We introduce an artificial intelligence approach to de novo design of molecules with desired physical or biological properties.
Abstract
We have devised and implemented a novel computational strategy for de novo design of molecules with desired properties termed ReLeaSE (Reinforcement Learning for Structural Evolution). On the basis of deep and reinforcement learning (RL) approaches, ReLeaSE integrates two deep neural networks—generative and predictive—that are trained separately but are used jointly to generate novel targeted chemical libraries. ReLeaSE uses simple representation of molecules by their simplified molecular-input line-entry system (SMILES) strings only. Generative models are trained with a stack-augmented memory network to produce chemically feasible SMILES strings, and predictive models are derived to forecast the desired properties of the de novo–generated compounds. In the first phase of the method, generative and predictive models are trained separately with a supervised learning algorithm. In the second phase, both models are trained jointly with the RL approach to bias the generation of new chemical structures toward those with the desired physical and/or biological properties. In the proof-of-concept study, we have used the ReLeaSE method to design chemical libraries with a bias toward structural complexity or toward compounds with maximal, minimal, or specific range of physical properties, such as melting point or hydrophobicity, or toward compounds with inhibitory activity against Janus protein kinase 2. The approach proposed herein can find a general use for generating targeted chemical libraries of novel compounds optimized for either a single desired property or multiple properties.
INTRODUCTION
The combination of big data and artificial intelligence (AI) was referred to by the World Economic Forum as the fourth industrial revolution that can radically transform the practice of scientific discovery (1). AI is revolutionizing medicine (2) including radiology, pathology, and other medical specialties (3). Deep learning (DL) technologies are beginning to find applications in drug discovery (4, 5) including areas of molecular docking (6), transcriptomics (7), reaction mechanism elucidation (8), and molecular energy prediction (9, 10).
The crucial step in many new drug discovery projects is the formulation of a well-motivated hypothesis for new lead compound generation (de novo design) or compound selection from available or synthetically feasible chemical libraries based on the available structure-activity relationship (SAR) data. The design hypotheses are often biased toward preferred chemistry (11) or driven by model interpretation (12). Automated approaches for designing compounds with the desired properties de novo have become an active field of research in the last 15 years (13–15). The diversity of synthetically feasible chemicals that can be considered as potential drug-like molecules was estimated to be between 1030 and 1060 (16). Great advances in computational algorithms (17, 18), hardware, and high-throughput screening technologies (19) notwithstanding, the size of this virtual library prohibits its exhaustive sampling and testing by systematic construction and evaluation of each individual compound. Local optimization approaches have been proposed, but they do not ensure the optimal solution, as the design process converges on a local or “practical” optimum by stochastic sampling or restricts the search to a defined section of chemical space that can be screened exhaustively (13, 20, 21).
Notably, a method for exploring chemical space based on continuous encodings of molecules was proposed recently (22). It allows efficient, directed gradient-based search through chemical space but does not involve biasing libraries toward special physical or biological properties. Another very recent approach for generating focused molecular libraries with the desired bioactivity using recurrent neural networks (RNNs) was proposed as well (23); however, properties of produced molecules could not be controlled well. An adversarial autoencoder was proposed (24) as a tool for generating new molecules with the desired properties; however, compounds of interest are selected by means of virtual screening of large libraries, not by designing novel molecules. Specifically, points from the latent space of chemical descriptors are projected to the nearest known molecule in the screening database, which are regarded as hit compounds.
Herein, we propose a novel method for generating chemical compounds with desired physical, chemical, and/or bioactivity properties de novo that is based on deep reinforcement learning (RL). RL is a subfield of AI, which is used to solve dynamic decision problems. It involves the analysis of possible actions and estimation of the statistical relationship between the actions and their possible outcomes, followed by the determination of a treatment regime that attempts to find the most desirable outcome. The integration of RL and neural networks dates back to the 1990s (25). However, with the recent advancement of DL, benefiting from big data, new powerful algorithmic approaches are emerging. There is a current renaissance of RL (26), especially when it is combined with deep neural networks, that is, deep RL. Most recently, RL was used to achieve superhuman performance in the game Go (27), which was considered an impossible task given the theoretical complexity of more than 10140 possible solutions (28). One may see an analogy with the complexity of chemical space exploration with an algorithm that avoids brute-force computing to examine every possible solution. Below, we describe the application of deep RL to the problem of designing chemical libraries with the desired properties and show that our approach termed ReLeaSE (Reinforcement Learning for Structural Evolution) affords a plausible solution to this problem.
The proposed ReLeaSE approach alleviates the deficiency of a small group of methodologically similar approaches discussed above. The most distinct innovative aspects of the approach proposed herein include the simple representation of molecules by their simplified molecular-input line-entry system (SMILES) strings only for both generative and predictive phases of the method and integration of these phases into a single workflow that includes a RL module. We demonstrate that ReLeaSE enables the design of chemical libraries with the desired physicochemical and biological properties. Below, we discuss both the algorithm and its proof-of-concept applications to designing targeted chemical libraries.
RESULTS
The general workflow for the ReLeaSE method (Fig. 1) includes two deep neural networks [generative (G) and predictive (P)]. The process of training consists of two stages. During the first stage, both models are trained separately with supervised learning algorithms, and during the second stage, the models are trained jointly with an RL approach that optimizes target properties. In this system, the generative model is used to produce novel chemically feasible molecules, that is, it plays a role of an agent, whereas the predictive model (that predicts the properties of novel compounds) plays the role of a critic, which estimates the agent’s behavior by assigning a numerical reward (or penalty) to every generated molecule. The reward is a function of the numerical property generated by the predictive model, and the generative model is trained to maximize the expected reward.
Fig. 1. The workflow of deep RL algorithm for generating new SMILES strings of compounds with the desired properties.
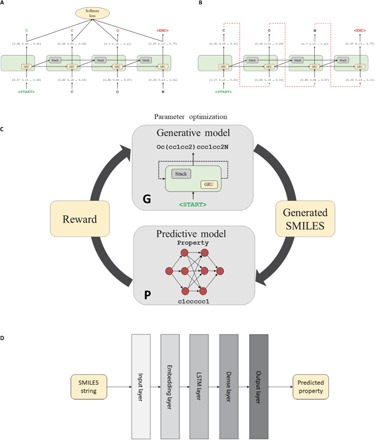
(A) Training step of the generative Stack-RNN. (B) Generator step of the generative Stack-RNN. During training, the input token is a character in the currently processed SMILES string from the training set. The model outputs the probability vector pΘ(at|st − 1) of the next character given a prefix. Vector of parameters Θ is optimized by cross-entropy loss function minimization. In the generator regime, the input token is a previously generated character. Next, character at is sampled randomly from the distribution pΘ(at| st − 1). (C) General pipeline of RL system for novel compound generation. (D) Scheme of predictive model. This model takes a SMILES string as an input and provides one real number, which is an estimated property value, as an output. Parameters of the model are trained by l2-squared loss function minimization.
RL formulation as applied to chemical library design
Both generative (G) and predictive (P) models are combined into a single RL system. The set of actions A is defined as an alphabet, that is, the entire collection of letters and symbols is used to define canonical SMILES strings that are most commonly used to encode chemical structures. For example, an aspirin molecule is encoded as [CC(═O)OC1═CC═CC═C1C(═O)O]. The set of states S is defined as all possible strings in the alphabet with lengths from zero to some value T. The state s0 with length 0 is unique and considered the initial state. The state sT of length T is called the terminal state, as it causes training to end. The subset of terminal states S* = {sT ∈ S} of S that contains all states sT with length T is called the terminal states set. Reward r(sT) is calculated at the end of the training cycle when the terminal state is reached. Intermediate rewards r(st), t < T are equal to zero. In these terms, the generative network G can be treated as a policy approximation model. At each time step t, 0 < t < T, G takes the previous state st − 1 as an input and estimates the probability distribution p(at | st − 1) of the next action. Afterward, the next action at is sampled from this estimated probability. Reward r(sT) is a function of the predicted property of sT using the predictive model P
| (1) |
where f is chosen depending on the task. Some examples of the functions f are provided in the computational experiment section. Given these notations and assumptions, the problem of generating chemical compounds with desired properties can be formulated as a task of finding a vector of parameters Θ of policy network G, which maximizes the expected reward
| (2) |
This sum iterates over the set S* of terminal states. In our case, this set is exponential, and the sum cannot be computed exactly. According to the law of large numbers, we can approximate this sum as a mathematical expectation by sampling terminal sequences from the model distribution
| (3) |
To estimate J(Θ), we sequentially sample at from the model G for t from 0 to T. The unbiased estimation for J(Θ) is the sum of all rewards in every time step, which, in our case, equals the reward for the terminal state as we assume that intermediate rewards are equal to 0. This quantity needs to be maximized; therefore, we need to compute its gradient. This can be done, for example, with the REINFORCE algorithm (29) that uses the approximation of mathematical expectation as a sum, which we provided in Eq. 3 and the following form
| (4) |
Therefore, the gradient of J(Θ) can be written down as
| (5) |
which gives an algorithm ∂ΘJ(Θ) estimation.
Neural network architectures
Model G (Fig. 1A) is a generative RNN, which outputs molecules in SMILES notation. We use a special type of stack-augmented RNN (Stack-RNN) (30) that has found success in inferring algorithmic patterns. In our implementation, we consider legitimate (that is, corresponding to chemically feasible molecules) SMILES strings as sentences composed of characters used in SMILES notation. The objective of Stack-RNN then is to learn hidden rules of forming sequences of letters that correspond to legitimate SMILES strings.
To generate a valid SMILES string, in addition to correct valence for all atoms, one must count ring opening and closure, as well as bracket sequences with several bracket types. Regular RNNs such as long short-term memory (LSTM) (31) and gated recurrent unit (GRU) (32) are unable to solve the sequence prediction problems because of their inability to count. One of the classical examples of sequences that cannot be properly modeled by regular RNNs are words from the Dyck language, where all open square brackets are matched with the respective closed ones (33). Formal language theory states that context-free languages, such as the Dyck language, cannot be generated by model without stack memory (34). As a valid SMILES string should at least be a sequence of all properly matched parentheses with multiple types of brackets, RNNs with an additional memory stack present a theoretically justified choice for modeling SMILES. Another weakness of regular RNNs is their inability to capture long-term dependencies, which leads to difficulties in generalizing to longer sequences (35). All of these features are required to learn the language of the SMILES notation. In a valid SMILES molecule, in addition to correct valence for all atoms, one must count ring opening and closure, as well as bracket sequences with several bracket types. Therefore, only memory-augmented neural networks such as Stack-RNN or Neural Turing Machines are the appropriate choice for modeling these sequence dependencies.
The Stack-RNN defines a new neuron or cell structure on top of the standard GRU cell (see Fig. 1A). It has two additional multiplicative gates referred to as the memory stack, which allow the Stack-RNN to learn meaningful long-range interdependencies. Stack memory is a differentiable structure onto and from which continuous vectors are inserted and removed. In stack terminology, the insertion operation is called PUSH operation and the removal operation is called POP operation. These traditionally discrete operations are continuous here, since PUSH and POP operations are permitted to be real values in the interval (0, 1). Intuitively, we can interpret these values as the degree of certainty with which some controller wishes to PUSH a vector v onto the stack or POP the top of the stack. Such an architecture resembles a pushdown automaton, which is a classic framework from the theory of formal languages, capable of dealing with more complicated languages. Applying this concept to neural networks provides the possibility to build a trainable model of the language of SMILES with correct syntaxes, proper balance of ring opening and closures, and correct valences for all elements.
The second model P is a predictive model (see Fig. 1D) for estimating physical, chemical, or biological properties of molecules. This property prediction model is a deep neural network, which consists of an embedding layer, an LSTM layer, and two dense layers. This network is designed to calculate user-specified property (activity) of the molecule taking a SMILES string as an input data vector. In a practical sense, this learning step is analogous to traditional quantitative structure–activity relationships (QSAR) models. However, unlike conventional QSAR, no numerical descriptors are needed, as the model distinctly learns directly from the SMILES notation as to how to relate the comparison between SMILES strings to that between target properties.
Generation of chemicals with novel structures
The generative network was trained with ~1.5 million structures from the ChEMBL21 database (please see Materials and Methods for technical details) (36); the objective of the training was to learn rules of organic chemistry that define SMILES strings corresponding to realistic chemical structures. To demonstrate the versatility of the baseline (unbiased) Stack-RNN, we generated over 1M compounds. All structures are available for download from the Supplementary Materials. Random examples of the generated compounds are shown in Fig. 2.
Fig. 2. A sample of molecules produced by the generative model.
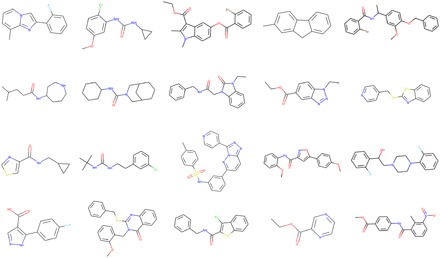
A known deficiency of approaches for de novo molecular design is frequent generation of chemically infeasible molecules (22, 37). To address this possible issue of concern, we have established that 95% of all generated structures were valid, chemically sensible molecules. The validity check was performed by the structure checker from ChemAxon (38). We compared the 1M de novo–generated molecules with those used to train the generative model from the ChEMBL database and found that the model produced less than 0.1% of structures from the training data set. Additional comparison with the ZINC15 database (39) of 320M synthetically accessible drug-like molecules showed that about 3% (~32,000 molecules) of de novo–generated structures could be found in ZINC. All ZINC IDs for the matching molecules are available in the Supplementary Materials.
To assess the importance of using a stack memory–augmented network as described in Materials and Methods, we compared several characteristics of chemical libraries generated by models developed either with or without stack memory. For this purpose, we trained another generative RNN with the same architecture but without using stack memory. Libraries were compared by the percentage of valid generated SMILES, internal diversity, and similarity of the generated molecules to those in the training data set (ChEMBL). The model without stack memory showed that only 86% of molecules in the respective library were valid (as evaluated by ChemAxon; cf. Materials and Methods) compared to 95% of molecules being valid in the library generated with stack memory network. As expected (cf. the justification for using stack memory augmented network in Materials and Methods), in the former library, syntactic errors such as open brackets, unclosed cycles, and incorrect bond types in SMILES strings were more frequent. On the basis of the analysis of respective sets of 10,000 molecules generated by each method (see Fig. 3A), the library obtained without stack memory showed a decrease in internal diversity of 0.2 units of the Tanimoto coefficient and yet a fourfold increase in the number of duplicates, from just about 1 to 5%. In addition, Fig. 3B shows that the number of molecules similar to the training data set (Ts > 0.85) for the library obtained without stack memory (28% of all molecules) is twice the number for the library obtained with stack memory (14%). These results highlight the advantages of using a neural network with memory for generating the highest number of realistic and predominantly novel molecules, which is one of the chief objectives of de novo chemical design.
Fig. 3. Performance of the generative model G, with and without stack-augmented memory.
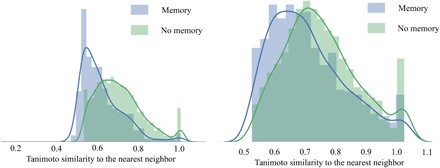
(A) Internal diversity of generated libraries. (B) Similarity of the generated libraries to the training data set from the ChEMBL database.
To further characterize the structural novelty of the de novo–generated molecules, we compared the content of the Murcko scaffolds (40) between the ChEMBL training set and the virtual library generated by our system. Murcko scaffolds provide a hierarchical molecular organization scheme by dividing small molecules into R groups, linkers, and frameworks (or scaffolds). They define the ring systems of a molecule by removing side chain atoms. We found that less than 10% of scaffolds in our library were present in ChEMBL. Overall, this analysis suggests that the generative Stack-RNN model did not simply memorize the training SMILES sequences but was indeed capable of generating extremely diverse yet realistic molecules as defined by the structure checker from ChemAxon.
In addition to passing the structure checker, an important requirement for de novo–generated molecules is their synthetic feasibility. To this end, we used the synthetic accessibility score (SAS) method (41), which relies on the knowledge extracted from known synthetic reactions and adds penalty for high molecular complexity. For ease of interpretation, SAS is scaled to be between 1 and 10. Molecules with high SAS values, typically above 6, are considered difficult to synthesize, whereas molecules with low SAS values are easily synthetically accessible. The distribution of SAS values calculated for 1M molecules generated by the ReLeaSE is shown in fig. S1. To illustrate the robustness of the de novo–generated chemical library, we compared its SAS distribution with that of the SAS values both for the full ChEMBL library (~1.5 million molecules) and for 1M random sample of molecules in ZINC. Similar to typical commercial vendor libraries, distribution of SAS for ReLeaSE is skewed toward more easily synthesizable molecules. Median SAS values were 2.9 for ChEMBL and 3.1 for both ZINC and ReLeaSE. More than 99.5% of de novo–generated molecules had SAS values below 6. Therefore, despite their high novelty, most generated compounds can be considered synthetically accessible.
Property prediction
For over more than 50 years of active development of the field, well-defined QSAR protocols and procedures have been established (42), including best practices for model validation, as reported in several highly cited papers by our group (42, 43). Any QSAR method can be generally defined as an application of machine learning (ML) and/or statistical methods to the problem of finding empirical relationships of the form y = ƒ(X1, X2,…,Xn), where y is the biological activity (or any property of interest) of molecules; X1, X2,…, Xn are calculated molecular descriptors of compounds; and ƒ is some empirically established mathematical transformation that should be applied to descriptors to calculate the property values for all molecules. Model validation is a critical component of model development; our approach to model validation in this study is described in Materials and Methods.
Building ML models directly from SMILES strings, which is a unique feature of our approach, completely bypasses the most traditional step of descriptor generation in QSAR modeling. In addition to being relatively slow, descriptor generation is nondifferentiable, and it does not allow a straightforward inverse mapping from the descriptor space back to molecules albeit a few approaches for such mapping (that is, inverse QSAR) have been proposed (44–46). For instance, one of the studies described above (22) used mapping from the point in a latent variable to real molecules represented by points most proximal to that point. In contrast, using neural networks directly on SMILES is fully differentiable, and it also enables direct mapping of properties to the SMILES sequence of characters (or strings). SMILES strings were previously used for QSAR model building (47, 48); however, in most cases, SMILES strings were used to derive string- and substring-based numerical descriptors (49). Note that, in our case, the ability to develop QSAR models using SMILES was critical for integrating property assessment (evaluative models) and de novo structure generation (generative models) into a single RL workflow, as described below.
In terms of external prediction accuracy, SMILES-based ML models also performed very well. For example, using fivefold cross-validation (5CV), we obtained the external model accuracy expressed as R2ext of 0.91 and root mean square error (RMSE) = 0.53 for predicting logP (see Materials and Methods). This compared favorably to a random forest model with DRAGON7 descriptors (R2ext = 0.90 and RMSE = 0.57). For the melting temperature (Tm) prediction, the observed RMSE of 35°C was the same as that predicted with the state-of-the-art consensus model obtained by using an ensemble of multiple conventional descriptor-based ML models (50), which afforded an RMSE of 35°C.
The following study was undertaken to evaluate the external predictive accuracy for novel compounds designed with the ReLeaSE method. We have identified more than 100 compounds that were not present in the training set from our library in the ChEMBL database. Then, we manually extracted their experimental logP or Tm data from the PubChem database. Multiple measurements were averaged. Final subsets were composed from about 20 molecules for each property. The comparison between predicted and experimental measurements yielded an RMSE of 0.9 for logP and ~42°C for Tm. This accuracy was slightly lower than that for the respective quantitative structure–property relationship (QSPR) model obtained with cross-validation. We consider the reasonable success of this exercise in property prediction for an external data set as additional evidence that our approach yields molecules with both desired and accurately predicted properties.
Generation of property value biased libraries with the RL system
To explore the utility of the RL algorithm in a drug design setting, we have conducted case studies to design libraries with three controlled target properties: (i) physical properties considered important for drug-like molecules, (ii) specific biological activity, and (iii) chemical complexity. For physical properties, we selected Tm and n-octanol/water partition coefficient (logP). For bioactivity prediction, we designed putative inhibitors of Janus protein kinase 2 (JAK2) with novel chemotypes. Finally, the number of benzene rings and the number of substituents (such as –OH, –NH2, –CH3–CN, etc.) were used as a structural reward to design novel chemically complex compounds. Figure 4 shows the distribution of predicted properties of interest in the training test molecules and in the libraries designed by our system. In all cases, we sampled 10,000 molecules by the baseline (no RL) generator and RL-optimized generative models and then calculated their properties with a corresponding predictive model. Values of the substructural features were calculated directly from the two-dimensional (2D) structure. Table 1 summarizes the analysis of generated molecules and the respective statistics.
Fig. 4. Property distributions for RL-optimized versus baseline generator model.
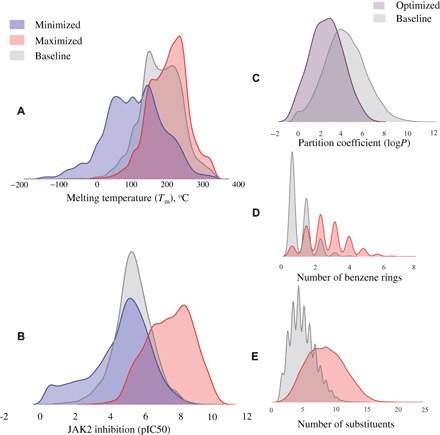
(A) Melting temperature. (B) JAK2 inhibition. (C) Partition coefficient. (D) Number of benzene rings. (E) Number of substituents.
Table 1. Comparison of statistics for generated molecular data sets.
| Property |
Valid molecules (%) |
Mean SAS |
Mean molar mass |
Mean value of target property |
Match with ZINC15 database (%) |
Match with ChEMBL database (%) |
|
| Tm | Baseline | 95 | 3.1 | 435.4 | 181 | 4.7 | 1.5 |
| Minimized | 31 | 3.1 | 279.6 | 137 | 4.6 | 1.6 | |
| Maximized | 53 | 3.4 | 413.2 | 200 | 2.4 | 0.9 | |
| Inhibition of JAK2 |
Baseline | 95 | 3.1 | 435.4 | 5.70 | 4.7 | 1.5 |
| Minimized | 60 | 3.85 | 481.8 | 4.89 | 2.5 | 1.0 | |
| Maximized | 45 | 3.7 | 275.4 | 7.85 | 4.5 | 1.8 | |
| LogP | Baseline | 95 | 3.1 | 435.4 | 3.63 | 4.7 | 1.5 |
| Range- optimized |
70 | 3.2 | 369.7 | 2.58 | 5.8 | 1.8 | |
| Number of benzene rings |
Baseline | 95 | 3.1 | 435.4 | 0.59 | 4.7 | 1.5 |
| Maximized | 83 | 3.15 | 496.0 | 2.41 | 5.5 | 1.6 | |
| Number of substituents |
Baseline | 95 | 3.1 | 435.4 | 3.8 | 4.7 | 1.5 |
| Maximized | 80 | 3.5 | 471.7 | 7.93 | 3.1 | 0.7 | |
Melting temperature
In this experiment, we set two goals: either to minimize or to maximize the target property. Upon minimization, the mean of the distribution in the de novo–generated library was shifted by 44°C, as compared to the training set distribution (Fig. 4A). The library of virtually synthesized chemicals included simple hydrocarbons such as butane, as well as polyhalogenated compounds such as CF2Cl2 and C6H4F2. The molecule with the lowest Tm = −184°C in the produced data set was CF4. This property minimization strategy was extremely effective, as it allowed for the discovery of molecules in the regions of the chemical space far beyond those of the training set of drug-like compounds. In the maximization regime, the mean of the Tm was increased by 20° to 200°C. As expected, the generated library indeed included substantially more complex molecules with the abundance of sulfur-containing heterocycles, phosphates, and conjugated double-bond moieties.
Designing a chemical library biased toward a range of lipophilicity (logP)
Compound hydrophobicity is an important consideration in drug design. One of the components of the famous Lipinski’s rule of five is that orally bioavailable compounds should have their octanol-water partition coefficient logP less than 5 (51). Thus, we endeavored to design a library that would contain compounds with logP values within a favorable drug-like range. The reward function in this case was defined as a piecewise linear function of logP with a constant region from 1.0 to 4.0 (see fig. S2). In other words, we set the goal to generate molecules according to a typical Lipinski’s constraint. As shown in Fig. 4C, we have succeeded in generating a library with 88% of the molecules falling within the drug-like region of logP values.
Inhibition of JAK2
In the third experiment, which serves as an example of the most common application of computational modeling in drug discovery, we have used our system to design molecules with the specific biological function, that is, JAK2 activity modulation. Specifically, we designed libraries with the goal of minimizing or maximizing negative logarithm of half maximal inhibitory concentration (pIC50) values for JAK2. While most of drug discovery studies are oriented toward finding molecules with heightened activity, bioactivity minimization is also pursued in drug discovery to mitigate off-target effects. Therefore, we were interested in exploring the ability of our system to bias the design of novel molecular structures toward any desired range of the target properties. JAK2 is a nonreceptor tyrosine kinase involved in various processes such as cell growth, development, differentiation, or histone modifications. It mediates essential signaling events in both innate and adaptive immunity. In the cytoplasm, it also plays an important role in signal transduction. Mutations in JAK2 have been implicated in multiple conditions such as thrombocythemia, myelofibrosis, or myeloproliferative disorders (52).
The reward functions in both cases (minimization and maximization) were defined as exponential functions of pIC50 (see fig. S2). The results of library optimization are shown in Fig. 4B. With minimization, the mean of predicted pIC50 distribution was shifted by about one pIC50 unit, and the distribution was heavily biased toward the lower ranges of bioactivity with 24% of molecules predicted to have practically no activity (pIC50 ≤ 4). In the activity maximization exercise, properties of generated molecules were more tightly distributed across the predicted activity range. In each case, our system virtually synthesized both known and novel compounds, with most de novo–designed molecules being novel compounds. The generation of known compounds (that is, not included in the training set) can be regarded as model validation. The system retrospectively discovered 793 commercially available compounds deposited in the ZINC database, which constituted about 5% of the total generated library. As many as 15 of them [exemplified by ZINC263823677 (http://zinc15.docking.org/substances/ZINC000263823677/) and ZINC271402431 (http://zinc15.docking.org/substances/ZINC000271402431/)] were actually annotated as possible tyrosine kinase inhibitors.
Substructure bias
Finally, we also performed two simple experiments mimicking the strategy of biased chemical library design where the designed library is enriched with certain user-defined substructures. We defined the reward function as the exponent of (i) the number of benzene rings (–Ph) and (ii) the total number of small group substituents. Among all case studies described, structure bias was found to be the easiest to optimize. The results of the library optimization study are shown in Fig. 4 (D and E). Furthermore, Fig. 5 illustrates the evolution of generated structures as the structural reward increases. We see that the model progresses toward generating increasingly more complex, yet realistic molecules with greater numbers of rings and/or substituents.
Fig. 5. Evolution of generated structures as chemical substructure reward increases.
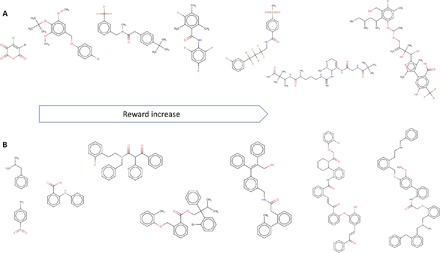
(A) Reward proportional to the total number of small group substituents. (B) Reward proportional to the number of benzene rings.
We expect that designing structurally biased libraries may be a highly desirable application of the ReLeaSE approach as researchers often wish to generate libraries enriched for certain privileged scaffold(s) and lead compound optimization (53). Conversely, the system also allows the avoidance of particular chemical groups or substructures (such as bromine or carboxyl group) that may lead to undesired compound properties such as toxicity. Finally, one could implement a certain substructure, or pharmacophore similarity, reward to explore additional chemical space.
Table 1 shows a decrease in the proportion of valid molecules after the optimization. We may explain this phenomenon by the weaknesses of predictive models P (see Fig. 1C) and the integration of predictive and generative models into a single design system. We presume that the generative model G tends to find some local optima of the reward function that correspond to invalid molecules, but the predictive model P assigns high rewards to these molecules. This explanation is also supported by the results of structure bias optimization experiments, as we did not use any predictive models in these experiments and the decrease in the proportion of valid molecules was insignificant. We also noticed that, among all experiments with predictive models, those with logP optimization showed the highest proportion of valid molecules and, at the same time, the predictive model for logP estimation had the highest accuracy R2 = 0.91 (see Materials and Methods). It is probably harder for the RL system to exploit the high-quality predictive model P and produce fictitious SMILES strings with predicted properties in the desired region.
Model analysis
Model interpretation is a highly significant component in any ML study. In this section, we demonstrate how Stack-RNN learns and memorizes useful information from the SMILES string as it is being processed. More specifically, we have manually analyzed neuron gate activations of the neural network as it processes the input data.
Figure 6 lists several examples of cells in neural networks with interpretable gate activations. In this figure, each line corresponds to activations of a specific neuron at different SMILES processing time steps by the pretrained baseline generative model. Each letter is colored according to the value of tanh activation in a cool-warm color map from dark blue to dark red, that is, from −1 to 1. We found that our RNN has several interpretable cells. These cells can be divided into two kinds of groups: chemically sensible groups, which activate in the presence of specific chemical groups or moieties, and syntactic groups, which keep tracks of numbers, bracket opening and closure, and even of SMILES string termination when the new molecule is generated. For instance, we saw cells reflecting the presence of a carbonyl group, aromatic groups, or NH moieties in heterocycles. We also observed that, in two of these three examples, there were counter cells that deactivate in the presence of the aforementioned chemical groups. Neural network–based models are notoriously uninterpretable (54), and most of the cells were indeed in that category. On the other hand, the possibility of even partial interpretation offered by this approach could be highly valuable for a medicinal chemist.
Fig. 6. Examples of Stack-RNN cells with interpretable gate activations.
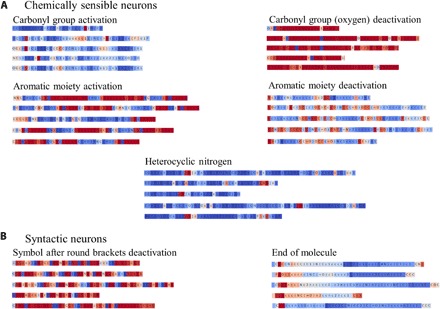
Color coding corresponds to GRU cells with hyperbolic tangent tanh activation function, where dark blue corresponds to the activation function value of −1 and red describes the value of the activation function of 1; the numbers in the range between −1 and 1 are colored using a cool-warm color map.
Visualization of new chemical libraries
To understand how the generative models populate chemical space with new molecules, we used t-distributed stochastic neighbor embedding (t-SNE) for dimensionality reduction (55). We selected data sets for three end points used in our case studies (Tm, logP, and JAK2) that were produced with corresponding optimized generative models G. For every molecule, we calculated a latent vector of representation from the feed-forward layer with a rectified linear unit (ReLU) activation function in the predictive model P for the respective property and constructed 2D projection using t-SNE. These projections are illustrated in Fig. 7. Every point corresponds to a molecule and is colored according to its property value.
Fig. 7. Clustering of generated molecules by t-SNE.
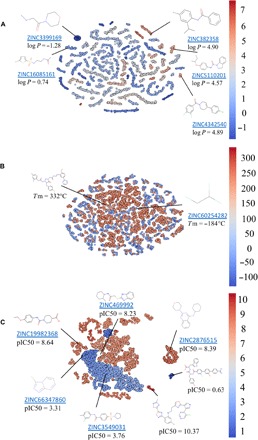
Molecules are colored on the basis of the predicted properties by the predictive model P, with values shown by the color bar on the right. (A and C) Examples of the generated molecules randomly picked from matches with ZINC database and property values predicted by the predictive model P. (A) Partition coefficient, logP. (B) Melting temperature, Tm (°C); examples show generated molecules with lowest and highest predicted Tm. (C) JAK2 inhibition, predicted pIC50.
For libraries generated to achieve certain partition coefficient distribution (Fig. 7A), we can observe well-defined clustering of molecules with similar logP values. In contrast, for Tm (Fig. 7B), there are no such clusters. High and low Tm molecules are intermixed together. This observation can be explained by the fact that Tm depends not only on the chemical structure of the molecule but also on intermolecular forces and packing in the crystal lattice. Therefore, plotting molecules in this neural net representation could not achieve good separation of high versus low Tm. In the case of the JAK2 model, we could observe two large nonoverlapping areas roughly corresponding to inactive (pIC50 < 6) and active (pIC50 ≥ 6) compounds. Inside these areas, molecules are typically clustered around multiple privileged scaffolds. Specifically for JAK2, we see an abundance of compounds with 1,3,5-triazine, 1,2,4-triazine, 5-methyl-1H-1,2,4-triazole, 7H-pyrrolo[2,3-d]pyrimidine, 1H-pyrazolo[3,4-d]pyrimidine, thieno-triazolo-pyrimidine, and other substructures. Overall, this approach offers a rapid way to visualize compound distribution in chemical space in terms of both chemical diversity and variability in the values of the specific prediction end point. Furthermore, joint embedding of both molecules in the training set and those generated de novo allows one to explore differences in the chemical space coverage by both sets and establish whether structurally novel compounds also have the desired predicted property of interest.
DISCUSSION
We have created and implemented a deep RL approach termed ReLeaSE for de novo design of novel chemical compounds with desired properties. To achieve this outcome, we combined two deep neural networks (generative and predictive) in a general workflow that also included the RL step (Fig. 1). The training process consists of two stages. In the first stage, both models were trained separately using supervised learning, and in the second stage, models were trained jointly with an RL method. Both neural networks use end-to-end DL. The ReLeaSe method does not rely on predefined chemical descriptors; the models are trained on chemical structures represented by SMILES strings only. This distinction differentiates this approach from traditional QSAR methods and simpler to both use and execute.
This method needs to be evaluated in the context of several previous and parallel developments elsewhere to highlight its unique innovative features. Our ReLeaSE method has benefited from the recent developments in the ML community as applied to natural language processing and machine translation. These new algorithms allow learning the mapping from an input sequence (for example, a sentence in one language) to an output sequence (that same sentence in another language). The entire input sentence represents an input vector for the neural network. The advantage of this approach is that it requires no handcrafted feature engineering.
Considering the use of similar approaches in chemistry, several comparable developments elsewhere should be discussed. RL approach for de novo molecular design was introduced in reference (37) as well. However, no data were provided to show that the predicted properties of molecular compounds are optimized. Instead of demonstrating the shift in distribution of biological activity values against dopamine receptor type 2 before and after the optimization, that study showed an increase in the fraction of the generated molecules, which are similar to those in training and test sets. This increase does not automatically mean that the generative model is capable of producing novel active compounds. In contrast, this result may indicate a model’s weaknesses in predicting novel valuable chemicals that are merely similar to the training set compounds, that is, the model is fitted to the training set but may have a limited ability to generate novel chemicals that are substantially different from the training set compounds. The generative model in references (23, 37) is a “vanilla” RNN without augmented memory stack, which does not have the capacity to count and infer algorithmic patterns (34). Another weakness of the approach described in reference (37), from our point of view, is the usage of a predictive model built with numerical molecular descriptors, whereas we propose a model that is essentially descriptor-free and naturally forms a coherent workflow together with the generative model. After the manuscript of our study was submitted for publication, a study by Jaques et al. (56) that used simple RNN and off-policy RL to generate molecules was published. However, in addition to low percentage (~30 to 35%) of valid molecules, in that study, the authors did not directly optimize any physical or biological properties but rather a proxy function that includes a SAS, drug-likeness, and a ring penalty.
It is important to highlight the critical element of using QSAR models as part of our approach as opposed to the traditional use of QSAR models for virtual screening of chemical libraries. The absolute majority of compounds generated de novo by the ReLeaSE method are novel structures as compared to the data sets used to train generative models, and any QSAR model could be used to evaluate their properties. However, one of our chief objectives was to develop a method that can tune not only structural diversity (cf. case study 1) but, most importantly, also bias the property (physical or biological) toward the desired range of values (case studies 2 and 3). The principal element of the ReLeaSE method as compared to traditional QSAR models is that QSAR models are implemented within the ReLEeaSE such as to put “pressure” on the generative model. Thus, although any QSAR model could evaluate properties of new chemicals, those built into our method are used directly for RL to bias de novo library design toward the desired property.
As a proof of principle, we tested our approach on three diverse types of end points: physical properties, biological activity, and chemical substructure bias. The use of flexible reward function enables different library optimization strategies where one can minimize, maximize, or impose a desired range to a property of interest in the generated compound libraries. As a by-product of these case studies, we have generated a data set of more than 1M of novel compounds. Here, we have focused on presenting the new methodology and its application for initial hit generation. However, ReLeaSE could also be used for lead optimization, where a particular privileged scaffold is fixed and only substituents are optimized. Our future studies will explore this direction.
Computational library design methods are often criticized for their inability to control synthetic accessibility of de novo–generated molecules (13). Computationally generated compounds are often quite complex; for instance, they may include exotic substituents. In many cases, these compounds may require multistep custom syntheses or could even be synthetically inaccessible with the current level of technology. In the pharmaceutical industry, the ease of synthesis of a prospective hit molecule is of primary concern as it strongly affects the cost of the manufacturing process required for the industrial-scale production. For all experiments in this paper, the synthetic accessibility of de novo–generated–focused libraries was estimated using the SAS (41). Distributions of SAS values are shown in fig. S3, and the medians of the SAS are listed in Table 1. This analysis shows that property optimization does not significantly affect synthetic accessibility of the generated molecules. The biggest shift of 0.75 for the distribution median was observed in the proof-of-concept study targeting the design of JAK2 inhibitors with minimized activity. Less than 0.5% of molecules had a high SAS of >6, which is an approximate cutoff for systems that are difficult to synthesize (41).
Obviously, it is technically feasible to include the SAS as an additional reward function; however, in our opinion, there are two main reasons as to why this is not desirable, at least with the current form of SAS. First, predicted SAS for newly generated molecules are practically independent of property optimization. Their distribution follows that from commercially available compounds. Second, “synthetic accessibility” is not a well-defined concept (57). In the process chemistry, it depends on multiple factors that determine the ease of synthesis of a particular molecule such as the availability of reagents, the number and difficulty of synthetic steps, the stability of intermediate products, the ease of their separation, reaction yields, etc. (58). In contrast, the most commonly used SAS method (also used in this work) is based on molecular complexity as defined by the number of substructures and molecular fragments (41). Therefore, optimizing SAS with RL as part of our approach would result in substantially reduced novelty of generated molecules and a bias toward substructures with low SAS used to train the model.
In summary, we have devised and executed a new strategy termed ReLeaSE for designing libraries of compounds with the desired properties that uses both DL and RL approaches. In choosing the abbreviation for the name of the method, we were mindful of one of the key meanings of the word “release,” that is, to “allow or enable to escape from confinement; set free.” We have conducted computational experiments that demonstrated the efficiency of the proposed ReLeaSE strategy in a single-task regime where each of the end points of interest is optimized independently. However, this system can be extended to afford multiobjective optimization of several target properties concurrently, which is the need of drug discovery where the drug molecule should be optimized with respect to potency, selectivity, solubility, and drug-likeness properties. Our future studies will address this challenge.
MATERIALS AND METHODS
Experimental data
The melting point data set was extracted from the literature (50). The PHYSPROP database (www.srcinc.com) was used to extract the octanol/water partition coefficient, logP for diverse set of molecules. Experimental IC50 and Ki data for compounds tested against JAK2 (CHEMBL ID 2971) were extracted from ChEMBL (36), PubChem (59), and the Eidogen-Sertanty Kinase Knowledgebase [KKB Q1 2017 (http://eidogen-sertanty.com/kinasekb.php)]. Compounds that had inconclusive IC50 values were considered unreliable and were not included in the modeling.
Data curation
Compiled data sets of compounds were carefully curated following the protocols proposed by Fourches et al. (60). Briefly, explicit hydrogens were added, and specific chemotypes such as aromatic and nitro groups were normalized using ChemAxon Standardizer. Polymers, inorganic salts, organometallic compounds, mixtures, and duplicates were removed. The modeling-ready curated data set contained 14,176 compounds for logP, 15,549 compounds for JAK2, and 47,425 compounds for Tm. All molecules were stored as normalized and canonicalized SMILES strings according to procedures developed elsewhere (61).
Property prediction models
We have built QSPR models for three different properties—Tm, logP, and pIC50 for JAK2. Curated data sets for all three end points were divided into training and training sets in 5CV fashion. In developing these QSPR models, we followed standard protocols and best practices for QSPR model validation (42). Specifically, it has been shown that multiple random splitting of data sets into training and test sets affords models of the highest stability and predictive power. Uniquely, models built herein did not use any calculated chemical descriptors; rather, SMILES representations were used. Each model consisted of an embedding layer transforming the sequence of discrete tokens (that is, SMILES symbols) into a vector of 100 continuous numbers, an LSTM layer with 100 units and tanh nonlinearity, one dense layer with 100 units and rectify nonlinearity function, and one dense layer with one unit and identity activation function. All three models were trained with the learning-rate decay technique until convergence. The resulting 5CV external accuracies of the models are shown in fig. S4.
Training for the generative model
In the first stage, we pretrained a generative model on a ChEMBL21 (36) data set of approximately 1.5 million drug-like compounds so that the model was capable of producing chemically feasible molecules (note that this step does not include any property optimization). This network had 1500 units in a GRU (32) layer and 512 units in a stack augmentation layer. The model was trained on a graphics processing unit (GPU) for 10,000 epochs. The learning curve is illustrated in fig. S5.
The generative model has two modes of processing sequences—training and generating. At each time step, in the training mode, the generative network takes a current prefix of the training object and predicts the probability distribution of the next character. Then, the next character is sampled from this predicted probability distribution and is compared to the ground truth. Afterward, on the basis of this comparison, the cross-entropy loss function is calculated, and parameters of the model are updated. At each time step, in generating mode, the generative network takes a prefix of already generated sequences and then, like in the training mode, predicts the probability distribution of the next character and samples it from this predicted distribution. In the generative model, we do not update the model parameters.
At the second stage, we combined both generative and predictive models into one RL system. In this system, the generative model plays the role of an agent, whose action space is represented by the SMILES notation alphabet, and state space is represented by all possible strings in this alphabet. The predictive model plays the role of a critic estimating the agent’s behavior by assigning a numerical reward to every generated molecule (that is, SMILES string). The reward is a function of the numerical property calculated by the predictive model. At this stage, the generative model is trained to maximize the expected reward. The entire pipeline is illustrated in Fig. 1.
We trained a Stack-RNN as a generative model. As mentioned above, for training, we used the ChEMBL database of drug-like compounds. ChEMBL includes approximately 1.5 million of SMILES strings; however, we only selected molecules with the lengths of SMILES string of fewer than 100 characters. The length of 100 was chosen because more than 97% of SMILES in the training data set had 100 characters or less (see fig. S6).
Stack-augmented recurrent neural network
This section describes the generative model G in more detail (30). We assume that the data are sequential, which means that they come in the form of discrete tokens, that is, characters. The goal is to build a model that is able to predict the next token conditioning on all previous tokens. The regular RNN has an input layer and a hidden layer. At time step t, the neural network takes the embedding vector of token number t from the sequence as an input and models the probability distribution of the next token given all previous tokens so that the next token can be sampled from this distribution. Information from all previously observed tokens is aggregated in the hidden layer. This can be written down as follows
| (6) |
where ht is a vector of hidden states, ht − 1 is the vector of hidden states from the previous time step, xt is the input vector at time step t, Wi is parameters of the input layers, Wh is a parameter of the hidden layer, and σ is the activation function.
The stack memory was used to keep the information and deliver it to the hidden layer at the next time step. A stack is a type of persistent memory, which can only be accessed through its topmost element. There are three operations supported by the stack: POP operation, which deletes an element from the top of the stack; PUSH operation, which puts a new element at the top of the stack; and NO-OP operation, which performs no action. The top element of the stack has value st[0] and is stored at position 0
| (7) |
where D is a 1 × m matrix and at = [at[PUSH], at[POP], at[NO − OP]] is a vector of stack control variables, which define the next operation to be performed. If at[POP] is equal to 1, then the value below is used to replace the top element of the stack. If at[PUSH] is equal to 1, then a new value will be added to the top, and all the rest values will be moved down. If at[NO − OP] is equal to 1, then the stack keeps the same value on top.
A similar rule is applied to the elements of the stack at a depth i > 0
| (8) |
Now, the hidden layer ht is updated as
| (9) |
where D is a matrix of size m × k and are the first k elements for the top of the stack at time step t − 1.
Supplementary Material
Acknowledgments
We wish to thank S. Cappuzzi for technical discussions and for proofreading the manuscript before submission. Funding: A.T. and O.I. acknowledge support from an Eshelman Institute for Innovation award and a U.S. Department of Defense Office of Naval Research grant (N00014-16-1-2311). A.T. also thanks the Russian Scientific Foundation (agreement no. 14-43-00024 from 1 October 2014) for its partial support of proof-of-concept studies on biasing chemical libraries toward structural diversity. O.I. acknowledges Extreme Science and Engineering Discovery Environment award DMR-110088, which is supported by NSF grant number ACI-1053575. M.P. acknowledges financial support from the Skolkovo Institute of Science and Technology. We gratefully acknowledge the support and hardware donation from NVIDIA Corporation and express our special gratitude to M. Berger. Author contributions: A.T. and O.I. devised and led this project and edited the final version of the manuscript. M.P. and O.I. developed and implemented the ReLeaSE method. M.P. wrote the manuscript with input from all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Code, data, and pretrained models are available from GitHub (https://github.com/isayev/ReLeaSE) and ZIP archive in the Supplementary Materials. We release a reference PyTorch implementation of our methods and provide iPython notebook examples explaining how to run the experiments described in our paper. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/7/eaap7885/DC1
Fig. S1. Distribution of SAS for the full ChEMBL21 database (~1.5 million molecules), random subsample of 1M molecules from ZINC15, and generated data set of 1M molecules with baseline generator model G.
Fig. S2. Reward functions.
Fig. S3. Distributions of SAS for all RL experiments.
Fig. S4. Distribution of residuals and predicted versus observed plots for predictive models.
Fig. S5. Learning curve for generative model.
Fig. S6. Distributions of SMILES’s string lengths.
REFERENCES AND NOTES
- 1.Gil Y., Greaves M., Hendler J., Hirsh H., Amplify scientific discovery with artificial intelligence. Science 346, 171–172 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Krittanawong C., Zhang H., Wang Z., Aydar M., Kitai T., Artificial intelligence in precision cardiovascular medicine. J. Am. Coll. Cardiol. 69, 2657–2664 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Chockley K., Emanuel E., The end of radiology? Three threats to the future practice of radiology. J. Am. Coll. Radiol. 13, 1415–1420 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Altae-Tran H., Ramsundar B., Pappu A. S., Pande V., Low data drug discovery with one-shot learning. ACS Cent. Sci. 3, 283–293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gawehn E., Hiss J. A., Schneider G., Deep learning in drug discovery. Mol. Inform. 35, 3–14 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Ragoza M., Hochuli J., Idrobo E., Sunseri J., Koes D. R., Protein–ligand scoring with convolutional neural networks. J. Chem. Inf. Model. 57, 942–957 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aliper A., Plis S., Artemov A., Ulloa A., Mamoshina P., Zhavoronkov A., Deep learning applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data. Mol. Pharm. 13, 2524–2530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Segler M. H. S., Waller M. P., Modelling chemical reasoning to predict and invent reactions. Chemistry 23, 6118–6128 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Smith J. S., Isayev O., Roitberg A. E., ANI-1: An extensible neural network potential with DFT accuracy at force field computational cost. Chem. Sci. 8, 3192–3203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schütt K. T., Arbabzadah F., Chmiela S., Müller K. R., Tkatchenko A., Quantum-chemical insights from deep tensor neural networks. Nat. Commun. 8, 13890 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnecke V., Boström J., Computational chemistry-driven decision making in lead generation. Drug Discov. Today 11, 43–50 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Macarron R., Critical review of the role of HTS in drug discovery. Drug Discov. Today 11, 277–279 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Schneider G., Fechner U., Computer-based de novo design of drug-like molecules. Nat. Rev. Drug Discov. 4, 649–663 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Mauser H., Guba W., Recent developments in de novo design and scaffold hopping. Curr. Opin. Drug Discov. Devel. 11, 365–374 (2008). [PubMed] [Google Scholar]
- 15.B. Sanchez-Lengeling, C. Outeiral, G. L. Guimaraes, A. Aspuru-Guzik, Optimizing distributions over molecular space. An Objective-Reinforced Generative Adversarial Network for Inverse-design Chemistry (ORGANIC) (2017); https://chemrxiv.org/articles/ORGANIC_1_pdf/5309668.
- 16.Polishchuk P. G., Madzhidov T. I., Varnek A., Estimation of the size of drug-like chemical space based on GDB-17 data. J. Comput. Aided Mol. Des. 27, 675–679 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Besnard J., Ruda G. F., Setola V., Abecassis K., Rodriguiz R. M., Huang X. P., Norval S., Sassano M. F., Shin A. I., Webster L. A., Simeons F. R., Stojanovski L., Prat A., Seidah N. G., Constam D. B., Bickerton G. R., Read K. D., Wetsel W. C., Gilbert I. H., Roth B. L., Hopkins A. L., Automated design of ligands to polypharmacological profiles. Nature 492, 215–220 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reker D., Schneider P., Schneider G., Multi-objective active machine learning rapidly improves structure–activity models and reveals new protein–protein interaction inhibitors. Chem. Sci. 7, 3919–3927 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipinski C., Hopkins A., Navigating chemical space for biology and medicine. Nature 432, 855–861 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Reker D., Schneider G., Active-learning strategies in computer-assisted drug discovery. Drug Discov. Today 20, 458–465 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Brown N., McKay B., Gilardoni F., Gasteiger J., A graph-based genetic algorithm and its application to the multiobjective evolution of median molecules. J. Chem. Inf. Comput. Sci. 44, 1079–1087 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Gómez-Bombarelli R., Wei J. N., Duvenaud D., Hernández-Lobato J. M., Sánchez-Lengeling B., Sheberla D., Aguilera-Iparraguirre J., Hirzel T. D., Adams R. P., Aspuru-Guzik A., Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 4, 268–276 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segler M. H. S., Kogej T., Tyrchan C., Waller M. P., Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent. Sci. 4, 120–131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadurin A., Aliper A., Kazennov A., Mamoshina P., Vanhaelen Q., Khrabrov K., Zhavoronkov A., The cornucopia of meaningful leads: Applying deep adversarial autoencoders for new molecule development in oncology. Oncotarget 8, 10883–10890 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.K. De Asis, J. F. Hernandez-Garcia, G. Z. Holland, R. S. Sutton, Multi-step reinforcement learning: A unifying algorithm, http://arxiv.org/abs/1703.01327 (2017).
- 26.Krakovsky M., Reinforcement renaissance. Commun. ACM. 59, 12–14 (2016). [Google Scholar]
- 27.Silver D., Huang A., Maddison C. J., Guez A., Sifre L., van den Driessche G., Schrittwieser J., Antonoglou I., Panneershelvam V., Lanctot M., Dieleman S., Grewe D., Nham J., Kalchbrenner N., Sutskever I., Lillicrap T., Leach M., Kavukcuoglu K., Graepel T., Hassabis D., Mastering the game of Go with deep neural networks and tree search. Nature 529, 484–489 (2016). [DOI] [PubMed] [Google Scholar]
- 28.van den Herik H. J., Uiterwijk J. W. H. M., van Rijswijck J., Games solved: Now and in the future. Artif. Intell. 134, 277–311 (2002). [Google Scholar]
- 29.Williams R. J., Simple statistical gradient-following algorithms for connectionist reinforcement learning. Mach. Learn. 8, 229–256 (1992). [Google Scholar]
- 30.A. Joulin, T. Mikolov, Inferring algorithmic patterns with stack-augmented recurrent nets, http://arxiv.org/abs/1503.01007) (2015).
- 31.Hochreiter S., Schmidhuber J., Long short-term memory. Neural. Comput. 9, 1735–1780 (1997). [DOI] [PubMed] [Google Scholar]
- 32.J. Chung, C. Gulcehre, K. Cho, Y. Bengio, Empirical evaluation of gated recurrent neural networks on sequence modeling, http://arxiv.org/abs/1412.3555 (2014).
- 33.T. Deleu, J. Dureau, Learning operations on a stack with Neural Turing Machines, http://arxiv.org/abs/1612.00827 (2016).
- 34.J. E. Hopcroft, J. D. Ullman, in Formal languages and their relation to automata (Addison-Wesley Longman Publishing, 1969), pp. 262. [Google Scholar]
- 35.E. Grefenstette, K. M. Hermann, M. Suleyman, P. Blunsom, Learning to transduce with unbounded memory, http://arxiv.org/abs/1506.02516 (2015).
- 36.Bento A. P., Gaulton A., Hersey A., Bellis L. J., Chambers J., Davies M., Krüger F. A., Light Y., Mak L., McGlinchey S., Nowotka M., Papadatos G., Santos R., Overington J. P., The ChEMBL bioactivity database: An update. Nucleic Acids Res. 42, D1083–D1090 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.M. Olivecrona, T. Blaschke, O. Engkvist, H. Chen, Molecular de novo design through deep reinforcement learning, http://arxiv.org/abs/1704.07555 (2017). [DOI] [PMC free article] [PubMed]
- 38.ChemAxon, MarvinSketch (2017); www.chemaxon.com/products/marvin/.
- 39.Irwin J. J., Sterling T., Mysinger M. M., Bolstad E. S., Coleman R. G., ZINC: A free tool to discover chemistry for biology. J. Chem. Inf. Model. 52, 1757–1768 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bemis G. W., Murcko M. A., The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 39, 2887–2893 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Ertl P., Schuffenhauer A., Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J. Cheminform. 1, 8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tropsha A., Best practices for QSAR model development, validation, and exploitation. Mol. Inform. 29, 476–488 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Cherkasov A., Muratov E. N., Fourches D., Varnek A., Baskin I. I., Cronin M., Dearden J., Gramatica P., Martin Y. C., Todeschini R., Consonni V., Kuz’min V. E., Cramer R., Benigni R., Yang C., Rathman J., Terfloth L., Gasteiger J., Richard A., Tropsha A., QSAR modeling: Where have you been? Where are you going to? J. Med. Chem. 57, 4977–5010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho S. J., Zheng W., Tropsha A., Rational combinatorial library design. 2. Rational design of targeted combinatorial peptide libraries using chemical similarity probe and the inverse QSAR approaches. J. Chem. Inf. Comput. Sci. 38, 259–268 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Brüggemann R., Pudenz S., Carlsen L., Sørensen P. B., Thomsen M., Mishra R. K., The use of Hasse diagrams as a potential approach for inverse QSAR. SAR QSAR Environ. Res. 11, 473–487 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Miyao T., Kaneko H., Funatsu K., Inverse QSPR/QSAR analysis for chemical structure generation (from y to x). J. Chem. Inf. Model. 56, 286–299 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Toropov A. A., Benfenati E., SMILES as an alternative to the graph in QSAR modelling of bee toxicity. Comput. Biol. Chem. 31, 57–60 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Haque I. S., Pande V. S., Walters W. P., SIML: A fast SIMD algorithm for calculating LINGO chemical similarities on GPUs and CPUs. J. Chem. Inf. Model. 50, 560–564 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ikebata H., Hongo K., Isomura T., Maezono R., Yoshida R., Bayesian molecular design with a chemical language model. J. Comput. Aided Mol. Des. 31, 379–391 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tetko I. V., Sushko Y., Novotarskyi S., Patiny L., Kondratov I., Petrenko A. E., Charochkina L., Asiri A. M., How accurately can we predict the melting points of drug-like compounds? J. Chem. Inf. Model. 54, 3320–3329 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lipinski C. A., Lombardo F., Dominy B. W., Feeney P. J., Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Kralovics R., Passamonti F., Buser A. S., Teo S. S., Tiedt R., Passweg J. R., Tichelli A., Cazzola M., Skoda R. C., A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 352, 1779–1790 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Welsch M. E., Snyder S. A., Stockwell B. R., Privileged scaffolds for library design and drug discovery. Curr. Opin. Chem. Biol. 14, 347–361 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.A. Nguyen, J. Yosinski, J. Clune, Deep neural networks are easily folled: High confidence predictions for unrecognizable images, in Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7 to 12 June 2015, pp. 427–436. [Google Scholar]
- 55.van der Maaten L. J. P., Hinton G. E., Visualizing high-dimensional data using t-sne. J. Mach. Learn. Res. 9, 2579–2605 (2008). [Google Scholar]
- 56.N. Jaques, S. Gu, D. Bahdanau, J. M. Hernández-Lobato, R. E. Turner, D. Eck, Sequence tutor: Conservative fine-tuning of sequence generation models with KL-control, in Proceedings of the 34th International Conference on Machine Learning, PMLR, International Convention Centre, Sydney, Australia, 6 to 11 August, 2017. [Google Scholar]
- 57.Lajiness M. S., Maggiora G. M., Shanmugasundaram V., Assessment of the consistency of medicinal chemists in reviewing sets of compounds. J. Med. Chem. 47, 4891–4896 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Zhang T. Y., Process chemistry: The science, business, logic, and logistics. Chem. Rev. 106, 2583–2595 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Wang Y., Bryant S. H., Cheng T., Wang J., Gindulyte A., Shoemaker B. A., Thiessen P. A., He S., Zhang J., PubChem BioAssay: 2017 update. Nucleic Acids Res. 45, D955–D963 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fourches D., Muratov E., Tropsha A., Trust, but verify: On the importance of chemical structure curation in cheminformatics and QSAR modeling research. J. Chem. Inf. Model. 50, 1189–1204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Boyle N. M., Towards a Universal SMILES representation - A standard method to generate canonical SMILES based on the InChI. J. Cheminform. 4, 22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/7/eaap7885/DC1
Fig. S1. Distribution of SAS for the full ChEMBL21 database (~1.5 million molecules), random subsample of 1M molecules from ZINC15, and generated data set of 1M molecules with baseline generator model G.
Fig. S2. Reward functions.
Fig. S3. Distributions of SAS for all RL experiments.
Fig. S4. Distribution of residuals and predicted versus observed plots for predictive models.
Fig. S5. Learning curve for generative model.
Fig. S6. Distributions of SMILES’s string lengths.


