Abstract
Allograft rejection remains a significant concern after all solid organ transplants. Although qualitative morphologic analysis with histologic grading of biopsy samples is the main tool employed for diagnosing allograft rejection, this standard has significant limitations in precision and accuracy that affect patient care. The use of endomyocardial biopsy to diagnose cardiac allograft rejection illustrates the significant shortcomings of current approaches for diagnosing allograft rejection. Despite disappointing interobserver variability, concerns about discordance with clinical trajectories, attempts at revising the histologic criteria and efforts to establish new diagnostic tools with imaging and gene expression profiling, no method has yet supplanted endomyocardial biopsy as the diagnostic gold standard. In this context, automated approaches to complex data analysis problems—often referred to as “machine learning”—represent promising strategies to improve overall diagnostic accuracy. By focusing on cardiac allograft rejection, where tissue sampling is relatively frequent, this review highlights the limitations of the current approach to diagnosing allograft rejection, introduces the basic methodology behind machine learning and automated image feature detection, and highlights the initial successes of these approaches within cardiovascular medicine.
The authors review “machine learning“ and “automated image feature detection“ to improve the diagnostic accuracy of endomyocardial biopsies to diagnose cardiac allograft rejection.
Cardiac allograft rejection (CAR) occurs in 30% to 40% of transplant recipients within the first year posttransplant,1-3 and carries an increased risk of both acute graft failure and reduced graft longevity. Because of the high morbidity of CAR when diagnosed after symptoms develop, surveillance endomyocardial biopsy (EMB) has been included in heart transplantation guidelines since 1990.4,5 Although EMB is the established gold standard for the diagnosis of CAR, the clinical utility of EMB using standard hematoxylin and eosin (H&E) histologic analysis is limited by marked interobserver variability and significant discordance between the histologic grade and clinical impression of CAR severity. These limitations result in undertreatment of important rejection events as well as overtreatment of less clinically important rejection events, both of which introduce a potential for patient harm. Although these concerns are particularly well characterized in cardiac allografts due to the frequency of EMB sampling, they are relevant to all solid organ transplants that periodically rely on tissue characterization to make important diagnoses.
Automated approaches to complex data analysis problems, often referred to as “machine learning,” (ML) represent promising strategies to improve overall diagnostic accuracy in solid organ transplants. To highlight the potential of ML to improve the histologic analysis of allograft biopsies, we review the limitations of the current diagnostic approach to transplant rejection, introduce the basic methodology behind ML and automated image feature detection, and highlight the initial successes of these approaches within cardiovascular and transplant medicine. Finally, this review discusses future applications for ML as a precision medicine tool to enable individualized management of solid organ transplant recipients. Based on all these considerations, we assert that applying ML technologies to allograft rejection is an opportunity whose time has come.
THE LIMITATIONS OF STANDARD OF CARE EMB FOR REJECTION SURVEILLANCE
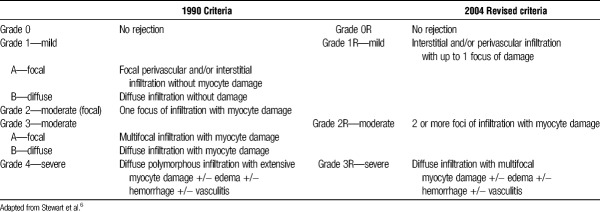
Because of the frequency and morbidity of CAR, heart transplant recipients are monitored with surveillance protocols that typically result in 12 or more scheduled EMBs in the first year posttransplant alone.4 This widespread use of tissue sampling led to the development of the 1990 International Society for Heart and Lung Transplantation (ISHLT) Working Formulation for the Standardization of Nomenclature in Heart Transplant to formalize the histologic grading of CAR in EMB samples.5 The histologic criteria outlined in this landmark publication called for light microscopy with H&E staining and relied on a qualitative examination for the presence of inflammatory cell infiltrates, the extent of infiltrates, and for signs of “myocyte damage” (Table 1). Although these criteria succeeded in implementing an international standard nomenclature and facilitating research, the qualitative and subjective nature of the morphologic grading scheme resulted in confusion and inconsistencies among users6-12 (Table 1).
TABLE 1.
1990 and 2004 Morphologic grading criteria for CAR
Attempts to revise the criteria were made in 19959 and 20017 before new formal consensus criteria were established in 2004. The ISHLT 2004 revised framework6 acknowledged a need for “further characterization of the nature of the inflammatory infiltrate and a definition of myocyte damage,” because there was widespread recognition that the vagaries of the language used in the 1990 scheme was a major contributor to the poor reliability and accuracy of CAR grading. However, despite significant exposition in the revised framework, qualitative and largely subjective language remains the foundation of histologic grading for CAR. How many inflammatory cells define an infiltrate? Is a larger infiltrate more important than a small one? How many focal infiltrates, or how large a single focus, before a sample is deemed “diffusely” infiltrated? How far away from a blood vessel does an infiltrate have to extend before it is no longer “perivascular”? What exactly is myocyte damage, and should damage without necrosis be differentiated from frank necrosis? The answers to these (and many other) questions may have important implications for the mechanism, severity, and treatment of rejection, but are not clearly addressed in the current rejection grading schema.
Poor Reliability and Diagnostic Accuracy
Because of the vagaries of the diagnostic criteria and the inherent subjectivity of traditional histologic analysis, the current diagnostic approach to CAR suffers from high interobserver variability and significant discordance between histologic and clinical impressions of rejection severity. Interobserver variability has been a widely recognized limitation of the morphologic assessment of CAR since the widespread adaption of the EMB procedure.3,10-14 In a study by Angelini et al,13 a combined κ statistic of 0.39 was calculated for grades assigned by the 18 study pathologists using the 2004 ISHLT criteria. Although this represented a small improvement over the 1990 criteria (κ = 0.31), this is still far from the degree of reproducibility one would expect from a gold standard test. In the Cardiac Allograft Rejection Gene Expression Observational II Study,10 concordance between a panel of 4 independent pathologists and local pathologists at study centers was examined. Although there was modest agreement between the 2 groups at 71% overall, there was a dismal agreement of 28.4% at the higher levels of rejection (grade 2R and higher) which typically result in major alterations of immunosuppression.
Issues with diagnostic accuracy have also plagued the current histologic approach to CAR. It has long been recognized that histologic rejection grade does not necessarily correlate with clinical findings of rejection as assessed by history/physical, echocardiography, and invasive hemodynamics. As far back as 1985, Greenberg et al15 noted that patients with and without biopsy evidence of rejection did not significantly differ in hemodynamic parameters measured by right heart catheterization. Larger studies by Frist et al16 in 1987 and Bolling et al17 in 1991 noted similar findings. Although these results were seen as supporting the continued use of EMB for CAR surveillance under the theory that prevention of a potential clinically important future rejection required early diagnosis histologically,12,17 one could also interpret these results as evidence of the poor positive predictive value of standard histologic grading when the disease of interest is clinically important rejection. In line with the latter interpretation, Klingenberg et al18 withheld treatment in a case series of 17 grade 3A (ISHLT 2004 grade 2R) rejections, all of whom experienced benign clinical courses with resolution of histologic rejection on subsequent biopsies. In the IMAGE trial, patients with 2R rejection based on retrospective review by a panel of expert pathologists who received no treatment (due to initial 1R grading) suffered no worse outcomes than patients initially and accurately diagnosed with 2R rejection who were treated per study protocol.19,20
Underdiagnosis due to poor negative predictive value also plagues the current diagnostic framework. Dandel et al21 examined 364 biopsies in 190 transplant patients to examine relationships between histologic EMB grades, clinical impression of rejection, and echocardiography data. There were 59 clinically important rejections in this study, and nearly half (49%) had histologically mild rejection on EMB (grade 1 or lower). Indeed, the concept of ‘biopsy negative rejection’ to describe cases of clear clinical rejection in the absence of histologic evidence of significant cellular rejection has been a source of concern and investigation for years.12,14,22,23 Also, although updates to ISHLT grading criteria in 2004 and the subsequent addition of more refined antibody-mediated rejection criteria have helped reduce the burden of biopsy negative rejection cases, there remains a number of false-negative EMBs in the setting of clinically important rejection.22,23 Although each of these examples represents a small case series of select patients, they suggest that the features currently used to assess rejection severity may not be optimal, and that new approaches to identifying other features might achieve better diagnostic accuracy.
Despite updates and revisions, there remain significant shortcomings with both the precision and accuracy of traditional EMB histologic analysis for diagnosing rejection. The ISHLT rejection grading framework has been invaluable for standardizing terminology, allowing for better study of heart transplant rejection on a population level over the past several decades. However, this same framework has clear limitations on an individual level, and looks increasingly outdated in a 21st century healthcare environment that is focused on quantifiable data and the delivery of personalized precision medicine. A new approach is needed to improve our ability to accurately and reliably diagnose and predict CAR.
IMPROVED PREDICTION AND PRECISION WITH ML
Traditional prediction modeling is based on regression analysis of a few selected clinical features that are thought to represent the important risk factors for a given disease or outcome. In the field of cardiovascular disease (CVD), risk factors such as age, hypertension, hyperlipidemia, smoking, and diabetes may be used, with the modeling process involving attempts to adjust the relative weights of these factors to provide the best prediction formula possible for a given cohort. Although a longstanding and well-established approach, risk models generated this way are used relatively sparingly in daily practice24 due to lack of external validation in diverse cohorts, modest concordance statistics, and poor performance on an individual patient level.25-28 For example, nearly half of incident myocardial infarctions (MI) will occur in patients who have 1 or no conventional risk factors and are thus not deemed to be at high risk by standard risk assessment tools.29,30 Findings like this highlight the significant shortcomings of traditional modeling with regards to making accurate predictions in complex biological disease processes.
The limitations underlying the classical approach to data analysis and risk prediction arise from several sources. Traditional prediction models rely on hand-picked variables with established independent strong risk associations and easily recognizable etiologic associations. The statistical regression modeling approaches that combine these few selected risk factors then make an implicit assumption that each risk factor is related in a linear fashion to the outcome of interest. Taken together, the traditional statistical approach oversimplifies the complex relationships present in many disease processes, which include large numbers of stronger and weaker risk factors, some with potentially unexpected and nonlinear interactions.28,31,32 Moreover, the scale and structure of complex modern data sets are not easily managed using traditional hands-on data analysis techniques. This results in a pragmatic but methodologically flawed pruning of data sets for easier human analysis, with a priori biases determining which variables are important enough to include in the model set and which are not. Although often necessary for traditional statistical modeling, this process limits the exploration and weighting of unexpected contributors to risk.
ML and Big Data
The term “big data” is frequently used to describe the large, complex, and often unstructured or semistructured data sets that arise in the digital age with widespread data capture and storage. The wealth of big data available in 21st century healthcare originating from diverse sources including the electronic health record, “omics” research, insurance databases, and wearable technologies, has led to increased interest in methods to better use data-rich resources and extract the maximum amount of clinically important information from them. Machine learning approaches have proven to be the most promising method of achieving this goal. Machine learning is a core component of artificial intelligence, contributing to computer programs that autonomously learn through experience to generate predictions. Over the past 2 decades, ML has progressed from a theoretical discipline, at the fringes of computer science, statistics and cognitive science, to a broadly applicable, widespread and commercially important technology. ML programs have diverse applications, ranging from weather prediction to credit card fraud detection, and from speech recognition to self-driving cars.
The advantages of advanced ML approaches over conventional data analysis and prediction modeling are a function of both the amount of data that can be processed and the manner in which data are analyzed. Although fundamentally any form of automated statistical modeling from basic linear regression to advanced deep learning neural networks can be described as a ML approach, advanced ML approaches are those which leverage modern computer processing power to perform more robust statistical analysis with a minimum of human input (and consequently, a minimum of a priori human bias). This allows for efficient processing of “big data” sources, exploiting the complex, nonlinear relationships that frequently arise in data-intensive fields while doing so in a comprehensive and unbiased fashion. In other words, advanced ML approaches assume less about the nature of the data to discover more and predict better.
ML in CVD and Transplant Medicine
Machine learning approaches to data analysis can be described generally as “supervised” or “unsupervised.” In supervised learning, the outcome of interest for each case in a data set is known and made available to the computer, allowing for the generated algorithm to correlate variables in the data set with the presence or absence of that outcome. Supervised learning is used for prediction, using a training set of manually labeled examples to train the algorithm (this is the “supervision” in supervised ML), then moving on to an unlabeled data set to validate predictive accuracy (Figure 1). For example, if the outcome of interest is CAR, a supervised ML approach would involve providing the algorithm with a labeled data set of cases with confirmed ‘rejection’ or confirmed ‘no rejection’ along with a number of potential variables that may be related to this outcome. Because the process of algorithm calibration is iterative, the algorithm can independently adapt as new data are added. As it learns from each consecutive case, a highly reliable and increasingly accurate prediction model is created. The limitations of supervised learning are often related to the specific method/algorithm used, with some prone to oversimplifications (eg, linear and logistic regressions), whereas others are more prone to overfitting (eg, decision trees and random forests), and still others are limited by high demands on processing power (eg, neural networks, support vector machines). However, as a general rule, the quality of the classification model generated with supervised learning is most dependent on the quality of the data set with regard to completeness and noise, as well as the accuracy and generalizability of the human-labeled data set used for training (Figure 1).
FIGURE 1.
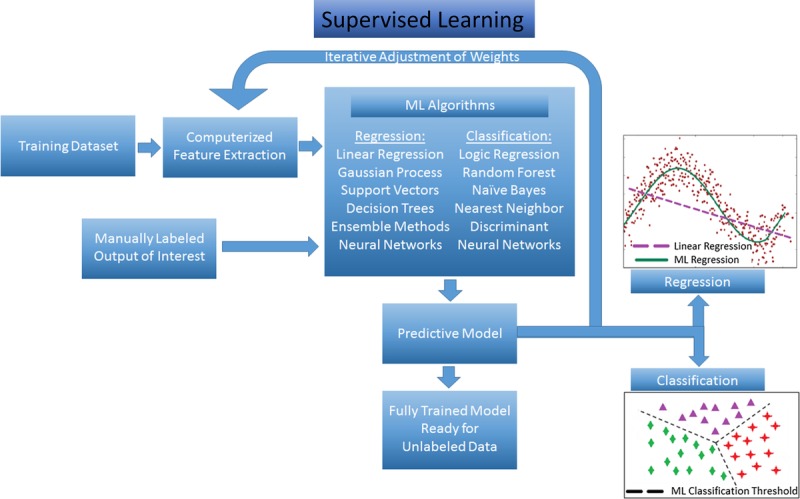
Supervised ML schematic. Training data set undergoes automated feature extraction, and this information along with manually labeled outcomes are fed into the selected ML algorithm. Many potential ML methods can be applied to data, depending on the desired output (regression for continuous outcomes, classification for categorical outcomes). After iterative adjustment of feature weighting to refine accuracy of predictive model based on the manually labeled outcomes, the algorithm is considered “trained” and ready for analysis of new unlabeled data.
Because of the availability of registry and electronic medical record data, supervised ML approaches to large and complex medical data sets have been the most widely implemented in CVD. Weng et al31 used a registry of family practices in the United Kingdom to analyze 30 variables of incident CVD events in 378 256 patients, with a neural network ML method predicting 7.6% more cardiovascular events in this cohort than the 2013 ACC/AHA risk model. Supervised learning ML approaches have been implemented to combine data from different sources, such as clinical data and annotated imaging findings to refine prediction as well. Motwani et al33 recently published the results of an ML algorithm to predict 5-year mortality in a cohort of 10 030 patients with suspected CAD who were referred for coronary CT angiogram based on 25 coronary CT angiogram parameters and 44 clinical parameters from the CONFIRM international registry. Their ML method using “boosted” decision trees resulted in an area under the receiver-operator characteristic curve (AUROC) of 0.79, which was far higher than the AUROCs obtained by traditional clinical or radiographic scores (Framingham Risk Score, 0.61; Segment Stenosis Score, 0.64; Segment Involvement Score, 0.64; or Modified Duke Index, 0.62). In transplant medicine, Yoo et al34 recently used an ensemble of ML models to predict long-term graft survival in a retrospective cohort of 3117 kidney transplant recipients in South Korea. The combined ML model using 33 patient-level variables achieved an index of concordance of 0.80, significantly outperforming a conventional Cox survival model using the same patient variables which achieved an index of concordance of only 0.60.
Unsupervised ML identifies groups and clusters within data rather than predicting an outcome from data. In unsupervised learning, the data set is examined for hidden patterns and groupings among the collected variables (Figure 2). Returning to examples within CAR, in a data set of patients with significant confirmed CAR containing a wide variety of clinical variables, an unsupervised ML approach may be able to identify distinct subgroups of CAR based on the clustering of certain variables. Female patients with a prior pregnancy and low ISHLT grades on EMB may form a cluster (perhaps representing antibody-mediated rejection cases?), or young patients longer than 1 year posttransplant with low tacrolimus levels may form another (noncompliance or lost insurance?). Through this process, groups for further characterization and investigation can be identified from large, complex data sets. An unsupervised approach is useful in heterogeneous populations and disease entities to identify phenotypes within a broader syndrome, and is commonly used in “omics” research. The limitations of unsupervised learning methods are that they tend to be quite sensitive to outliers and noisy data and that they inherently do not provide “answers” in the form of a diagnosis or classification or prediction. Shah et al35 effectively used an unsupervised ML approach to identify pheno-group clusters within a cohort of 397 patients with heart failure with preserved ejection fraction. After identifying 3 strong clusters based on clinical, demographic, and echocardiographic data, a supervised ML approach was then used to compare different phenogroups based on clinically important outcomes of interest such as hospitalization (Figure 2).
FIGURE 2.

Unsupervised ML schematic. As with supervised learning, a training data set undergoes automated feature extraction, with this information being analyzed by the selected ML algorithm. Unlike supervised learning, there are no manually labeled outputs to “train” and refine prediction. Instead, data are analyzed for intrinsic patterns, and through iterative adjustment input data, increasingly refined cutoffs for distinct clusters within the data can be generated.
The ability of ML approaches to perform unbiased analysis of “big data,” to boost predictive power, and to identify hidden structures in data have already begun to impact CVD and transplant research. Also, with dozens of open source ML frameworks available and major companies such as Google, Amazon, IBM, and Microsoft offering professional consultative ML services, the role of ML methods in research and patient care will continue to grow. With concurrent investments in medical informatics and precision medicine, potential cost savings due to improved disease forecasting with ML methods could be as high as US $300 billion in the United States alone according to a recent report by the McKinsey Global Institute.36 However, massive data set interrogation is only 1 aspect of ML, and even more novel and exciting applications for computerized data analysis are likely to be realized as ever more complex ML methods are introduced to medical practice and research.
QUANTITATIVE IMAGE ANALYSIS, DEEP LEARNING, AND NEURAL NETWORKS
Although the ML approaches discussed so far can identify relationships in large data sets in a versatile and comprehensive manner, there remains a fundamental bias common to all of them. This bias is a reliance on traditional data—those pieces of transcribable information deliberately compiled for the sake of future quantitative analysis. Constraining analysis to only the categorical and continuous variables that were already recognized as data worth collecting imposes a fundamental limitation on the ability of computer algorithms to identify truly novel relationships and predictors. Implementing ML approaches on raw and nontraditional data therefore opens up the possibility of discovering new quantifiable relationships, features, and patterns in places where humans typically do not think to look for them. This includes analysis of diverse sources of raw, noisy, and unconventional information.
This field can be classified into 2 basic approaches—the older handcrafted feature analysis and the newer deep learning feature analysis. In general, handcrafted feature analysis refers to automated image segmentation and analysis that relies on a base set of manually labeled features. This approach has the potential to expand on the base feature set by quantifying and uncovering intensity and positional relationships, but is fundamentally defined by the human-labeled features that serve as a primer. Deep learning, on the other hand, is the latest development in the field, and through a neural network approach turns the work of feature detection completely over to the software that applies filters and transformations to uncover quantifiable relationships within an image.
There are strengths and weaknesses to both approaches. Handcrafted features provide transparency due to the human-labeled features that serve as the basis of the method, and this fact makes the features uncovered and predictions made more readily interpretable and explainable due to a foundation built upon things that the programmer already knows are relevant. Of course, this basis on what the programming team chooses to label introduces potential bias and may limit truly novel feature identification. This method is also labor intensive, because manual image labeling takes significant upfront effort and time. Moreover, if insufficient domain-specific information is known about the images being analyzed, then it will constrain the ability to manually label features of relevance that serve as the basis for a handcrafted approach. These limitations are the reason deep learning approaches have undergone rapid development and dissemination in recent years. Deep learning neural networks have their own limitations, requiring larger training sets and often being described as a “black box” due to the difficulty the programming team often has in understanding exactly why the algorithm makes the predictions it makes. However, the lack of upfront labor, as well as the removal of a priori bias, results in algorithms that are incredibly powerful and frequently provide more accurate predictions than their handcrafted counterparts.
Deep Learning Neural Networks
Deep learning with convolutional neural networks (CNNs) is the ML approach behind cutting edge applications, like computer speech recognition, natural language processing, and computer vision/image analysis, each of which represent disciplines in which unconventional raw data (a sound, a letter, or a pixel) comprise the input data of interest. For example, in deep learning for image analysis, each pixel that comprises a digital image is a quantifiable piece of input data. Digital images are in actuality a matrix of numbers with position within the matrix corresponding to position in space and different numerical values corresponding to brightness/color. In a CNN, this matrix of numbers is passed through layers of interconnected artificial neurons (“nodes”) which perform filtering operations (“convolutions”) and compressing/aggregating operations (“pooling”) to identify the often obscure fundamental features of importance (“primitives”) for predicting the desired output of interest. The weighting of these computer-identified primitive image features is adjusted iteratively as more data are analyzed based on their relative value in accurately classifying and predicting the outcome, ultimately creating a comprehensive feature map that captures complex, nonlinear relationships in a way which maximizes predictive power (Figure 3). For a more detailed and comprehensive review of deep learning and neural networks, we refer readers to reviews by Janowczyk et al37 and Lecun et al,38 whereas for a more rigorous technical overview, we refer readers to Schmidhuber39 (Figure 3).
FIGURE 3.
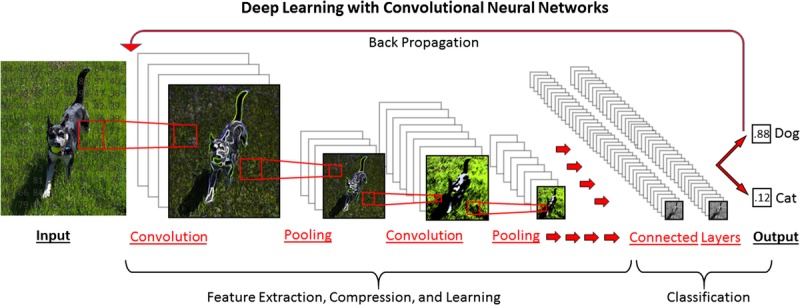
Deep learning schematic with a CNN, designed to classify an input image as either a dog or a cat. The initial digital image is analyzed as a matrix of numerical values corresponding to position and color. This matrix is analyzed through a series of “convolutions” in which filters are applied (sharpening, Gaussian blurring, edge detection etc.) to extract underlying features. Pooling steps compress the data, reducing the features to their base components. After a user-determined number of convolutions and pooling steps have been performed, all of the identified features are combined in “fully connected” layers for final predictive model generation. By a process of “back propagation,” weights of different features are iteratively adjusted to improve prediction when a supervised learning method is used.
Quantitative Image Analysis in Medical Research
With an unparalleled ability to identify novel features of predictive importance, quantitative image analysis has begun to fulfill some of its potential as a translational technology in the field of digital pathology. Although the human brain has an undeniably powerful pattern recognition capacity, there are inevitably unrecognized patterns and unquantified variables in histologic samples that make this discipline an ideal place to use handcrafted feature analysis and, more recently, deep learning neural networks. Work in this regard has made the greatest headway in the tissue-rich field of oncology, in particular in breast cancer research. Beck et al40 applied an earlier method of digital image analysis and manually identified features along with an ML classifier to analyze tissue microarrays of breast cancer biopsies. Using 248 local samples for algorithm training before validating on an external set of 328 samples, the “computerized pathologist (C-Path)” algorithm identified 6642 unique morphologic features in these microarrays, creating a predictive model based on these morphologic features and the outcome of interest, 5-year survival. The ML-derived risk score was significantly associated with 5-year survival independent of any other clinical, genetic or molecular factor. Moreover, the C-path algorithm was able to accurately stratify risk of mortality within each conventional morphologic tumor grade, identifying higher and lower risk patients that would otherwise be homogeneously labeled. When bootstrapping analysis was performed on the 6642 morphologic features that made up the C-path risk score, 11 strong and completely novel morphologic predictors of mortality were identified. Interestingly, 3 of these features pertained to the stroma rather than the neoplastic cellular regions of the tissue, whereas an additional 7 were relational features between neoplastic cells and stroma representing larger-scale tissue architectural morphology. Unexpected findings like these that have inspired the use of even more powerful deep learning morphologic classifiers in the field of digital medical image analysis.
Cruz-Roa et al41 recently performed a similar analysis using a state-of-the-art deep learning CNN on 349 breast cancer biopsies to determine the presence of invasive ductal adenocarcinoma. The CNN algorithm performed well at this task, with the most notable finding being a true-negative rate of 99.64% suggesting great potential for this technology to be used as a decision support and screening tool to save pathologists time on reading negative biopsies. Notably, while the study by Beck et al in 2011 analyzed tissue microarrays taken from biopsy samples due to limitations in computing power, the study by Cruz-Roa et al performed nearly 5 years later was able to perform whole-slide image analysis, reducing the risk of sampling bias which could be a critical limitation when analyzing biopsy samples for evidence of invasive carcinoma. Arevalo et al42 demonstrated an unsupervised CNN algorithm with a 98.1% classification accuracy for basal cell carcinoma from histopathology, significantly outperforming other state-of-the-art methods for tumor identification. In similar experiments beyond oncology, Gulshan et al43 used deep learning CNNs to analyze digitized images to demonstrate an impressive AUROC of 0.99 for the diagnosis of diabetic retinopathy on a validation set of nearly 12 000 funduscopic images. Taken together, these experiments demonstrate the promise of deep feature ML methods to analyze complex, raw, and unconventional digital image data, generating not only robust prediction and discrimination tools, but also discovery tools with the ability to shed new light on disease processes at the morphologic level.
FUTURE DIRECTIONS
Despite the progress and promise deep learning feature identification has already shown this decade, these techniques have yet to make an impact on CVD or transplant medicine. Recently, a team of researchers from the University of Pennsylvania and Case Western Reserve University presented a first-in-heart deep learning CNN for the classification of failing versus nonfailing hearts based on histologic samples from full thickness biopsies taken at the time of heart explant or VAD placement44 (Figure 4). This deep learning algorithm achieved a sensitivity of 99% and specificity of 94%, far exceeding the performance of 2 expert pathologists. The highly accurate performance of this CNN not only serves to support the theory that deep learning methods can be used for cardiac tissue analysis, but also suggests that some of the perceived limitations of morphometric analysis based on random biopsies may be not be so important when a deep learning approach is used. Fundamentally, biopsy procedures are limited by the potential for sampling error. The classic histologic lesion of ischemic cardiomyopathy—areas of dense replacement fibrosis—should occur in some locations and not others within the myocardium, and because of this heterogeneity, should impose an intrinsic limitation on the degree of diagnostic accuracy achievable when analyzing random biopsies. And yet, the failing/nonfailing CNN classifier described above performed at a very high sensitivity on a cohort containing many ischemic cardiomyopathy patients, suggesting that although the most recognizable histologic feature of a disease process may be patchy, other more homogenously distributed features are present that a deep learning CNN can use to generate strong predictions. Nevertheless, this work serves only as a proof of concept highlighting the potential of the deep learning method in cardiac tissue. To fulfill the translational promise of advanced ML, for tissue analysis, what is needed is an important diagnostic challenge with well-documented limitations and a clear need for enhanced predictive accuracy. Ideally this disease process will have plenty of raw, complex primary data that has proved resistant to quantifiable and reproducible analyses. In this context, the diagnosis of allograft rejection appears to be an ideal candidate for such research (Figure 4).45
FIGURE 4.
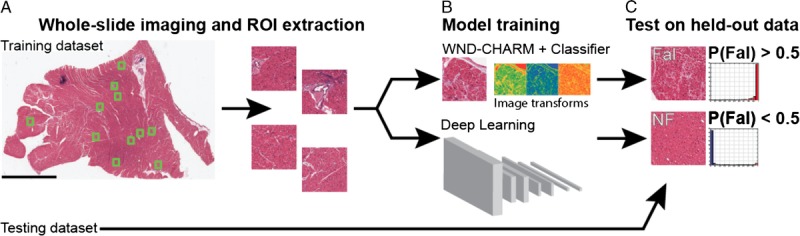
Deep learning workflow for classification of Fal and NF based on selected ROIs from heart tissue samples. Fal, failing; NF, nonfailing; ROIs, regions of interest. Courtesy of Nirschl et al.45
The development of deep-learning algorithms within transplant rejection should focus initially on those areas where the current approach is most obviously falling short. For example, high-throughput decision support systems using deep-learning CNNs could be developed relatively easily to use the ISHLT grading framework in a way that adds value to the standard pathologist interpretation. Given the somewhat arbitrary nature of discrete rejection grade cutoffs and the significant and well-documented inter-reader variability, a CNN that produces a discrete ISHLT grade is probably not the right approach. Instead, the CNN can provide a probability for each rejection grade from 0R to 3R that complements the pathologist's interpretation. Not all EMBs resulting in a common rejection grade look the same, and it is likely that understanding the variability within each grade has value. By providing probability outputs instead of all-or-nothing classifications, this CNN would be able to define a biopsy sample as, for example, having majority 1R character but also with a significant (albeit minority) amount of 2R character. This information can be used to assist the pathologist in labeling borderline cases and may also be useful to the transplant physicians in planning treatment changes or follow-up biopsy schedules.
Alternatively, deep-learning CNNs could be developed that leave the ISHLT grading framework behind, training the CNN on EMB samples that are classified not by their ISHLT grade but based on the clinical impression of rejection severity. Through this approach, the CNN would identify tissue-level features that correlate strongly with clinically important rejection. Such approaches would be more likely to identify variant subtypes of allograft rejection: cellular, antibody-mediated, and combined rejections. Here again, outputs could be graded based on both severity and the degree of diagnostic certainty.
In considering the role of deep learning neural networks in CVD and rejection surveillance in particular, it is important to recognize not only their power as predictors but also their potential as tools of discovery. Although commonly referred to as “black boxes” because of the perceived opacity of the logic and dimensional reduction processes going on within a neural network, the accuracy of this criticism is increasingly being called into question as new approaches to validate and interrogate the inner workings of these powerful algorithms are developed. Recently, Sundararajan et al46 used a novel “Integrated Gradients” method to uncover the features of highest prognostic importance behind a series of deep learning image classifiers, including the Gulshan diabetic retinopathy classifier discussed previously. With application of the integrated gradients, an output image corresponding to the input image but highlighting the specific patterns and areas associated with the choice of diagnosis by the classifier can be produced. This work suggests that it is possible to have the best of both worlds—a fully unbiased, nonlinear, automated deep-learning prediction system along with the transparent feature identification for user reassurance or hypothesis generation. Leveraging deep learning feature identification to diagnose and predict episodes of CAR is certainly a valuable pursuit, but from a transplant medicine standpoint, prediction is only 1 part of a larger clinical problem. If researchers can “see” inside of the machine and apply domain-specific knowledge to the features discovered through unbiased deep learning analysis, better histopathologic grading schemes can be developed and a better understanding of the mechanisms of rejection achieved. This in turn could lead to earlier and better targeted therapeutics to disrupt the rejection process. Additionally, some demonstration of biological plausibility, via explicit identification of discrete features with a potential etiologic role in CAR, will likely be necessary before widespread adaption of CNN classifiers into clinical practice can be expected.
Finally, it is important to recognize that the potential of ML in rejection surveillance can extend beyond conventional H&E analysis alone. Deep-learning image analysis has the potential to provide additional value when paired with more sophisticated microscopy techniques, such as multiplex immunostaining or electron microscopy.47-49 Moreover, because not every important process and predictor of rejection is necessarily evident on morphologic analysis alone, there is great opportunity for synergy when ML techniques for analyzing morphologic data are combined with techniques for analyzing other forms of patient-level data. Whether incorporating broad electronic medical record laboratory and demographic data with deep learning analysis of morphologic samples,47 incorporating Allomap or Allosure results, or exploring deeper histogenomic and histoproteomic relationships,50 combined analytic approaches have the potential to offer more robust prediction in the clinical realm and more comprehensive mechanistic study in the research realm.
CONCLUSIONS
In this article, we have reviewed the well-studied and widely recognized limitations of conventional histologic analysis of EMB samples for the diagnosis of CAR. These limitations include simplistic and vague feature identification criteria, significant interobserver variability, and a concerningly high rate of potentially misleading “false” positive and negative histology results. These limitations lead to confusion among providers and potential patient harm through both undertreatment of important rejection events and overtreatment of less clinically significant rejection events. For more than 30 years, the limitations of conventional EMB analysis for CAR have been recognized, and despite multiple attempts at revising the histologic criteria and a multitude of efforts to establish new diagnostic tools with imaging51-53 and gene expression profiling,20,54,55 no method has yet supplanted EMB as the diagnostic gold standard. The failure of alternative approaches to make a major impact on rejection surveillance is a complex issue and beyond the scope of this review, but it bares mentioning that validating any new method in this field will be inherently limited by the failings of the standard they attempt to compare themselves with. Proving the worth of noninvasive rejection surveillance methods through comparison to an established standard which suffers from poor inter-rater agreement and questionable accuracy is a setup for failure. Because these issues have cemented tissue diagnosis as the diagnostic standard in rejection going forward (regardless of what complimentary imaging and gene profiling techniques evolve to reduce the frequency of tissue samples in the future), the onus is on the transplant community to develop more advanced morphologic analysis tools to bring the field up to the standards of 21st century precision medicine. Deep ML feature identification could provide invaluable decision support to pathologists in the diagnosis of CAR, ensuring a reliable standard output, improving diagnostic accuracy, and uncovering previously unrecognized histologic patterns and features of potential diagnostic or prognostic importance. By providing a better standard of diagnosis, these methods can also reinvigorate research into complementary and alternative methods of rejection surveillance. A significant investment in the development of advanced computer-assisted morphologic classifiers would be consistent with the innovative and pioneering research spirit that has defined advanced heart failure and transplant medicine and would keep the field at the leading edge of this century's push toward technologically integrative precision medicine.
Footnotes
This publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number TL1TR001880, the National Cancer Institute of the National Institutes of Health under award numbers (R21CA179327-01, R21CA195152-01, U24CA199374-01, the National Institute of Diabetes and Digestive and Kidney Diseases under award number R01DK098503-02, the National Heart Lung and Blood Institute under award number R01-HL105993, the DOD Prostate Cancer Synergistic Idea Development Award (PC120857); the DOD Lung Cancer Idea Development New Investigator Award (LC130463), the DOD Prostate Cancer Idea Development Award; the Case Comprehensive Cancer Center Pilot Grant, the VelaSano Grant from the Cleveland Clinic the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University, the I-Corps@Ohio Program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or other grant providers.
E.G.P. declares no conflicts of interest.
A.M. is the cofounder and stakeholder in Ibris Inc., a cancer diagnostics company. A.M. is an equity holder and has technology licensed to both Elucid Bioimaging and Inspirata Inc. A.M. is a scientific advisory consultant for Inspirata Inc. Part of his research work has been sponsored by Philips Healthcare and he has NIH-funded projects in collaboration with PathCore Inc. and Inspirata. He also sits on the Tumor Modeling advisory board at Astrazeneca Inc and at Inspirata Inc. K.B.M. holds research grants from Thoratec Corporation, Merck and Glaxo-Smith-Kline, and serves as a scientific consultant/advisory board member for Janssen, Merck, Pfizer and Glaxo-Smith-Kline.
E.G.P. is the primary author, participated in writing. A.M. participated in writing. K.B.M. is the corresponding author and participated in writing.
REFERENCES
- 1.Eisen HJ, Tuzcu EM, Dorent R, et al. Everolimus for the prevention of allograft rejection and vasculopathy in cardiac-transplant recipients. N Engl J Med. 2003;349:847–858. [DOI] [PubMed] [Google Scholar]
- 2.Kobashigawa JA, Miller LW, Russell SD, et al. Tacrolimus with mycophenolate mofetil (MMF) or sirolimus vs. cyclosporine with MMF in cardiac transplant patients: 1-year report. Am J Transplant. 2006;6:1377–1386. [DOI] [PubMed] [Google Scholar]
- 3.Patel JK, Kobashigawa JA. Should we be doing routine biopsy after heart transplantation in a new era of anti-rejection? Curr Opin Cardiol. 2006;21:127–131. [DOI] [PubMed] [Google Scholar]
- 4.Costanzo MR, Dipchand A, Starling R, et al. The International Society of Heart and Lung Transplantation Guidelines for the care of heart transplant recipients. J Heart Lung Transplant. 2010;29:914–956. [DOI] [PubMed] [Google Scholar]
- 5.Billingham ME, Cary NR, Hammond ME, et al. A working formulation for the standardization of nomenclature in the diagnosis of heart and lung rejection: Heart Rejection Study Group. The International Society for Heart Transplantation. J Heart Transplant. 1990;9:587–593. [PubMed] [Google Scholar]
- 6.Stewart S, Winters GL, Fishbein MC, et al. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J Heart Lung Transplant. 2005;24:1710–1720. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez ER. International Society for Heart and Lung Transplantation. The pathology of heart transplant biopsy specimens: revisiting the 1990 ISHLT working formulation. J Heart Lung Transplant. 2003;22:3–15. [DOI] [PubMed] [Google Scholar]
- 8.Winters GL. The challenge of endomyocardial biopsy interpretation in assessing cardiac allograft rejection. Curr Opin Cardiol. 1997;12:146–152. [DOI] [PubMed] [Google Scholar]
- 9.Winters GL, Marboe CC, Billingham ME. The International Society for Heart and Lung Transplantation grading system for heart transplant biopsy specimens: clarification and commentary. J Heart Lung Transplant. 1998;17:754–760. [PubMed] [Google Scholar]
- 10.Crespo-Leiro MG, Zuckermann A, Bara C, et al. Concordance among pathologists in the second Cardiac Allograft Rejection Gene Expression Observational Study (CARGO II). Transplantation. 2012;94:1172–1177. [DOI] [PubMed] [Google Scholar]
- 11.Marboe CC, Billingham M, Eisen H, et al. Nodular endocardial infiltrates (Quilty lesions) cause significant variability in diagnosis of ISHLT Grade 2 and 3A rejection in cardiac allograft recipients. J Heart Lung Transplant. 2005;24(7 Suppl):S219–S226. [DOI] [PubMed] [Google Scholar]
- 12.Fishbein MC, Kobashigawa J. Biopsy-negative cardiac transplant rejection: etiology, diagnosis, and therapy. Curr Opin Cardiol. 2004;19:166–169. [DOI] [PubMed] [Google Scholar]
- 13.Angelini A, Andersen CB, Bartoloni G, et al. A web-based pilot study of inter-pathologist reproducibility using the ISHLT 2004 working formulation for biopsy diagnosis of cardiac allograft rejection: the European experience. J Heart Lung Transplant. 2011;30:1214–1220. [DOI] [PubMed] [Google Scholar]
- 14.Patel J, Kittleson M, Rafiei M, et al. The natural history of biopsy negative rejection after heart transplantation. J Heart Lung Transplant. 2012;31:S235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenberg ML, Uretsky BF, Reddy PS, et al. Long-term hemodynamic follow-up of cardiac transplant patients treated with cyclosporine and prednisone. Circulation. 1985;71:487–494. [DOI] [PubMed] [Google Scholar]
- 16.Frist WH, Stinson EB, Oyer PE, et al. Long-term hemodynamic results after cardiac transplantation. J Thorac Cardiovasc Surg. 1987;94:685–693. [PubMed] [Google Scholar]
- 17.Bolling SF, Putnam JB, Jr, Abrams GD, et al. Hemodynamics versus biopsy findings during cardiac transplant rejection. Ann Thorac Surg. 1991;51:52–55. [DOI] [PubMed] [Google Scholar]
- 18.Klingenberg R, Koch A, Schnabel PA, et al. Allograft rejection of ISHLT grade >/=3A occurring late after heart transplantation—a distinct entity? J Heart Lung Transplant. 2003;22:1005–1013. [DOI] [PubMed] [Google Scholar]
- 19.Kobashigawa JA. The search for a gold standard to detect rejection in heart transplant patients: are we there yet? Circulation. 2017;135:936–938. [DOI] [PubMed] [Google Scholar]
- 20.Pham MX, Teuteberg JJ, Kfoury AG, et al. Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med. 2010;362:1890–1900. [DOI] [PubMed] [Google Scholar]
- 21.Dandel M, Hummel M, Meyer R, et al. Left ventricular dysfunction during cardiac allograft rejection: early diagnosis, relationship to the histological severity grade, and therapeutic implications. Transplant Proc. 2002;34:2169–2173. [DOI] [PubMed] [Google Scholar]
- 22.Tang Z, Kobashigawa J, Rafiei M, et al. The natural history of biopsy-negative rejection after heart transplantation. J Transplant. 2013;2013:236720 doi: 10.1155/2013/236720. Published December 18, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veiga Barreiro A, Crespo Leiro M, Doménech García N, et al. Severe cardiac allograft dysfunction without endomyocardial biopsy signs of cellular rejection: incidence and management. Transplant Proc. 2004;36:778–779. [DOI] [PubMed] [Google Scholar]
- 24.Hobbs FD, Jukema JW, Da Silva PM, et al. Barriers to cardiovascular disease risk scoring and primary prevention in Europe. QJM. 2010;103:727–739. [DOI] [PubMed] [Google Scholar]
- 25.Wilson PW, D'Agostino RB, Levy D, et al. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–1847. [DOI] [PubMed] [Google Scholar]
- 26.Cooney MT, Dudina AL, Graham IM. Value and limitations of existing scores for the assessment of cardiovascular risk: a review for clinicians. J Am Coll Cardiol. 2009;54:1209–1227. [DOI] [PubMed] [Google Scholar]
- 27.Damen JA, Hooft L, Schuit E, et al. Prediction models for cardiovascular disease risk in the general population: systematic review. BMJ. 2016;16:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krittanawong C, Zhang H, Wang Z, et al. Artificial intelligence in precision cardiovascular medicine. J Am Coll Cardiol. 2017;69:2657–2664. [DOI] [PubMed] [Google Scholar]
- 29.Ridker PM, Danielson E, Fonseca FA, et al. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N Engl J Med. 2008;359:2195–2207. [DOI] [PubMed] [Google Scholar]
- 30.Canto JG, Kiefe CI, Rogers WJ, et al. Number of coronary heart disease risk factors and mortality in patients with first myocardial infarction. JAMA. 2011;306:2120–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weng SF, Reps J, Kai J, et al. Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS One. 2017;12:e0174944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science. 2015;349:255–260. [DOI] [PubMed] [Google Scholar]
- 33.Motwani M, Dey D, Berman DS, et al. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur Heart J. 2017;38:500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoo KD, Noh J, Lee H, et al. A machine learning approach using survival statistics to predict graft survival in kidney transplant recipients: a multicenter cohort study. Sci Rep. 2017;7:8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah SJ, Katz DH, Selvaraj S, et al. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation. 2015;131:269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bughin J, Hazan E, Ramaswamy S, et al. Artificial Intelligence: The Next Digital Frontier? McKinsey Global Institute. 2017; Discussion Paper June 2017:24. http://www.mckinsey.com/business-functions/mckinsey-analytics/our-insights/how-artificial-intelligence-can-deliver-real-value-to-companies.
- 37.Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J Pathol Inform. 2016;7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. [DOI] [PubMed] [Google Scholar]
- 39.Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015;61:85–117. [DOI] [PubMed] [Google Scholar]
- 40.Beck AH, Sangoi AR, Leung S, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3:108ra113. [DOI] [PubMed] [Google Scholar]
- 41.Cruz-Roa A, Gilmore H, Basavanhally A, et al. Accurate and reproducible invasive breast cancer detection in whole-slide images: a deep learning approach for quantifying tumor extent. Sci Rep. 2017;7:46450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arevalo J, Cruz-Roa A, Arias V, et al. An unsupervised feature learning framework for basal cell carcinoma image analysis. Artif Intell Med. 2015;64:131–145. [DOI] [PubMed] [Google Scholar]
- 43.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. [DOI] [PubMed] [Google Scholar]
- 44.Nirschl JJ, Janowczyk A, Peyster EG, et al. Deep learning classifier to predict cardiac failure from whole-slide H&E images. Lab Invest. 2017;97:532A–533A. [Google Scholar]
- 45.Nirschl JJ, Janowczyk A, Peyster EG, et al. A deep-learning classifier identifies patients with clinical heart failure using whole-slide images of H&E tissue. PLoS One. 2018;13: e0192726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sundararajan M, Taly A, Yan Q. Axiomatic Attribution for Deep Networks. Published Mar 4, 2017. Revised June 13, 2017. doi: arXiv:1703.01365 [cs.LG].
- 47.Madabhushi A, Lee G. Image analysis and machine learning in digital pathology: challenges and opportunities. Med Image Anal. 2016;33:170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhargava R, Madabhushi A. Emerging themes in image informatics and molecular analysis for digital pathology. Annu Rev Biomed Eng. 2016;18:387–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martel AL, Hosseinzadeh D, Senaras C, et al. An image analysis resource for cancer research: piip-pathology image informatics platform for visualization, analysis, and management. Cancer Res. 2017;77:e83–e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee G, Singanamalli A, Wang H, et al. Supervised multi-view canonical correlation analysis (sMVCCA): integrating histologic and proteomic features for predicting recurrent prostate cancer. IEEE Trans Med Imaging. 2015;34:284–297. [DOI] [PubMed] [Google Scholar]
- 51.Butler CR, Thompson R, Haykowsky M, et al. Cardiovascular magnetic resonance in the diagnosis of acute heart transplant rejection: a review. J Cardiovasc Magn Reson. 2009;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butler CR, Savu A, Bakal JA, et al. Correlation of cardiovascular magnetic resonance imaging findings and endomyocardial biopsy results in patients undergoing screening for heart transplant rejection. J Heart Lung Transplant. 2015;34:643–650. [DOI] [PubMed] [Google Scholar]
- 53.Wu YL, Ye Q, Sato K, et al. Noninvasive evaluation of cardiac allograft rejection by cellular and functional cardiac magnetic resonance. JACC Cardiovasc Imaging. 2009;2:731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deng MC, Eisen HJ, Mehra MR, et al. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. 2006;6:150–160. [DOI] [PubMed] [Google Scholar]
- 55.Crespo-Leiro MG, Stypmann J, Schulz U, et al. Clinical usefulness of gene-expression profile to rule out acute rejection after heart transplantation: CARGO II. Eur Heart J. 2016;37:2591–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]