Infection by the opportunistic pathogen Pseudomonas aeruginosa is the leading cause of morbidity and mortality seen in CF patients. Poor patient prognosis correlates with the genotypic and phenotypic change of the bacteria from a typical nonmucoid to a mucoid form in the CF lung, characterized by the overproduction of alginate. The expression of this exopolysaccharide is under the control an alternate sigma factor, AlgT/U, that is regulated posttranslationally by a series of proteases. A better understanding of this regulatory phenomenon will help in the development of therapies targeting alginate production, ultimately leading to an increase in the length and quality of life for those suffering from CF.
KEYWORDS: mucoid conversion, Prc/AlgO/Tsp, RseP/MucP/YaeL, σE, σ22, AlgT/U, cystic fibrosis, sigma factors, sigma-22, sigma-E, anti-sigma factor
ABSTRACT
The progression of cystic fibrosis (CF) from an acute to a chronic disease is often associated with the conversion of the opportunistic pathogen Pseudomonas aeruginosa from a nonmucoid form to a mucoid form in the lung. This conversion involves the constitutive synthesis of the exopolysaccharide alginate, whose production is under the control of the AlgT/U sigma factor. This factor is regulated posttranslationally by an extremely unstable process and has been commonly attributed to mutations in the algT (algU) gene. By exploiting this unstable phenotype, we isolated 34 spontaneous nonmucoid variants arising from the mucoid strain PDO300, a PAO1 derivative containing the mucA22 allele commonly found in mucoid CF isolates. Complementation analysis using a minimal tiling path cosmid library revealed that most of these mutants mapped to two protease-encoding genes, algO, also known as prc or PA3257, and mucP. Interestingly, our algO mutations were complemented by both mucP and algO, leading us to delete, clone, and overexpress mucP, algO, mucE, and mucD in both wild-type PAO1 and PDO300 backgrounds to better understand the regulation of this complex regulatory mechanism. Our findings suggest that the regulatory proteases follow two pathways for regulated intramembrane proteolysis (RIP), where both the AlgO/MucP pathway and MucE/AlgW pathway are required in the wild-type strain but where the AlgO/MucP pathway can bypass the MucE/AlgW pathway in mucoid strains with membrane-associated forms of MucA with shortened C termini, such as the MucA22 variant. This work gives us a better understanding of how alginate production is regulated in the clinically important mucoid variants of Pseudomonas aeruginosa.
IMPORTANCE Infection by the opportunistic pathogen Pseudomonas aeruginosa is the leading cause of morbidity and mortality seen in CF patients. Poor patient prognosis correlates with the genotypic and phenotypic change of the bacteria from a typical nonmucoid to a mucoid form in the CF lung, characterized by the overproduction of alginate. The expression of this exopolysaccharide is under the control an alternate sigma factor, AlgT/U, that is regulated posttranslationally by a series of proteases. A better understanding of this regulatory phenomenon will help in the development of therapies targeting alginate production, ultimately leading to an increase in the length and quality of life for those suffering from CF.
INTRODUCTION
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen capable of thriving in a variety of environments. Due to its ability to adapt and infect, P. aeruginosa is commonly found in immunocompromised patients as well as those with cystic fibrosis (CF). In about 80% of patients with CF, P. aeruginosa can be found in the respiratory tract, leading to multiple complications and ultimate deterioration of the lung (1). One of the mechanisms of survival of P. aeruginosa in the lungs of CF patients is its ability to overproduce alginate, a capsule-like linear copolymer composed of β-d-mannuronic acid and α-l-guluronic acid (2). Following colonization of the CF lung, P. aeruginosa mucoid colonies emerge characterized by a phenotypic change from an Alg− nonmucoid state to an Alg+ mucoid colony morphology (3, 4). Although the selective pressures that drive the conversion of P. aeruginosa from a nonmucoid to a mucoid state are not fully understood, it has been shown that exposure of nonmucoid strains to activated polymorphonuclear leukocytes (PMNs) or low levels of hydrogen peroxide can result in mucA defective mucoid variants (5). Other authors have reported that antimicrobial peptides (6), carbon, nitrogen, or phosphorus starvation (7), or estradiol (8) could also induce mutations in mucA leading to mucoidy. The presence of mucoid colonies is indicative of the onset of chronic infection and a poor patient prognosis (9).
The master regulator of alginate production is σ22, also known as AlgT/U (10). It presents 65% similarity to the sigma factor σE, responsible for the stress-mediated response in Escherichia coli (11, 12). AlgT/U is sequestered in the cytoplasm by the negative regulator MucA, known as the anti-sigma factor, located across the inner membrane (13–15). The mucA gene, in the algT (algU) mucABCD operon (16), has been identified as the major site for mutations present in mucoid strains isolated from the CF lung, with such mutations usually localized to the 3′ end of the gene (17, 18). Many of these mutations, which cause alterations in the C terminus of the protein, affect the interaction of MucB with MucA in the periplasmic space, causing degradation of MucA by sequential proteases and leading to the release of AlgT/U, resulting in the overexpression of alginate (14, 15, 19).
The production of alginate is regulated by the degradation of MucA in a manner similar to that for the E. coli σE stress response pathway (20, 21). Sequential proteolytic cleavage of MucA (E. coli RseA) is initiated by the accumulation of envelope proteins, such as MucE containing the C-terminal amino acid sequence WVF (Fig. 1) (20). This signature sequence, along with C-terminal signatures YVF, LVF, WIF, and WVW, is necessary to interact with the intramembrane-cleaving (I-CLiP) site 1 serine protease AlgW. The MucE-AlgW interaction is required for AlgW, the E. coli DegS homolog, to perform primary cleavage of MucA (20, 22). This cleavage liberates MucB from the carboxyl terminus of MucA, allowing the site 2 zinc metalloprotease MucP to further degrade MucA (20, 21). MucP shows 63% similarity to the RseP/YaeL protease in E. coli that is responsible for cleavage of RseA within the inner membrane (21, 23). The residual MucA protein, located in the cytoplasmic space, is then finally degraded by the protease complex ClpXP, allowing activation of AlgT/U; this process is aided by the SspA protein (24, 25).
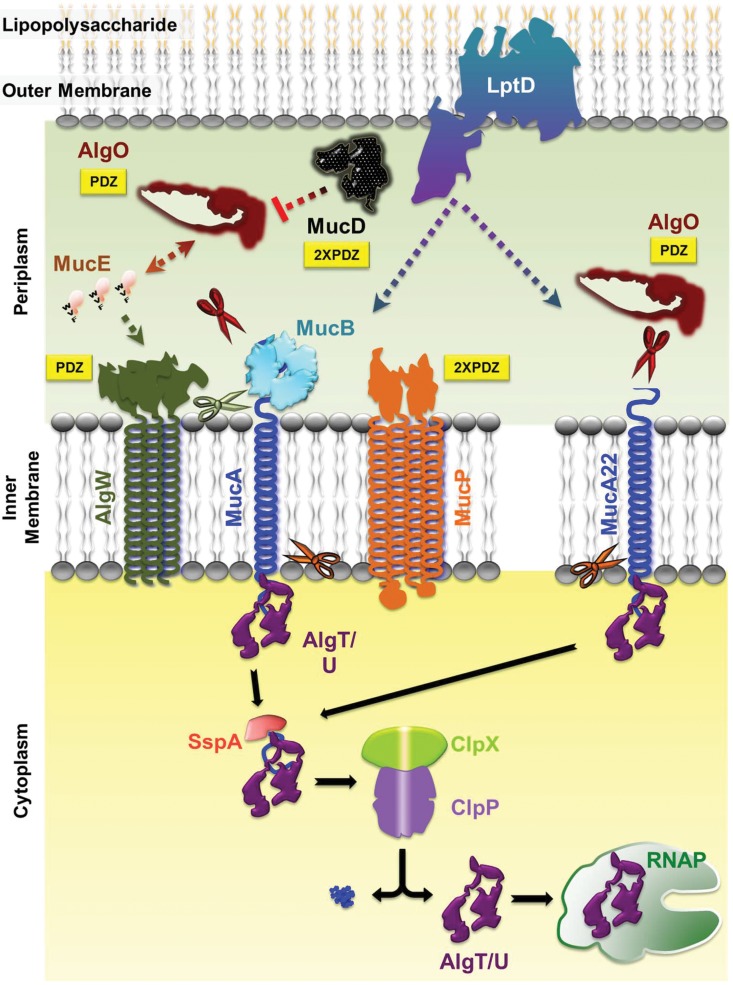
FIG 1.
Regulated intramembrane proteolysis pathway of P. aeruginosa. The C-terminal WVF motif of MucE indirectly activates the AlgW protease to cleave MucA. This cleavage requires the removal of the MucB protein from the MucA C terminus. Subsequent cleavage of MucA is performed by the AlgO and MucP proteases, which releases the AlgT/U sigma factor. Further processing by SspA, ClpX, and ClpP removes the remaining MucA fragment, allowing AlgT/U to interact with RNA polymerase (RNAP) and begin transcription of the alginate pathway genes. This process is negatively regulated in the periplasmic space by MucD. In the mucoid mucA22 mutant, the truncated C terminus of the protein is not bound by MucB, allowing for cleavage by the AlgO protease (see the text for details). The cleaved MucA22 protein is then processed by SspA, ClpX, and ClpP. Both wild-type and MucA22 pathways also undergo regulation by the LptD outer membrane protein (46). AlgO, MucD, AlgW, and MucP all contain PDZ domains (yellow boxes) involved in protein-protein interactions. Scissors indicate proteolytic cleavage of MucA or MucA22 by AlgW (green), AlgO (burgundy), and MucP (orange).
In addition to the proteases AlgW, MucP, and ClpXP, involved in the degradation of MucA, two other proteases have been shown to take part in the regulation of alginate production. The serine protease MucD presents 57% similarity and 39% identity to the E. coli protein HtrA (DegP) (25–27). MucD has been shown to be a negative regulator of alginate production as well as a positive regulator of high-temperature stress (26). It has been postulated that MucD regulates alginate production by removing accumulated proteins such as MucE that can activate proteolytic degradation of MucA (20). The second protease shown to be involved in alginate production is the serine protease AlgO (also known as Prc) (28, 29). AlgO shows 42% similarity to the E. coli periplasmic protease Prc (Tsp), shown to be involved in stress response and processing of carboxyl termini of target proteins (29–31). AlgO does not share similar functions with E. coli Prc and has no effect on antibiotic resistance or heat shock response (29). However, its involvement in alginate production is clear, since loss of AlgO in an Alg+ mucA mutant strain renders the strain nonmucoid (28, 29).
In this study, we further characterize the remaining suppressors of alginate production (sap) mutants isolated from P. aeruginosa strain PDO300 (5, 29). These suppressors were isolated by growing a defined PAO1mucA22 mucoid strain (PDO300) without aeration, a stressful condition that selects for mutations that revert the metabolically demanding mucoid condition to a nonmucoid phenotype. The majority of these mutations were mapped to either algT (algU) or to one of two protease-encoding genes known to be involved in the regulation of alginate production, leading us to further explore the role of these proteases in alginate regulation. This study explores the potential regulatory cascade involving proteases using precise in-frame deletions and overexpression of the various genes known to be involved in alginate regulation.
RESULTS
Characterization of sap mutants by cosmid complementation.
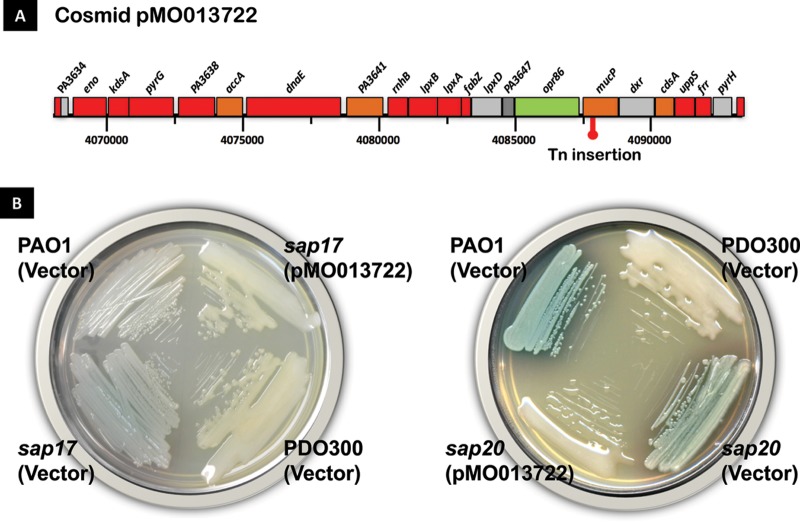
Previously, 34 nonmucoid revertants (sap mutants) were isolated by growing a defined PAOmucA22 mutant (Alg+ PDO300) without aeration, a condition for this facultative aerobe that selects for nonmucoid revertants (29). Fourteen sap mutants were found to harbor unidentified mutations in algT (algU), and the three remaining mutants are currently being characterized. DNA sequence analysis had previously mapped the mutation in another sap mutant, sap22, to be in PA3257 (29), which encodes the periplasmic protease AlgO (29), also known as Tsp/Prc (28). To identify mutations in the remaining 16 sap mutants, each strain was complemented with the PAO1 minimal tiling path (MTP) library which had been previously divided into four pools (29). The mucoid phenotype was restored in several mutants. Two sap strains, sap17 and sap20 strains, were chosen for further characterization, since both were complemented with an identical cosmid (pMO013722) containing PAO1 genomic sequences from coordinates 4068269 to 409343, encompassing a total of 21 open reading frames (ORFs) (Fig. 2A and B).
FIG 2.
Complementation of sap17 and sap20 mutants with cosmid pMO013722 on LBTc plates. (A) The P. aeruginosa genes found in cosmid pMO013722 (corresponding to nucleotides 4068269 to 409343 of the PAO1 genome) are shown. The red symbol indicates the position of the EZ::Tn transposon insertion. (B) Phenotype of wild-type, mutant, and complemented strains inoculated on LB plates. The parental PDO300 strain displays a mucoid phenotype which is lost in the sap mutants but restored upon transformation of pMO013722. The prototypic PAO1 strain, transformed with the empty vector plasmid pLAFR3, is included as a nonmucoid control. Cells were streaked on LB plates containing tetracycline and were incubated at 37°C for 24 h.
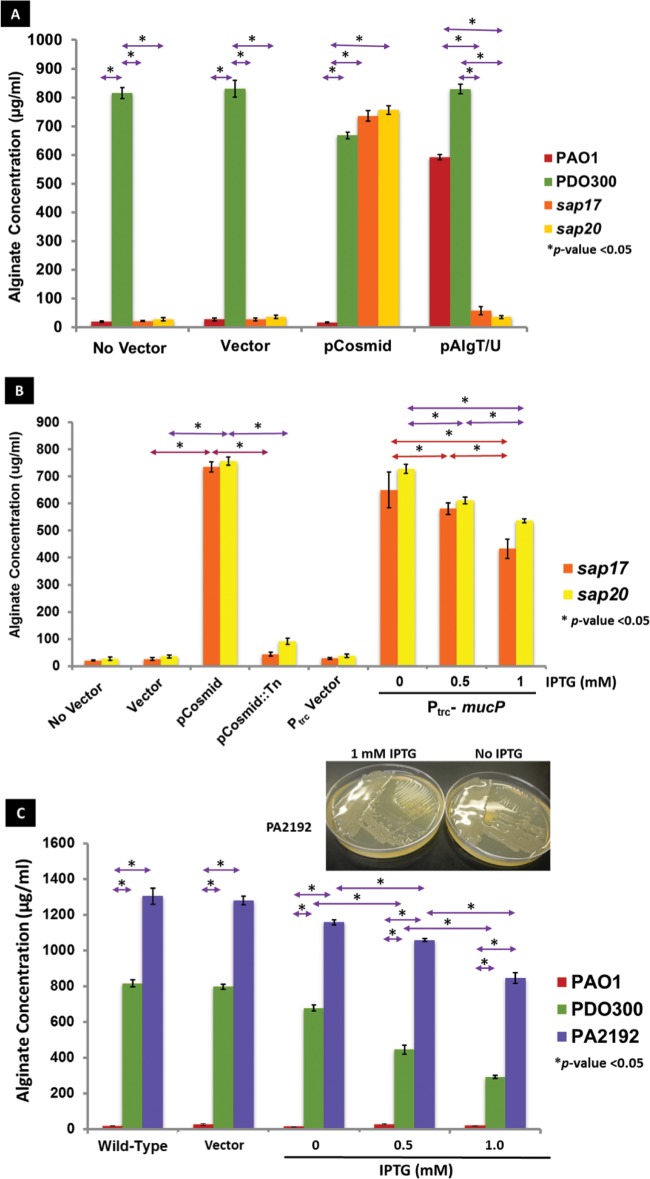
To map the ORF in the cosmid responsible for restoration of the mucoid phenotype, the cosmid was subjected to transposon (Tn) mutagenesis using EZ::TN (32, 33). The mutagenized library was introduced into sap17 and sap20 strains, and transconjugants were screened for loss of the mucoid phenotype, indicating a Tn disruption of the complementing gene on the cosmid. The mutagenized cosmid was then isolated from P. aeruginosa and introduced into E. coli for further identification. The gene responsible for the loss of the mucoid phenotype was mapped to PA3649 (mucP) by sequencing of the Tn junction, which indicated an insertion between nucleotides 599 and 600 of the mucP coding sequence (Fig. 2A; see also Fig. S1 in the supplemental material). The restoration of the mucoid phenotype was further confirmed by alginate quantification using the carbazole assay (34). PAO1, sap17, and sap20 strains containing the empty vector pLAFR3 produced negligible amounts of alginate as well as PAO1 without vector (Fig. 3A). The introduction of an algT (algU)-expressing plasmid induced alginate production in PAO1 but not in the sap17 and sap20 mutants, confirming the initial observations that these particular sap mutants were not complemented by AlgT/U (29) (Fig. 3A). PDO300 and the two sap mutants possessing pMO013722 produced significant amounts of alginate compared to the nonmucoid controls (Fig. 3A), and these levels were reduced in the sap mutants containing the transposon-mutagenized cosmid (Fig. 3B).
FIG 3.
Alginate levels in P. aeruginosa strains. Assays were performed in triplicate, and statistics were calculated using the two-tailed Student t test; asterisks indicate P values of <0.05 (A) Alginate levels of wild-type (PAO1), mucoid (PDO300), sap17, and sap20 overnight LB cultures were measured using the uronic acid assay standardized with purified sodium alginate, expressed in micrograms of alginate per milliliter of culture supernatant. Strains lacked plasmid (No Vector), contained plasmid pLAFR3 (Vector), contained pMO013722 (pCosmid), or contained pJG293 (pAlgT/U) as indicated. (B) sap mutants containing pLAFR3 (Vector), pMO013722 (pCosmid), pMO013722 containing the transposon insertion (pCosmid::Tn), pMF54 (Ptrc Vector), and pLVF54 containing mucP (Ptrc-mucP) were grown overnight in LB medium containing carbenicillin (except the no vector control) and induced at the indicated IPTG concentrations. (C) Wild-type PAO1 and mucoid strains PDO300 and PA2192 were transformed with the mucP-containing pLVF52 plasmid and grown in LB broth containing carbenicillin with increasing concentrations of IPTG. The inset image indicates the mucoid phenotype of pLVF52-transformed PA2192 in the presence and absence of 1 mM IPTG.
Complementation of sap17 and sap20 strains with mucP.
To confirm that the restoration of the mucoid phenotype in sap17 and sap20 mutants was due to mucP, a plasmid carrying mucP under the control of the Ptrc promoter (pMucP) was introduced into these strains. The mucoid phenotype was restored in both sap mutants, whereas complementation with the vector control remained nonmucoid (Fig. S1).
It was observed that after the introduction of pMO013722 into PDO300, the alginate levels were slightly reduced compared to those of the original mucoid strain (Fig. 3A). Similarly, introducing pMucP into sap17 and sap20 strains increased the alginate levels, but these levels decreased with increasing concentrations of isopropyl-β-d-thiogalactopyranoside (IPTG) (Fig. 3B). In fact, the highest levels of alginate in the pMucP-complemented mutants were found at an IPTG concentration of 0 mM, which still produces small amounts of MucP due to the inherent leakiness of the Ptrc promoter (35). This suggests that an optimal amount of MucP is required for alginate production, and levels that exceed this amount produce less alginate in the complemented strain. To determine if this phenotype was strain dependent, the pMucP plasmid was introduced into a CF P. aeruginosa mucoid strain, PA2192 (36). This clinical isolate contains a more stable mucA mutation that is different from the one found in PDO300 (37). The PA2192 parental strain's alginate levels were measured to be 1,303 μg ml−1, while in PA2192(pMucP) at 1 mM IPTG, the levels were significantly decreased to 746 μg ml−1 (Fig. 3C). Thus, overexpression of mucP suppresses alginate production in the mucoid clinical isolate as well as in PDO300. The pMucP plasmid was also introduced into PAO1 as a negative control. PAO1 (pMucP) remained strictly nonmucoid, suggesting that the effect of overexpression of the mucP gene was specific to strains carrying mucA mutations only (Fig. 3C).
Mapping the sap17 and sap20 mutations.
Our results suggested that the sap17 and sap20 strains harbored a mutation within mucP. Sequence analysis of the sap20 strain revealed a single-nucleotide deletion of a guanine (G) at nucleotide 1174, resulting in a frameshift mutation after amino acid (aa) 391 (mucP392) and resulting in premature termination at codon 437 (Fig. S2). Interestingly, no mutation was found in the mucP gene in the sap17 mutant.
sap17 strain harbors a mutation in algO.
The absence of mutations in the mucP gene in the sap17 strain suggested that MucP could overcome the defect by a compensatory mutation elsewhere. Since it is known that multiple proteases play a key role in the degradation of MucA (20), it was hypothesized that sap17 had a mutation in a different protease involved in alginate regulation, and overproduction of MucP compensated for such a mutation. A mutation in the gene encoding the AlgO protease was previously characterized in a sap mutant (29), leading us to consider that our cloned mucP plasmid is able to complement AlgO. To test this hypothesis, pAlgO, a plasmid encoding the AlgO protease, was introduced into the sap17 strain. The presence of pAlgO restored the mucoid phenotype, suggesting a defect in algO is causing the phenotype observed in the sap17 mutant (data not shown).
Analysis of other sap mutants.
To determine if the remaining 14 sap mutants harbored mutations in algO or mucP, pAlgO and pMucP were introduced into the sap mutant strains and screened for the restoration of the mucoid phenotype. The mucoid phenotype was rescued in the presence of pAlgO in 11 of the mutants and in all 14 mutants in the presence of pMucP (data not shown). The algO and mucP genes in the remaining mutants were sequenced, revealing four novel mutations in algO and three novel mutations in mucP (Fig. S2 and S3).
Analysis of the algO alleles.
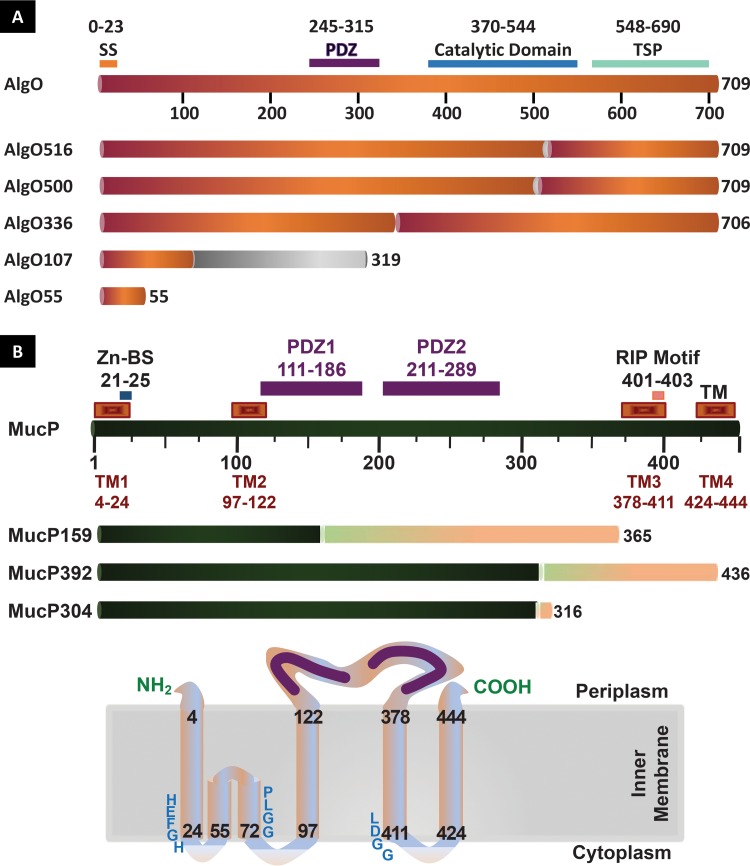
AlgO, previously referred to as Prc or PA3257, is a 698-aa periplasmic protein with a predicted molecular weight of 78.2 kDa (29). The protein possesses a signal sequence from residues 1 to 23 and contains a PDZ domain involved in protein-protein interactions at positions 245 to 315, an S41 peptidase domain at positions 370 to 544, and a tail-specific protease C-terminal domain at positions 548 to 690 (Fig. 4A). Sequence analysis revealed the mutation in the sap16, sap17, sap23, sap24, and sap25 strains was due to a substitution of an adenine (A) for a cytosine (C) at nucleotide 1499 of algO (Fig. S3). The mutation in this allele (algO500) corresponds to a glutamine (Q)-to-proline (P) substitution at amino acid 500 (Fig. S3), placing the mutation in the peptidase catalytic domain (Fig. 4A) (29). In addition, analysis of the sap46 and sap47 mutations revealed a guanine (G)-to-A substitution at nucleotide 1515, creating allele algO516 and resulting in an amino acid change at residue 516 from a glycine (G) to aspartic acid (D) (Fig. S3), also within the catalytic domain of this protein (Fig. 4A).
FIG 4.
Linear representations of the AlgO (A) and MucP (B) proteins and their mutant variants. Signal sequence (SS), zinc-binding (Zn-BS), protein-protein association (PDZ), transmembrane (TM), catalytic and tail-specific protease (TSP), and RIP motif domains are indicated. Changes in the sequence due to frameshifting or single-amino-acid changes are indicated in grey (AlgO) and pink (MucP), and the specific deletion in AlgO336 is indicated as a gap in the sequence. The positioning of MucP in the inner membrane is indicated at the bottom of panel B. The zinc-binding and RIP metalloprotease motifs are indicated as HEFGH and LDGG, respectively. The C1N and MRE β-loop domains (62) are situated between TM1 and TM2, respectively. The conserved motif of the MRE β-loop (63) is indicated as PLGG.
The mutations in sap26, sap27, and sap31 strains were mapped to the same position as sap22 (29), where an insertion of a thymine (T) nucleotide at position 321 led to a frameshift mutation occurring after amino acid 106 and early termination of the protein at codon 320 (algO107) (Fig. S3). In the case of the sap32 strain, a deletion of nine nucleotides between positions 1005 and 1013 in algO was identified, changing the amino acid sequence at residue 336 and resulting in an in-frame deletion of three amino acids (algO336) (Fig. S3). This change maps to a region between the PDZ domain and the catalytic domain, suggesting that this intervening region is important for AlgO activity (Fig. 4A). The sap42 mutation was found to be due to a G-T substitution at nucleotide 163, resulting in a stop codon at residue 55 (algO55) (Fig. 4A and Fig. S3).
sap20, sap27, sap30, and sap36 strains harbor mutations in mucP.
PA3649 (mucP) is a 1,353-bp ORF encoding a 48-kDa protein which shows 63% identity to the E. coli inner membrane metalloprotease RseP/YaeL (38, 39). TMpred analysis (40) of the 450-aa MucP sequence reveals the presence of four possible transmembrane helices and one membrane-associated β-loop domain (Fig. 4B). A conserved HEXXH metalloprotease zinc-binding motif (41) is found at positions 21 to 25, and the regions between amino acids 111 to 190 and 211 to 287 correspond to PDZ binding domains implicated in protein-protein interactions and C-terminus processing (42, 43). An RIP motif (38) is located between amino acids 401 and 403. The mucP sequence in the sap21 strain showed the presence of the same mutation as that found in the sap20 strain. This mutation corresponds to a single-nucleotide guanine (G) deletion at nucleotide 1174, resulting in a frameshift mutation starting after codon 391 and premature termination at codon 437 (mucP392) (Fig. 4B and Fig. S2), mapping to the final transmembrane domain of the protein. The sap30 sequence revealed a nucleotide insertion of a cytosine (C) at nucleotide 475 of mucP, leading to a frameshift mutation after residue 158 and early termination of the protein at codon 366 (mucP159) (Fig. 4B and Fig. S2). The mutation in the sap36 strain was found to be due to a seven-nucleotide deletion between positions 910 and 917 of mucP, resulting in a frameshift mutation starting after codon 303 and a premature termination of the protein at codon 317 (mucP304) (Fig. 4B and Fig. S2).
Roles of AlgO and MucP in alginate production.
The observation that the majority of our sap mutants mapped in either algO or mucP, plus the fact that MucP overproduction could compensate for the loss of either AlgO or MucP function, led us to examine the role of all of the proteases and regulatory factors of the RIP pathway of P. aeruginosa using a holistic approach. Precise in-frame deletions of genes encoding proteases and regulatory factors were constructed in PAO1 and PDO300. In addition, the algO, mucP, and mucD protease-encoding genes, along with the regulatory factor-encoding mucE gene, were cloned in plasmids with a controllable promoter. This permitted the controlled expression of each gene in wild-type and deletion strains, demonstrating which factors were required for inducing mucoidy (Alg+) in PAO1 and what led to the loss of mucoidy (Alg−) in PDO300.
MucE-induced mucoidy in PAO1 requires AlgO and AlgW.
The results of the complementation analysis in PAO1 showed that the deletion of mucE, algO, and algW did not alter the nonmucoid phenotype of the parental PAO1 strain (Table 1). No change was observed when vector pMucD, pAlgO, pAlgW, or pMucP was introduced into either of these deletion mutants. Only the presence of the pMucE plasmid resulted in a mucoid phenotype when the plasmid was induced in either the wild-type or in the mucE deletion background, suggesting that increased amounts of MucE was the cause of this phenotype and required the presence of either AlgO or AlgW. This suggests that MucE works alongside AlgO to activate AlgW's cleavage of MucA.
TABLE 1.
Alginate phenotype in mutant strains
| Strain and genotype | Phenotype ofa: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No-vector control | Vector |
pMucE |
pMucD |
pAlgO |
pMucP |
||||||
| NON | IND | NON | IND | NON | IND | NON | IND | NON | IND | ||
| PAO1 | |||||||||||
| Wild type | − | − | − | − | + | − | − | − | − | − | − |
| ΔmucE | − | − | − | − | + | − | − | − | − | − | − |
| ΔalgO | − | − | − | − | − | − | − | − | − | − | − |
| ΔalgW | − | − | − | − | − | − | − | − | − | − | − |
| PDO300 | |||||||||||
| Wild type | + | + | + | + | + | + | + | + | + | + | + |
| ΔmucE | + | + | + | + | + | − | − | + | N/G | + | N/G |
| ΔalgO | − | − | − | − | + | − | − | + | N/G | + | N/G |
| ΔalgW | − | − | − | − | − | − | − | + | + | + | + |
NON, noninduced; IND, induced with 1 mM IPTG; N/G, no growth; +, mucoid; −, nonmucoid.
AlgO and MucP can overcome loss of mucoidy in PDO300 ΔalgO and ΔalgW strains.
In PDO300 (Table 1), the Alg+ phenotype was retained when all plasmids were introduced into the strain, and no change was observed in the presence or absence of IPTG induction. Deletion of algO or algW abolished the mucoid phenotype of PDO300, which was rescued by the presence of algO or mucP on a complementing plasmid (Table 1). In addition, the overexpression of mucE could rescue the mucoid phenotype in algO deletion mutants but not in algW deletion mutants. This shows that both AlgO and AlgW are required for mucoidy, presumably to perform the initial cleavage of MucA; overexpression of mucP may overcome this by allowing second-site cleavage to occur independently of AlgW/AlgO-mediated first-site cleavage. The overproduction of MucE can also overcome the nonmucoid phenotype of the PDO300 ΔalgO strain, most likely by increased activation of AlgW proteolytic activity.
In the mucE deletion background, the nonmucoid phenotype was only observed when pMucD was present, whether induced or not. Interestingly, induction of pAlgO and pMucP in the mucE mutant inhibited growth in these strains. This phenotype was also observed in the algO deletion strain, suggesting that the overproduction of these proteins in these backgrounds interferes with some essential cellular process. In this strain, the mucoid phenotype was only observed in the presence of the induced pMucE plasmid or with the uninduced pAlgO or pMucP plasmid. Taken together, these results show that in PDO300, mucoidy requires the presence of AlgO and AlgW, but this requirement can be overcome by the overproduction of the AlgO or MucP protease, or, in certain cases (algO deletion mutant), MucE. In addition, the loss of MucE, coupled with the slight overexpression of mucD, can also inhibit mucoidy in the PDO300 strain, confirming MucD's negative regulatory role in this process (26). This also suggests that the target of MucD is not MucE and is more likely to be a protein involved in the later stages of RIP.
DISCUSSION
Alginate overproduction by P. aeruginosa in the CF lung has been delineated as the leading cause of morbidity and mortality in these patients. Therefore, it is crucial to further understand the molecular mechanisms of alginate regulation. MucA, the anti-sigma factor responsible for regulation of the sigma factor AlgT/U, has been identified as the major site of mutations in mucoid strains isolated from the CF lung (5, 15). These mutations lead to instability in MucA, allowing AlgT/U to promote the expression of the biosynthetic operon, resulting in the overproduction of alginate (14). This study explores the regulatory pathway leading to the degradation of MucA, which leads to the release of AlgT/U. An isogenic PAO1 derivative was used which is constitutively Alg+ due to the replacement of the mucA gene with mucA22, a common allele found in mucoid CF isolates (5). Thirty-four nonmucoid revertants were isolated after growing PDO300 under low-oxygen tension; 14 were found to be in algT (algU), three were uncharacterized, and the remaining 17 mutants were analyzed in this study. Four of these mutants mapped to mucP and the remaining 13 mapped to algO (see Fig. S1 and S2 in the supplemental material). These 13 algO mutants were complemented in trans by plasmids expressing either algO or mucP. The mucP mutants were complemented by a mucP-expressing plasmid; the pAlgO plasmid failed to complement these strains, suggesting MucP acts downstream of AlgO. Similar results were observed where a majority of mutations that suppressed the mucA22 mutation mapped to algT (algU) or algO (28, 52). However, these authors did not observe any suppressor mutations that mapped to mucP, most likely due to the small number of mutants obtained in their study.
Interestingly, two of the mucP alleles identified here involved a string five Gs. The ubiquitous mucA allele, mucA22, found in most clinical CF isolates (40%), is also due to a loss of a G in a homopolymeric tract of 5 Gs that is prone to single-nucleotide deletion (5, 17, 53–55). The mucP sequence has five such homopolymeric tracts, all of them at the 3′ end of the gene. It is becoming increasingly recognized that prokaryotes have evolved to use these homopolymeric tracts, which mutate at a higher frequency, for the efficient inactivation of genes; these poly(G) tracts tend to occur at the 3′ ends of coding regions (56). P. aeruginosa PAO1 has two copies of nine-nucleotide poly(G) tracts and 706 copies of the 6-G tract. This is also the first report where the second-site alginate suppressor mutations were mapped to mucP in P. aeruginosa.
The mucP-complemented sap17 and sap20 mutants were quantified for alginate production with the original cosmid, the transposon-inserted cosmid, and a plasmid expressing mucP under IPTG control (Fig. 3B). Strains that had an observable mucoid phenotype produced significantly higher levels of alginate than nonmucoid strains. Interestingly, IPTG-induced overproduction of MucP produced slightly less alginate than noninduced plasmids (Fig. 3B), suggesting the increased levels of MucP are slightly inhibitory although still greater than that observed in the vector and nonvector controls. A similar phenomenon was observed when pMucP was expressed in the P. aeruginosa PA2192 clinical CF isolate (Fig. 3C), suggesting that this observation is not strain dependent. Further analysis is required to determine if this is a regulatory effect or simply due to growth inhibition due to MucP or AlgT/U overproduction in the mucoid strains.
Our results suggest that MucP is acting downstream in the regulatory pathway and can overcome the loss of AlgO found in most of the sap mutant strains. To further elucidate the regulatory cascade of alginate production, several precise in-frame deletions were constructed of the various players involved in MucA proteolysis. Although the specific signal that initiates the cascade that results in the mucoid phenotype has not been well characterized, MucA degradation occurs via RIP (20, 25, 28). This proteolytic cascade occurs in a manner similar to that of degradation of RseA in E. coli involved in the σE stress response (20). In E. coli three proteases, DegS (P. aeruginosa AlgW), RseP (P. aeruginosa MucP), and ClpXP (P. aeruginosa ClpXP), degrade RseA (P. aeruginosa MucA) in a sequential manner that results in the release of σE (P. aeruginosa AlgT/U). Current literature suggests that MucE interacts with AlgW, a transmembrane serine protease, and activates this protein to initiate cleavage of MucA (20, 22). These are based on experiments showing that both MucE and AlgW are necessary for the cleavage of MucA in vitro. Our results agree with previous literature showing that overexpression of mucE in PAO1 causes mucoidy and mucE overexpression in the algW deletion mutant does not (Table 1) (20). This is consistent with the requirement of AlgW and MucE in the initial degradation of MucA. However, we also observed the loss of the mucoid phenotype when mucE was overexpressed in the PAOΔalgO strain. This is different from what was observed by Qiu et al. (20), who did not see suppression of mucE-overexpressed mucoidy in algO mutant backgrounds. It is possible that the AlgO requirement we observed was due to lower levels of MucE expression in our strains, whereas the very high levels of MucE produced by Qiu et al. allow it to overcome this requirement. We therefore suggest that MucE does not interact directly with AlgW but rather in concert with AlgO to expose MucB-bound MucA for cleavage by AlgW.
In the Alg+ mucA22 mutant strain PDO300, deletion of the mucE coding sequence did not result in loss of the mucoid phenotype, suggesting that other factors are required to help initiate MucA degradation or that MucE does not interact with AlgW in maintaining mucoidy in this mucA mutant. Other signaling motifs besides those found in MucE have been shown to activate the degradation cascade (20), showing that there is still much to be elucidated. Another phenotype observed was that overexpression of mucE in the nonmucoid PDOΔalgO strain restored the mucoid phenotype to that of PDO300. Although contradictory to the results seen for the PAOΔalgO strain, this could be explained by the fact that in a PDO300 background, the altered MucA protein has a reduced interaction with MucB which, in turn, alleviates AlgO from its role with MucE to expose MucA, allowing it to perform differently in this mucA mutant strain. MucE alone might be enough to expose MucA for cleavage or may interact with another factor to help activate MucA degradation. After the introduction of pMucD into the mucoid PDOΔmucE strain, a loss of the mucoid phenotype was observed. It has been suggested that the target of MucD is MucE, which acts to remove the WVF signaling peptide to prevent the activation of AlgW and, ultimately, release of AlgT/U. Our results suggest that the target of MucD is elsewhere in the periplasm, as a change in phenotype in the absence of MucE was observed when MucD was overproduced. In fact, even slight overproduction of MucD was sufficient to block mucoidy in all of the mutants we constructed. One possible target of MucD is AlgO, as MucD overproduction mimics the phenotype obtained in PDO300 algO mutants.
Overproduction of AlgO and MucP in mucE and algO deletion mutants resulted in growth inhibition. The precise nature of this inhibition is still under study. Since one of the proteins is periplasmic and the other is membrane bound, one cannot say that the effect is solely due to the disruption of the cell membrane due to MucP accumulation. It is more likely that the overproduction of these factors either competes or interferes with an essential process in mucoid strains. It is also noteworthy that AlgO overexpression in the PDO300 algO deletion mutant inhibits growth, while overexpression in the parental algO+ strain does not. It is possible that deletion of algO from the chromosome removes some cis-acting regulatory factor that can lead to altered expression of some other factor, resulting in growth inhibition. The algO coding sequence does contain an AmrZ binding site (57), which plays a role in the regulation of motility and alginate genes in P. aeruginosa. The removal of the AmrZ site from algO may affect the expression of the adjacent PA3256 (putative oxidoreductase) or PA3258 (unknown function) gene. Further studies on this effect, including the isolation of suppressor mutants, may help us elucidate this process in more detail.
Here, we propose two parallel pathways of MucA degradation and alginate regulation in P. aeruginosa (Fig. 1). In PAO1, the alginate pathway is tightly regulated, and MucA needs to be degraded by the sequential action of proteases. In response to certain stresses in the periplasm, the proteolytic cascade is indirectly activated by MucE activating AlgW for MucA cleavage, leading to cleavage by AlgO after its cleavage site is exposed by some other unknown factor. Finally, MucP performs an N-terminal cleavage, allowing the rest of MucA to release AlgT/U, activating transcription. While MucE/AlgW action occurs before the action of AlgO and MucP, both pathways are individually regulated by MucD in the periplasm, as overexpression of MucD shuts off alginate production in all PDO300 single deletion mutants. With appropriate signals, the two can be initiated and act sequentially to release AlgT/U. In PAO1, deletion of mucD releases its inhibitory effect on MucE and the unknown factor and activates both pathways, leading to mucoidy (26). While overexpression of mucE can turn PAO1 mucoid, overexpression of algO and mucP cannot because the AlgO cleavage site is not accessible when MucA is intact.
In the clinical scenario, as expected with strain PDO300, when MucA is truncated and more susceptible for cleavage, although the MucE/AlgW pathway is still required, it can be easily bypassed by overexpressing the AlgO/MucP pathway because MucA is more readily accessible for cleavage by AlgO once the cleavage site is exposed. This could explain why, after algT (algU) (41%), most of the sap mutants have mutations in algO (38%) or mucP (12%). Hence, the activation of both AlgW and AlgO pathways may require divergent or overlapping signals to regulate alginate production in new infections as well as in mucA-mutated chronic strains. With PAO1, alginate production is turned on by mutations in mucA to generate the mucoid phenotype and the mucoid colonies are turned off by mutations in genes in the AlgO/MucP pathway, which is more prominent in mucoid strains.
This complex regulatory framework of membrane-bound and periplasmic proteins reveals that alginate production is not a simple process in P. aeruginosa. Recent work from our laboratory (46) indicating the additional involvement of an outer membrane protein, LptD, in restoring mucoidy to algO and algW mutants has added an extra layer of complexity to this medically important pathway, clearly indicating the need for continued research to understand the exact role of the critical elements involved in the events leading to mucoid development and maintenance in P. aeruginosa-infected CF patients.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and primers.
The P. aeruginosa and E. coli strains and plasmids that were used are listed in Table 2. E. coli strains were grown on Luria-Bertani (LB) medium and, when required, were supplemented with tetracycline (Tc; 20 μg ml−1), chloramphenicol (Cm; 10 μg ml−1), ampicillin (Ap; 100 μg ml−1), kanamycin (Km; 20 μg ml−1), or gentamicin (Gm; 15 μg ml−1). P. aeruginosa strains were grown on LB or on LB-PIA agar. LB-PIA agar was composed of a 1:1 mixture of LB agar and Pseudomonas isolation agar (PIA) (14). Antibiotics for P. aeruginosa, when required, were Tc (100 μg ml−1), Gm (75 μg ml−1), and carbenicillin (Cb; 150 μg ml−1). All cultures were grown at 37°C unless stated otherwise. Primers (Table 2) used were all synthesized by Integrated DNA Technologies (Coralville, IA).
TABLE 2.
List of strains, plasmid and primers
| Strain, plasmid, or primer | Relevant genotype, phenotype, and/or sequence | Reference or source |
|---|---|---|
| Escherichia coli | ||
| DH5α | ϕ80lacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 deoR Δ(lacZYA-argF)U169 | Invitrogen (Bethesda Research Laboratories) |
| MTP216 | Tcr, PAO1 fragment 4068269–4093431 (pMO013722) | 44 |
| Pseudomonas aeruginosa | ||
| PA2192 | Alg+ mucA180 | 36 |
| PAO1 | Alg− prototypic strain, nonmucoid | 45 |
| PDO300 | Alg+ mucA22, constitutively mucoid (PAOmucA22) | 5 |
| PKM800 | Alg− sap22 mucA22 alg107-1 | 29 |
| PKM816 | Alg− sap26 mucA22 alg107-2 | This study |
| PKM817 | Alg− sap27 mucA22 alg107-3 | 46 |
| PKM818 | Alg− sap31 mucA22 alg107-4 | This study |
| PKM819 | Alg− sap42 mucA22 algO55-1 | This study |
| PKM820 | Alg− sap32 mucA22 algO336-1 | This study |
| PKM821 | Alg− sap16 mucA22 algO500-1 | This study |
| PKM822 | Alg− sap17 mucA22 algO500-2 | This study |
| PKM823 | Alg− sap23 mucA22 algO500-3 | This study |
| PKM824 | Alg− sap24 mucA22 algO500-4 | This study |
| PKM825 | Alg− sap25 mucA22 algO500-5 | This study |
| PKM826 | Alg− sap46 mucA22 algO516-1 | This study |
| PKM827 | Alg− sap47 mucA22 algO516-2 | This study |
| PKM828 | Alg− sap30 mucA22 mucP159-1 | This study |
| PKM829 | Alg− sap36 mucA22 mucP304-1 | This study |
| PKM830 | Alg− sap20 mucA22 mucP392-1 | This study |
| PKM831 | Alg− sap21 mucA22 mucP392-2 | This study |
| PKM832 | Alg− ΔalgO | PAOΔalgO (46) |
| PKM833 | Alg− mucA22 ΔalgO | PDOΔalgO (46) |
| PKM834 | Alg− ΔalgW | PAOΔalgW (46) |
| PKM835 | Alg− mucA22 ΔalgW | PDOΔalgW (46) |
| PKM836 | Alg− ΔmucE | PAOΔmucE; this study |
| PKM837 | Alg+ mucA22 ΔmucE | PDOΔmucE; this study |
| Plasmids | ||
| pCD10 | Apr Kmr; pCRII-TOPO with 300-bp fragment containing mucE | This study |
| pCD11 | Apr Kmr; pCRII-TOPO with 1,400-bp fragment containing mucD | This study |
| pCD12 | Apr Kmr; pCRII-TOPO with 1,400-bp fragment containing algW | This study |
| pCD13 | Apr Kmr; pCRII-TOPO with 2,000-bp fragment containing algO | This study |
| pCD20 | Apr/Cbr; pMF54 with 300-bp fragment containing mucE | pMucE; this study |
| pCD21 | Apr/Cbr; pMF54 with 1,400-bp fragment containing mucD | pMucD; this study |
| pCD22 | Apr/Cbr; pMF54 with 1,400-bp fragment containing algW | pAlgW; this study |
| pCD23 | Apr/Cbr; pMF54 with 2,000-bp fragment containing algO | pAlgO; this study |
| pCRII-TOPO | Apr Kmr; colE1 lacZa | Invitrogen |
| pJG293 | Tcr Hgr Kmr pAlgT/U oriT (RK2-Tra+)::Tn501-250 | 29 |
| pLAFR3 | Tcr; cosmid vector derived from pLAFR1; IncP1 λ cos+ rlx | 47 |
| pLVF48 | Apr Kmr; pCRII-TOPO with 1,400-bp fragment containing mucP | This study |
| pLVF52 | Apr/Cbr; pMF54 with 1,400-bp fragment containing mucP | pMucP; this study |
| pLVF60 | pMO013722 (mucP::Tn5-200) | This study |
| pME6030 | Tcr oriV pVS1 oriV P15A oriT | 48 |
| pMF54 | Apr/Cbr ori ColE1-SF oriT (RK2) Ptrc, lacIq | 49 |
| pMO013722 | Tcr PAO1 fragment 4068269–4093431 | 44 |
| pRK2013 | Kmr colE1 RK2− Tra+ | 50 |
| pRK600 | Cmr colE1 RK2− Tra+ | 51 |
| Primers | ||
| cos-1 | 5′-CGCCCTCTGGTAAGGTTG-3′ | 44 |
| CC_algO_F | 5′-CTCGACACGACCTTCGATACCTTGC-3′ | This study |
| CC_algO_R | 5′-TTTATGACGCTCCCGCTGAACGCTAGC-3′ | This study |
| CC_MucD_F | 5′-TCTAGAAAGAAGGAGATATACATGCATACCCTAAAACG-3′ | This study |
| CC_MucD_R | 5′-AAGCTTGCCGGCTTATTCGGCCAGCTT-3′ | This study |
| CC_mucE_F | 5′-TTTTCTAGAAAGAAGGAGATATACATGGGTTTCCGGCCAGTTAGCC-3′ | This study |
| CC_mucE_R | 5′-AAGCTTTCGCGTTCAAAACACCCAG-3′ | This study |
| LF_mucE_UF (P1) | 5′-GGAATTGTGAGCGGATAACAATTTCACACAGGAAACAGCTGAAGCGCTCGTAGAGATATT-3′ | This study |
| LF_mucE_UR (P) | 5′-CTATTTCAGATTCGTCGCGTCTAGTTAGCTAGCATGGCTACGACTCCTTGATAGG-3′ | This study |
| LF_mucE_DF (P2) | 5′-CCTATCAAGGAGTCGTAGCCATGCTAGCTAACTAGACGCGACGAATCTGAAATAG-3′ | This study |
| LF_mucE_DR (P2) | 5′-CCAGGCAAATTCTGTTTTATCAGACCGCTTCTGCGTTCTGATCGCGGCTTGCAACGAGTA-3′ | This study |
| LF_MucP_F | 5′-CCATGGTTTAAAGAAGGAGATATACCATGAGTGCGCTTTACATGATCG-3′ | This study |
| LF_MucP_R | 5′-AAGCTTCTACAGACGACTCAGATCGTTGACC-3′ | This study |
| KAN-2 FP-1 | 5′-ACCTACAACAAAGCTCTCATGAACC-3′ | EpiCenter |
| KAN-2 RP-1 | 5′-GCAATGTAACATCAGAGATTTTGAG-3′ | EpiCenter |
DNA manipulations.
All molecular techniques were performed according to current molecular biology protocols. DNA sequencing was done using the BigDye Terminator v. 3.1 cycle sequencing kit (Applied Biosystems, Carlsbad, CA) and analyzed on an Applied Biosystems 3100 genetic analyzer. To ensure high fidelity, the top strand was sequenced twice, while the bottom strand was done once (standard two-plus-one sequencing). Genomic DNA was isolated from Pseudomonas strains with the PureLink genomic DNA minikit (Invitrogen Life Technologies, Grand Island, NY) using the manufacturer's directions.
Isolation and analysis of sap mutants.
The suppressors of alginate production (sap) mutants were isolated by growing PDO300 under low-oxygen tension and selecting for nonmucoid revertants (29).
Triparental mating.
Plasmids and cosmids were conjugated into P. aeruginosa via triparental mating using helper plasmids pRK2013 and pRK600 (50, 58). Conjugants were selected on LB-PIA plates containing Tc or Cb.
Construction of deletion mutants.
An unmarked algO null mutant of P. aeruginosa was generated by allelic replacement using primers CC_AlgO_P1(F) and CC_AlgO_P1(R) (to generate the upstream product P1, flanked by HindIII and NheI sites) and primers CC_AlgO_P2(F) and CC_AlgO_P2(R) (to generate the downstream product P2, flanked by NheI and BamHI sites) using AccuPrime high-fidelity Taq DNA polymerase (Invitrogen Life Technologies, Grand Island, NY). After sequencing to ensure the absence of mutations, P1 and P2 were spliced together and subcloned into a P. aeruginosa nonreplicative plasmid, pEXG2 (59), as a HindIII-BamHI fragment and moved into PAO1 and PDO300 by conjugation to generate single crossovers (46, 60). Double-crossover mutants were selected for their sucrose resistance and gentamicin sensitivity, indicating loss of the plasmid.
For deletion mutants of algW and mucE in PAO1 and PDO300, deletion constructs in pMQ30 were constructed via the yeast system of double-stranded gap repair and homologous recombination (61). The P1 region of algW was generated using the primers HK_AlgW_P1_F and HK_AlgW_P1_R. The P2 region was generated using primers HK_AglW_P2_F and HK_AlgW_P2_R (46). The P1 region of mucE was constructed using primers LF_mucE_UF(P1) and LF_mucE_UR(P1). Primers LF_mucE_DF(P2) and LF_mucE_DR(P2) were used to amplify the P2 region (Table 2). The extracted plasmid constructs were sequenced to ensure the absence of mutations and transformed into E. coli DH5α, which then served as the donor strain in the triparental mating as previously described. The presence of the gene deletion in all mutants was confirmed using standard molecular methods (PCR, restriction analysis of amplicons, and DNA sequencing of the locus).
Construction of complementation clones.
The algO (PA3257), mucE (PA4033), and mucD (PA0766) open reading frames (ORFs) were amplified from PAO1 genomic DNA using primers containing a ribosomal binding site (RBS) and flanked by XbaI and HindIII, as shown in Table 1. The mucP gene (PA3649) was amplified with primers carrying NcoI and HindIII restriction sites. The amplicons were cloned into pCR2.1 TOPO using the TA cloning technique. After confirming the absence of mutations by sequencing, each ORF was subcloned into pMF54 downstream of the LacI-regulatable Ptrc promoter (49). Expression of these genes was induced by plating the strains on LB media containing ampicillin and 2 mM IPTG.
Amplification of the mucP and algO loci from the sap strains for sequencing.
Genomic DNA was prepared from the sap strains and from the parental PAO1 and PDO300 strains. Primers LF_MucP_F and LF_MucP_R were used to amplify mucP, and CC_algO_F and CC_algO_R were used to amplify algO; amplicons were amplified for sequencing using the AccuPrime Taq DNA high-fidelity polymerase (Invitrogen Life Technologies, Grand Island, NY). Primer sequences are listed in Table 2.
Alginate assay.
Alginate concentrations in overnight culture supernatants were measured after extensive dialysis using a colorimetric carbazole assay for uronic acids (34). To determine the alginate concentration, a set of standards was made with sodium alginate (Sigma, St. Louis, MO). The alginate concentration was expressed as milligrams per milliliter of supernatant.
Cosmid DNA identification.
Cosmids from a P. aeruginosa minimal tiling path array (44), obtained from Paul Phibbs, East Carolina University, were extracted using the PureLink quick plasmid miniprep kit (Invitrogen Life Technologies, Grand Island, NY) by following the manufacturer's instructions. Individual cosmids were then identified by sequencing the insert junction using the cos-1 primer (Table 2).
Transposon mutagenesis and characterization.
To identify the ORF of interest, the complementing cosmid was mutagenized using the EZ::TN transposon kit (Epicentre, Madison, WI) according to the manufacturer's protocol, with slight modifications, followed by transformation into TOP10 electrocompetent cells (Invitrogen, Carlsbad, CA) (29). Sequencing of the transposon insertion was performed as described above using the KAN-2 FP-1 and KAN-2 RP-1 primers (Table 2).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the NIH National Institute of Allergy and Infectious Diseases (NIAID) 1R15Al111210 (to K.M. and H.K.) and the NIH National Institute of General Medical Sciences (NIGMS) T34 GM08368 (to L.F.) and R25 GM061347 (to C.D., I.L., and R.T.S.).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Paul Phibbs, East Carolina University, for supplying the P. aeruginosa minimal tiling path cosmid library. We thank members of the Mathee laboratory, particularly Supurna Dhar, for comments on and assistance with the manuscript.
This paper is dedicated to the late Robert J. Smiddy.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00215-18.
REFERENCES
- 1.Hoiby N. 2011. Recent advances in the treatment of Pseudomonas aeruginosa infections in cystic fibrosis. BMC Med 9:32. doi: 10.1186/1741-7015-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Remminghorst U, Rehm BH. 2006. Bacterial alginates: from biosynthesis to applications. Biotechnol Lett 28:1701–1712. doi: 10.1007/s10529-006-9156-x. [DOI] [PubMed] [Google Scholar]
- 3.Evans LR, Linker A. 1973. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J Bacteriol 116:915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Govan JR, Harris GS. 1986. Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol Sci 3:302–308. [PubMed] [Google Scholar]
- 5.Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JI, Jensen P, Johnsen AH, Givskov M, Ohman DE, Molin S, Hoiby N, Kharazmi A. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145(Part 6):1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- 6.Limoli DH, Rockel AB, Host KM, Jha A, Kopp BT, Hollis T, Wozniak DJ. 2014. Cationic antimicrobial peptides promote microbial mutagenesis and pathoadaptation in chronic infections. PLoS Pathog 10:e1004083. doi: 10.1371/journal.ppat.1004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terry JM, Pina SE, Mattingly SJ. 1991. Environmental conditions which influence mucoid conversion Pseudomonas aeruginosa PAO1. Infect Immun 59:471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chotirmall SH, Smith SG, Gunaratnam C, Cosgrove S, Dimitrov BD, O'Neill SJ, Harvey BJ, Greene CM, McElvaney NG. 2012. Effect of estrogen on pseudomonas mucoidy and exacerbations in cystic fibrosis. N Engl J Med 366:1978–1986. doi: 10.1056/NEJMoa1106126. [DOI] [PubMed] [Google Scholar]
- 9.Henry RL, Mellis CM, Petrovic L. 1992. Mucoid Pseudomonas aeruginosa is a marker of poor survival in cystic fibrosis. Pediatr Pulmonol 12:158–161. doi: 10.1002/ppul.1950120306. [DOI] [PubMed] [Google Scholar]
- 10.Wozniak DJ, Ohman DE. 1994. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol 176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hershberger CD, Ye RW, Parsek MR, Xie ZD, Chakrabarty AM. 1995. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative sigma factor (sigma E). Proc Natl Acad Sci U S A 92:7941–7945. doi: 10.1073/pnas.92.17.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lonetto M, Gribskov M, Gross CA. 1992. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol 174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie ZD, Hershberger CD, Shankar S, Ye RW, Chakrabarty AM. 1996. Sigma factor-anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol 178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathee K, McPherson CJ, Ohman DE. 1997. Posttranslational control of the algT (algU)-encoded sigma22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J Bacteriol 179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schurr MJ, Yu H, Martinez-Salazar JM, Boucher JC, Deretic V. 1996. Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol 178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin DW, Holloway BW, Deretic V. 1993. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J Bacteriol 175:1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anthony M, Rose B, Pegler MB, Elkins M, Service H, Thamotharampillai K, Watson J, Robinson M, Bye P, Merlino J, Harbour C. 2002. Genetic analysis of Pseudomonas aeruginosa isolates from the sputa of Australian adult cystic fibrosis patients. J Clin Microbiol 40:2772–2778. doi: 10.1128/JCM.40.8.2772-2778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boucher JC, Yu H, Mudd MH, Deretic V. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect Immun 65:3838–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowen DW, Deretic V. 2000. Membrane-to-cytosol redistribution of ECF sigma factor AlgU and conversion to mucoidy in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Mol Microbiol 36:314–327. doi: 10.1046/j.1365-2958.2000.01830.x. [DOI] [PubMed] [Google Scholar]
- 20.Qiu D, Eisinger VM, Rowen DW, Yu HD. 2007. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 104:8107–8112. doi: 10.1073/pnas.0702660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood LF, Ohman DE. 2009. Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol 72:183–201. doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- 22.Cezairliyan BO, Sauer RT. 2009. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol Microbiol 72:368–379. doi: 10.1111/j.1365-2958.2009.06654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akiyama Y, Kanehara K, Ito K. 2004. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J 23:4434–4442. doi: 10.1038/sj.emboj.7600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu D, Eisinger VM, Head NE, Pier GB, Yu HD. 2008. ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiology 154:2119–2130. doi: 10.1099/mic.0.2008/017368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandey S, Martins KL, Mathee K. 2016. Posttranslational regulation of antisigma factors of RpoE: a comparison between the Escherichia coli and Pseudomonas aeruginosa systems. In de Bruijn FJ (ed), Stress and environmental regulation of gene expression and adaptation in bacteria. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 26.Boucher JC, Martinez-Salazar J, Schurr MJ, Mudd MH, Yu H, Deretic V. 1996. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J Bacteriol 178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood LF, Ohman DE. 2006. Independent regulation of MucD, an HtrA-like protease in Pseudomonas aeruginosa, and the role of its proteolytic motif in alginate gene regulation. J Bacteriol 188:3134–3137. doi: 10.1128/JB.188.8.3134-3137.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reiling SA, Jansen JA, Henley BJ, Singh S, Chattin C, Chandler M, Rowen DW. 2005. Prc protease promotes mucoidy in mucA mutants of Pseudomonas aeruginosa. Microbiology 151:2251–2261. doi: 10.1099/mic.0.27772-0. [DOI] [PubMed] [Google Scholar]
- 29.Sautter R, Ramos D, Schneper L, Ciofu O, Wassermann T, Koh CL, Heydorn A, Hentzer M, Hoiby N, Kharazmi A, Molin S, Devries CA, Ohman DE, Mathee K. 2012. A complex multilevel attack on Pseudomonas aeruginosa algT/U expression and AlgT/U activity results in the loss of alginate production. Gene 498:242–253. doi: 10.1016/j.gene.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hara H, Yamamoto Y, Higashitani A, Suzuki H, Nishimura Y. 1991. Cloning, mapping, and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J Bacteriol 173:4799–4813. doi: 10.1128/jb.173.15.4799-4813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silber KR, Keiler KC, Sauer RT. 1992. Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C termini. Proc Natl Acad Sci U S A 89:295–299. doi: 10.1073/pnas.89.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goryshin IY, Jendrisak J, Hoffman LM, Meis R, Reznikoff WS. 2000. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat Biotechnol 18:97–100. doi: 10.1038/72017. [DOI] [PubMed] [Google Scholar]
- 33.Withers TR, Yin Y, Yu HD. 2014. Identification and characterization of a novel inhibitor of alginate overproduction in Pseudomonas aeruginosa. Pathog Dis 70:185–188. doi: 10.1111/2049-632X.12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain S, Ohman DE. 1998. Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J Bacteriol 180:634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balzer S, Kucharova V, Megerle J, Lale R, Brautaset T, Valla S. 2013. A comparative analysis of the properties of regulated promoter systems commonly used for recombinant gene expression in Escherichia coli. Microb Cell Fact 12:26. doi: 10.1186/1475-2859-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pier GB, Matthews WJ Jr, Eardley DD. 1983. Immunochemical characterization of the mucoid exopolysaccharide of Pseudomonas aeruginosa. J Infect Dis 147:494–503. doi: 10.1093/infdis/147.3.494. [DOI] [PubMed] [Google Scholar]
- 37.Colbert B. 2017. Identification of alginate-regulating genes in the clinical cystic fibrosis isolate of Pseudomonas aeruginosa PA2192. Undergraduate honors thesis 76 Florida International University, Department of Biological Sciences, Miami, FL: bioRxiv 319004 doi: 10.1101/319004. [DOI] [Google Scholar]
- 38.Kanehara K, Akiyama Y, Ito K. 2001. Characterization of the yaeL gene product and its S2P-protease motifs in Escherichia coli. Gene 281:71–79. doi: 10.1016/S0378-1119(01)00823-X. [DOI] [PubMed] [Google Scholar]
- 39.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 40.Hofmann KSW. 1993. TMbase–a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler 374:166. [Google Scholar]
- 41.Damron FH, Yu HD. 2011. Pseudomonas aeruginosa MucD regulates the alginate pathway through activation of MucA degradation via MucP proteolytic activity. J Bacteriol 193:286–291. doi: 10.1128/JB.01132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. 1997. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 43.Harris BZ, Lim WA. 2001. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci 114:3219–3231. [DOI] [PubMed] [Google Scholar]
- 44.Huang B, Whitchurch CB, Croft L, Beatson SA, Mattick JS. 2000. A minimal tiling path cosmid library for functional analysis of the Pseudomonas aeruginosa PAO1 genome. Microb Comp Genom 5:189–203. doi: 10.1089/omi.1.2000.5.189. [DOI] [PubMed] [Google Scholar]
- 45.Holloway BW, Morgan AF. 1986. Genome organization in Pseudomonas. Annu Rev Microbiol 40:79–105. doi: 10.1146/annurev.mi.40.100186.000455. [DOI] [PubMed] [Google Scholar]
- 46.Pandey S, Delgado C, Kumari H, Florez L, Mathee K. Outer-membrane protein LptD (PAO595) plays a role in the regulation of alginate synthesis in Pseudomonas aeruginosa. J Med Microbiol, in press. doi: 10.1099/jmm.0.000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedman AM, Long SR, Brown SE, Buikema WJ, Ausubel FM. 1982. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene 18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 48.Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, Wade J, Walsh U, O'Gara F, Haas D. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol Plant Microbe Interact 13:232–237. doi: 10.1094/MPMI.2000.13.2.232. [DOI] [PubMed] [Google Scholar]
- 49.Franklin MJ, Chitnis CE, Gacesa P, Sonesson A, White DC, Ohman DE. 1994. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J Bacteriol 176:1821–1830. doi: 10.1128/jb.176.7.1821-1830.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Lorenzo V, Herrero M, Jakubzik U, Timmis KN. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in Gram-negative eubacteria. J Bacteriol 172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeVries CA, Ohman DE. 1994. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J Bacteriol 176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spencer DH, Kas A, Smith EE, Raymond CK, Sims EH, Hastings M, Burns JL, Kaul R, Olson MV. 2003. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J Bacteriol 185:1316–1325. doi: 10.1128/JB.185.4.1316-1325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bragonzi A, Wiehlmann L, Klockgether J, Cramer N, Worlitzsch D, Doring G, Tummler B. 2006. Sequence diversity of the mucABD locus in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology 152:3261–3269. doi: 10.1099/mic.0.29175-0. [DOI] [PubMed] [Google Scholar]
- 55.Yoon SS, Coakley R, Lau GW, Lymar SV, Gaston B, Karabulut AC, Hennigan RF, Hwang SH, Buettner G, Schurr MJ, Mortensen JE, Burns JL, Speert D, Boucher RC, Hassett DJ. 2006. Anaerobic killing of mucoid Pseudomonas aeruginosa by acidified nitrite derivatives under cystic fibrosis airway conditions. J Clin Investig 116:436–446. doi: 10.1172/JCI24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Orsi RH, Bowen BM, Wiedmann M. 2010. Homopolymeric tracts represent a general regulatory mechanism in prokaryotes. BMC Genomics 11:102. doi: 10.1186/1471-2164-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones CJ, Newsom D, Kelly B, Irie Y, Jennings LK, Xu B, Limoli DH, Harrison JJ, Parsek MR, White P, Wozniak DJ. 2014. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog 10:e1003984. doi: 10.1371/journal.ppat.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finan TM, Kunkel B, De Vos GF, Signer ER. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol 167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. 2005. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schweizer HP, Hoang TT. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15–22. doi: 10.1016/0378-1119(95)00055-B. [DOI] [PubMed] [Google Scholar]
- 61.Shanks RM, Kadouri DE, MacEachran DP, O'Toole GA. 2009. New yeast recombineering tools for bacteria. Plasmid 62:88–97. doi: 10.1016/j.plasmid.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akiyama K, Hizukuri Y, Akiyama Y. 2017. Involvement of a conserved GFG motif region in substrate binding by RseP, an Escherichia coli S2P protease. Mol Microbiol 104:737–751. doi: 10.1111/mmi.13659. [DOI] [PubMed] [Google Scholar]
- 63.Akiyama K, Mizuno S, Hizukuri Y, Mori H, Nogi T, Akiyama Y. 2015. Roles of the membrane-reentrant beta-hairpin-like loop of RseP protease in selective substrate cleavage. Elife 4:e08928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.