Abstract
Colon cancer is the main cause of cancer mortality worldwide. Its poor prognosis is mainly ascribed to high recurrence rates. Identifying novel prognostic biomarkers and therapeutic key points for management is crucial and important. Long non-coding RNAs (lncRNAs) are a class of RNAs, which have various roles in carcinogenicity and molecular mechanisms. The lncRNA small nucleolar RNA host gene 1 (SNHG1) contributes to the promotion of tumor development, however, the connections between SNHG1 and colon cancer are still unclear. The aim of the present study was to investigate the clinical significance, the biological functions, and the potential mechanism of SNHG1 in colon cancer. In the present study, we referred to the Oncomine database and used RT-qPCR to determine that SNHG1 expression was significantly higher both in colon cancer tissues and cancerous cell lines than in normal samples. Cell functional experiments were performed after knockdown of SNHG1, including Cell Counting Kit-8 assay, colony formation assay, Transwell® assay, and flow cytometric analyses of cell apoptosis, which suggested that SNHG1 stimulated colon cancer cell proliferation, promoted cell invasion and migration, and inhibited apoptosis. Immunohistochemical staining and western blotting experiments revealed that in colon cancer cells with SNHG1 knockdown, β-catenin, c-Myc and cyclin D1 protein levels were decreased, while E-cadherin was increased, which suggested that SNHG1 promoted colon cancer cell proliferation, migration and invasion through the Wnt/β-catenin signaling pathway. Our results indicated that SNHG1 and its interrelated components may be future therapeutic targets of carcinoma of the colon.
Keywords: lncRNA, SNHG1, colon cancer, proliferation, invasion, Wnt/β-catenin signaling pathway
Introduction
Colon cancer has a high incidence and mortality rate, and it is the second most common malignancy in the Western world (1). Despite recent improvements in treatments, early diagnostic techniques are still required, and tumor cell metastasis and postoperative recurrence remain challenges. Thus, early detection and diagnosis is vital for prognosis, and has great potential to reduce the burden of this disease (2). Long non-coding RNAs (lncRNAs) are a class of RNAs whose transcripts are over 200 nucleotides in length, which do not have a protein coding capacity (3). Studies have shown the diversity and complexity of lncRNA functions and mechanisms. The functions of lncRNAs have been ascertained in many cancers, including gastric cancer, hepatocellular carcinoma, cervical and lung cancer (4–9). For example, increased lncRNA-ATB [lncRNA activated by transforming growth factor β (TGF-β)], a mediator of TGF-β signaling, induced epithelial-mesenchymal transition (EMT) and invasion, and indicated hepatocellular carcinoma metastases and poor prognoses, in addition to promoting organ colonization of disseminated tumor cells (4). In non-small cell lung cancer (NSCLC), smoking history, infiltration degree, lymph node metastasis, and distant metastasis are all related with the expression of lncRNA AFAP1-AS1, and a high expression of lncRNA AFAP1-AS1 may be associated with the short survival times of NSCLC patients (6). Dysregulation and aberrant expression are important causes of tumorigenesis, and promote relevant tumor progression. Notably, the roles that lncRNAs play are vital in nearly every aspect of tumor biology. However, the clinical prognostic significance and potential functions of lncRNAs are still not understood in colon tumorigenesis.
The lncRNA small nucleolar RNA host gene 1 (SNHG1) (Gene ID: 23642, HGNC: 32688, MIM: 603222, Location: 11q12.3, Exon count: 11) has been reported to play a role in cancer carcinogenesis. Cui et al revealed that in NSCLC, SNHG1 indicated a poor prognosis and promoted NSCLC development via the SNHG1/miR-101-3p/SOX9/Wnt/β-catenin axis (10). Yan et al further revealed that SNHG1 directly bound miR-338 and promoted esophageal carcinoma cell growth by alleviating cell apoptosis of CST3 cells caused by miR-338 (11). Li et al determined that SNHG1 acted as a competing endogenous (ce) RNA for miR-199a-3p in prostate cancer, by inhibiting the activity of miR-199a-3p, and reducing the suppression of CDK7 by miR-199a-3, thus promoting cell proliferation and cell cycle progression in prostate cancer (12). However, studies in colon cancer on SNHG1 expression, biological function, and tumor correlation mechanisms are few. We referred to the Oncomine database, and determined the expression of SNHG1 in human normal colon tissues, cancerous colon tissues, and colon cancerous cell lines. We then used cell function tests to identify potential molecular mechanisms in cells before and after knockdown of SNHG1, to determine if SNHG1 influenced colon cancer cell proliferation, apoptosis, migration, and invasion. In addition, possible pathways involved in the mechanism of colon cancer carcinogenesis were suggested.
Materials and methods
Database and patient tissue samples
Oncomine is a bioinformatics database of abundant DNA microarrays used in collecting, standardizing, and as an analyzing platform, aimed at facilitating the discovery of the functions from genome-wide expression analysis (13,14). We used the Oncomine database (www.oncomine.org) to identify differentially expressed genes between colon cancer tissues and normal tissues. By searching ‘Gene: SNHG1’; ‘Analysis Type: cancer vs. normal analysis’; ‘Cancer Type: colorectal cancer’; and setting ‘P value: all’; ‘Fold Change: all’; ‘Gene rank: all’; ‘Sample Type: Clinical Specimen’; and ‘Data Type: mRNA’, we obtained seven useable studies as follows: three out of seven related to the colon, and 1,352 samples in total. We analyzed SNHG1 differential expression in these three datasets between normal and colon cancer tissues, then used GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS version 19.0 (SPSS, Inc., Chicago, IL, USA) to analyze the statistical differences. We also randomly acquired 13 colon cancer patient tissues, who underwent surgical treatment at the First Affiliated Hospital of Chongqing Medical University from January 2015 to December 2016. None of the patients involved in this study had undergone radiation or chemotherapy prior to surgery. Every excised colon tissue, no matter if cancerous or adjacent normal colon tissue, was immediately stored in liquid nitrogen, and saved at the Chongqing Medical University Laboratory. The histopathological features of cells in these tissue samples were observed by pathologists at Chongqing Medical University who used standard methods to diagnose colon cancer. There were no obvious tumor cells in adjacent non-cancerous tissues, which were included as standard control samples. The use of the human tissues was approved by the Ethics Committee of Chongqing Medical University (Chongqing, China).
Cell culture
The colon cancer cell lines HCT-116, Caco-2 and HT-29, were obtained from the Molecular Tumor and Epigenetics Laboratory at the First Affiliated Hospital of Chongqing Medical University. All the cell lines were cultured in RPMI-1640 medium (Hyclone™; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Grand Island, NY, USA), 100 mg/ml streptomycin and 100 µg/ml penicillin (both from Beyotime Institute of Biotechnology, Beijing, China), in an incubator at 37°C with 5% CO2. The medium was changed each day and cells were subcultured every 2–3 days.
RNA extraction, reverse transcription and real-time quantitative PCR (RT-qPCR)
RNAiso Plus (Takara Bio USA, Inc., Mountain View, CA, USA) was used to isolate total RNA from all tissues and cell lines following the manufacturer's instructions. All RNA concentrations and purities were examined with ultraviolet spectrometry (ND-1000 spectrophotometer; NanoDrop Technologies, Wilmington, DE, USA). The reverse-transcribed extracted RNA was produced from cDNA using the GoScript™ Reverse Transcription System (Promega Corp., Sunnyvale, CA, USA) according to the manufacturer's instructions, and RT-qPCR was used to compare SNHG1 expression. RT-qPCR primer sequences were designed by Takara Bio, Inc. (Shiga, Japan) as follows: SNHG1: 5′-TTGCTGCCTTTCTTACATGATCC-3′ (sense primer), 5′-AGACACGAAGTGGAGTTATGGGAA-3′ (antisense primer). β-actin was employed as an internal control. The β-actin primer sequences were 5′-TCCTGTGGCATCCACGAAACT-3′ (sense primer) and 5′-GAAGCATTTGCGGTGGACGAT-3′ (antisense primer). The concentration of primers was 10 µmol/l. The RT-qPCR steps were in accordance to the standard SYBR-Green (BioTool, Kirchberg, Switzerland) instructions and were performed on a Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The thermal cycler protocol was as follows: step 1, repeated once, 50°C for 2 min; step 2, repeated once, 95°C for 10 min; step 3, repeated 40 times, 95°C for 15 sec, and 60°C for 1 min; step 4, repeated once, 95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec.
Plasmid construction, transfection, G418 selection
To assess if SNHG1 influenced colon cancer cell biological functions, we acquired plasmid pRNAT-U6.1/Neo-vector and pRNAT-U6.1/Neo-sh-SNHG1, which were synthesized by Guangzhou RiboBio, Co., Ltd. (Guangzhou, China), and transfected them into colon cancer cells. Sequences were as follows: GATCCCGAGGACATCAGAAGGTGAATTGATATCCGT TCACCTTCTGATGTCCTCTTTTTTCCAAA (forward) and AGCTTTTGGAAAAAAGAGGACATCAGAAGGTGAAC GGATATCAATTCACCTTCTGATGTCCTCGG (reverse). Regarding the transfections, we plated the cells (Caco-2 and HCT-116 cells) into 6-well plates; transfection was performed at 40% cell density, at room temperature. Initially 5 µl Lipofectamine™ 2000 reagent (Invitrogen; Thermo Fisher Sientific, Shanghai, China) was mixed with 500 µl RPMI-1640 medium (Hyclone; GE Healthcare Life Sciences) for 5 min, then 4 µg plasmid (vector or sh-SNHG1) (Guangzhou RiboBio, Co., Ltd.) with 500 µl RPMI-1640 was added for 5 min. Two 500-µl samples were added to 1 ml of the mixture for 20 min, then 1 ml of RPMI-1640 medium was added to a total 2 ml final volume. The samples were added into every well at 40% cell density. After culturing the dilution at 37°C containing 5% CO2 for 4–6 h, 2 ml of medium with 10% FBS was used to replace the medium. Then, 48 h later, we used fluorescence microscopy to observe plasmid transfection efficiency. When the efficiency reached 30–40%, G418 (Amresco, LLC, Solon, OH, USA) was added into the medium (G418 at 300 µg/ml for Caco-2 cells and 400 µg/ml for HCT-116 cells) to select stably transfected cells. The medium containing G418 was changed every 3 days until the non-transfected cells died. To further confirm that the plasmids were successfully transfected into colon cancer cells after G418 selection, RT-qPCR was performed to detect SNHG1 differential expression between the vector group and the sh-SNHG1 cell group. The RT-qPCR in this step was performed as aforementioned.
Cell proliferation assays [Cell Counting Kit-8 (CCK-8) and clone formation assays]
To assess cell proliferation before and after SNHG1 knockdown, the cells (HCT-116 and Caco-2) were divided into the HCT-116-vector, HCT-116-sh-SNHG1, Caco-2-vector and Caco-2-sh-SNHG1 groups. They were plated at a density of 5×103 cells/well in triplicate in 96-well plates. The cells were all stably transfected. After culturing at 0, 24, 48 and 72 h, 10 µl of CCK-8 reagent (Dojindo Molecular Technologies, Rockville, MD, USA), was added to each well for 2 h and the OD (optical density) at 450 nm was recorded for growth density. The absorbances at 0, 24, 48 and 72 h were plotted. By comparing each point-in-time OD value the statistical differences between the vector group and sh-SNHG1 groups were identified.
In another colony formation assay study, the cells (HCT-116 and Caco-2 cells) were divided into the HCT-116-vector, HCT-116-sh-SNHG1, Caco-2-vector and Caco2-sh-SNHG1 groups. Each cell group was plated at 5×102 cells/well in 6-well plates, and cultured in RPMI-1640 medium with 10% FBS. G418 was added (G418 300 µg/ml for Caco-2 and 400 µg/ml for HCT-116 cells) and exchanged for fresh medium every 3 days until single colonies (≥50 cells) formed. The number of colonies in each group was counted by eyes and ImageJ version 2.1.4.7 software (National Institutes of Health, Bethesda, MD, USA) after fixing with paraformaldehyde and staining with crystal violet. We conducted statistical analyses to compare the proliferation differences between the vector and experimental cell groups.
Flow cytometric analyses of the cell cycle and apoptosis assays
Stably transfected HCT-116 and Caco-2 cells were cultured at 37°C in 5% CO2 for 48–72 h until 70% confluence, then harvested with trypsin without EDTA, harvested by centrifugation (120 × g, 5 min), resuspended in 1 ml of phosphate-buffered saline (PBS), and 5×106 cells from each group were used. Binding buffer (500 µl) was used to resuspend the cells, then 5 µl of Annexin V-fluorescein isothiocyanate (Annexin V-FITC) and 5 µl of propidium iodide were added and incubated without light at room temperature for 5–15 min, and finally analyzed by flow cytometry. The Annexin V-EGFP Apoptosis Detection kit was purchased from Beyotime Institute of Biotechnology.
Transwell® migration and invasion assays
The migration ability of the cells (HCT-116 and Caco-2 cells) was assessed using 24-well Transwell (Corning 3422; Corning Inc., Corning, NY, USA) plates. Medium (700 µl) containing 10% FBS was added into the lower chamber, and 200 µl of serum-free medium with 6×104 transfected cells were added to the upper chamber. After 48 h, the cells still remaining in the upper chamber were discarded; cells migrating to the lower chamber were fixed with paraformaldehyde and stained by hematoxylin and eosin.
The invasion assays required the stimulation of cancer cells to migrate in the 24-well Transwell plates. Therefore, Matrigel (BD Biosciences, San Jose, CA, USA) was diluted 1:7 with serum-free medium at 4°C according to the manufacturer's instructions. Then, 100 µl of the diluted Matrigel was added into the upper chamber at 37°C until the Matrigel had solidified. Subsequently, 700 µl of medium containing 10% FBS was added to the lower chamber, and 100 µl of serum-free medium with 8×104 stably transfected cells were added to the upper chamber (on the surface of the Matrigel). Each sample was plated in triplicate. After incubation for 48 h, the stained cells were counted under a light microscope (magnification, ×200).
Western blotting
RIPA lysis buffer and phenylmethanesulfonyl fluoride were used to extract cell proteins, and the protein concentrations were determined using a BCA protein assay kit (all from Beyotime Institute of Biotechnology). Protein (40 µg) was separated by electrophoresis (5% stacking gel and 12% separating gel) and transferred onto PVDF membranes (EMD Millipore, Billerica, MA, USA). Non-specific binding was blocked with 5% evaporated milk and the membranes were reacted with primary antibodies overnight at 4°C. The primary antibodies were all obtained from Cell Signaling Technology (Danvers, MA, USA) and included anti-E-cadherin (1:500 dilution; cat. no. 3195), anti-β-catenin (1:500 dilution; cat. no. 8480), anti-cyclin D1 (1:500 dilution; cat. no. 2978) and anti-c-Myc (1:500 dilution; cat. no. 13987). The secondary antibodies (1:3,000 dilution; cat. no. 7074; Cell Signaling Technology) were incubated for 2 h. They were decided by the corresponding primary antibodies. After washing the blots with TBST three times, the target proteins were detected using BeyoECL Plus reagent (Beyotime Institute of Biotechnology, Shanghai, China) using a FUSION FX imager instrument (Vilber Lourmat, Marne-la-Vallée, France). The electrophoresis and blotting were assessed on the basis of each target protein's molecular weight.
Immunohistochemistry
We obtained 20 paired colon cancer and corresponding normal paraffin sections from Chongqing Medical University from January 2015 to December 2016. Sections were placed in an oven at 60°C for 2 h, dewaxed with water and rinsed with PBS (pH 7.4) 3 times. Then, the sections were dehydrated through a series of concentration gradient ethanol (100, 95 90, 80 and 70%). Subsequently, we added 3% hydrogen peroxide to each section and incubated the sections for 10 min at room temperature. Then, the sections were rinsed with PBS. The primary mouse monoclonal antibody E-cadherin (1:100 dilution; cat. no. WL00941), β-catenin (1:100 dilution; cat. no. WL0962a), cyclin D1 (1:100 dilution; cat. no. WL01435a) and c-Myc (1:100 dilution; cat. no. WL01781) all purchased from Wanleibio (Beijing, China) were added to sections and incubated at 4°C overnight. The following morning, we washed the sections 3 times with PBS, and the corresponding secondary antibody (80 µl) was added to each section. The sections were then incubated for 30 min at room temperature. 3,3′-Diaminobenzidine tetrahydrochloride (DAB) was used to stain the sections and hematoxylin was used to counterstain the cell membrane and cytoplasm for 15 sec. The sections were then immediately rinsed with water. Finally, the sections were treated with a series of gradient alcohol, xylene and neutral gum in order to be dehydrated and sealed. We used PBS instead of a primary antibody as a blank control, and known positive staining as a positive control. Immunohistochemical staining of the sections was observed under light microscope by three pathologists independently.
Assessment of immunohistochemistry
The evaluations were as follows: i) β-catenin: strong nuclear staining and cytoplasmic staining of >10% of cells was considered aberrant positive expression in colon cancer tissues (15); ii) E-cadherin: a membranous stain of >60% of cells was considered to be a score 3+; a score 2+ was considered as a stain of 20–60% of cells; a stain of <20% of cells was considered as a score 1+; a score 0 was used for a 0% stain. A score of 3+ was classified as having positive expression, and all others were classified as having decreased expression (16); iii) c-Myc: strong nuclear staining of >25% of cells was considered positive expression (17); iv) cyclin D1: nuclear staining of >5% of cells was considered as positive expression, while negative expression was considered as a stain of <5% of cells (18).
Statistical analysis
We used SPSS, version 19.0 software (IBM Corp., Armonk, NY, USA), ImageJ version 2.1.4.7 software (National Institutes of Health, Bethesda, MD, USA) and GraphPad Prism version 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA) to perform statistical analyses. The independent samples t-test was applied to intra-group differentiation. The Chi-squared test analysis was used to assess clinicopathological relationships. Repeated measures analysis of variance (ANOVA) was used for several groups. P<0.05 was considered to indicate a statistically significant difference.
Results
SNHG1 expression is upregulated in colon cancer tissues compared to adjacent non-cancerous tissues, and is higher in colon cancer cells than in normal cells
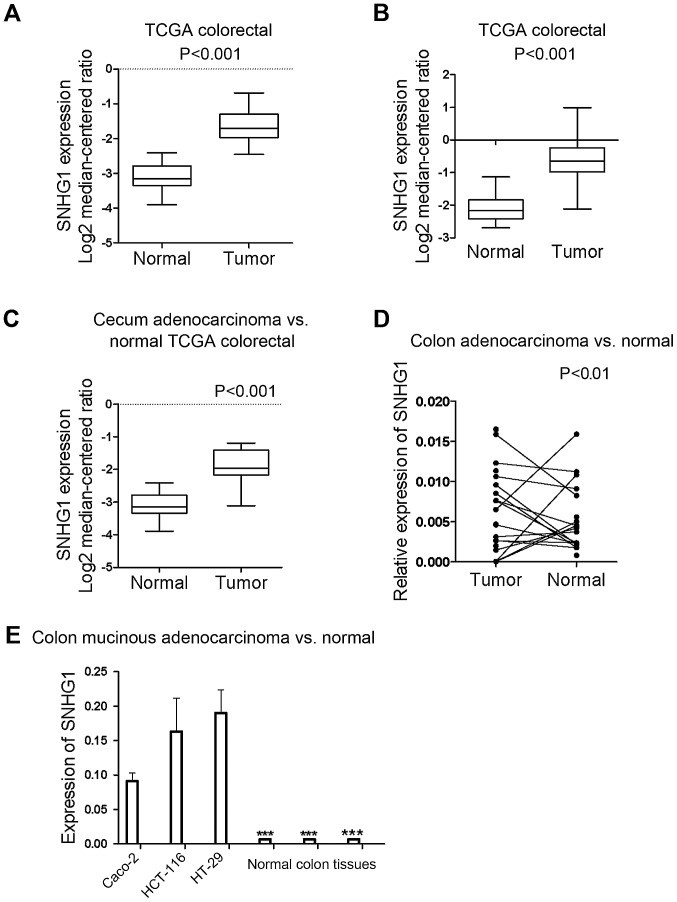
All the samples identified from the Oncomine database revealed that the expression of SNHG1 in colon cancer tissues was significantly higher than in normal colon tissues (P<0.05; P-value for the median-ranked analysis: 2.31E-19). The sample microarrays of three datasets (TCGA colorectal: cecum adenocarcinoma vs. normal; colon adenocarcinoma vs. normal; and colon mucinous adenocarcinoma vs. normal) exhibited respective fold changes between cancer and normal tissues of 2.697-fold (Fig. 1A; P<0.001), 2.806-fold (Fig. 1B; P<0.001), and 2.241-fold (Fig. 1C; P<0.001), respectively. All the dataset microarray results revealed that SNHG1 expression in colon cancer samples was at least 2-fold higher than the normal sample levels.
Figure 1.
Statistical analysis results of the expression of SNHG1 in 3 datasets obtained from the Oncomine database, and the expression of SNHG1 in 13 pairs of patient tissues and 3 cell lines. (A) TCGA colorectal: cecum adenocarcinoma vs. normal: 2.697-fold (P<0.001); (B) colon adenocarcinoma vs. normal: 2.806-fold (P<0.001); (C) colon mucinous adenocarcinoma vs. normal: 2.241-fold (P<0.001). (D) Except for 2 colon cancer patients, all of the colon cancer cases indicated a significantly higher expression of SNHG1 in cancer tissues compared with adjacent non-cancer tissues (P<0.01). (E) SNHG1 was upregulated in HCT-116, Caco-2, HT-29 cell lines than in human normal colon tissues (***P<0.001).
To further ascertain SNHG1 expression differences in patient tissues, RT-qPCR was used to examine SNHG1 expression in 13 pairs of colon cancer patient tissues compared with adjacent non-cancer tissues. Since SNHG1 belongs to lnc-RNAs and does not encode a protein, in tissues and cells it can be measured and analyzed by RT-qPCR. The results revealed that except for 2 colon cancer patients, all of the colon cancer cases indicated a significantly higher expression of SNHG1 in cancer tissues compared with adjacent non-cancer tissues (Fig. 1D; P<0.01). Table I contains the clinicopathological characteristics of the 13 patients with colon cancer.
Table I.
Clinicopathological characteristics of the 13 patients with colon cancer.
| Sex | Age | Weight | Smoking | TNM stage | Differentiation | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female | Male | (years) | (kg) | Yes | No | I | II | III | IV | Middle | Middle-low | Low | |
| Number | 7/13 | 6/13 | 56±14 | 55±7 | 4/13 | 9/13 | 0 | 7 | 4 | 2 | 9 | 3 | 1 |
The differences in the expression of SNHG1 not only existed in the tissues, but also in different colon cancer cell lines. RT-qPCR was performed on three colon cancer cell lines (HCT-116, HT29 and Caco-2) and three control colon tissue sample. We found that SNHG1 was upregulated in HCT-116, Caco-2 and HT-29 cells compared with the human normal colon tissue (Fig. 1E; P<0.001).
In summary, SNHG1 was upregulated in colon cancer tissues and cell lines compared to normal tissues and cells. SNHG1 may therefore be an oncogene playing a role in the promotion of colon cancer development, as has also been reported in NSCLC and prostate cancer.
Recombinant plasmid is successfully transfected
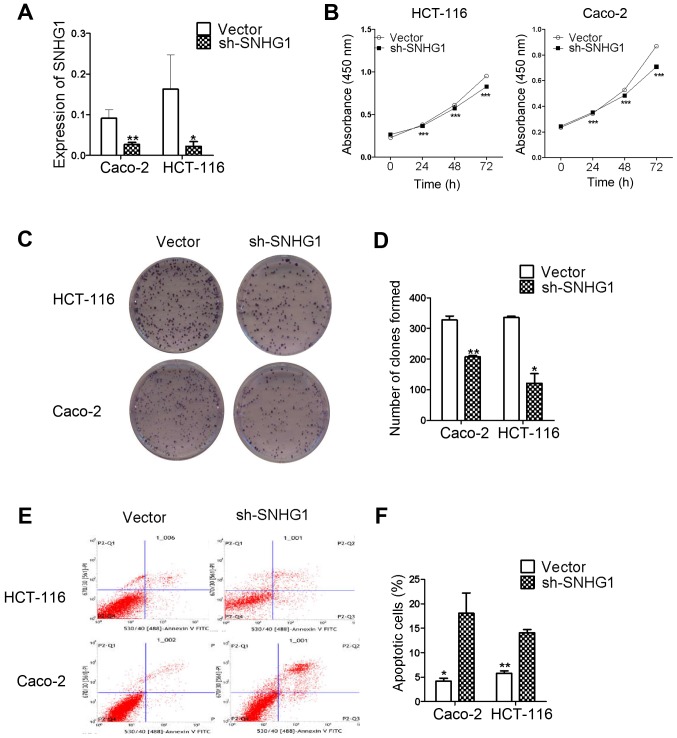
The transfection of recombinant plasmids into colon cancer cells was used to observe if SNHG1 downregulation influenced cell biological functions. To verify transfection success, fluorescence cell counting was used to examine plasmid transfection efficiency, and the results revealed ~30–40% efficiency (Fig. 2). We added G418 to select stably transfected cells, and RT-qPCR detected plasmid transfection in the stably transfected cells. The results revaled that SNHG1 was significantly decreased in Caco-2 and HCT-116 cells transfected with pRNAT-U6.1/Neo-sh-SNHG1 when compared with the pRNAT-U6.1/Neo-vector transfected cells (Fig. 3A; Caco-2, P<0.01; HCT-116, P<0.05). These results indicated that recombinant plasmid transfection decreased SNHG1.
Figure 2.
Flourescence counting roughly examined plasmid transfection efficiency and it was revealed to be ~30–40%.
Figure 3.
Plasmid transfection results and a series of cell function experiments including CCK-8, colony formation, apoptosis assay between the sh-SNHG1 group and the vector. (A) The plasmid pRNAT-U6.1/Neo-sh-SNHG1 and pRNAT-U6.1/Neo-vector transfection results were detected by RT-qPCR, and they revealed that SNHG1 was significantly decreased in cells (Caco-2 and HCT-116) transfected with pRNAT-U6.1/Neo-sh-SNHG1 than those with pRNAT-U6.1/Neo-vector (Caco-2: **P<0.01; HCT-116: *P<0.05). These results indicated that the recombinant plasmid had interfered with SNHG1 successfully. (B) The result of the CCK-8 assay. Cells proliferated slower at 24, 48 and 72 h in the sh-SNHG1 group than the vector group. (C) The different number of colonies in the vector and the sh-SNHG1 group in Caco-2 and HCT-116 cell lines. (D) The statistical analysis of the colony formation assay. The sh-SNHG1 group formed fewer colonies compared with the vector (Caco-2: **P<0.01; HCT-116: *P<0.05). (E) Cell apoptosis images detected by combining flow cytometry and Annexin V-FITC/PI staining. (F) The statistical analysis of E. The apoptosis rate in the vector group was lower than the sh-SNHG1 group (Caco-2: *P<0.05; HCT-116: **P<0.01).
CCK-8 and colony formation assays reveal that SNHG1 promotes colon cancer cell proliferation
The CCK-8 and colony formation assay results confirmed that SNHG1 influenced CaCo-2 and HCT-116 cell proliferation in the sh-SNHG1 transfected group. The CCK-8 results presented in Table II, revealed that cell proliferation was slower at 24, 48 and 72 h in the sh-SNHG1 group compared to the vector group. The results were also revealed in Fig. 3B for both Caco-2 and HCT-116 cells (Caco-2, P<0.001; HCT-116, P<0.001).
Table II.
Effect of recombinant plasmids on cell proliferation.
| A, Effect of recombinant plasmids on HCT-116 cell proliferation (means ± SD) | ||
|---|---|---|
| Time (h) | Vector group | sh-SNHG1 group |
| 0 | 0.2277±0.0020 | 0.2644±0.0015 |
| 24 | 0.3796±0.0123 | 0.3655±0.0193 |
| 48 | 0.6093±0.0179 | 0.5722±0.0571 |
| 72 | 0.9510±0.0089 | 0.8267±0.0136 |
| B, Effect of recombinant plasmids on Caco-2 cell proliferation (means ± SD) | ||
| Time (h) | Vector group | sh-SNHG1 group |
| 0 | 0.2341±0.0091 | 0.2438±0.0057 |
| 24 | 0.3420±0.0112 | 0.3514±0.0051 |
| 48 | 0.5264±0.0131 | 0.4845±0.0146 |
| 72 | 0.8685±0.0106 | 0.7076±0.0260 |
The colony formation assay results further ascertained the proliferation abilities of Caco-2 and HCT-116 cells. Fig. 3C revealed the colony formation results of the vector and sh-SNHG1-transfected groups in the two cell lines. Similar to the results presented in Fig. 3D, the Caco-2 vector group vs. the sh-SNHG1 group was 328±23 vs. 207±6 (P<0.01), and the HCT-116 vector group vs. the sh-SNHG1 group was 336±7 vs. 122±54 (P<0.05), respectively, which confirmed that colon cancer cell proliferation was decreased in the sh-SNHG1 group. The results from both assays demonstrated that SNHG1 played a role in promoting colon cancer cell proliferation.
Flow cytometry: the effects of SNHG1 on cell apoptosis
Flow cytometric analyses were used to identify whether SNHG1 was associated with colon cancer cell apoptosis. As revealed in Fig. 3E and F, the apoptosis level in the Caco-2 vector group vs. the sh-SNHG1 group was 4.18±1.03 vs. 18.07±7.19 (P<0.05), and the HCT-116 vector group vs. the sh-SNHG1 group was 5.78±0.83 vs. 14.08±1.14 (P<0.01). These data further demonstrated that SNHG1 reduced colon cancer cell apoptosis in both HCT-116 and Caco-2 cells.
Transwell assay results reveal that SNHG1 promotes colon cancer cell migration and invasion abilities
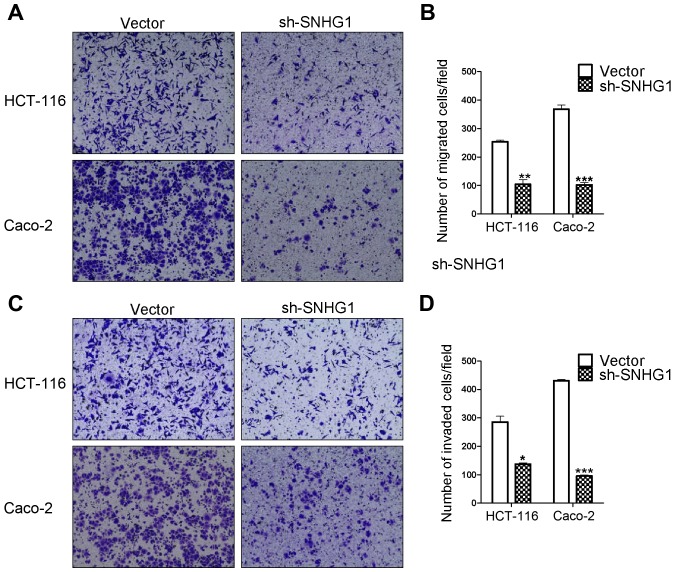
Transwell migration and invasion assays are based on the medium concentration difference between the upper and lower side of the chambers. Cells migrate from a nutrient-poor side chamber to a nutrient-rich side chamber (the lower side). The migration assay results revealed that the HCT-116 sh-SNHG1 group cell counts (104±29 cells) were significantly decreased compared to the HCT-116 vector group (254±8 cells) (P<0.01), in addition, the cell counts for the Caco-2 sh-SNHG1 group were 102±13 vs. the CacCo-2 vector group 368±25 (P<0.001). The invasion experiment results from the HCT-116 sh-SNHG1 group vs. the vector group were 137±8 cells vs. 285±35 (P<0.05) and the CacCo-2 sh-SNHG1 group vs. the vector group were 95±7 vs. 430±9 (P<0.001). These results were in agreement with the images displayed in Fig. 4A-D, which revealed that HCT-116- and Caco-2-vector transfected cells had greater migration and invasion abilities than the sh-SNHG1-transfected cells, and further suggested the important carcinogenenic role of SNHG1 in colon cancer cell migration and invasion.
Figure 4.
Transwell® assays of the migration and invasion in Caco-2 and HCT-116 cells. (A) The migration assay in Caco-2 and HCT-116 cells. (B) The statistical analysis of (A). The migration ability in the vector group was higher than that of the sh-SNHG1 group (HCT-116: **P<0.01; Caco-2: ***P<0.001). (C) The invasion assay in Caco-2 and HCT-116 cells. (D) The statistical analysis of (C). The invasion ability in the vector group was higher than that of the sh-SNHG1 group (HCT-116: *P<0.05; Caco-2: ***P<0.001).
Key markers of SNHG1 in cell metastatic diffusion and proliferation
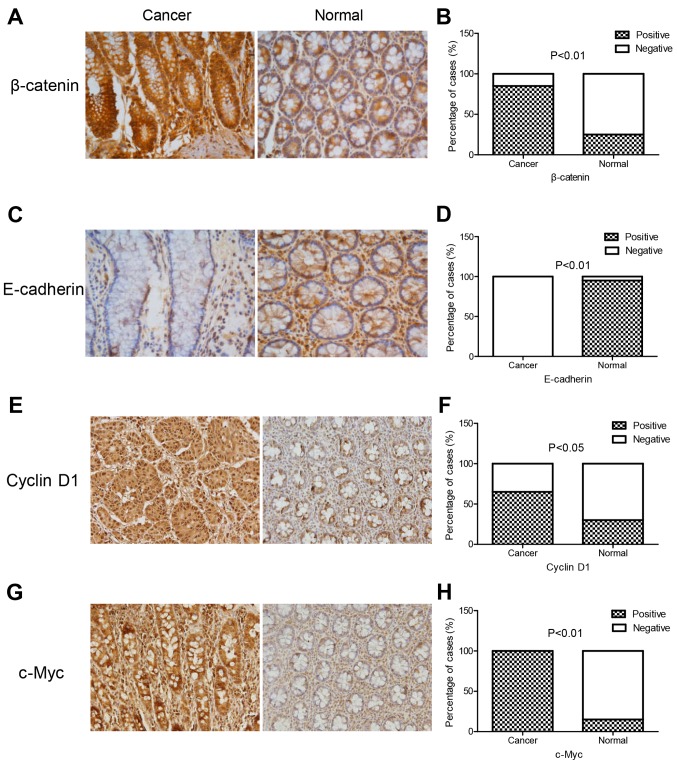
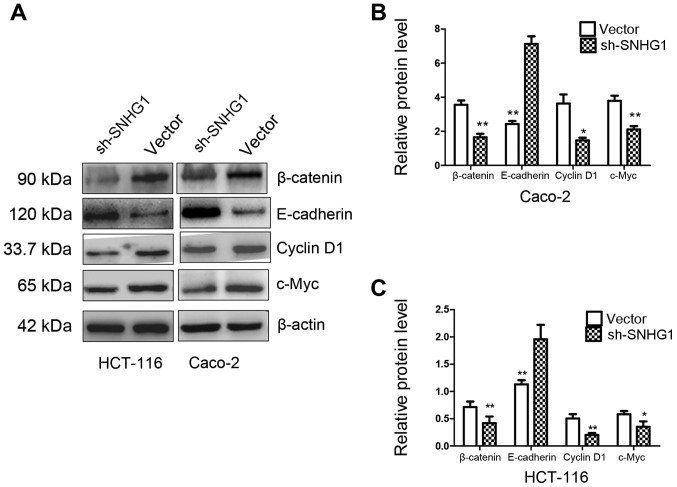
The Transwell results confirmed that SNHG1 could facilitate colon cancer cell invasion and migration. The CCK-8 and colony assays revealed that SNHG1 could promote colon cancer cell proliferation. We next performed immunohistochemistry and western blot analysis to explore the underlying mechanisms. E-cadherin, β-catenin, cyclin D1 and c-Myc proteins were detected in these experiments. The immunohistochemistry experiment revealed that the expression of β-catenin, cyclin D1 and c-Myc in colon cancer tissues was significantly higher than that in normal tissues, while E-cadherin expression was lower in colon cancer tissues compared to normal tissues (Fig. 5; Table III). In addition, western blotting indicated that both in HCT-116 and Caco-2 cell lines with sh-SNHG1 transfection, proteins β-catenin, cyclin D1 and c-Myc were significantly decreased, while E-cadherin exhibited a higher expression than the vector group (Fig. 6; P<0.05, P<0.01). These results implied that the trends observed while investigating the expression of E-cadherin, β-catenin, cyclin D1 and c-Myc in the sh-SNHG1 group corresponded to the trends in colon normal tissues, which further confirmed the carcinogenic characteristics of SNHG1 and suggested that the Wnt/β-catenin signaling pathway may participate in SNHG1-induced carcinogenesis.
Figure 5.
Immunohistochemical staining of E-cadherin, β-catenin, cyclin D1 and c-Myc proteins in colon cancer tissues and normal tissues. The expression of β-catenin, cyclin D1, and c-Myc in colon cancer tissues was significantly higher than that in normal tissues, while E-cadherin expression was lower. (A, C, E and G) β-catenin, E-cadherin, cyclin D1 and c-Myc protein expression in colon cancer tissues and normal tissues, and (B, D, F and H) the quantitative analysis of A, C, E and G).
Table III.
Expression of β-catenin, E-cadherin, cyclin-D1 and c-Myc in colon cancer tissues and colon normal tissues.
| β-catenin %(n/total) | E-cadherin %(n/total) | Cyclin D1 %(n/total) | c-Myc %(n/total) | |||||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | |
| Cancer | 85 (17/20) | 15 (3/20) | 0 (0/20) | 100 (20/20) | 65 (13/20) | 35 (7/20) | 100 (20/20) | 0 (0/20) |
| Normal | 25 (5/20) | 75 (15/20) | 95 (19/20) | 5 (1/20) | 30 (6/20) | 70 (14/20) | 15 (3/20) | 85 (17/20) |
| P-value | P<0.01 | P<0.01 | P<0.05 | P<0.01 | ||||
Figure 6.
Western blot analysis of E-cadherin, β-catenin, cyclin D1 and c-Myc protein expression in the sh-SNHG1 group and the vector group. (A) β-catenin, cyclin D1, c-Myc were significantly decreased while E-cadherin was significantly increased in the sh-SNHG1 group compared to the vector goup in HCT-116 and Caco-2 cell lines. (B and C) The statistical quantitative analysis of (A). *P<0.05, **P<0.01.
Discussion
Colon cancer is one of the most common gastrointestinal malignancies and the third leading cause of cancer-related mortality among males and females worldwide (19,20). The high rate of relapse and unfavorable prognoses suggest a need for novel molecular biomarkers for the diagnosis and therapeutic treatment of colon cancer to increase disease survival (21). lncRNAs have specific expressions in tissues and cell lines (22), and may have uses in the dysregulation of cancer cells, including epigenetic effects, biological effects, and other potential antitumor properties (23,24). SNHG1 is a novel biomarker that may play a role in lung cancer, hepatocellular carcinoma, and neuroblastoma. Its mechanism of action has been reported (25–28). SNHG1 has a cancer-inducing role on different target genes, but its regulatory mechanism in colon cancer is unknown.
We used a database (Oncomine) to identify differentially expressed genes in colon cancer tissues vs. normal tissues, and it was determined that SNHG1 expression was statistically higher in cancer than in normal tissues. SNHG1 expression in cancer vs. normal tissues was at least 2-fold higher, and this was in agreement with the microarray GSE31737 analyses (19). However, database or chip accession is always an initial determination, therefore we then verified SNHG1 expression in colon cancer tissues and normal tissues using RT-qPCR, and found that except for 2 colon cancer patients, all of the colon cancer cases indicated a significantly higher expression of SNHG1 in cancer tissues compared with adjacent non-cancer tissues, which was consistent with the Oncomine database and statistically significant. However, this study lacked sufficient tissue samples from patients to analyze and correlate the clinical characteristics of patients, such as age, sex, tumor size, or aggressive stages, therefore more studies with a larger patient population are necessary for further investigation. Normal colon tissues consist of various normal colon cells, thus we also compared the expression of SNHG1 between three colon cancer cell lines (HCT-116, Caco-2 and HT-29 cells) with normal colon tissues. It was revealed that SNHG1 was upregulated in colon cancer cells, which implied that SNHG1 may be a biomarker for colon cancer. This result was in agreement with the SNHG1 levels in other cancers, such as in esophageal and lung cancer (10,11). Knockdown of SNHG1 was used to investigate SNHG1 biological functions, and the results revealed that SNHG1 promoted cell proliferation and colony formation, favored cell migration and invasion abilities, but reduced cell apoptosis in colon cancer cells. These results demonstrated that SNHG1 played an oncogenic role in colon cancer, and that suppressing SNHG1 expression and interfering with its function may be possible new therapeutic approaches.
The canonical WNT pathway (Wnt/β-catenin signaling) participates in a variety of cellular processes including embryonic development, maintenance of tissue homeostasis, and cancer pathogenesis, among which β-catenin is the key regulatory factor (29). In Homo sapiens, β-catenin is a multifunctional protein (30) consisting of 781 amino acids. When the Wnt pathway is inactivated, the destruction complex, composed of CSNK1A1, AXIN1, GSK3b and APC phosphorylates serine residues and stabilizes β-catenin ubiquitination. Once the Wnt pathway is activated, the stabilization of β-catenin leads to its translocation from the cytoplasm to the nucleus, causing its association with high-mobility group domain factors such as T-cell factor (Tcf)/lymphocyte enhancer factor, resulting in transcriptional activation of target genes leading to aberrant cell proliferation, differentiation, migration, and invasion to causing carcinogenesis and poor prognoses (30). β-catenin is a pivotal protein in Wnt/β-catenin signaling (31).
In a recent study, lncRNA SNHG1 and Wnt/β-catenin signaling were linked to carcinogenesis. SNHG1 promoted NSCLC progression via miR-101-3p and SOX9 activating the Wnt/β-catenin signaling pathway (10). In our study, SNHG1 promoted colon cancer cell aberrant proliferation, migration, and invasion. In addition, a significant decrease of β-catenin in the sh-SNHG1 group was observed compared to the vector group. This suggested that β-catenin played a role in the molecular mechanism of SNHG1 action, and the Wnt/β-catenin signaling pathway may be involved. Previous studies have described β-catenin as part of the E-cadherin complex (32), and this specific complex may link the actin cytoskeleton to accommodate cell adhesive abilities, stabilize integrity, and alter cellular functions. E-cadherin loss is a critical factor of EMT, a fundamental process that facilitates tumor cell migration and invasion into surrounding tissues during tumor metastasis (33,34). If E-cadherin lacks enough β-catenin its target genes or other affiliated proteins may be affected and this could cause carcinogenesis, thus, E-cadherin and β-catenin expression may have opposite effects (29). Our western blotting results revealed that E-cadherin expression was significantly higher in the sh-SNHG1 group than the vector group. β-catenin decreased after SNHG1 was downregulated, which was in agreement with our Transwell assay results which revealed that SNHG1 inhibition induced decreased invasion and migration abilities of colon cancer cells. This further suggested that in the vector-transfected colon cancer cells, knockdown of SNHG1 influenced EMT and the Wnt/β-catenin pathway to reduce the migration and invasion of colon cancer cells. In summary, SNHG1 led to colon cancer invasion and migration possibly via the Wnt/β-catenin pathway.
To further study this pathway, we detected c-Myc and cyclin D1, the downstream targets of the Wnt/β-catenin pathway (35,36), which are involved in Wnt and β-catenin signaling and are known as cell cycle modulators showing accumulated expression during accelerated cell proliferation (37,38). Our western blot analyses revealed that cyclin D1 and c-Myc decreased in Caco-2 and HCT-116 cells with SNHG1 downregulation; which was also in agreement with our CCK-8 and colony formation assays. SNHG1 reduced E-cadherin but increased β-catenin protein levels, cyclin D1 and c-Myc expression, supporting our hypothesis that SNHG1 promoted colon cancer cell migration, invasion, and proliferation possibly via the Wnt/β-catenin pathway. Concurrently, the immunohistochemical experiment revealed that the expression of β-catenin, cyclin D1 and c-Myc in colon cancer tissues was significantly higher than in normal tissues, while E-cadherin expression was lower, which implied that the trends of those proteins in normal colon tissues corresponded to the trends of sh-SNHG1 group in the western blots. Combining the results of two experiments further confirmed the oncogenic role of SNHG1. Further investigations such as examining the phosphorylation of key proteins, and inhibiting or positively stimulating specific pathways, and detecting more sufficient efficacious pathway signaling factors should be performed in the future.
The data demonstrated that SNHG1 functioned as an oncogene in colon cancer and may act via the Wnt/β-catenin pathway to promote carcinogenesis. SNHG1 could be a potential predictor for colon cancer patient prognosis and may be a therapeutic target.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Author's contributions
ZJ and HY designed the study. HY, SW, YJK and CW performed the experiments. HY and SW wrote the paper. YJK, CW, YX, YZ and ZJ received and edited the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All experimental protocols were approved by the Chongqing Medical University Ethics Committee (Chongqing, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Negm OH, Hamed MR, Schoen RE, Whelan RL, Steele RJ, Scholefield J, Dilnot EM, Kumara Shantha HM, Robertson JF, Sewell HF. Human blood autoantibodies in the detection of colorectal cancer. PLoS One. 2016;11:e0156971. doi: 10.1371/journal.pone.0156971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 3.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 4.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Peng W, Fan H. Long non-coding RNA PANDAR correlates with poor prognosis and promotes tumorigenesis in hepatocellular carcinoma. Biomed Pharmacother. 2015;72:113–118. doi: 10.1016/j.biopha.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Deng J, Liang Y, Liu C, He S, Wang S. The up-regulation of long non-coding RNA AFAP1-AS1 is associated with the poor prognosis of NSCLC patients. Biomed Pharmacother. 2015;75:8–11. doi: 10.1016/j.biopha.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Li L, Zhang L, Zhang Y, Zhou F. Increased expression of LncRNA BANCR is associated with clinical progression and poor prognosis in gastric cancer. Biomed Pharmacother. 2015;72:109–112. doi: 10.1016/j.biopha.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Fu Z, Ji C, Gu P, Xu P, Yu N, Kan Y, Wu X, Shen R, Shen Y. Systematic gene microarray analysis of the lncRNA expression profiles in human uterine cervix carcinoma. Biomed Pharmacother. 2015;72:83–90. doi: 10.1016/j.biopha.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Yang Q, Xu E, Dai J, Liu B, Han Z, Wu J, Zhang S, Peng B, Zhang Y, Jiang Y. A novel long noncoding RNA AK001796 acts as an oncogene and is involved in cell growth inhibition by resveratrol in lung cancer. Toxicol Appl Pharmacol. 2015;285:79–88. doi: 10.1016/j.taap.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/β-catenin signaling pathway. Oncotarget. 2017;8:17785–17794. doi: 10.18632/oncotarget.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan Y, Fan Q, Wang L, Zhou Y, Li J, Zhou K. LncRNA Snhg1, a non-degradable sponge for miR-338, promotes expression of proto-oncogene CST3 in primary esophageal cancer cells. Oncotarget. 2017;8:35750–35760. doi: 10.18632/oncotarget.16189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Zhang Z, Xiong L, Guo C, Jiang T, Zeng L, Li G, Wang J. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;487:146–152. doi: 10.1016/j.bbrc.2017.03.169. [DOI] [PubMed] [Google Scholar]
- 13.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jalving M, Heijink DM, Koornstra JJ, Boersma-van Ek W, Zwart N, Wesseling J, Sluiter WJ, de Vries EG, Kleibeuker JH, de Jong S. Regulation of TRAIL receptor expression by β-catenin in colorectal tumours. Carcinogenesis. 2014;35:1092–1099. doi: 10.1093/carcin/bgt484. [DOI] [PubMed] [Google Scholar]
- 16.Rashed HE, Hussein S, Mosaad H, Abdelwahab MM, Abdelhamid MI, Mohamed SY, Mohamed AM, Fayed A. Prognostic significance of the genetic and the immunohistochemical expression of epithelial-mesenchymal-related markers in colon cancer. Cancer Biomark. 2017;20:107–122. doi: 10.3233/CBM-170034. [DOI] [PubMed] [Google Scholar]
- 17.Ji L, Wei Y, Jiang T, Wang S. Correlation of Nrf2, NQO1, MRP1, cmyc and p53 in colorectal cancer and their relationships to clinicopathologic features and survival. Int J Clin Exp Pathol. 2014;7:1124–1131. [PMC free article] [PubMed] [Google Scholar]
- 18.Senol S, Ceyran AB, Kösemetin D, Gobanoglu B, Aydin D, Duran EA, Leblebici M. Immunohistochemical profile of tumor pathways and prognostic significance in colon adenocarcinomas. J Environ Pathol Toxicol Oncol. 2017;36:29–41. doi: 10.1615/JEnvironPatholToxicolOncol.2017016530. [DOI] [PubMed] [Google Scholar]
- 19.Weijun C, Hailu W, Yushu Z, Zhenqiu C. Study on expression of long non-coding RNA in colon carcinoma. Linchuang Zhongliuxue Zazhi. 2013;18:882–886. [Google Scholar]
- 20.Xu X, Chen R, Li Z, Huang N, Wu X, Li S, Li Y, Wu S. MicroRNA-490-3p inhibits colorectal cancer metastasis by targeting TGFβR1. BMC Cancer. 2015;15:1023. doi: 10.1186/s12885-015-2032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agüero F, Murta-Nascimento C, Gallén M, Andreu-García M, Pera M, Hernández C, Burón A, Macià F. Colorectal cancer survival: Results from a hospital-based cancer registry. Rev Esp Enferm Dig. 2012;104:572–577. doi: 10.4321/S1130-01082012001100004. [DOI] [PubMed] [Google Scholar]
- 22.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, Hall JA, Miraldi ER, Ng CP, Rigo F, et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015;528:517–522. doi: 10.1038/nature16193. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Zhang M, Wang W, Li T, Yu X, Zhu Y, Ding F, Li D, Yang T. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother. 2016;80:73–79. doi: 10.1016/j.biopha.2016.02.036. [DOI] [PubMed] [Google Scholar]
- 26.You J, Fang N, Gu J, Zhang Y, Li X, Zu L, Zhou Q. Non-coding RNA small nucleolar RNA host gene 1 promote cell proliferation in nonsmall cell lung cancer. Indian J Cancer. 2014;7:99–102. doi: 10.4103/0019-509X.154092. [DOI] [PubMed] [Google Scholar]
- 27.Chaudhry MA. Small nucleolar RNA host genes and long non-coding RNA responses in directly irradiated and bystander cells. Cancer Biother Radiopharm. 2014;29:135–141. doi: 10.1089/cbr.2013.1574. [DOI] [PubMed] [Google Scholar]
- 28.Sahu D, Hsu CL, Lin CC, Yang TW, Hsu WM, Ho SY, Juan HF, Huang HC. Co-expression analysis identifies long noncoding RNA SNHG1 as a novel predictor for event-free survival in neuroblastoma. Oncotarget. 2016;7:58022–58037. doi: 10.18632/oncotarget.11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenbluh J, Wang X, Hahn WC. Genomic insights into WNT/β-catenin signaling. Trends Pharmacol Sci. 2014;35:103–109. doi: 10.1016/j.tips.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui J, Zhou X, Liu Y, Tang Z, Romeih M. Wnt signaling in hepatocellular carcinoma: Analysis of mutation and expression of beta-catenin, T-cell factor-4 and glycogen synthase kinase 3-beta genes. J Gastroenterol Hepatol. 2003;18:280–287. doi: 10.1046/j.1440-1746.2003.02973.x. [DOI] [PubMed] [Google Scholar]
- 31.Prasetyanti PR, Zimberlin CD, Bots M, Vermeulen L, Melo FS, Medema JP. Regulation of stem cell self-renewal and differentiation by Wnt and Notch are conserved throughout the adenoma-carcinoma sequence in the colon. Mol Cancer. 2013;12:126. doi: 10.1186/1476-4598-12-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greco A, De Virgilio A, Rizzo MI, Pandolfi F, Rosati D, de Vincentiis M. The prognostic role of E-cadherin and β-catenin overexpression in laryngeal squamous cell carcinoma. Laryngoscope. 2016;126:E148–E155. doi: 10.1002/lary.25736. [DOI] [PubMed] [Google Scholar]
- 33.Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- 34.Stoyianni A, Goussia A, Pentheroudakis G, Siozopoulou V, Ioachim E, Krikelis D, Golfinopoulos V, Cervantes A, Bobos M, Fotsis T, et al. Immunohistochemical study of the epithelial-mesenchymal transition phenotype in cancer of unknown primary: Incidence, correlations and prognostic utility. Anticancer Res. 2012;32:1273–1281. [PubMed] [Google Scholar]
- 35.MacDonald BT, Tamai K, He X. Wnt/β-catenin signaling: Components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vlad A, Röhrs S, Klein-Hitpass L, Müller O. The first five years of the Wnt targetome. Cell Signal. 2008;20:795–802. doi: 10.1016/j.cellsig.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 37.Prensner JR, Chen W, Han S, Iyer MK, Cao Q, Kothari V, Evans JR, Knudsen KE, Paulsen MT, Ljungman M, et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16:900–908. doi: 10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto K, Lee BJ, Li C, Dubois RL, Hobeika E, Bhagat G, Zha S. Early B-cell-specific inactivation of ATM synergizes with ectopic CyclinD1 expression to promote pre-germinal center B-cell lymphomas in mice. Leukemia. 2015;29:1414–1424. doi: 10.1038/leu.2015.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.