Abstract
Mitochondria play a critical role in the cardiomyocyte physiology by generating majority of the ATP required for the contraction/relaxation through oxidative phosphorylation (OXPHOS). Aging is a major risk factor for cardiovascular diseases (CVD) and mitochondrial dysfunction has been proposed as potential cause of aging. Recent technological innovations in Seahorse XFe24 Analyzer enhanced the detection sensitivity of oxygen consumption rate and proton flux to advance our ability study mitochondrial function. Studies of the respiratory function tests in the isolated mitochondria have the advantages to detect specific defects in the mitochondrial protein function and evaluate the direct mitochondrial effects of therapeutic/pharmacological agents. Here, we provide the protocols for studying the respiratory function of isolated murine cardiac mitochondria by measuring oxygen consumption rate using Seahorse XFe24 Analyzer. In addition, we provide details about experimental design, measurement of various respiratory parameters along with interpretation and analysis of data.
Keywords: Cardiac mitochondria, Respiratory control ratio, Oxygen consumption rate
Introduction
Mitochondria, the power houses of the cells, generate ATP through oxidation of NADH and FADH2. Transfer of electrons from NADH to the oxygen through electron transport chain (ETC) is coupled with proton transfer across the inner mitochondrial membrane generating membrane potential. ATP synthase makes use of the proton gradient to generate ATP from ADP. The process of electron transfer to oxygen coupled with the formation of ATP is termed oxidative phosphorylation (OXPHOS). Reactive oxygen species (ROS) formed within the ETC, particularly at complex I and III, are considered to regulate cellular proliferation, differentiation, and apoptosis (Nunnari and Suomalainen 2012). ROS generated from the mitochondria can lead to organelle dysfunction, contributing to age-related degenerative diseases (Nunnari and Suomalainen 2012).
Aging is the main risk factor for cardiovascular diseases (CVD) (North and Sinclair 2012). Notably, mitochondrial dysfunction plays a critical role in the cardiovascular aging (Dai et al. 2012). It has been hypothesized that prolonged exposure to ROS causes mitochondrial dysfunction during aging (Dai et al. 2012) by inducing mtDNA mutations and oxidizing the mitochondrial proteins and lipids. Previous studies reported decreased complex I and III activities, state III respiration, and increase ROS generation in mitochondria from aged hearts (Dai et al. 2012). In addition, increased oxidative damage to mitochondrial proteins and DNA also has been shown in aged hearts and is proposed to play a key role cardiovascular aging (Herrero and Barja 2001; Martin-Fernandez and Gredilla 2016). Furthermore, mice containing transgenic mitochondrial targeted catalase (mCAT mice) were found to have longer lifespan and protected from cardiovascular abnormalities observed with the aging (Dai et al. 2009). Interestingly, mitochondria from aged mCAT mice showed reduced aging-induced oxidative damage and DNA deletions (Dai et al. 2009). Similarly, the importance of mitochondria in cardiovascular aging is further supported by observations from mice with targeted mutations in mitochondrial polymerase gamma (Polgm/m), enzyme involved in proof-reading of mtDNA (Trifunovic et al. 2004). These mice exhibited increased oxidative damage in cardiac mitochondria with increased cardiomyocyte apoptosis (Dai et al. 2010). They exhibit cardiomyopathy with worsened age-related systolic and diastolic function (Dai et al. 2010). In addition, the role of impaired mitochondrial function in aging is further supported by partial restoration of cardiac dysfunction in these mice when targeted with mCAT (Dai et al. 2010). Moreover, consistent with the observations in transgenic mice, mitochondrial-targeted antioxidant proteins like SS-31 have been reported to ameliorate various age-related cardio and cerebrovascular diseases (Dai et al. 2012; Tarantini et al. 2018). Furthermore, mitochondrial dysfunction is also known to play a key role in heart failure, myocardial infarction, and cardiac ischemia-reperfusion injury (Lesnefsky et al. 2017; Rosca and Hoppel 2013). Both mitochondrial and nuclear DNA mutations of ETC and OXPHOS proteins have been associated with various cardiomyopathies (Meyers et al. 2013). Considering the significance of mitochondrial function in the cardiovascular aging, in vitro high throughput functional tests of isolated cardiac mitochondria could identify the specific functional deficits and underlying mechanisms, and provide screening tools for potential therapeutic agents.
Seahorse XFe24 is high throughput technique for determination of mitochondrial respiratory function by simultaneously measuring oxygen consumption rate (OCR) and extracellular proton flux in the adherent cells. Recently, methodologies have been developed to measure the OCR in the isolated mitochondria from various organs and tissue biopsies (Boutagy et al. 2015; Rogers et al. 2011). OCR measurements in the isolated mitochondria allow the researchers to determine the direct effects of pharmacological agents on the mitochondrial respiration by excluding the potential cellular signaling mechanisms influencing the mitochondrial function. In the present study, we optimized the method to measure respiratory function in the isolated cardiac mitochondria from mouse using seahorse XFe24. Using this optimized protocol, researchers can measure the effects of various therapeutic agents or antioxidants or transgenic mice on respiration in isolated mitochondria in vitro. Furthermore, studies may be designed to understand the impact of interventions like calorie restriction and exercise on cardiovascular aging. Thus, there are numerous research applications in cardiovascular aging for the proposed protocols to test respiratory function in isolated mitochondria.
First, measurements of respiration in isolated mitochondria from mouse liver and rat heart tissues utilizing XFe24 were reported in 2011 (Rogers et al. 2011). Similarly, Das et al. reported standardized coupling and electron flow assays and identified respiratory defects in the isolated cardiac mitochondria from aged mice (Das and Muniyappa 2013). However, previous reports lack the details necessary for isolating mitochondria, designing the functional assay, and analyzing the various aspects of mitochondrial respiration. Here, we report the detailed steps for isolating and purifying the mitochondria from the mouse heart which could be easily adapted for mitochondrial isolation from other tissues. We also provided the experimental design and analysis of data for assaying mitochondrial respiration function. We standardized our protocol with lower reaction volumes than the previously reported and validated our protocol with respiratory measurements made in isolated cardiac mitochondria from the mice. Thus, the present manuscript provides the first detailed protocol to make the respiratory functional measurements in the isolated mouse cardiac mitochondria using Seahorse XFe24 Analyzer.
Materials and methods
Animal procedures and protocols were performed in accordance with institutional and federal guidelines. All procedures from this study have been approved by the Institution of Animal Care and Use Committee (IACUC) at Tulane University School of Medicine.
Mitochondrial isolation
Mitochondrial isolation protocol was standardized in our laboratory and developed from previously published protocols (Kristian 2010; Sordahl 1984). Perform all the steps on ice.
Isolation of mitochondria from mouse hearts
Anesthetize the mouse with isoflurane (VETOne, ID, USA). Tape the arms down on paper towels. Make an incision in the mouse at the abdomen and cut the skin to the top of the thorax. Use tweezers to lift the sternum and cut away the diaphragm and the ribcage to expose the heart.
Remove the heart and place in 5 mL of mitochondrial isolation medium (IM) on ice. Press with force to eject the blood from the chambers. Remove the aorta and atrium from the heart and cut the ventricles into three to four large pieces.
Transfer the tissue to a separate petri dish with fresh IM (5 mL), and chop into fine pieces with small scissors.
Transfer the minced tissue into the glass homogenizer along with the IM (5 mL). Perform 15 strong strokes with 4 turns on ice; then, transfer the homogenate into a 30-mL polycarbonate centrifuge tube. Care should be taken to prevent the negative pressure during lifting of the glass rod.
Centrifuge the homogenate at 27,000×g for 10 min at 4 °C (DuPont Instruments, DE, USA).
Discard the supernatant and gently resuspend the pellet in 5 mL IM by mixing 10–20 times with a 5-mL pipette.
Centrifuge the solution at 500×g for 5 min at 4 °C (Eppendorf, Hamburg, Germany).
Transfer the supernatant to a separate 30-mL polycarbonate centrifuge tube, and centrifuge at 10,000×g for 6 min at 4 °C.
During this spin, prepare the required amount of 40, 24, and 15% Percoll solutions diluting 100% Percoll in IM. Add 3.5 mL of 24% Percoll to a new centrifuge tube. Add 4.5 mL of 40% Percoll slowly at the bottom of the centrifuge tube, beneath the 24% Percoll. A visible interface should be seen between the concentration gradient layers.
Remove the supernatant and resuspend the pellet in 3.5 mL of 15% Percoll (Mix gently 10–20 times each sequentially with 5- and 1-mL pipette). Slowly layer this mitochondrial suspension on top of the 24% Percoll. Visible interfaces should be observed between the three Percoll gradient solutions.
Centrifuge at 30,000×g for 8 min at 4 °C. Mitochondrial layer will be observed between the 24 and 40% Percoll layers. Collect the mitochondrial layer using a 5-mL BD syringe with spinal needle (approximately 3–5 mL suspension). Transfer the mitochondrial suspension into a new centrifuge tube with 5 mL of IM. Gently mix the suspension 10–20 times using the 5-mL pipette.
Centrifuge the mitochondrial suspension at 16,000×g for 10 min at 4 °C.
A loose pellet (or small multiple pellets) will be observed at the bottom of the tube. Use 5mL-, 1mL-, and 200-μL pipettes to remove the supernatant and leave some supernatant above the pellet without disturbing the pellet. Add 0.5 mL of BSA diluted in IM (10 mg/mL) to the pellet. Tap the tube to dislodge the pellet into the BSA solution. Add 4.5 mL of IM and gently mix the suspension 10–20 times using the 5-mL pipette.
Centrifuge the solution at 7000×g for 10 min at 4 °C. Discard the supernatant. Remove the supernatant and resuspend the mitochondria in 50 μL of IM without BSA. Gently mix the pellet suspension 10–20 times using the 100-μL pipette. Take required amount of the mitochondrial solution for protein quantification. Immediately double the mitochondrial suspension using IM (with BSA) and gently mix with pipette. Keep the mitochondria on ice to be used within 4 h.
Mitochondrial protein quantification
Use the Pierce™ BCA protein assay kit (Thermofisher, MA, USA) to quantify mitochondrial protein. Add 10 μL of BSA standards (0 to 2000 μg/mL of BSA) to the 96-well plate.
Add 10 μL of diluted mitochondrial suspensions (IM without BSA) to the 96-well plate. Choose the dilutions to be in the range of the standard curve (approximately 5 to 20 times dilution).
To each well, add 200 μL of BCA reagent made by combining assay reagents A and B (Thermofisher, MA, USA) at 50:1 ratio and incubate the plate for 15–20 min at 37 °C.
After incubation, measure the absorbance at 595 nm using the plate reader (BMG Labtech, Ortenberg, Germany). Deduct the background absorbance of the IM from the sample’s absorbance (similarly diluted ones) and calculate the protein concertation using the dilution factor.
Seahorse assay
- Preparation of the XFe24 calibration plate:
- The day before the assay, add 1 mL of XFe24 calibration buffer to the wells of XFe24 sensor cartridge, cover the edges tightly with the parafilm, and incubate overnight at 37 °C in a non-CO2 incubator.
- On the day of assay, prepare the desired amount of MAS with pyruvate and malate (10 and 2 mmol/L) and adjust the pH to 7.4. Also, prepare the desired amount of ADP and adjust the pH to 7.4. Dilute the XFe assay drugs in the required concentrations from the stock solutions using MAS with pyruvate/malate: ADP (50 mmol/L); oligomycin (50 μmol/L); FCCP (50 μmol/L); antimycin A/rotenone (100 μmol/L/20 μmol/L).
- After or during the protein quantification of mitochondrial samples, load the XFe24 sensor cartridge injection ports with different drugs as follows. Compounds and volumes added to each port: port A: ADP (22 μL); port B: oligomycin (24 μL); port C: FCCP (26 μL); and port D: Antimycin A and rotenone (28 μL). After the injection, mitochondria will be exposed to a 10× diluted concentration of ADP and drugs: Final concentrations of drugs in the well will be ADP (5 mmol/L); oligomycin (5 μmol/L); FCCP (5 μmol/L); antimycin A (10 μmol/L); and rotenone (2 μmol/L).
- Design and start the assay protocol in the Seahorse XFe24 Analyzer. Load the sensor cartridge into the analyzer for the calibration (calibration takes approximately 20 to 30 min) (protocol given in Table 1).
- Preparation and loading of mitochondria to the cell culture microplate:
- Dilute mitochondria to the desired final concentration (5 μg/50 μL) using the MAS with pyruvate/malate.
- Load 50 μL of MAS with pyruvate/malate (no mitochondria) to blank wells A1, C3, B4, and D6.
- Load 50 μL of the mitochondrial suspension (5 μg of protein) to each of the remaining 20 wells.
- Centrifuge the cell culture microplate at 2000×g for 20 min at 4 °C.
- After the centrifugation, carefully add 150 μL of MAS with pyruvate/malate to each well and incubate in a non-CO2 incubator for 10 min (At this step, we use the drugs that we are interested to test by diluting the stock solutions in MAS with pyruvate/malate).
- Exchange the calibration plate with the cell culture microplate once the Seahorse XFe24 Analyzer finished the calibration.
Table 1.
Seahorse XFe24 assay protocol for isolated cardiac mitochondria from mouse and rat
| Command | Time (min) | Compound |
|---|---|---|
| Equilibration | 12 | |
| Mix | 1 | |
| Wait | 3 | |
| Measure | 3 | |
| Mix | 1 | |
| Wait | 3 | |
| Measure | 3 | |
| Inject port A | ADP | |
| Mix | 1 | |
| Measure | 6 | |
| Inject port B | Oligomycin | |
| Mix | 1 | |
| Measure | 3–6 | |
| Inject port C | FCCP | |
| Mix | 1 | |
| Measure | 3 | |
| Inject port D | Antimycin A + rotenone | |
| Mix | 1 | |
| Measure | 6 |
Seahorse data analysis
Open the assay results file in the “overview” window (Wave 2.3.0).
Check for the OCR values of blank wells individually by clicking on the each well. Remove any blank value if it has higher positive value and significantly affects the OCR values of the sample wells.
Select the “standard error of mean” instead of “standard deviation” for OCR graphs.
Export data in the Excel file and arrange the data in new sheet according to the treatment groups for different measurements (basal 1, basal 2, ADP, oligomycin, FCCP, antimycin A/rotenone).
Determine the average of all basal OCR values of the control group. Use this control mean value to normalize the OCR values of all wells and express them as percentage of control. We use this approach to account for the day-to-day and plate-to-plate variations in the OCR values of the assays.
Arrange the relative OCR data from each plate that belong to same experiment (run on the same day or on multiple days) in the new Excel sheet. Take the average of the two basal respiration readings as state II. Represent the OCR values following treatment with ADP, oligomycin, and FCCP as state III, state IVO, and state IIIu, respectively. Use this data for statistical analysis to determine the significant differences between the treatments or groups.
Calculate the respiratory control ratio (RCR) from state III/state IVO. As mid-point OCR values underestimate the RCR, the highest OCR point in state III and the lowest point in the state IVO from the point-point data are used to calculate RCR values. As treatments with drugs or compounds can alter the RCR, RCR from the mitochondria from the control group is used as the indicator of the quality of the mitochondrial preparation. Typically, RCR values obtained by the above method of calculation vary based on the substrates used, ADP concentration, and the quality of the mitochondrial preparations.
Typically, five technical replicates (wells) for each group per treatment are recommended. Further, experiments from 5 to 10 mice for each group is recommended for accuracy of the observations.
For representative images, point-point or mid-point data plots of a single experiment may be generated using the Agilent Seahorse analysis software. In addition, cumulative data from all the experiments may be processed offline and graphs can be generated by plotting software such as GraphPad Prism (Figs. 1, 2, and 3).
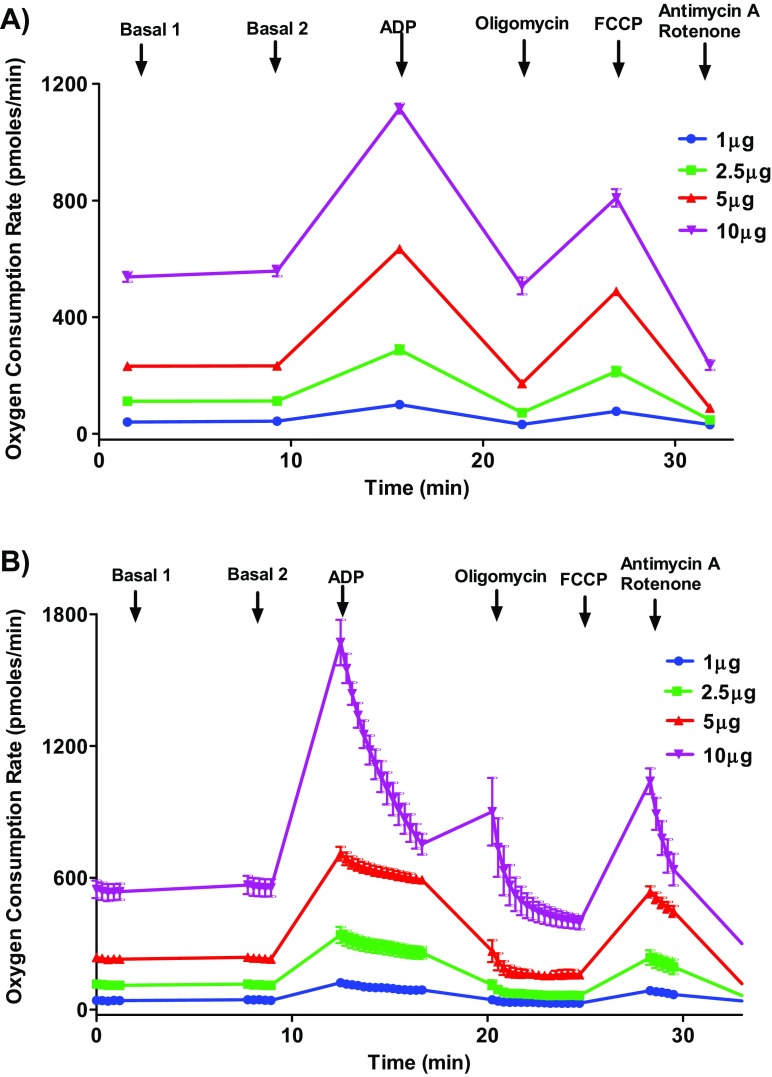
Fig. 1.
Optimizing protein concentration for isolated mouse cardiac mitochondrial oxygen consumption rate measurements. Isolated mouse (C57BL/6J) cardiac mitochondria at various concentrations (1, 2.5, 5, and 10 μg protein/well) were treated with ADP (5 mmol/L), oligomycin (5 μmol/L), FCCP (5 μmol/L), and antimycin (10 μmol/L)/rotenone (2 μmol/L) combination in the presence of pyruvate (10 mmol/L) and malate (2 mmol/L) and OCR was measured. Five-microgram mitochondria provide optimal responses to the ADP and FCCP. High protein concentrations can yield poor responses by depleting the oxygen levels, substrates, and ADP. a Representative plot of point-to-point OCR data. b Representative plot of mid-point OCR. Data expressed as mean ± SEM (n = 5 wells/group)
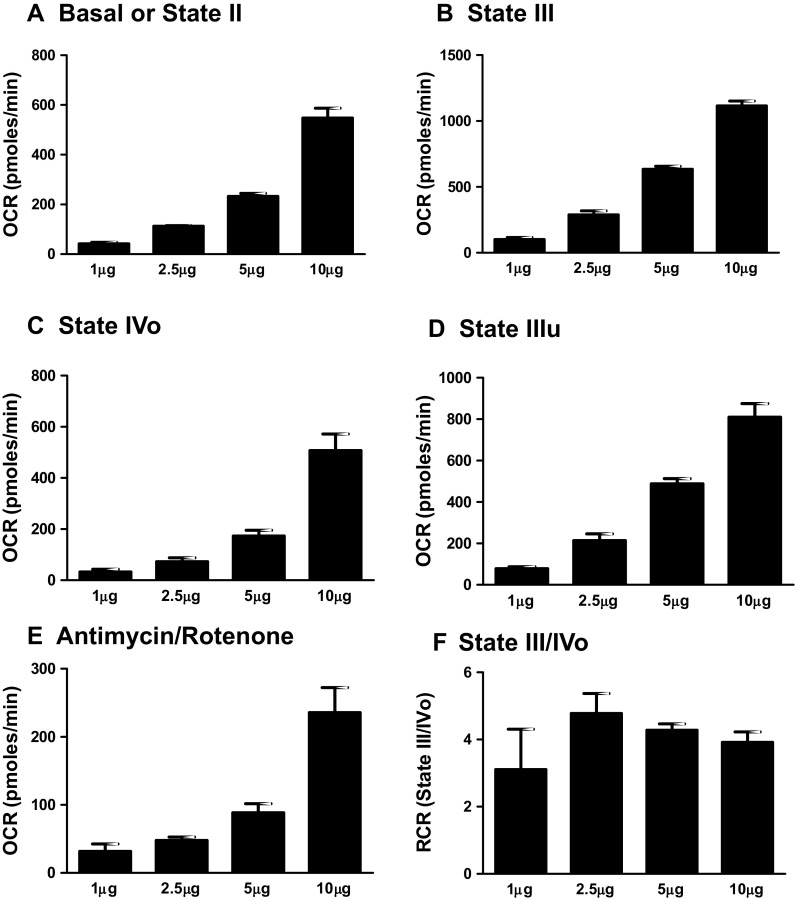
Fig. 2.
Optimizing protein concentration for isolated mouse mitochondrial oxygen consumption rate measurements. Isolated mouse (C57BL/6J) cardiac mitochondria at various concentrations (1, 2.5, 5, and 10 μg protein/well) were treated with ADP (5 mmol/L), oligomycin (5 μmmol/L), FCCP (5 μmmol/L), and antimycin (10 μmmol/L)/rotenone (2 μmmol/L) combination in the presence of pyruvate (10 mmol/L) and malate (2 mmol/L) and OCR was measured. a State II or basal respiration. b State III respiration. c State IVO respiration. d State IIIu respiration. e Antimycin/rotenone. f Respiratory control ratio (state III/IVO). Dose-dependent increase in various respiratory parameters was observed with various protein concentrations. We use 5-μg mitochondria per well for the assays. High protein concentrations can yield poor responses by depleting the oxygen levels, substrates, and ADP. RCR values were higher than four indicating the good quality of isolated mitochondria. Data expressed as mean ± SEM (n = 5 wells/group)
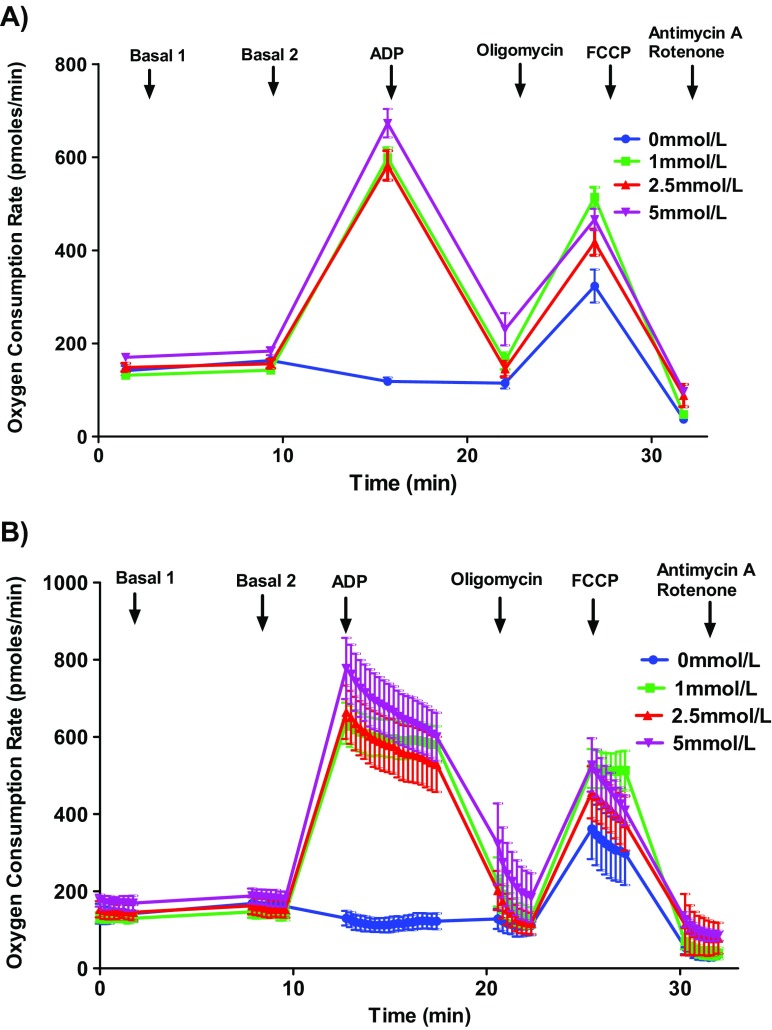
Fig. 3.
Optimizing ADP concentration for mouse mitochondrial oxygen consumption rate measurements. Isolated mouse (C57BL/6J) cardiac mitochondria (5 μg protein/well) were treated different ADP concentrations (0, 1, 2.5, and 5 mmol/L, 5 wells/concentrations), oligomycin (5 μmol/L), FCCP (5 μmol/L), and antimycin (10 μmol/L)/rotenone (2 μmol/L) in the presence of pyruvate (10 μmol/L) and malate (2 mmol/L) and OCR was measured. Selected ADP concentrations not altered the state III response dose-dependently. We use 5 mmol/L ADP for the assays. a Representative plot of point-to-point OCR data. b Representative plot of mid-point OCR data. c State III respiration. Data expressed as mean ± SEM (n = 5 wells/group)
Notes
Mitochondrial isolation
This isolation procedure and volumes of reagents are standardized for isolation of mitochondria from one mouse heart. Pooled isolations from multiple hearts may need relative increase in reagent volumes with gentle homogenization of one mouse heart at a time.
Step 7 can be repeated and the pooled supernatant can be used in step 8 for higher mitochondrial yields.
Prepare the required amount of fresh IM and 100% Percoll every week and store them at 4 °C. Check for the pH before starting the mitochondrial isolation and adjust the pH with 1 N KOH. Note that mitochondrial solutions contain high potassium representing intracellular potassium concentration. We observed poor mitochondrial yield and quality even with 2-week-old 100% Percoll solution (stored at 4 °C).
Try to follow the suggested number of pipetting steps when resuspending the mitochondrial pellet at various steps. A high number of pipetting steps can physically damage the mitochondrial membranes. Tissue pieces may be observed at step 6 but they are discarded in step 8. While resuspending the mitochondrial pellet at step 8, even after pipetting gently for 20 times, we observed visible clumps of mitochondria. However, these clumps of mitochondria do not affect the mitochondrial yield as these pellets will reach the same layer as the individual mitochondria during the Percoll gradient centrifugation.
Prepare 40, 24, and 15% Percoll dilutions fresh every day. Vortex the solutions for at least 30–60 s after mixing the IM and 100% Percoll in the required quantities.
Low amount of mitochondrial layer between the 40 and 24% Percoll layers indicates a poor quality of 100% Percoll solution. During the collection of the mitochondrial layer, care should be taken so that the material in the 40% Percoll layer or in the interphase from 15 and 24% Percoll (which may contain nuclei and other cell organelles) is not drawn out contaminating the mitochondrial layer.
Mix the mitochondrial solution gently (10–20 times with 100-μL/1-mL pipette) whenever it is used (either for protein assay or for diluting and loading the mitochondria for the XFe24 assay). Squirting the mitochondria through narrow pipette tip or pipetting with force may rupture the mitochondria.
Seahorse XFe24 experiment
Overnight incubation of the cartridges in the calibration buffer at 37 °C in a non-CO2 incubator is highly recommended.
After the completion of the run, monitor the pH changes by selecting pH on Y2-axis in the overview mode. It helps to find out about any abnormal changes in pH during the assay. Ideally, the pH should be between 7 and 7.5 throughout each measurement during the assay (depends on the pH of the MAS used).
Observe the plate for mitochondrial attachment both after the microplate centrifugation step and after assay completion. OCR measurements from the wells with excessive attachment of mitochondria to the wall prior to or after the assay could be abnormal and may be represented as outlier.
Prepare 2× MAS and adjust pH to 7.4 using potassium hydroxide store at 4 °C. Use 2× MAS to prepare pyruvate, malate stocks along with ADP. Prepare 250 mmol/L pyruvate and malate stock solutions in the MAS and adjust the pH to 7.4. Make aliquots of 10 to 20 mL and store them at − 20 °C. Thaw an aliquot and store at 4 °C and use it for preparing MAS with pyruvate/malate (pH should be adjusted).
Prepare the ADP solution (50 or 100 mmol/L depending on the desired concentration) in MAS with pyruvate and malate and adjust the pH to 7.4 with KOH.
Prepare the stock solutions of drugs in 95% alcohol (10 mmol/L for oligomycin, FCCP and antimycin, and 2 mmol/L for rotenone) and aliquot them (25 or 50 μL) and store them in − 20 °C. Use fresh aliquots to prepare the working solutions. Prepare the working drug solutions (oligomycin, FCCP, and antimycin/rotenone) in required quantities freshly every day in the pH-adjusted MAS with pyruvate and malate (checking pH of drug solutions is not required).
During data analysis, always consider both mid-point and point-to-point OCR data for assay standardization and for identifying any well with “outlier” OCR measurements.
For OCR measurements after ADP supplementation, it is ideal to select the optimal ADP concentration and the measurement time such that the increased OCR is steady or exhibits a minimal decrease throughout the measurement period. We recommend 3 to 6 min of measuring time with an ADP concentration ranging from 1 to 5 mmol/L.
Though previous studies suggested basal OCR values between 100 and 200 pmol/min and state III (after ADP addition) within 1600–1800 pmol/min, in our experiments, even basal OCR between 300 and 400 pmol/min showed robust responses to drugs as long as the state III respiration does not cross 1800 pmol/min. As day-to-day protein quantity and quality may vary, basal OCR values may be less than 100 (sometimes between 40 and 60 pmol/min), which may result in poor responses to drugs or increase the technical variation.
As the entire experiment, including mitochondrial isolation and Seahorse assay, takes a minimum of 6 to 8 h (for two Seahorse assays), prior planning and reagent preparation are required for consistent results. As such, we always run two Seahorse assays with isolated mitochondria (takes 3 to 4 h total time). Typically, we use the second plate of isolated mitochondria for replicating the first plate.
As previous studies reported the existence of two different mitochondrial populations in cardiomyocytes depending on the position in the cells: intrafibromyolar (IFM) and subsarcolemmal (SSM) mitochondria. We predict our isolation protocol contains both types of mitochondrial populations, so any conclusions drawn must be applied to the whole cardiac mitochondria instead of a specific population.
Materials
Isolation medium, pH 7.4
Sucrose (70 mmol/L, S3-500, Fisher Scientific, PA, USA)
Mannitol (210 mmol/L, M4125, Sigma-Aldrich, MO, USA)
HEPES (5 mmol/L, BP310-500, Fisher Scientific, PA, USA)
EGTA (1 mmol/L, E-4370, Sigma-Aldrich, MO, USA)
Fatty acid-free bovine serum albumin (BSA) (0.5%, A7030, Sigma-Aldrich, MO, USA)
Mitochondrial assay buffer, pH 7.4
Sucrose (70 mmol/L)
Mannitol (210 mmol/L)
HEPES (2 mmol/L)
EGTA (1 mmol/L)
Magnesium chloride (5 mmol/L, M9272, Sigma-Aldrich, MO, USA)
Potassium dihydrogen phosphate (10 mmol/L, P5655, Sigma-Aldrich, MO, USA)
Fatty acid-free bovine serum albumin (0.2%)
Sodium pyruvate (10 mmol/L, P2256, Sigma-Aldrich, MO, USA)
D-Malic acid (2 mmol/L, 46940-U, Sigma-Aldrich, MO, USA)
Mitochondrial assay buffer, pH 7.4 without BSA
Percoll solution, pH 7.4
Percoll (17-0891-01, GE Healthcare, IL, USA)
Sucrose (70 mmol/L)
Mannitol (210 mmol/L)
HEPES (5 mmol/L)
EGTA (1 mmol/L)
Seahorse assay chemicals
Adenosine 5′-diphosphate sodium salt (A2754, Sigma-Aldrich, MO, USA)
Oligomycin (495455—10mg, EMD Millipore, Darmstadt, Germany)
FCCP (15218, Cayman Chemicals, MI, USA)
Antimycin A (A8674, Sigma-Aldrich, MO, USA)
Rotenone (2 μmmol/L, R8875, Sigma-Aldrich, MO, USA)
Funding information
This work was supported by the National Institutes of Health grants NS094834 (P.V. Katakam: funded by National Institute of General Medical Sciences, NIGMS, and National Institute of Neurological Disorders and Stroke, NINDS) and DK107694 (R. Satou: National Institute of Diabetes and Digestive and Kidney Diseases, NIDDK), American Heart Association National Center Scientist Development grant (14SDG20490359 to P.V. Katakam), American Heart Association Greater Southeast Affiliate Predoctoral Fellowship grant (16PRE27790122 to V.N. Sure), American Heart Association Greater Southeast Affiliate Scientist Development Grant (17SDG33410366 to Ibolya Rutkai), and LACaTS (supported in part by U54 GM104940 from the National Institute of General Medical Sciences of the National Institutes of Health, which funds the Louisiana Clinical and Translational Science Center to Ibolya Rutkai).
Compliance with ethical standards
Animal procedures and protocols were performed in accordance with institutional and federal guidelines. All procedures from this study have been approved by the Institution of Animal Care and Use Committee (IACUC) at Tulane University School of Medicine.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Boutagy NE, Rogers GW, Pyne ES, Ali MM, Hulver MW, and Frisard MI (2015) Using isolated mitochondria from minimal quantities of mouse skeletal muscle for high throughput microplate respiratory measurements. J Visualized Exp : JoVE, 53216 [DOI] [PMC free article] [PubMed]
- Dai DF, Chen T, Wanagat J, Laflamme M, Marcinek DJ, Emond MJ, Ngo CP, Prolla TA, Rabinovitch PS. Age-dependent cardiomyopathy in mitochondrial mutator mice is attenuated by overexpression of catalase targeted to mitochondria. Aging Cell. 2010;9:536–544. doi: 10.1111/j.1474-9726.2010.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DF, Rabinovitch PS, Ungvari Z. Mitochondria and cardiovascular aging. Circ Res. 2012;110:1109–1124. doi: 10.1161/CIRCRESAHA.111.246140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DF, Santana LF, Vermulst M, Tomazela DM, Emond MJ, MacCoss MJ, Gollahon K, Martin GM, Loeb LA, Ladiges WC, Rabinovitch PS. Overexpression of catalase targeted to mitochondria attenuates murine cardiac aging. Circulation. 2009;119:2789–2797. doi: 10.1161/CIRCULATIONAHA.108.822403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das KC, Muniyappa H. Age-dependent mitochondrial energy dynamics in the mice heart: role of superoxide dismutase-2. Exp Gerontol. 2013;48:947–959. doi: 10.1016/j.exger.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A, Barja G. Effect of aging on mitochondrial and nuclear DNA oxidative damage in the heart and brain throughout the life-span of the rat. J Am Aging Assoc. 2001;24:45–50. doi: 10.1007/s11357-001-0006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristian T (2010) Isolation of mitochondria from the CNS. Current protocols in neuroscience Chapter 7, Unit 7.22 [DOI] [PMC free article] [PubMed]
- Lesnefsky EJ, Chen Q, Tandler B, Hoppel CL. Mitochondrial dysfunction and myocardial ischemia-reperfusion: implications for novel therapies. Annu Rev Pharmacol Toxicol. 2017;57:535–565. doi: 10.1146/annurev-pharmtox-010715-103335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Fernandez B, Gredilla R. Mitochondria and oxidative stress in heart aging. Age (Dordr) 2016;38:225–238. doi: 10.1007/s11357-016-9933-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers DE, Basha HI, Koenig MK. Mitochondrial cardiomyopathy: pathophysiology, diagnosis, and management. Tex Heart Inst J. 2013;40:385–394. [PMC free article] [PubMed] [Google Scholar]
- North BJ, Sinclair DA. The intersection between aging and cardiovascular disease. Circ Res. 2012;110:1097–1108. doi: 10.1161/CIRCRESAHA.111.246876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW, Brand MD, Petrosyan S, Ashok D, Elorza AA, Ferrick DA, Murphy AN. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS One. 2011;6:e21746. doi: 10.1371/journal.pone.0021746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosca MG, Hoppel CL. Mitochondrial dysfunction in heart failure. Heart Fail Rev. 2013;18:607–622. doi: 10.1007/s10741-012-9340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordahl LA (1984) In Methods in studying cardiac membranes. D. NS, ed. (Boca Raton, Florida: CRC Press), pp. 65–74
- Tarantini S, Valcarcel-Ares NM, Yabluchanskiy A, Fulop GA, Hertelendy P, Gautam T, Farkas E, Perz A, Rabinovitch PS, Sonntag WE, Csiszar A, Ungvari Z. Treatment with the mitochondrial-targeted antioxidant peptide SS-31 rescues neurovascular coupling responses and cerebrovascular endothelial function and improves cognition in aged mice. Aging Cell. 2018;17:e12731. doi: 10.1111/acel.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]