer Res. 2012;72:2970–2979. doi: 10.1158/0008-5472.CAN-11-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rebecca VW, Amaravadi RK. Emerging strategies to effectively target autophagy in cancer. Oncogene. 2016;35:1–11. doi: 10.1038/onc.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorburn A, Thamm DH, Gustafson DL. Autophagy and cancer therapy. Mol Pharmacol. 2014;85:830–838. doi: 10.1124/mol.114.091850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang M, Zeng P, Kang R, Yu Y, Yang L, Tang D, Cao L. S100A8 contributes to drug resistance by promoting autophagy in leukemia cells. PLoS One. 2014;9:e97242. doi: 10.1371/journal.pone.0097242. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Ajabnoor GM, Crook T, Coley HM. Paclitaxel resistance is associated with switch from apoptotic to autophagic cell death in MCF-7 breast cancer cells. Cell Death Dis. 2012;3:e260. doi: 10.1038/cddis.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qadir MA, Kwok B, Dragowska WH, To KH, Le D, Bally MB, Gorski SM. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat. 2008;112:389–403. doi: 10.1007/s10549-007-9873-4. [DOI] [PubMed] [Google Scholar]
- 41.Sun WL, Chen J, Wang YP, Zheng H. Autophagy protects breast cancer cells from epirubicin-induced apoptosis and facilitates epirubicin-resistance development. Autophagy. 2011;7:1035–1044. doi: 10.4161/auto.7.9.16521. [DOI] [PubMed] [Google Scholar]
- 42.Guo JY, Teng X, Laddha SV, Ma S, Van Nostrand SC, Yang Y, Khor S, Chan CS, Rabinowitz JD, White E. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev. 2016;30:1704–1717. doi: 10.1101/gad.283416.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo JY, White E. Autophagy, metabolism, and cancer. Cold Spring Harb Symp Quant Biol. 2016;81:73–78. doi: 10.1101/sqb.2016.81.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab. 2017;25:1037–1043. doi: 10.1016/j.cmet.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rangwala R, Chang YC, Hu J, Algazy KM, Evans TL, Fecher LA, Schuchter LM, Torigian DA, Panosian JT, Troxel AB. Combined MTOR and autophagy inhibition: Phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1391–1402. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao D, Wang P, Zhang J, Fu L, Ouyang L, Wang J. Deconvoluting the relationships between autophagy and metastasis for potential cancer therapy. Apoptosis. 2016;21:683–698. doi: 10.1007/s10495-016-1237-2. [DOI] [PubMed] [Google Scholar]
- 48.Debnath J. Detachment-induced autophagy during anoikis and lumen formation in epithelial acini. Autophagy. 2008;4:351–353. doi: 10.4161/auto.5523. [DOI] [PubMed] [Google Scholar]
- 49.Herrero-Martin G, Hoyer-Hansen M, Garcia-Garcia C, Fumarola C, Farkas T, López-Rivas A, Jäättelä M. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 2009;28:677–685. doi: 10.1038/emboj.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: An awakening field. Nat Rev Cancer. 2014;14:611–622. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ojha R, Bhattacharyya S, Singh SK. Autophagy in cancer stem cells: A potential link between chemoresistance, recurrence, and metastasis. Biores Open Access. 2015;4:97–108. doi: 10.1089/biores.2014.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu Z, Luo RZ, Lu Y, Zhang X, Yu Q, Khare S, Kondo S, Kondo Y, Yu Y, Mills GB, et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest. 2008;118:3917–3929. doi: 10.1172/JCI35512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maes H, Kuchnio A, Peric A, Moens S, Nys K, De Bock K, Quaegebeur A, Schoors S, Georgiadou M, Wouters J, et al. Tumor vessel normalization by chloroquine independent of autophagy. Cancer Cell. 2014;26:190–206. doi: 10.1016/j.ccr.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 54.Zhao X, Fang Y, Yang Y, Qin Y, Wu P, Wang T, Lai H, Meng L, Wang D, Zheng Z, et al. Elaiophylin, a novel autophagy inhibitor, exerts antitumor activity as a single agent in ovarian cancer cells. Autophagy. 2015;11:1849–1863. doi: 10.1080/15548627.2015.1017185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou J, Dong D, Cheng R, Wang Y, Jiang S, Zhu Y, Fan L, Mao X, Gui Y, Li Z, et al. Aberrant expression of KPNA2 is associated with a poor prognosis and contributes to OCT4 nuclear transportation in bladder cancer. Oncotarget. 2016;7:72767–72776. doi: 10.18632/oncotarget.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takada T, Tsutsumi S, Takahashi R, Ohsone K, Tatsuki H, Suto T, Kato T, Fujii T, Yokobori T, Kuwano H. KPNA2 over-expression is a potential marker of prognosis and therapeutic sensitivity in colorectal cancer patients. J Surg Oncol. 2016;113:213–217. doi: 10.1002/jso.24114. [DOI] [PubMed] [Google Scholar]
- 57.White E. Autophagy and p53. Cold Spring Harb Perspect Med. 2016;6:a026120. doi: 10.1101/cshperspect.a026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J, Zhang C, Hu W, Feng Z. Tumor suppressor p53 and its mutants in cancer metabolism. Cancer Lett. 2015;356:197–203. doi: 10.1016/j.canlet.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu Y, Xiao L, Liu Y, Wang H, Li H, Zhou Q, Pan J, Lei B, Huang A, Qi S. MIR517C inhibits autophagy and the epithelial-to-mesenchymal (-like) transition phenotype in human glioblastoma through KPNA2-dependent disruption of TP53 nuclear translocation. Autophagy. 2015;11:2213–2232. doi: 10.1080/15548627.2015.1108507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maiuri MC, Galluzzi L, Morselli E, Kepp O, Malik SA, Kroemer G. Autophagy regulation by p53. Curr Opin Cell Biol. 2010;22:181–185. doi: 10.1016/j.ceb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 61.Galluzzi L, Morselli E, Kepp O, Maiuri MC, Kroemer G. Defective autophagy control by the p53 rheostat in cancer. Cell Cycle. 2010;9:250–255. doi: 10.4161/cc.9.2.10493. [DOI] [PubMed] [Google Scholar]
- 62.O'Brate A, Giannakakou P. The importance of p53 location: Nuclear or cytoplasmic zip code? Drug Resist Updat. 2003;6:313–322. doi: 10.1016/j.drup.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 63.Kim IS, Kim DH, Han SM, Chin MU, Nam HJ, Cho HP, Choi SY, Song BJ, Kim ER, Bae YS, Moon YH. Truncated form of importin alpha identified in breast cancer cell inhibits nuclear import of p53. J Biol Chem. 2000;275:23139–23145. doi: 10.1074/jbc.M909256199. [DOI] [PubMed] [Google Scholar]
- 64.Kau TR, Silver PA. Nuclear transport as a target for cell growth. Drug Discov Today. 2003;8:78–85. doi: 10.1016/S1359-6446(02)02562-X. [DOI] [PubMed] [Google Scholar]
- 65.Jamali T, Jamali Y, Mehrbod M, Mofrad MR. Nuclear pore complex: Biochemistry and biophysics of nucleocytoplasmic transport in health and disease. Int Rev Cell Mol Biol. 2011;287:233–286. doi: 10.1016/B978-0-12-386043-9.00006-2. [DOI] [PubMed] [Google Scholar]
- 66.Dickmanns A, Kehlenbach RH, Fahrenkrog B. Nuclear pore complexes and nucleocytoplasmic transport: From structure to function to disease. Int Rev Cell Mol Biol. 2015;320:171–233. doi: 10.1016/bs.ircmb.2015.07.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article. Still, more details about the datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.