Abstract
Inhibition of mTOR signaling using rapamycin has been shown to increase lifespan and healthspan in multiple model organisms; however, the precise mechanisms for the beneficial effects of rapamycin remain uncertain. We have previously reported that rapamycin delays senescence in human cells and that enhanced mitochondrial biogenesis and protection from mitochondrial stress is one component of the benefit provided by rapamycin treatment. Here, using two models of senescence, replicative senescence and senescence induced by the presence of the Hutchinson-Gilford progeria lamin A mutation, we report that senescence is accompanied by elevated glycolysis and increased oxidative phosphorylation, which are both reduced by rapamycin. Measurements of mitochondrial function indicate that direct mitochondria targets of rapamycin are succinate dehydrogenase and matrix alanine aminotransferase. Elevated activity of these enzymes could be part of complex mechanisms that enable mitochondria to resume their optimal oxidative phosphorylation and resist senescence. This interpretation is supported by the fact that rapamycin-treated cultures do not undergo a premature senescence in response to the replacement of glucose with galactose in the culture medium, which forces a greater reliance on oxidative phosphorylation. Additionally, long-term treatment with rapamycin increases expression of the mitochondrial carrier protein UCP2, which facilitates the movement of metabolic intermediates across the mitochondrial membrane. The results suggest that rapamycin impacts mitochondrial function both through direct interaction with the mitochondria and through altered gene expression of mitochondrial carrier proteins.
Electronic supplementary material
The online version of this article (10.1007/s11357-018-0030-2) contains supplementary material, which is available to authorized users.
Keywords: Senescence, Cardiac fibroblasts, Oxidative phosphorylation, Alanine aminotransferase, Rapamycin, Aging
Introduction
Cardiac fibroblasts represent the largest population of cells in the human heart. As with most fibroblast connective tissue cells, cardiac fibroblasts synthesize and degrade the extracellular matrix that provide the structural support for the heart. By sensing changes in their extracellular milieu, the cardiac fibroblasts are able to regulate the cell-derived extracellular matrix scaffolding and secrete growth factors and cytokines facilitating adaption of the heart to normal and pathological stimuli (Souders et al. 2009). Cardiac fibroblasts also have roles in angiogenesis, neoplasia, and even electrical signaling and there is an evidence that senescent cells accumulate with age in the heart (Chimenti et al. 2003), although the impact of senescence specifically within the cardiac fibroblast population during aging requires further investigation.
Cellular senescence was initially defined by Hayflick and Moorhead as a cellular aging process that limits the number of cell divisions that somatic cells can undergo in culture (Hayflick and Moorhead 1961). However, it is now clear that senescence is also a stress response pathway parallel to apoptosis that can be activated by multiple stressors including oxidative stress, genotoxic stress, telomere attrition, and oncogene conversion (Campisi 2013). More recently, evidence has emerged that mitochondrial stress induces senescence, even in the absence of overt DNA (Nacarelli et al. 2016; Wiley et al. 2016). In terms of the relevance of senescence to aging of the organism, targeting senescent cells has been reported to provide protection in both progeroid models (Baker et al. 2011) and during normal aging (Baker et al. 2016), while the identification of senescent cells in multiple tissues as a function of age further demonstrates the importance of senescence in the aging process (Herbig et al. 2006; Jeyapalan and Sedivy 2008).
One of the key players in establishing replicative, genotoxic stress and oncogene-induced senescence is an increase in reactive oxygen species (ROS) (Colavitti and Finkel 2005; Lu and Finkel 2008; Nair et al. 2015). The generation of intracellular ROS appears to be a much earlier event than the onset of other senescent phenotypes as shown in a time-progression analysis of replicative senescence in human fibroblast (Kim et al. 2013). Mitochondria are a major source of intracellular ROS production and a significant increase in mitochondrial superoxide anion has been reported with increasing population doublings and replicative senescence in human fibroblasts, mesenchymal stem cells, and vascular smooth muscle cells (Bielak-Zmijewska et al. 2014; Estrada et al. 2013; Lerner et al. 2013; Nacarelli et al. 2016; Nacarelli et al. 2015; Passos et al. 2013; Passos et al. 2007; Wiley et al. 2016). Elevated non-phosphorylating mitochondrial respiration has been demonstrated in replicative, oxidative stress and oncogene-mediated senescence (Hutter et al. 2004; Kaplon et al. 2013; Nacarelli et al. 2015; Quijano et al. 2012). Not only are mitochondria potential sources of ROS, but also potential targets of ROS. During replicative senescence of human fibroblasts, evidence of ROS-induced oxidative damage was detected within the mitochondria (Ahmed et al. 2010), suggesting that the mitochondria are both a ROS source and a target during the senescence transition. Yet, aging-related decline in organismal metabolic rate refers not only to generation of abnormalities but also to “normal” mitochondrial changes which are adaptive to the aging processes. These changes include shifts in oxidative metabolites, rewiring in routes of carbon in the TCA cycle, and activity of mitochondrial enzymes and transporters. An increase in mitochondrial pyruvate oxidation, TCA cycle, and respiration has been observed in senescence induced through oncogene activation (Kaplon et al. 2013; Quijano et al. 2012), genotoxic stress (Wang et al. 2016), mitochondrial stress (Nacarelli et al. 2016), and in replicative senescence (Nacarelli et al. 2016; Takebayashi et al. 2015). Importantly, it has been demonstrated that senescence-mediated acceleration of mitochondria oxidative metabolism is accompanied by elevated glycolysis. Seminal studies on the metabolism of cellular senescence show that both glucose consumption and lactate production are elevated during replicative aging and senescence (Bittles and Harper 1984; Goldstein et al. 1982; Moiseeva et al. 2009; Dorr et al. 2013; Liao et al. 2014; Wang et al. 2016; James et al. 2015; Takebayashi et al. 2015). Thus, although in an essentially irreversible growth arrest, senescent cells exhibit a highly active metabolism that is a key characteristic of the senescent phenotype, although the precise mechanisms of balancing of different energetic pathways, including glycolysis and oxidative phosphorylation, are not fully understood.
Rapamycin, an inhibitor of mTOR metabolic and growth control pathway used as part of immunosuppressive therapy (Kennedy and Lamming 2016), has been intensively employed both as an investigational tool and a medicinal agent. It has been shown that rapamycin extends lifespan in multiple organisms including yeast, C. elegans, Drosophila, and mice, and improves late life cardiac function in mice (Dai et al. 2014); Miller et al. 2011). Rapamycin also provides benefit in settings of mitochondrial dysfunction such as mutations in the NDUFS4 mitochondrial subunit which models Leigh syndrome (Johnson et al. 2013), and in a lamin A/B null model (Ramos et al. 2012). We and other laboratories have reported that rapamycin delays replicative senescence in both normal human cells and cells from Hutchinson-Gilford progeria syndrome (HGPS) patients as well as reduces the production of the senescence-associated secretory profile, known as the SASP (Cao et al. 2011b; Laberge et al. 2015; Lerner et al. 2013; Wiley et al. 2016). These studies demonstrate that rapamycin can delay or prevent senescence; however, a greater understanding of the metabolic reprogramming of the senescent cells and the potential impact of interventions to delay senescence, such as rapamycin, are needed to facilitate the development of targeted interventions that will modify metabolism and provide benefits associated with a reduced burden of senescent cells. In the present study, we have examined the mitochondrial oxidative phosphorylation processes both on intact cells and isolated organelles and the impact of rapamycin treatment in two settings of senescence, human cardiac fibroblasts grown to replicative senescence, and senescence induced by a mutant lamin A protein which underlies Hutchinson-Gilford progeria syndrome. This multidimensional approach enabled us to explore the potential mitochondrial targets of rapamycin which could be a part of mechanisms involved in senescence-mediated metabolic adaptation.
Materials and methods
Cell culture and cell culture reagents
Human cardiac fibroblast cells (ScienCell Inc. Carlsbad, CA) and HGPS patient-derived fibroblasts (Coriell Institute, Camden, NJ) were cultivated in minimal essential medium (MEM) supplemented with 10% fetal bovine serum, 1% l-glutamine, 1% MEM vitamins, and 1% MEM non-essential amino acids according to standard culture protocol for lifespan analysis of human diploid fibroblasts (Cristofalo and Charpentier 1980), with or without the addition of 1 nM rapamycin. Rapamycin was present in the cultures continuously. The HGPS fibroblasts, HGFDN0178 and AG01972, were obtained from the Hutchinson-Gilford Foundation and Coriell Research Institute respectively while the unaffected adult skin fibroblasts, HGFDFN168 and HGMDFN090, were obtained from the Hutchinson-Gilford Foundation. Progerin- and lamin-expressing cells were generated through retroviral introduction of cDNA constructs producing GFP-fusion proteins generously deposited in the Addgene site by Dr. Thomas Misteli (Scaffidi and Misteli 2008). Atypical nuclei were scored as any nuclei which exhibited abnormal morphology, including folding of the nuclear lamina and invaginations in the nuclear membrane. For galactose studies, cultures were transferred to Dulbecco’s modified eagle medium (DMEM) containing either 10 or 20 mM galactose and followed for lifespan according to standard protocols for lifespan analysis as above.
Cellular respiration, extracellular acidification, and glucose uptake measurements
Oxygen consumption and extracellular acidification rates were measured on a Seahorse XF24 Bioanalyzer (Seahorse Bioscience, North Billerica, MA), using the XF Cell Mito Stress Test and XF Mito Fuel Flex Test kits. Cells were seeded at a density of 15,000 to 25,000 cells per well in an XF24 microplate. Plates were placed in the bioanalyzer pre-loaded with the sensor cartridge containing oligomycin, carbonilcyanide p-triflouromethoxyphenylhydrazone (FCCP), and rotenone/antimycin A for the Mito Stress Test kit or UK5099(8 μM), BPTES (10 μM), and etomoxir (15 μM) for the Mito Fuel Flex Test kit. Respiration and acidification rates were assessed at least in triplicates as outlined in published methods (Hill et al. 2012; Dranka et al. 2011). In the inhibitory analysis, the rates of oxidation were calculated based on the percentage of oxygen consumption upon treatment with UK5099, an inhibitor of pyruvate oxidation, and with sequential addition of BPTES/etomoxir to interfere the glutamine/palmitate oxidation. The values were normalized to the number of cells harvested in each well at the completion of assay and counted using a Guava EasyCyte Mini (Guava Technologies, Hayward, CA). Glucose uptake was determined by measuring of glucose level in the media using an ACCU-CHEK glucometer (Roche, Basel, Switzerland). The difference in glucose concentration in the stock culture media and cultured cell media was normalized to cell number using a Guava EasyCyte Mini for cell counts.
Isolation of mitochondria from cell cultures
Mitochondria were isolated by differential centrifugation. The cell pellet rinsed with ice-cold PBS (without Ca+2/Mg+2) was re-suspended in hypotonic solution (100 mM sucrose, 10 mM MOPS, pH 7.2, 1 mM EGTA) to let cells swell for 10 min on ice according to Panov (Panov et al. 2005). The suspension was homogenized in a Teflon-glass homogenizer by gentle circular strokes following immediate dilution with hypertonic solution (1.25 M sucrose, 10 mM MOPS, pH 7.2), 1 ml per 10 ml cell suspension, to restore the buffer isotonicity. The mix was diluted with three volumes of isolation buffer (75 mM mannitol, 225 mM sucrose, 10 mM MOPS, pH 7.2, 1 mM EGTA, 0.1% fatty acid-free BSA). Cellular debris were sedimented at 980g, 4 °C for 5 min. Supernatant containing mitochondria was further centrifuged at 10,300g, 4 °C, for 20 min. The resulting crude mitochondria pellet was rinsed by centrifugation at the same parameters in 10 ml of MiR05 respiration buffer (110 mM sucrose, 60 mM K-lactobionate, 20 mM HEPES, pH 7.2, 1 mM KH2PO4, 3 mM MgCl2x6H2O, 0.5 mM EGTA, 20 mM taurine, 0.1% fatty acid-free BSA) (Gnaiger et al. 2000). The final pellet was re-suspended in 100 μl MiR05 by vortexing and stored on ice in a refrigerator. The mitochondria protein was determined by the Bradford protein assay.
Measurement of oxidative phosphorylation of isolated mitochondria
The isolated mitochondria respiration was analyzed by high-resolution respirometry at 37 °C in a two-chamber respirometer OROBOROS Oxygraph-2K (Innsbruck, Austria) (Gnaiger 2008; Gnaiger et al. 1995; Pesta and Gnaiger 2012). The OROBOROS DatLab software was used for data acquisition and analysis. Each 2-ml chamber was loaded with 0.2 mg mitochondria protein. To assess different respirometric complexes, we applied sequential stimulation of mitochondria with 15 μM ADP (to assess state 4 respiration) following 2 mM ADP (to assess state 3 respiration) in the presence of metabolic substrates, namely 10 mM glutamate, 10 mM pyruvate, 10 mM succinate, and 15 μM palmitoyl-l-carnitine all in the presence of 2 mM malate along with inhibitors of NADH dehydrogenase (1 μg/ml rotenone) and adenine nucleotide transporter (5 μM carboxyatractilozide).
Transaminase assay
The activity of alanine aminotransferase (ALT) was measured in isolated mitochondria of the same preparation used for respirometric analysis using commercial assay kit according to the manufacturer protocol (BioAssay Systems, Hayward, CA). The 0.1 mg/ml mitochondria protein was used per sample, and both control and rapamycin treated went through one freezing-thawing cycle. Colorimetric detection of enzyme activity is based on quantification of pyruvate produced by ALT as a function of decrease of absorbance of associated NADH.
Immunoblotting and immunoprecipitation
Cell protein extracts were prepared by extracting with RIPA buffer containing a protease inhibitor cocktail (Sigma-Aldrich) and protein concentration was quantified using a bicinchoninic acid (BCA) assay (Pierce Biotechnology). Western blot analysis was performed using 15 to 30 μg of protein extracts run on SDS-PAGE and transferred onto nitrocellulose (Biorad) membranes. Blots were incubated with antibodies specific for beta-actin (A2066 Sigma-Aldrich) and progerin (EMD Millipore).
Statistical analysis
Statistical analyses were performed with GraphPad Prism version 5.03 for Windows (GraphPad Software, San Diego, CA). The statistical differences between control and rapamycin-treated cells and isolated mitochondria were estimated by paired, two-tailed Student’s t test and were considered significant at p < 0.05.
Results
Cell respirometric and glycolytic activities
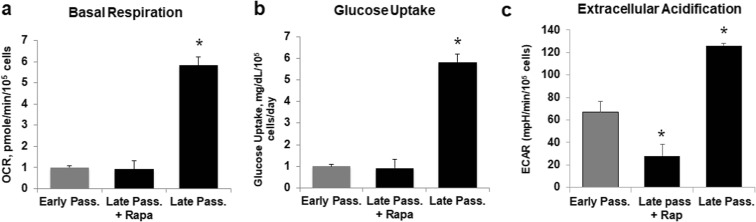
We examined the metabolic state of senescent (late passage greater than 90% lifespan completed) cardiac fibroblasts relative to early passage cardiac fibroblasts by monitoring oxygen consumption and extracellular acidification rates as a measure of activity of oxidative phosphorylation (OxPhos) and glycolysis respectively. The matched sets of cells had been cultured in the presence or absence of 1 nM rapamycin beginning at mid-lifespan (population doubling 27) and maintained until the control cultures began to approach senescence. Consistent with our previous observations, the senescent cells exhibited a higher basal oxygen consumption rate, i.e., OxPhos, up to 6 pmol/min/105cells vs 1 pmol/min/105cells in early passage cells, and increased glycolysis 5.5 vs 1 mg/dl/105cells, along with associated medium acidification (Fig. 1). Long-term treatment of cells with rapamycin corrected all three processes back to the early passage cells.
Fig. 1.
Late passage human fibroblasts are relatively glycolytic. Seahorse bioanalyzer measurements of basal respiration (a), glucose uptake (b), and extracellular acidification (c) in early and late passage (> 90% lifespan completed) human cardiac fibroblasts. The values were normalized to cell numbers in each well. Samples were measured in quintuplicate and results are representative of at least two independent experiments ± SD. The bars with an asterisk represent values that are significantly (*, p < 0.05) differ from the late passage ones
Cell oxidation of pyruvate and glutamine/palmitate
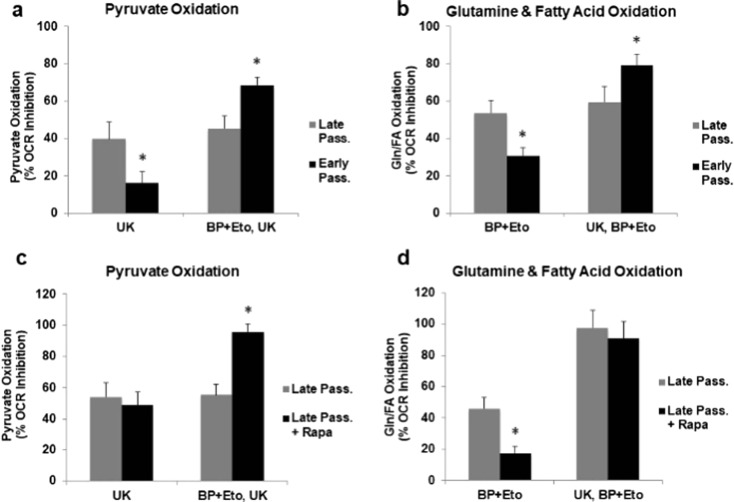
To explore the other energetic routes, we examined the ability of the late and early passage cells to maintain OxPhos via utilization of pyruvate and glutamine/palmitate. This was achieved through sequential addition of inhibitors of the corresponding mitochondrial pyruvate transporter (UK5099), glutaminase (BPTES), and palmitate transporter (etomoxir). About 40–50% inhibition of oxygen consumption by paired inhibitors indicates relative addiction of senescent cells to these three metabolites (Fig. 2a, b). There is a greater inhibition in early passage cells following the additional exposure to either BPTES/etomoxir in the presence of UK5099 (Fig. 2a) or UK5099 in the presence of BPTES/etomoxir (Fig. 2b). This suggests that the early passage cells are able to divert utilization when first challenged with the inhibitor. Interestingly, the late passage cells that went through the long-term exposure to 1 nM rapamycin exhibit an elevated oxidative capacity (Fig. 2c, d).
Fig. 2.
Distinct rates of pyruvate and glutamine/palmitate oxidation by early and late passage human fibroblasts and the effect of rapamycin. a The rates of pyruvate-mediated respiration were calculated as the percentage of inhibition of oxygen consumption by specific pyruvate transporter inhibitor UK5099. The rates of oxygen consumption obtained via sequential addition of first the glutamine and palmitate oxidation inhibitors, BPTES and etomoxir, and then UK5099 represent the capacity of pyruvate oxidation. b The rates of glutamine/palmitate oxidation were determined in the presence of corresponding inhibitors BPTES and etomoxir, while the capacity was determined following first the addition of pyruvate inhibitor UK5099 and then BPTES/etomoxir. c, d Effect of rapamycin on capacity of late passage cells to oxidize pyruvate and glutamine/palmitate in late passage fibroblasts maintained in the presence or absence of 1 nM rapamycin. Values are generated as in b through the addition of inhibitors and calculating the percent inhibition of oxygen consumption. In all cases, graphs represent the values of one of at least two independent experiments measured in triplicates and presented as mean ± SD; *p < 0.05 vs late passage
Oxidative phosphorylation and transaminase activities of isolated mitochondria
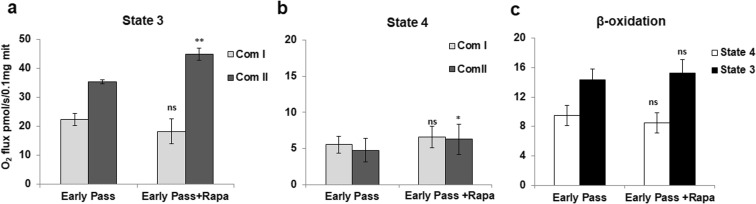
To address the mechanisms of rapamycin-mediated metabolic reprogramming, we studied bioenergetic processes in isolated mitochondria of cardiac fibroblasts. To assess the direct mitochondria targets of rapamycin, we applied higher concentrations of rapamycin, 100 nM, 500 nM, and 5 μM, to the isolated mitochondria. The choice of these concentrations was made based on a large body of studies utilizing rapamycin for cell treatment (Kumar and Lombard 2016; Ramanathan and Schreiber 2009). We found that higher concentrations do not immediately alter respiration, but rather require at least 1 h of pre-incubation to develop detectable mitochondria effects. Figure 3 presents quantitative data of complex I and II and beta-oxidation respiration of mitochondria isolated from cardiac fibroblasts.
Fig. 3.
Oxidative phosphorylation activity of cardiac mitochondria. (a, b) Complex I-mediated oxidation of 10 mM glutamate and 2 mM malate does not change upon mitochondria incubation with 100 nM rapamycin. The complex II-mediated succinate oxidation significantly elevated in the presence of rapamycin in both active and resting states 3 and 4 correspondingly. (c) Oxidation of 15 μM palmitoyl-l-carnitine did not change noticeably, although the trend was observed in every experiment performed. Data are mean ± SD; *p < 0.04, **p < 0.003
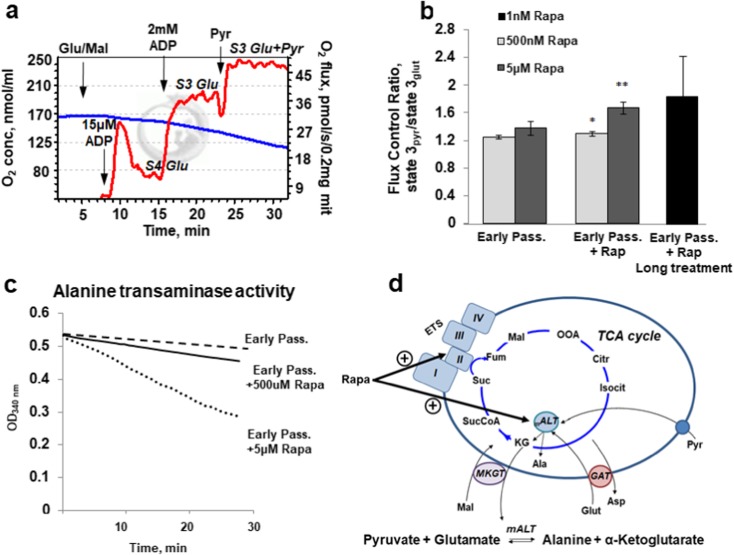
As seen from the data, rapamycin does not affect glutamate/malate oxidation in complex I (Fig. 3(a, b)). Similarly, pyruvate/malate oxidation is not affected (data not shown). Succinate oxidation is elevated in the presence of 100 nM rapamycin in both active state 3 to 44.9 ± 2.2 vs 35.4 ± 0.8 pmol/s/0.1 mg mitochondria protein and resting state 4 to 22.3 ± 2.1 vs 18.2 ± 4.2 pmol/s/0.1 mg mitochondria protein. To assess the complex II activity, rotenone was used prior to addition of succinate to demonstrate the direct targeting of succinate dehydrogenase by rapamycin. Complex I- and II-mediated changes of oxidation are not profound but statistically significant. Oxidation of palmitoyl-l-carnitine, at least in our experimental setting, was shown to be not affected, remaining in the presence of rapamycin at 15.3 ± 1.8 pmol/s/0.1 mg mitochondrial protein. To mimic in vivo heart physiological condition, we challenged the mitochondria with different substrates given over glutamate- and malate-stimulated respiration. In recent work on rat heart mitochondria, the importance of the use of tissue-specific substrate mixtures for physiological data has been described (Panov 2018). Addition of pyruvate over glutamate and malate increased the state 3 respiration of isolated mitochondria. The data presented in Fig. 4a, b demonstrates the original respirometric record of this protocol and the quantitative data of the effect of rapamycin on oxidative phosphorylation rates.
Fig. 4.
Combinatory oxidation of glutamate, malate, and pyruvate by isolated cardiac mitochondria. a The original oxygraphic protocol applied to study of oxidation of the substrate combination. Addition of 15 μM of ADP enabled to evaluate state 4 and addition of 2 mM ADP to read out the maximal state 3 respiration rates. Sequential addition of 10 mM glutamate and 2 mM malate and then 10 mM pyruvate gave higher rates of oxygen consumption than the glutamate and pyruvate alone. b Quantitative flux control ratios were obtained as state 3pyr/state 3glut. Pre-incubation of mitochondria with rapamycin for 1 h dose-dependently elevated glutamate/malate/pyruvate oxidation. Elevation of pyruvate oxidation over glutamate/malate was also observed in mitochondria isolated from cells long treated with rapamycin for 14 days. Data are mean ± SD; *p < 0.044, **p < 0.0021. c Alanine aminotransferase activity also dose-dependently elevates in the presence of rapamycin. d The schematics demonstrates transamination of pyruvate and glutamate by mitochondria matrix alanine aminotransferase and the formed α-ketoglutarate further converts in TCA cycle to succinate and activates succinate dehydrogenase-dependent respiration. Abbreviations: ETS, electron transport system; mALT, mitochondria alanine aminotransferase; MKGT, malate-α-ketoglutarate transporter; GAT, glutamate-aspartate transporter
Interestingly, pretreatment with rapamycin caused dose-dependent elevation of pyruvate oxidation, while oxidation of glutamate and malate without pyruvate was not sensitive to rapamycin (Fig. 3(a, b)). Long-term treatment with 1 nM rapamycin for 14 days also elevates glutamate/malate/pyruvate oxidation, although shown to be statistically non-significant (Fig. 3b, black bar). These results led us to examine the activity of mitochondria alanine aminotransferase (mALT), the enzyme that promotes conversion of glutamate and pyruvate to α-ketoglutarate and alanine. Figure 4d shows the chain of reactions that involves transamination and further metabolizes α-ketoglutarate to succinate. Enzymatic activity of mALT was shown to be elevated also in a dose-dependent way in the presence of rapamycin (Fig. 4c).
We also examined a gene expression dataset of long-term rapamycin-treated cultures which we have generated to compare the impact of methionine restriction and rapamycin (Azar et al. 2018) for differential expression which could relate to the metabolic changes we observe here. This analysis revealed multiple changes in steady-state mRNA levels for transcripts related to mitochondrial function. Among these transcripts was the uncoupling protein 2 (UCP2) mRNA (adjusted p < 0.001). We verified the differential expression of UCP2 using PCR analysis of mRNA levels and by immunoblot for protein levels (Fig. 5S).
Fig. 5.
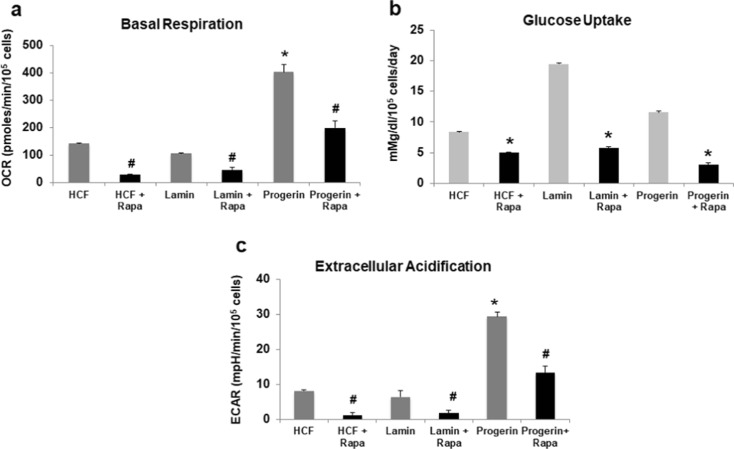
Progerin expression induces a glycolytic phenotype which is reversed by rapamycin. Measurements of basal respiration (a), glucose uptake (b), and extracellular acidification (ECAR) (c) in human cardiac fibroblasts (HCFs) expressing lamin A or progerin maintained with or without 1 nM rapamycin. Oxygen consumption rates were determined using a Seahorse bioanalyzer. Oxygen consumption and extracellular acidification rates were normalized to cell number. Data presented as mean ± SD (n = 2–3, in triplicates). The differences are shown to be statistically significant with p < 0.05 in comparison to the late passage + rapamycin values
Rapamycin alters metabolism of lamin A mutant cells
In order to examine modulation of metabolism by rapamycin during senescence, we examined the effect of rapamycin on the metabolic state in cells expressing a mutant form of lamin A, which underlies Hutchinson-Gilford progeria syndrome (DeBusk 1972; Goldman et al. 2004). Rapamycin is known to extend the replicative lifespan of patient-derived HGPS fibroblasts potentially through an increase in autophagy (Cao et al. 2011a). Human fibroblasts expressing the mutant form of lamin A, progerin, displayed nuclear atypia consistent with the known impact of the lamin A mutation (Fig. 1S). We also verified that rapamycin rescues cells from premature senescence induced by progerin expression (Supplemental Materials, Fig. 2S). Human fibroblast cells expressing progerin exhibited an increase in basal oxygen consumption, glucose uptake, and extracellular acidification (Fig. 5a–c), while rapamycin treatment reduced all of these parameters in a manner similar to the effects in the late passage cells (black bars, Fig. 5a–c).
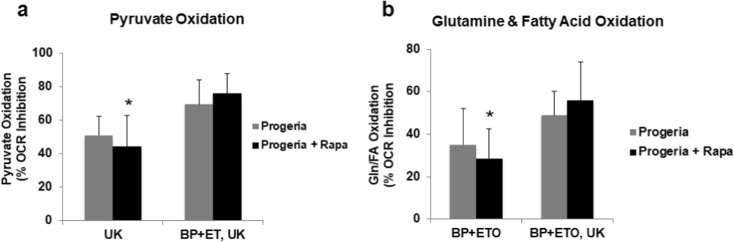
We next examined the impact of rapamycin treatment on the ability of progeria cells to maintain oxygen consumption rates following a block in the utilization of either pyruvate or glutamine and palmitate through the sequential addition of corresponding inhibitors UK5099, BPTES, and etomoxir. Although not as robust as the impact on normal fibroblasts, this analysis suggests that rapamycin-treated cultures had an enhanced capacity to utilize either pyruvate or glutamine/fatty acids (Fig. 6a, b).
Fig. 6.
Rapamycin improves metabolic flexibility in human fibroblast cells expressing progerin. Measurements of the capacity for oxidizing pyruvate (a) or glutamine/palmitate (b) in late passage fibroblasts maintained in the presence or absence of 1 nM rapamycin. Measurements were made as in Fig. 2. The data were normalized to cell number in each well. Data presented as mean ± SD (n = 2, in triplicates). The differences are shown to be statistically significant with p < 0.05 in comparison to the late passage values
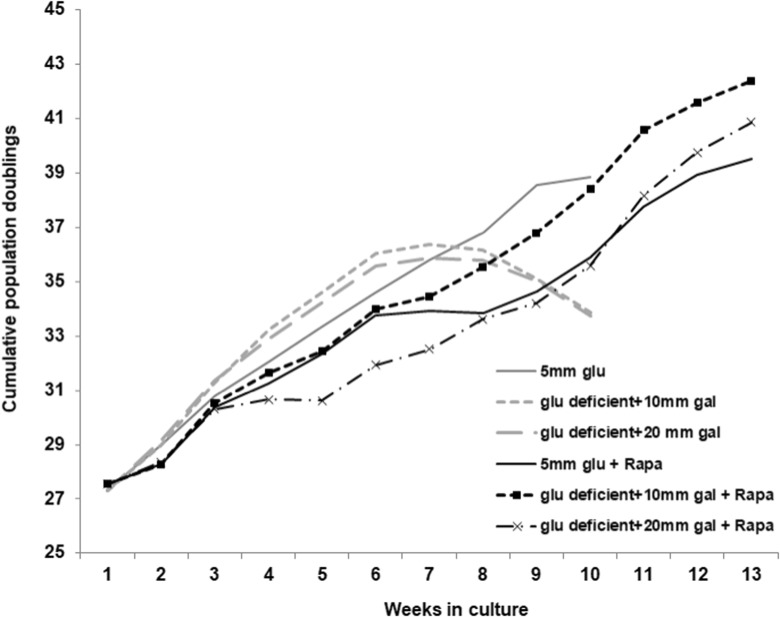
In order to directly test whether rapamycin treatment allows human fibroblasts to resist senescence induced by a metabolic stress, we grew the normal cardiac fibroblasts in glucose-free culture media supplemented with either 10 or 20 mM galactose. Due to the additional phosphorylation events required to process galactose for use in glycolysis (Holden et al. 2003), cells are relatively more reliant on oxidative phosphorylation when grown in galactose-containing medium. We found that the normal human fibroblasts undergo a premature senescence when provided galactose instead of glucose as a source of energy (Fig. 7, gray lines compared to gray dashed lines). Consistent with our interpretation of an enhanced metabolic flexibility, rapamycin provided protection against galactose-induced premature senescence (rapamycin-treated cultures are indicated by black lines in Fig. 7).
Fig. 7.
Rapamycin prevents premature senescence induced by galactose. Human cardiac fibroblasts maintained with or without 1 nM rapamycin were transferred to glucose-free medium supplemented with either 10 or 20 mM galactose at a population doubling of 27.5. Cumulative population doublings were calculated as described in “Materials and methods” section
Discussion
We have employed oxygen consumption rates as an indirect measure to gain insight into the metabolic state of senescent cells and to examine the impact of rapamycin on metabolism as it relates to senescence. First, we found that senescent cells are relatively reliant on both glycolysis and OxPhos (Fig. 1a, b). The parallel activity of both energetic routs is not inherently controversial. To compensate for their mitochondrial dysregulation, senescence cells could employ mechanisms which have been observed in tissues with high energy demand. For example, muscle cells are capable of converting glycolytic lactate to pyruvate to be further oxidized by mitochondria (Elustondo et al. 2013). In our senescent cell measurements, the elevated lactate production (Fig. 1c) is in support of this possibility. Second, we found that when challenged by inducing a block on specific metabolic pathways, senescent cells are less able to shift between carbon sources to maintain oxygen consumption, compared to early passage cells (Fig. 2). Whether the change in metabolism is a cause or consequence of senescence is not clear. However, it appears that rapamycin both reduces the dependence on glycolysis and enhances the ability to utilize other respiratory metabolites. It appears that the metabolic rejuvenation of senescence cells through long-term treatment with a low (nM) dose of rapamycin may occur via reprogramming of pyruvate and glutamate oxidation, and possibly fatty acids (Fig. 2). To better understand this metabolic reprogramming and to explore the mitochondria targets of rapamycin, we examined the direct effects of rapamycin on mitochondria isolated from cardiac fibroblasts. These oxygraph studies demonstrated that short-term treatment with rapamycin (1 h) causes elevation of complex II but not complex I activity both in active and resting states of respiration (Fig. 3(a, b)). Interestingly, similar effects of rapamycin on succinate dehydrogenase activity have been reported in studies on drosophila (Villa-Cuesta et al. 2014). The rates of beta-oxidation of cardiac fibroblasts mitochondria had a tendency to increase in the presence of rapamycin, although not statistically significant (Fig. 3(c)). Palmitoyl-l-carnitine was added alone and in combination with glutamate, malate, and pyruvate. The importance of application of hybrid substrates has been demonstrated on rat heart mitochondria (Panov 2018). Following that experimental approach, we added pyruvate to mitochondria-oxidizing glutamate and malate. The moderate rates of glutamate/malate-stimulated oxygen consumption elevated upon addition of pyruvate. Interestingly, rapamycin was able to further increase the state 3 respiration higher than in control mitochondria. The increase of oxygen consumption when both glutamate and pyruvate are present could be due to active transamination supported by mitochondria alanine aminotransferase (ALT). The enzymatic analysis confirmed the ability of rapamycin to activate ALT (Fig. 4c). As a result of transamination, pyruvate and glutamate metabolize to alanine and α-ketoglutarate with further conversion of latter to succinate. The operation of α-ketoglutarate dehydrogenase in TCA cycle that converts α-ketoglutarate to succinyl-CoA and then to succinate could be a rescue mechanism that enables mitochondria functionality based on substrate-level phosphorylation (Chinopoulos et al. 2010). The data on ALT activity demonstrating elevated production of α-ketoglutarate by rapamycin which further proceeds to generation of succinate in TCA is in agreement with the increased activity of succinate dehydrogenase promoted by rapamycin (Fig. 3(a, b)). Thus, as shown on the schematics on Fig. 4d, we revealed at least two potential direct mitochondria targets of rapamycin, succinate dehydrogenase and alanine aminotransferase. In addition, our long-term treatment of cells with rapamycin reveals potentially indirect mitochondrial targets of rapamycin. For example, we have performed gene expression profiling of long-term rapamycin-treated cultures (Azar et al. 2018) and find several changes related to mitochondrial metabolism. Among the induced set of genes is the uncoupling protein 2, which displays elevated transcript and protein levels following prolonged treatment with rapamycin (Supplementary Materials, Fig. 5S). Biochemical analysis has demonstrated that UCP2 acts as a transporter of 4 carbon, Krebs cycle intermediates and participates in the regulation of substrate phosphorylation (Vozza et al. 2014). The intact cell data corroborated by those obtained with isolated mitochondria enabled us to explore mitochondria constituents that could be involved in aging-mediated dysfunctions and be corrected by rapamycin. The observed changes in UCP2 are consistent with this data and indicate that altered gene expression of proteins related to mitochondrial function is impacted by rapamycin. These results extend our previous observations on the impact of rapamycin on NFE2L2 (NRF2) signaling (Lerner et al. 2013) to enhance mitochondrial biogenesis as well as clearance.
In addition to cardiac fibroblasts, we also examined the metabolic state of HGPS patient-derived skin fibroblasts relative to skin fibroblasts from unaffected individuals. HPGS fibroblasts exhibited a glycolytic phenotype similar to the progerin-expressing HCFs (Supplementary Materials, Fig. 3S) and rapamycin provided a similar shift towards oxidative metabolism (Supplementary Materials, Fig. 4S).
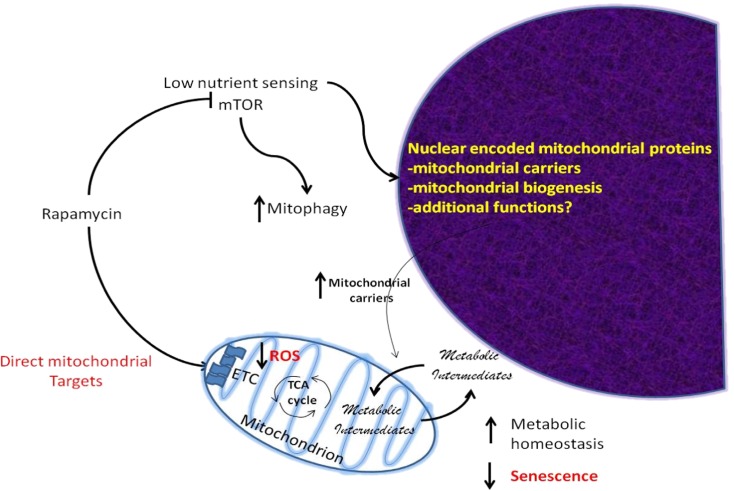
Interestingly, when cells are forced to rely upon oxidative phosphorylation by replacing glucose with galactose in the culture medium rapamycin treatment allows cells to maintain proliferative potential which suggests protection against metabolic stress. Galactose requires conversion to glucose-1-phosphate via the Leloir pathway to enter metabolism (Holden et al. 2003), which places an extra metabolic demand on the cells and a relative reliance on oxidative phosphorylation for ATP production. In terms of the relationship to senescence, replacement of glucose with micromolar concentrations of galactose has been shown to induce senescence in human fibroblasts, while cells harboring a Val32Met mutation in galactokinase display accelerated senescence (Elzi et al. 2016). Thus, these results and our observations support the concept that metabolic imbalance can induce senescence and that rapamycin improves metabolic homeostasis to prevent senescence caused by metabolic catastrophe (Fig. 8).
Fig. 8.
Model of metabolic responses to rapamycin improving metabolic homeostasis. Rapamycin interacts directly with the mitochondria to provide enhanced complex II function and inhibits the mTOR complex which impacts nuclear-encoded mitochondrial genes including mitochondrial carrier proteins which shuttle metabolic intermediates across the mitochondrial membrane providing greater metabolic flexibility to meet the demand for ATP and metabolic precursors and reduced ROS production. This increased flexibility prevents metabolic catastrophe, which can result in senescence (Nacarelli and Sell 2017)
In the literature, there are numerous links between mitochondrial function and senescence. For example, complete disruption of mitochondrial function induces senescence (Wiley et al. 2016) through an AMPK-dependent manner, while low levels of mitochondrial stress induce senescence in an mTOR-dependent manner (Nacarelli et al. 2016) and depletion of mitochondria in senescent cells leads to a reduction in SASP production (Correia-Melo et al. 2016). Proteomic analysis suggests that HGPS also involves mitochondrial dysfunction (Peinado et al. 2011; Rivera-Torres et al. 2013). In this regard, it is interesting to note that an improved metabolism has recently been reported in LMNA −/− mice treated with rapamycin (Liao et al. 2016). In this case, the authors find an improved balance between lipolysis and glucose utilization which allows the LMNA −/− mice to survive in the face of profound metabolic dysfunction. These results are consistent with previous reports suggesting a mitochondrial defect in Hutchinson-Gilford progeria and our observations regarding the impact of rapamycin on metabolic flexibility. It is interesting to note that both rapamycin treatment and low IGF-1 levels also provide enhanced ability to maintain metabolic balance when challenged by a high fat diet (Chang et al. 2009; Lorenzini et al. 2014, Salmon et al. 2015).
Based on our studies, we believe that rapamycin increases metabolic flexibility defined as the ability to utilize multiple carbon sources to satisfy metabolic demands, and that this flexibility averts a metabolic stress contributing to the lifespan extension that is provided by rapamycin treatment, both in cell culture and in the organism. This effect appears to be mediated both by direct interaction of rapamycin with the mitochondria and through increased expression of mitochondrial transporters such as UCP2.
Electronic supplementary material
(DOCX 355 kb)
Acknowledgments
The support of the Drexel Aging Initiative is appreciated.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s11357-018-0030-2) contains supplementary material, which is available to authorized users.
References
- Ahmed EK, Rogowska-Wrzesinska A, Roepstorff P, Bulteau AL, Friguet B. Protein modification and replicative senescence of WI-38 human embryonic fibroblasts. Aging Cell. 2010;9:252–272. doi: 10.1111/j.1474-9726.2010.00555.x. [DOI] [PubMed] [Google Scholar]
- Azar A, Lawrence I, Jofre S, Mell J, Sell C. Distinct patterns of gene expression in human cardiac fibroblasts exposed to rapamycin treatment or methionine restriction. Ann N Y Acad Sci. 2018;18(1):95–105. doi: 10.1111/nyas.13566. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, Saltness RA, Jeganathan KB, Verzosa GC, Pezeshki A, Khazaie K, Miller JD, van Deursen JM. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielak-Zmijewska A, Wnuk M, Przybylska D, Grabowska W, Lewinska A, Alster O, Korwek Z, Cmoch A, Myszka A, Pikula S, Mosieniak G, Sikora E. A comparison of replicative senescence and doxorubicin-induced premature senescence of vascular smooth muscle cells isolated from human aorta. Biogerontology. 2014;15:47–64. doi: 10.1007/s10522-013-9477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittles AH, Harper N. Increased glycolysis in ageing cultured human diploid fibroblasts. Biosci Rep. 1984;4:751–756. doi: 10.1007/BF01128816. [DOI] [PubMed] [Google Scholar]
- Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K, Graziotto J, Blair C, Mazzulli J, Erdos M, Krainc D, Collins F. Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson-Gilford progeria syndrome cells. Sci Transl Med. 2011;3:89ra58. doi: 10.1126/scitranslmed.3002346. [DOI] [PubMed] [Google Scholar]
- Cao K, Graziotto JJ, Blair CD, Mazzulli JR, Erdos MR, Krainc D, Collins FS. Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson-Gilford progeria syndrome cells. Sci Transl Med. 2011;3:89ra58. doi: 10.1126/scitranslmed.3002346. [DOI] [PubMed] [Google Scholar]
- Chang GR, Wu YY, Chiu YS, Chen WY, Liao JW, Hsu HM, Chao TH, Hung SW, Mao FC. Long-term administration of rapamycin reduces adiposity, but impairs glucose tolerance in high-fat diet-fed KK/HlJ mice. Basic Clin Pharmacol Toxicol. 2009;105:188–198. doi: 10.1111/j.1742-7843.2009.00427.x. [DOI] [PubMed] [Google Scholar]
- Chimenti C, Kajstura J, Torella D, Urbanek K, Heleniak H, Colussi C, Di Meglio F, Nadal-Ginard B, Frustaci A, Leri A, Maseri A, Anversa P. Senescence and death of primitive cells and myocytes lead to premature cardiac aging and heart failure. Circ Res. 2003;93:604–613. doi: 10.1161/01.RES.0000093985.76901.AF. [DOI] [PubMed] [Google Scholar]
- Chinopoulos C, Gerencser AA, Mandi M, Mathe K, Töröcsik B, Doczi J, Turiak L, Kiss G, Konràd C, Vajda S, Vereczki V, Oh RJ, Adam-Vizi V. Forward operation of adenine nucleotide translocase during F0F1-ATPase reversal: critical role of matrix substrate-level phosphorylation. FASEB J. 2010;24:2405–2416. doi: 10.1096/fj.09-149898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavitti R, Finkel T. Reactive oxygen species as mediators of cellular senescence. IUBMB Life. 2005;57:277–281. doi: 10.1080/15216540500091890. [DOI] [PubMed] [Google Scholar]
- Correia-Melo C, Marques FD, Anderson R, Hewitt G, Hewitt R, Cole J, Carroll BM, Miwa S, Birch J, Merz A, Rushton MD, Charles M, Jurk D, Tait SW, Czapiewski R, Greaves L, Nelson G, Bohlooly-Y M, Rodriguez-Cuenca S, Vidal-Puig A, Mann D, Saretzki G, Quarato G, Green DR, Adams PD, von Zglinicki T, Korolchuk VI, Passos JF (2016) Mitochondria are required for pro-ageing features of the senescent phenotype EMBO J 35:724–742 [DOI] [PMC free article] [PubMed]
- Cristofalo VJ, Charpentier R (1980) A standard procedure for cultivating human diploid fibroblastlike cells to study cellular aging. J Tissue Cult Methods 6:117–121
- Dai DF, Karunadharma PP, Chiao YA, Basisty N, Crispin D, Hsieh EJ, Chen T, Gu H, Djukovic D, Raftery D, Beyer RP, MacCoss MJ, Rabinovitch PS. Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell. 2014;13:529–539. doi: 10.1111/acel.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBusk FL. The Hutchinson-Gilford progeria syndrome. Report of 4 cases and review of the literature. J Pediatr. 1972;80:697–724. doi: 10.1016/S0022-3476(72)80229-4. [DOI] [PubMed] [Google Scholar]
- Dörr JR, Yu Y, Milanovic M, Beuster G, Zasada C, Däbritz JH, Lisec J, Lenze D, Gerhardt A, Schleicher K, Kratzat S, Purfürst B, Walenta S, Mueller-Klieser W, Gräler M, Hummel M, Keller U, Buck AK, Dörken B, Willmitzer L, Reimann M, Kempa S, Lee S, Schmitt CA. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature. 2013;501:421–425. doi: 10.1038/nature12437. [DOI] [PubMed] [Google Scholar]
- Dranka BP, Zielonka J, Kanthasamy AG, Kalyanaraman B (2011) Alterations in bioenergetic function induced by Parkinson’s disease mimetic compounds: lack of correlation with superoxide generation. J Neurochemistry 122(5):941–951 [DOI] [PMC free article] [PubMed]
- Elustondo PA, White AE, Hughes ME, Brebner K, Pavlov E, Kane DA. Physical and functional association of lactate dehydrogenase (LDH) with skeletal muscle mitochondria. J Biol Chem. 2013;288:25309–25317. doi: 10.1074/jbc.M113.476648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzi DJ, Song M, Shiio Y. Role of galactose in cellular senescence. Exp Gerontol. 2016;73:1–4. doi: 10.1016/j.exger.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Estrada JC, Torres Y, Benguría A, Dopazo A, Roche E, Carrera-Quintanar L, Pérez RA, Enríquez JA, Torres R, Ramírez JC, Samper E, Bernad A. Human mesenchymal stem cell-replicative senescence and oxidative stress are closely linked to aneuploidy. Cell Death Dis. 2013;4:e691. doi: 10.1038/cddis.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnaiger E (2008) Polarographic oxygen sensors, the oxygraph and high-resolution respirometry to assess mitochondrial function in Mitochondrial dysfunction in drug-induced toxicity D.J.a.W. Y, ed. (Wiley):27–352
- Gnaiger E, Kuznetsov A, Schneeberger S (2000) Mitochondria in the cold. In: Heldmaier G, Klingenspor M, editors Life in the Cold. Berlin, Heidelberg, New York: Springer;:431–442
- Gnaiger E, Steinlechner-Maran R, Mendez G, Eberl T, Margreiter R. Control of mitochondrial and cellular respiration by oxygen. J Bioenerg Biomembr. 1995;27:583–596. doi: 10.1007/BF02111656. [DOI] [PubMed] [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, Collins FS. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein S, Ballantyne SR, Robson AL, Moerman EJ. Energy metabolism in cultured human fibroblasts during aging in vitro. J Cell Physiol. 1982;112:419–424. doi: 10.1002/jcp.1041120316. [DOI] [PubMed] [Google Scholar]
- Hayflick L, Moorhead P. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- Hill BG, Benavides GA, Lancaster Jr. JR, Ballinger S, Dell’Italia L, Zhang J, Darley-Usmar VM (2012) Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol Chem 393(12):1485–1512 [DOI] [PMC free article] [PubMed]
- Holden HM, Rayment I, Thoden JB. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J Biol Chem. 2003;278:43885–43888. doi: 10.1074/jbc.R300025200. [DOI] [PubMed] [Google Scholar]
- Hutter E, Renner K, Pfister G, Stockl P, Jansen-Durr P, Gnaiger E. Senescence-associated changes in respiration and oxidative phosphorylation in primary human fibroblasts. The Biochemical journal. 2004;380:919–928. doi: 10.1042/bj20040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James EL, Michalek RD, Pitiyage GN, de Castro AM, Vignola KS, Jones J, Mohney RP, Karoly ED, Prime SS, Parkinson EK. Senescent human fibroblasts show increased glycolysis and redox homeostasis with extracellular metabolomes that overlap with those of irreparable DNA damage, aging, and disease. J Proteome Res. 2015;14:1854–1871. doi: 10.1021/pr501221g. [DOI] [PubMed] [Google Scholar]
- Jeyapalan J, Sedivy J. Cellular senescence and organismal aging. Mech Ageing Dev. 2008;129:467–541. doi: 10.1016/j.mad.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Yanos ME, Kayser EB, Quintana A, Sangesland M, Castanza A, Uhde L, Hui J, Wall VZ, Gagnidze A, Oh K, Wasko BM, Ramos FJ, Palmiter RD, Rabinovitch PS, Morgan PG, Sedensky MM, Kaeberlein M. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science. 2013;342:1524–1528. doi: 10.1126/science.1244360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplon J, Zheng L, Meissl K, Chaneton B, Selivanov VA, Mackay G, van der Burg SH, Verdegaal EM, Cascante M, Shlomi T, Gottlieb E, Peeper DS. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498:109–112. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Lamming DW. The mechanistic target of rapamycin: the grand ConducTOR of metabolism and aging. Cell Metab. 2016;23:990–1003. doi: 10.1016/j.cmet.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Byun HO, Jee BA, Cho H, Seo YH, Kim YS, Park MH, Chung HY, Woo HG, Yoon G. Implications of time-series gene expression profiles of replicative senescence. Aging Cell. 2013;12:622–634. doi: 10.1111/acel.12087. [DOI] [PubMed] [Google Scholar]
- Kumar S, Lombard DB (2016) Finding Ponce de Leon’s pill: challenges in screening for anti-aging molecules. F1000Res 5 [DOI] [PMC free article] [PubMed]
- Laberge RM, Sun Y, Orjalo AV, Patil CK, Freund A, Zhou CSC, Davalos AR, Wilson-Edell KA, Liu S, Limbad C, Demaria M, Li P, Hubbard GB, Ikeno Y, Javors M, Desprez PY, Benz CC, Kapahi P, Nelson PS, Campisi J. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat Cell Biol. 2015;17:1049–1061. doi: 10.1038/ncb3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner C, Bitto A, Pulliam D, Nacarelli T, Konigsberg M, Van Remmen H, Torres C, Sell C (2013) Reduced mammalian target of rapamycin activity facilitates mitochondrial retrograde signaling and increases life span in normal human fibroblasts. Aging Cell 12(6):966–977 [DOI] [PMC free article] [PubMed]
- Liao C-Y, Anderson SS, Chicoine NH, Mayfield JR, Academia EC, Wilson JA, Pongkietisak C, Thompson MA, Lagmay EP, Miller DM, Hsu Y-M, McCormick MA, O'Leary MN, Kennedy BK. Rapamycin reverses metabolic deficits in lamin A/C-deficient mice. Cell Rep. 2016;17:2542–2552. doi: 10.1016/j.celrep.2016.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao EC, Hsu YT, Chuah QY, Lee YJ, Hu JY, Huang TC, Yang PM, Chiu SJ. Radiation induces senescence and a bystander effect through metabolic alterations. Cell Death Dis. 2014;5:e1255. doi: 10.1038/cddis.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzini A, et al. Mice producing reduced levels of insulin-like growth factor type 1 display an increase in maximum, but not mean, lifespan. Aging Cell. 2014;69(4):410–419. doi: 10.1093/gerona/glt108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Finkel T. Free radicals and senescence. Exp Cell Res. 2008;314:1918–1922. doi: 10.1016/j.yexcr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, Harrison DE, Astle CM, Baur JA, Boyd AR, de Cabo R, Fernandez E, Flurkey K, Javors MA, Nelson JF, Orihuela CJ, Pletcher S, Sharp ZD, Sinclair D, Starnes JW, Wilkinson JE, Nadon NL, Strong R. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. the Journals of Gerontology Series A Biological Sciences and Medical Sciences. 2011;66:191–201. doi: 10.1093/gerona/glq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseeva O, Bourdeau V, Roux A, Deschenes-Simard X, Ferbeyre G. Mitochondrial dysfunction contributes to oncogene-induced senescence. Mol Cell Biol. 2009;29:4495–4507. doi: 10.1128/MCB.01868-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacarelli T, Azar A, Sell C. Aberrant mTOR activation in senescence and aging: a mitochondrial stress response? Exp Gerontol. 2015;68:66–70. doi: 10.1016/j.exger.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacarelli T, Azar A, Sell C. Mitochondrial stress induces cellular senescence in an mTORC1-dependent manner. Free Radic Biol Med. 2016;95:133–154. doi: 10.1016/j.freeradbiomed.2016.03.008. [DOI] [PubMed] [Google Scholar]
- Nacarelli T, Sell C. Targeting metabolism in cellular senescence, a role for intervention. Mol Cell Endocrinol. 2017;455:83–92. doi: 10.1016/j.mce.2016.08.049. [DOI] [PubMed] [Google Scholar]
- Nair RR, Bagheri M, Saini DK. Temporally distinct roles of ATM and ROS in genotoxic-stress-dependent induction and maintenance of cellular senescence. J Cell Sci. 2015;128:342–353. doi: 10.1242/jcs.159517. [DOI] [PubMed] [Google Scholar]
- Panov AV. Synergistic oxidation of fatty acids, glucose and amino acids metabolites by isolated rat heart mitochondria EC. Cardiology. 2018;5:198–208. [Google Scholar]
- Panov AV, Lund S, Greenamyre JT. Ca2+-induced permeability transition in human lymphoblastoid cell mitochondria from normal and Huntington's disease individuals. Mol Cell Biochem. 2005;269:143–152. doi: 10.1007/s11010-005-3454-9. [DOI] [PubMed] [Google Scholar]
- Passos JF, Miwa S, von Zglinicki T. Measuring reactive oxygen species in senescent cells. Methods Mol Biol. 2013;965:253–263. doi: 10.1007/978-1-62703-239-1_17. [DOI] [PubMed] [Google Scholar]
- Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, Peters H, Wappler I, Birket MJ, Harold G, Schaeuble K, Birch-Machin MA, Kirkwood TBL, von Zglinicki T. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 2007;5:e110. doi: 10.1371/journal.pbio.0050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado JR et al. (2011) Proteomic profiling of adipose tissue from Zmpste24−/− mice, a model of lipodystrophy and premature aging, reveals major changes in mitochondrial function and vimentin processing Mol Cell Proteomics 10:M111 [DOI] [PMC free article] [PubMed]
- Pesta D, Gnaiger E. High-resolution respirometry: OXPHOS protocols for human cells and permeabilized fibers from small biopsies of human muscle. Methods Mol Biol. 2012;810:25–58. doi: 10.1007/978-1-61779-382-0_3. [DOI] [PubMed] [Google Scholar]
- Quijano C, Cao L, Fergusson MM, Romero H, Liu J, Gutkind S, Rovira II, Mohney RP, Karoly ED, Finkel T. Oncogene-induced senescence results in marked metabolic and bioenergetic alterations. Cell Cycle. 2012;11:1383–1392. doi: 10.4161/cc.19800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A, Schreiber SL. Direct control of mitochondrial function by mTOR. Proc Natl Acad Sci U S A. 2009;106:22229–22232. doi: 10.1073/pnas.0912074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos FJ, Chen SC, Garelick MG, Dai DF, Liao CY, Schreiber KH, MacKay VL, An EH, Strong R, Ladiges WC, Rabinovitch PS, Kaeberlein M, Kennedy BK. Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci Transl Med. 2012;4:144ra103. doi: 10.1126/scitranslmed.3003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Torres J, Acín-Perez R, Cabezas-Sánchez P, Osorio FG, Gonzalez-Gómez C, Megias D, Cámara C, López-Otín C, Enríquez JA, Luque-García JL, Andrés V. Identification of mitochondrial dysfunction in Hutchinson-Gilford progeria syndrome through use of stable isotope labeling with amino acids in cell culture. J Proteome. 2013;91:466–477. doi: 10.1016/j.jprot.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Salmon AB, Lerner C, Ikeno Y, Motch Perrine SM, McCarter R, Sell C (2015) Altered metabolism and resistance to obesity in long-lived mice producing reduced levels of IGF-I. Am J Physiol Endocrinol Metab 308(7):E545-53 [DOI] [PMC free article] [PubMed]
- Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol. 2008;10(4):452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souders CA, Bowers SLK, Baudino TA (2009) Cardiac fibroblast: the renaissance cell. Circ Res 105:1164–1176 [DOI] [PMC free article] [PubMed]
- Takebayashi S, Tanaka H, Hino S, Nakatsu Y, Igata T, Sakamoto A, Narita M, Nakao M. Retinoblastoma protein promotes oxidative phosphorylation through upregulation of glycolytic genes in oncogene-induced senescent cells. Aging Cell. 2015;14:689–697. doi: 10.1111/acel.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa-Cuesta E, Holmbeck MA, Rand DM. Rapamycin increases mitochondrial efficiency by mtDNA-dependent reprogramming of mitochondrial metabolism in Drosophila. J Cell Sci. 2014;127:2282–2290. doi: 10.1242/jcs.142026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vozza A, Parisi G, De Leonardis F, Lasorsa FM, Castegna A, Amorese D, Marmo R, Calcagnile VM, Palmieri L, Ricquier D, Paradies E, Scarcia P, Palmieri F, Bouillaud F, Fiermonte G. UCP2 transports C4 metabolites out of mitochondria, regulating glucose and glutamine oxidation. Proc Natl Acad Sci U S A. 2014;111:960–965. doi: 10.1073/pnas.1317400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu Y, Zhang R, Zhang F, Sui W, Chen L, Zheng R, Chen X, Wen F, Ouyang HW, Ji J. Apoptotic transition of senescent cells accompanied with mitochondrial hyper-function. Oncotarget. 2016;7:28286–28300. doi: 10.18632/oncotarget.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley CD, Velarde MC, Lecot P, Liu S, Sarnoski EA, Freund A, Shirakawa K, Lim HW, Davis SS, Ramanathan A, Gerencser AA, Verdin E, Campisi J. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab. 2016;23:303–314. doi: 10.1016/j.cmet.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 355 kb)