We explored the relationship between macrophage glycolysis and replication of an intracellular bacterial pathogen, Legionella pneumophila. Previous studies demonstrated that a glycolysis inhibitor, 2-deoxyglucose (2DG), blocks replication of L. pneumophila during infection of macrophages, leading to speculation that L. pneumophila may exploit macrophage glycolysis. We isolated L. pneumophila mutants resistant to the inhibitory effect of 2DG in macrophages, identifying a L. pneumophila hexose-phosphate transporter, UhpC, that is required for bacterial sensitivity to 2DG during infection. Our results reveal how a bacterial transporter mediates the direct antimicrobial effect of a toxic metabolite. Moreover, our results indicate that neither induction nor impairment of host glycolysis inhibits intracellular replication of L. pneumophila, which is consistent with a view of L. pneumophila as a metabolic generalist.
KEYWORDS: 2-deoxyglucose, Legionella pneumophila, macrophages, metabolism
ABSTRACT
Toll-like receptor (TLR) stimulation induces a pronounced shift to increased glycolytic metabolism in mammalian macrophages. We observed that bone marrow-derived macrophages (BMMs) increase glycolysis in response to infection with Legionella pneumophila, but the role of host macrophage glycolysis in terms of intracellular L. pneumophila replication is not currently understood. Treatment with 2-deoxyglucose (2DG) blocks L. pneumophila replication in mammalian macrophages but has no effect on bacteria grown in broth. In addition, we found that 2DG had no effect on bacteria grown in amoebae. We used a serial enrichment strategy to reveal that the effect of 2DG on L. pneumophila in macrophages requires the L. pneumophila hexose-phosphate transporter UhpC. Experiments with UhpC-deficient L. pneumophila revealed that mutant bacteria are also resistant to growth inhibition following treatment with phosphorylated 2DG in broth, suggesting that the inhibitory effect of 2DG on L. pneumophila in mammalian cells requires 2DG phosphorylation. UhpC-deficient L. pneumophila replicates without a growth defect in BMMs and protozoan host cells and also replicates without a growth defect in BMMs treated with 2DG. Our data indicate that neither TLR signaling-dependent increased macrophage glycolysis nor inhibition of macrophage glycolysis has a substantial effect on intracellular L. pneumophila replication. These results are consistent with the view that L. pneumophila can employ diverse metabolic strategies to exploit its host cells.
IMPORTANCE We explored the relationship between macrophage glycolysis and replication of an intracellular bacterial pathogen, Legionella pneumophila. Previous studies demonstrated that a glycolysis inhibitor, 2-deoxyglucose (2DG), blocks replication of L. pneumophila during infection of macrophages, leading to speculation that L. pneumophila may exploit macrophage glycolysis. We isolated L. pneumophila mutants resistant to the inhibitory effect of 2DG in macrophages, identifying a L. pneumophila hexose-phosphate transporter, UhpC, that is required for bacterial sensitivity to 2DG during infection. Our results reveal how a bacterial transporter mediates the direct antimicrobial effect of a toxic metabolite. Moreover, our results indicate that neither induction nor impairment of host glycolysis inhibits intracellular replication of L. pneumophila, which is consistent with a view of L. pneumophila as a metabolic generalist.
INTRODUCTION
Mammalian macrophages and dendritic cells exhibit dramatic metabolic remodeling in response to pathogen-associated molecular patterns detected by pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs) (1–3). PRR-stimulated macrophages display a characteristic “Warburg”-type metabolism with elevated levels of glycolysis that can be measured through increased uptake of glucose and increased secretion of lactate (4). Increased glycolysis is thought to allow macrophages to rapidly process carbon from glucose and glutamine to generate biomolecules such as cytokines, chemokines, and other inflammatory mediators during the acute immune response to infection (1, 5, 6). In addition to providing for the energetic and biosynthetic requirements of macrophages during an immune response to pathogens, it is increasingly clear that the metabolic changes that occur in macrophages can also directly influence the ability of intracellular pathogens to survive and replicate within macrophages as host cells (7–9).
The intracellular bacterial pathogen Legionella pneumophila naturally replicates in protozoan host amoebae. L. pneumophila is an accidental pathogen of mammals and can cause a severe pneumonia in humans, called Legionnaires' disease, via infection of lung macrophages (10–12). Upon uptake by phagocytosis, L. pneumophila employs a type IV secretion system (Dot/Icm) to translocate effector proteins into the host cell cytosol, a subset of which hijack host trafficking machinery and allow the bacteria to establish a replicative vacuole (13, 14). While flagellin produced by wild-type (WT) L. pneumophila can result in restriction of bacterial replication via activation of the NAIP/NLRC4 inflammasome in infected mouse cells, L. pneumophila that lack flagellin (ΔflaA) replicate to high levels in mouse bone marrow-derived macrophages (BMMs) (15–21).
Here, we report that BMMs infected with L. pneumophila display a characteristic increase in glycolysis, similar to observations noted in a recent report (22). We speculated that increased host cell glycolysis may benefit L. pneumophila because the glycolysis inhibitor 2-deoxyglucose (2DG) has been reported to block L. pneumophila replication in mammalian cells, though the mechanism(s) underlying this effect remains obscure (23–25). It has been suggested that 2DG acts on host cells during L. pneumophila infection by modulating autophagy, inhibiting uptake of bacteria by interfering with phagocytosis, or altering the intracellular metabolic environment (22–25). Consistent with the notion that 2DG acts via an effect on host cells, the inhibitory effect of 2DG on L. pneumophila is specific to infection of mammalian cells, since 2DG has no effect on bacterial replication in broth (23), and we observed that 2DG does not inhibit bacterial growth in protozoan host cells. However, the idea that inhibition of macrophage glycolysis affects intracellular replication of L. pneumophila is complicated by the observation that another glycolysis inhibitor, sodium fluoride, did not similarly affect bacterial growth in macrophages (23). Indeed, we tested multiple independent glycolysis-restricting strategies in macrophages and did not observe inhibitory effects on intracellular L. pneumophila replication. Since 2DG is frequently used to investigate metabolic aspects of immunity, including recent investigations of the metabolic relationships between intracellular pathogens and host cells (8, 9), including L. pneumophila (22), a better understanding the effects of 2DG in these systems is needed.
To distinguish whether the inhibitory effect of 2DG acts through modulating host macrophage metabolism or instead acts more directly on L. pneumophila during infection of macrophages, we used a serial enrichment approach to identify L. pneumophila mutants with the ability to replicate in 2DG-treated macrophages. This approach revealed that the L. pneumophila hexose-phosphate transporter UhpC is required for bacterial sensitivity to 2DG during infection of macrophages. We went on to demonstrate that L. pneumophila UhpC is required for sensitivity to phosphorylated 2DG (2DG-6-phosphate [2DGP]). These results suggest that phosphorylation of 2DG and the subsequent bacterial uptake of 2DGP from host macrophages via UhpC is the mechanism by which 2DG treatment restricts L. pneumophila replication in macrophages. The isolation of a 2DG-resistant uhpC mutant allowed us to assess for the first time whether intracellular replication of L. pneumophila is affected by 2DG-mediated inhibition of host glycolysis. We found that L. pneumophila can replicate without a growth defect in 2DG-treated macrophages with severely impaired glycolysis. Overall, our results indicate that host cell glycolysis has little effect on L. pneumophila intracellular replication and are consistent with the view that L. pneumophila is a generalist that encodes redundant systems to capture metabolites from its environment.
RESULTS
L. pneumophila-infected macrophages upregulate glycolysis.
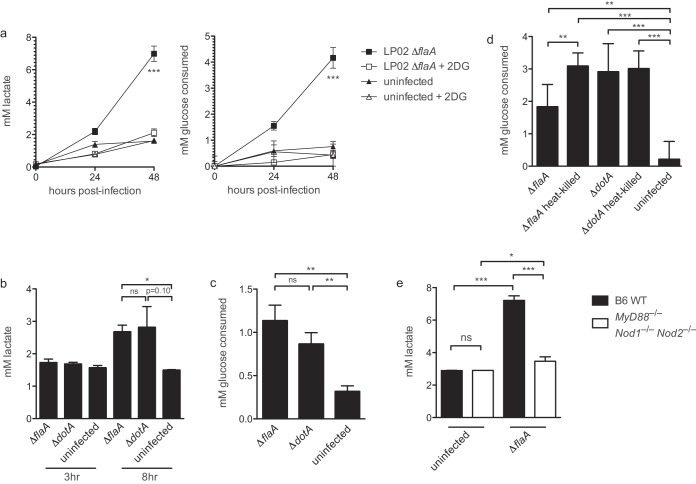
To determine whether macrophages increase glycolysis during infection with L. pneumophila, we measured the lactate levels in the supernatant of BMMs infected with LP02 ΔflaA L. pneumophila. In comparison to culture media containing uninfected BMMs, we observed increased lactate in culture media of infected BMMs over time (Fig. 1a). Infected BMMs also consumed an increased amount of glucose over the course of infection, suggesting that infected BMMs are increasing glycolysis in response to infection (Fig. 1a). Increased secretion of lactate and consumption of glucose by L. pneumophila-infected BMMs was largely blocked by the addition of the glycolysis inhibitor 2DG, reinforcing the idea that L. pneumophila infection triggers increased glycolysis in host macrophages (Fig. 1a). To determine whether an intact bacterial type IV secretion system is required for increased BMM glycolysis during infection with L. pneumophila, we measured the lactate levels in the supernatant of BMMs infected with LP01 ΔflaA L. pneumophila or Dot/Icm-deficient LP01 ΔdotA L. pneumophila at 3 and 8 h postinfection. At 8 h postinfection, we observed increased lactate secretion by BMMs infected with both strains (Fig. 1b). Increased secretion of lactate at 8 h corresponded with increased uptake of glucose by BMMs infected with both LP01 ΔflaA and LP01 ΔdotA L. pneumophila strains (Fig. 1c). These results suggest that bacterial replication is not required for increased glycolysis in BMMs infected with L. pneumophila. Furthermore, triggering of increased glycolysis did not require live bacteria, since exposure to live and heat-killed LP02 ΔflaA and ΔdotA L. pneumophila had similar effects on macrophage glucose consumption (Fig. 1d). Decreased glucose consumption by BMMs infected with live versus heat-killed LP02 ΔflaA L. pneumophila likely reflects increased host cell death observed under conditions with actively replicating bacteria. Lactate secretion was not induced to the same extent in infected BMMs derived from mice lacking the PRRs MyD88 and nucleotide-binding oligomerization domain-containing proteins 1 and 2 (MyD88−/− Nod1−/− Nod2−/− triply deficient mice) compared to WT BMMs (Fig. 1e). MyD88−/− Nod1−/− Nod2−/− triply deficient mice fail to activate TLR or NOD signaling in response to L. pneumophila (26). In sum, these results indicate that increased glycolysis in BMMs in response to infection by L. pneumophila is independent of bacterial replication or bacterial metabolism and relies on intact host macrophage TLR/NOD signaling.
FIG 1.
Increased glycolysis in BMMs during L. pneumophila infection. (a) Lactate levels and glucose consumption over 48 h in culture supernatant from uninfected BMMs (derived from wild-type C57BL/6 mice unless otherwise indicated) and BMMs infected with LP02 ΔflaA with or without 1.0 mM 2DG added 1 h postinfection (MOI = 0.05). (b) Lactate levels in culture supernatant from uninfected BMMs and BMMs infected with LP01 ΔflaA or LP01 ΔdotA L. pneumophila (MOI = 2.0). (c) Glucose consumption by uninfected BMMs or BMMs infected with LP01 ΔflaA or LP01 ΔdotA L. pneumophila (MOI = 2.0) at 8 h postinfection. Levels were calculated by measuring the change in glucose concentration at 8 h relative to 0-h controls. (d) Glucose consumed at 24 h postinfection by uninfected BMMs or BMMs infected with live and heat-killed LP02 ΔflaA and LP02 ΔdotA L. pneumophila (MOI = 1.0). The levels were calculated by measuring the change in glucose concentration at 24 h relative to 0-h controls. (e) Lactate levels in culture supernatant from uninfected wild-type BMMs and MyD88−/− Nod1−/− Nod2−/− BMMs and infected BMMs of both genotypes at 48 h postinfection with LP02 ΔflaA L. pneumophila (MOI = 0.05). Data are representative of at least three independent experiments. In panel a, symbols and error bars represent means ± the standard deviations from three technical replicates. Curves in panel a depicting BMMs infected with LP02 ΔflaA are significantly distinct from other curves in each graph (P < 0.001, two-way analysis of variance [ANOVA]). For histograms, bars and error bars represent means ± the standard deviations from at least three technical replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant (two-tailed Student t test).
2DG blocks L. pneumophila replication in WT and Atg5−/− macrophages but not in broth or protozoan host cells.
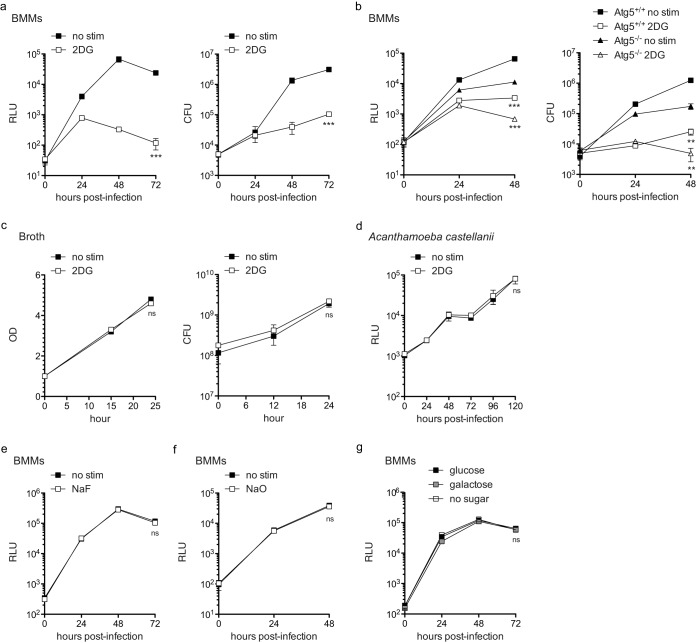
To test the effect of blocking glycolysis on L. pneumophila replication in BMMs, we treated BMMs infected with of LP02 ΔflaA expressing a luminescence cassette containing lux genes from P. luminescens (27) (LP02 ΔflaA lux) with 2DG at 1 h postinfection. In accord with previous observations (23–25), we found that L. pneumophila replication was potently inhibited by the addition of 2DG to the infection culture media (Fig. 2a). Inhibition of L. pneumophila replication in 2DG-treated BMMs was not due to increased host death in response to 2DG since BMM cell viability was not adversely affected by 0.2 mM 2DG up to 48 h (data not shown). Matsuda et al. proposed that induction of host autophagy by 2DG was responsible for the restriction of L. pneumophila replication in BMMs (25). To test this, we infected Atg5-deficient BMMs with L. pneumophila and treated infected cultures with 2DG. ATG5 is considered essential for autophagy (28). While we observed lower levels of L. pneumophila replication in Atg5−/− BMMs, 2DG-dependent restriction was unaffected (Fig. 2b). This result indicated that 2DG is unlikely to restrict L. pneumophila replication in BMMs via increased host autophagy. In addition, 2DG did not have an adverse effect on L. pneumophila growth in broth, in agreement with a previous report (Fig. 2c) (23), and also did not restrict L. pneumophila growth in a laboratory model of its natural protozoan host cells Acanthamoeba castellanii (Fig. 2d). These data suggest that 2DG inhibition of L. pneumophila replication requires a mammalian host cell and does not rely on induction of host autophagy.
FIG 2.
2DG restricts L. pneumophila replication in BMMs independent of Atg5, and perturbation of macrophage glycolysis via other inhibitors or glucose substitution does not affect intracellular replication of L. pneumophila in macrophages. (a) LP02 ΔflaA lux L. pneumophila replication measured by light emission (relative light units [RLU]) detected via luminometer (left) or CFU (right) in unstimulated BMMs (no stim) or in the presence of 0.2 mM 2DG (MOI = 0.01). (b) LP02 ΔflaA lux L. pneumophila replication (RLU and CFU) in Atg5+/+ BMMs (derived from LysMCre+/+ C57BL/6 mice) or Atg5−/− BMMs (derived from LysMCre+/+ Atg5fl/fl C57BL/6 mice) with or without 0.2 mM 2DG (MOI = 0.01). (c) LP02 ΔflaA L. pneumophila replication in broth with or without 0.2 mM 2DG from a starting OD of 1.0 as measured by OD (left) and CFU (right). (d) LP02 ΔflaA (pJB908) (thyA+) lux L. pneumophila replication in A. castellanii with or without 0.2 mM 2DG as measured by light emission (RLU). **, P < 0.01; ***, P < 0.001; ns, not significant (comparing growth in the presence of 2DG to that with no stimulation by two-way ANOVA). Symbols and error bars represent means ± the standard deviations from at least three technical replicates. (e) LP02 ΔflaA lux L. pneumophila replication (RLU) in WT BMMs with or without 1.0 mM sodium fluoride (NaF; MOI = 0.05). (f) LP02 ΔflaA lux L. pneumophila replication (RLU) in WT BMMs with or without 1.25 mM sodium oxamate (NaO; MOI = 0.05). (g) LP02 ΔflaA lux L. pneumophila replication (RLU) in WT BMMs in culture conditions containing 11.11 mM glucose or 11.11 mM galactose in the absence of glucose or in the absence of any sugar (no sugar). In each graph, no curves differ significantly (ns, not significant) as assessed by two-way ANOVA. Data are representative of at least two independent experiments.
Treatment with other inhibitors of glycolysis, substitution of glucose for galactose, and removal of glucose from cell culture media do not affect intracellular replication of L. pneumophila in macrophages.
Previous work found that 2DG but not another glycolysis inhibitor, sodium fluoride (NaF), had an inhibitory effect on L. pneumophila replication in macrophages (23). To further investigate whether host macrophage glycolysis inhibition negatively affects L. pneumophila replication during infection, we perturbed glycolysis using NaF, a lactate dehydrogenase inhibitor sodium oxamate (NaO), or by altering the availability of glucose during infection of wild-type BMMs with LP02 ΔflaA lux L. pneumophila. In agreement with Ogawa et al., we did not observe an effect of NaF on intracellular replication of L. pneumophila (Fig. 2e) (23). Similarly, NaO did not have an inhibitory effect on L. pneumophila replication (Fig. 2f). Replacement of glucose in the infection media with galactose, a sugar known to inhibit the transition to increased glycolysis in macrophages during bacterial infection (29), or removal of glucose from the infection media had no inhibitory effect on L. pneumophila replication in BMMs (Fig. 2g). These results indicate that L. pneumophila is able to replicate in host macrophages exhibiting a wide range of glycolytic states and that host glycolysis per se does not affect bacterial replication. Further, these data strongly suggest that the inhibitory effect of 2DG on L. pneumophila growing in macrophages is not due to the effect of 2DG on macrophage glycolysis.
Mutations in hexose-phosphate transporter gene uhpC allow L. pneumophila to replicate in 2DG-treated macrophages.
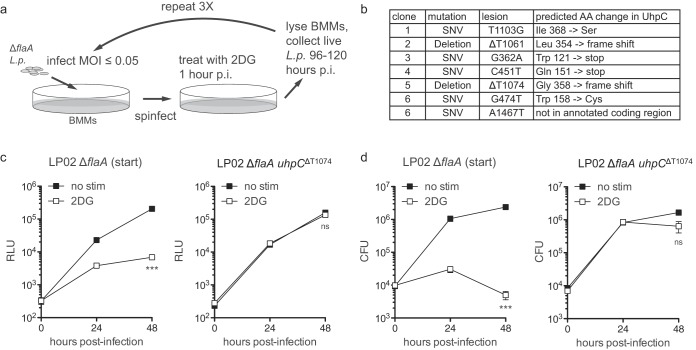
2DG-mediated inhibition of L. pneumophila replication in macrophages may act through changes in host macrophage metabolism or could result from a more direct effect on L. pneumophila during infection of macrophages. To determine whether L. pneumophila has the ability to overcome the replication block imposed by 2DG during infection of macrophages, we serially passaged LP02 ΔflaA lux L. pneumophila in BMMs treated with 2DG (Fig. 3a). After three rounds of enrichment in 2DG-treated BMMs, we were able to purify several independent clones of LP02 ΔflaA lux L. pneumophila with the ability to replicate without a growth defect in 2DG-treated BMMs. Whole-genome sequencing of six independent clones of L. pneumophila with the ability to replicate in 2DG-treated BMMs revealed that each clone harbored mutations in a single shared gene, the hexose-phosphate transporter uhpC (NCBI Gene ID 19834118) (Fig. 3b). Several of the mutations observed in uhpC were predicted to introduce premature stop codons or frameshifts for the translated UhpC protein, suggesting that mutations resulting in nonfunctional UhpC protect L. pneumophila from 2DG-mediated restriction in BMMs (Fig. 3b). Analysis of an isolated clone from the serial enrichment experiment, a thymidine deletion at position 1074 of uhpC (LP02 ΔflaA uhpCΔT1074 lux), demonstrated that compared to its parental strain, LP02 ΔflaA uhpCΔT1074 lux L. pneumophila replicates in 2DG-treated BMMs without a growth defect (Fig. 3c and d). These results suggest that L. pneumophila sensitivity to 2DG during infection of BMMs depends on interaction of 2DG or a 2DG-derived molecule directly with L. pneumophila via the bacterial protein UhpC.
FIG 3.
Enrichment of uhpC-mutant L. pneumophila following serial infection of BMMs treated with 2DG. (a) Strategy for serial passaging of LP02 ΔflaA lux L. pneumophila in 2DG-treated BMMs. L.p., Legionella pneumophila; p.i., postinfection. (b) Table of mutations in uhpC and corresponding predicted amino acid (AA) changes in UhpC protein observed in independent clones of LP02 ΔflaA lux L. pneumophila with the ability to replicate in 2DG-treated BMMs. SNV, single nucleotide variant. (c) LP02 ΔflaA lux (starting strain before the serial enrichment experiment) and LP02 ΔflaA uhpCΔT1074 lux replication in BMMs with or without 0.5 mM 2DG as measured by light emission (RLU). (d) LP02 ΔflaA lux and LP02 ΔflaA uhpCΔT1074 lux replication in BMMs with or without 0.5 mM 2DG as measured by CFU. ***, P < 0.001; ns, not significant (comparing growth in the presence of 2DG to that with no stimulation by two-way ANOVA). Data are representative of at least two independent experiments. Symbols and error bars represent means ± the standard deviations from at least three technical replicates.
ΔuhpC L. pneumophila is resistant to 2DG-mediated growth inhibition in BMMs and does not display a growth defect in broth or in protozoan host cells.
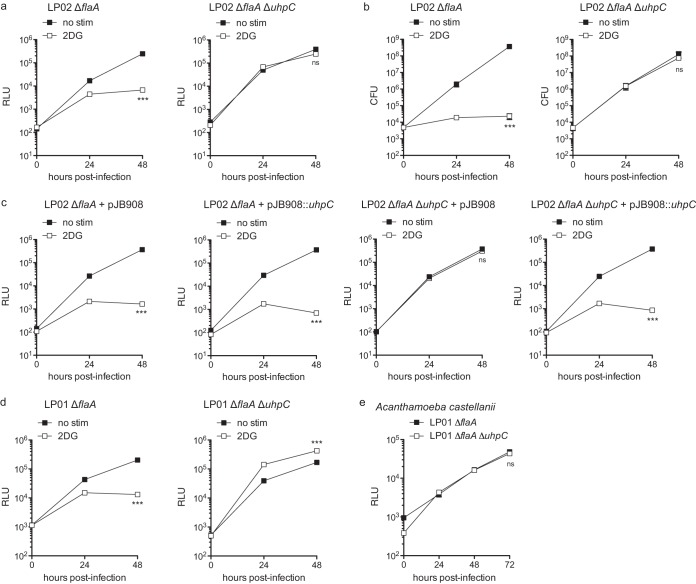
To confirm that loss of functional UhpC is responsible for the rescue of L. pneumophila replication in 2DG-treated BMMs, we generated in-frame deletions of uhpC in LP02 ΔflaA and LP01 ΔflaA strains of L. pneumophila. Compared to LP02 ΔflaA, LP02 ΔflaA ΔuhpC was insensitive to the inhibitory effect of 2DG during replication in BMMs (Fig. 4a and b). To verify that L. pneumophila uhpC is responsible for 2DG sensitivity, we transformed LP02 ΔflaA lux and LP02 ΔflaA ΔuhpC lux with the expression vector pJB908, which rescues the thymidine auxotrophy of LP02 L. pneumophila (30), or pJB908 expressing uhpC cloned from WT LP02 L. pneumophila. We observed that pJB908::uhpC conferred 2DG sensitivity to LP02 ΔflaA ΔuhpC during infection of BMMs (Fig. 4c). LP01 ΔflaA ΔuhpC did not display a defect in replication during infection of BMMs or A. castellanii (Fig. 4d and e). All ΔuhpC strains of L. pneumophila grow without defect in broth (data not shown). These results indicate that functional UhpC is responsible for 2DG-mediated inhibition of growth in BMMs. Notably, L. pneumophila does not appear to require functional UhpC to replicate in any laboratory conditions tested, including BMMs treated with high levels of 2DG.
FIG 4.
In-frame deletion of uhpC rescues 2DG-mediated inhibition of L. pneumophila replication in BMMs. (a) LP02 ΔflaA lux and LP02 ΔflaA ΔuhpC lux L. pneumophila replication in BMMs with or without 1.0 mM 2DG as measured by light emission (RLU). (b) LP02 ΔflaA and LP02 ΔflaA ΔuhpC L. pneumophila replication in BMMs with or without 1.0 mM 2DG as measured by CFU. (c) Replication of LP02 ΔflaA lux and LP02 ΔflaA ΔuhpC lux L. pneumophila complemented with pJB908 or pJB908 expressing wild-type (LP02) uhpC in BMMs with or without 0.5 mM 2DG as measured by light emission (RLU). (d) LP01 ΔflaA lux and LP01 ΔflaA ΔuhpC lux L. pneumophila replication in BMMs with or without 1.0 mM 2DG as measured by light emission (RLU). (e) LP01 ΔflaA lux and LP01 ΔflaA ΔuhpC lux L. pneumophila replication in Acanthamoeba castellanii as measured by light emission (RLU). ***, P < 0.001; ns, not significant (comparing growth curves by two-way ANOVA). Data are representative of at least two independent experiments. Symbols and bars represent means ± the standard deviations from at least three technical replicates.
2DG-phosphate inhibits wild-type but not ΔuhpC L. pneumophila replication in broth.
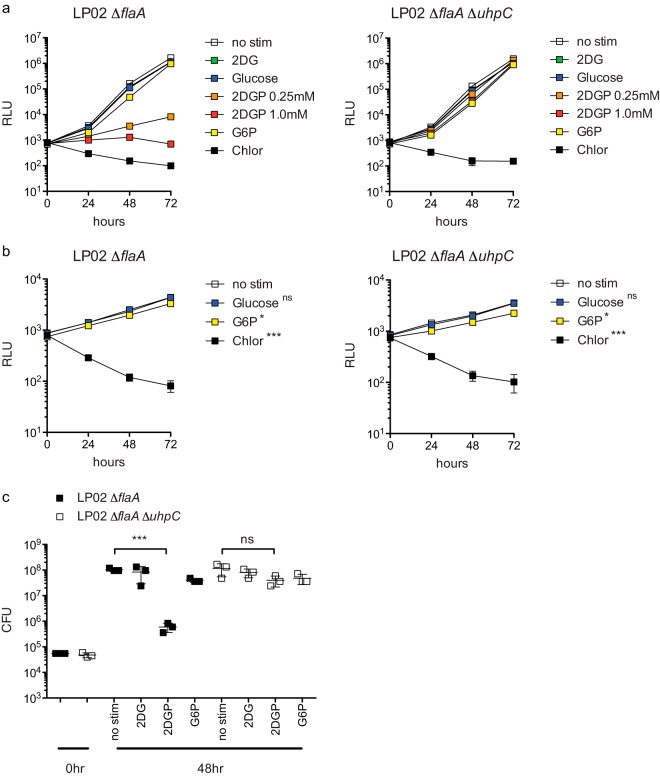
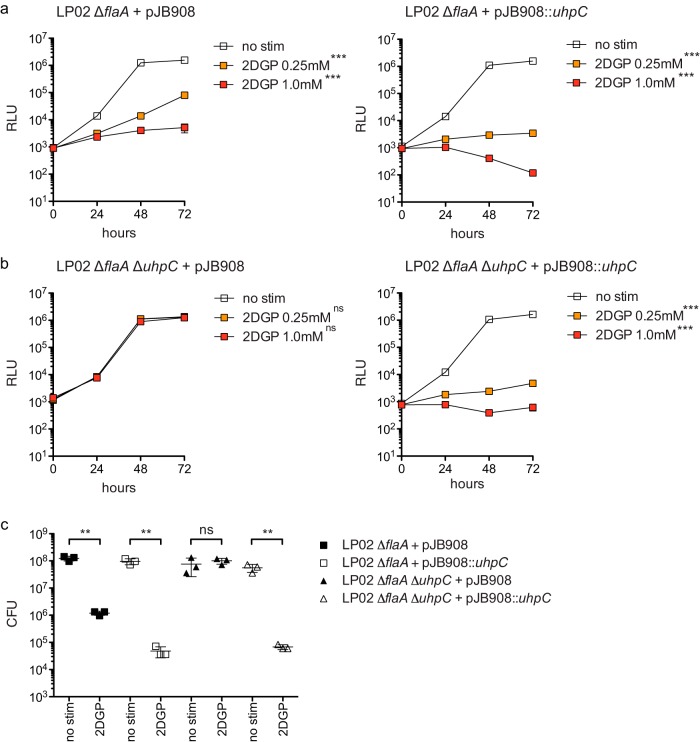
Given that the predicted function of L. pneumophila UhpC is to transport hexose-phosphates and that 2DG is itself a hexose sugar analog, we hypothesized that the inhibitory effect of 2DG in BMMs requires phosphorylation to produce 2DG 6-phosphate (2DGP). In support of this hypothesis, we saw that LP02 ΔflaA lux displayed a profound growth defect in broth in the presence of 2DGP but not 2DG (Fig. 5a). The inhibitory effect of 2DGP on L. pneumophila growth in broth was largely uhpC-dependent, as growth of LP02 ΔflaA ΔuhpC lux was not potently inhibited by the presence of 2DGP in broth (Fig. 5a). We observed that the addition of glucose had no effect on L. pneumophila replication in replete broth or in broth containing a low level of cysteine (Fig. 5a and b). Given that UhpC is a predicted hexose-phosphate transporter, we tested whether L. pneumophila growth in replete broth or cysteine-limited broth could be enhanced by phosphorylated glucose (glucose-6-phosphate [G6P]) and whether this effect depended on UhpC. However, we observed that the addition of G6P did not increase the ability of either LP02 ΔflaA or LP02 ΔflaA ΔuhpC lux L. pneumophila to replicate in either replete or cysteine-limited conditions and, in fact, slightly decreased the bacterial replication of both strains (Fig. 5a and b). We verified that 2DGP inhibited the growth of LP02 ΔflaA lux but not LP02 ΔflaA ΔuhpC lux L. pneumophila growth in broth by measuring CFU at 48 h (Fig. 5c). Complementation of LP02 ΔflaA ΔuhpC lux with pJB908::uhpC restored sensitivity 2DGP in broth (Fig. 6). These results demonstrate that L. pneumophila UhpC is required for sensitivity to 2DGP and strongly suggest that the inhibitory effect of 2DG on L. pneumophila replication in macrophages is mediated by phosphorylated 2DG.
FIG 5.
Restriction of L. pneumophila replication by 2DGP in broth is uhpC dependent. (a) LP02 ΔflaA lux and LP02 ΔflaA ΔuhpC lux L. pneumophila growth in replete broth in the presence of no stimulation (no stim), 1.0 mM 2DG, 1.0 mM glucose, 0.25 mM 2DGP, 1.0 mM 2DGP, 1.0 mM G6P, and 10.0 μg/ml chloramphenicol (Chlor) as measured by light emission (RLU). Growth under all stimulation conditions differs from growth in no stimulation conditions (P < 0.05 by two-way ANOVA with Bonferroni posttest to control for multiple comparisons). (b) LP02 ΔflaA lux and LP02 ΔflaA ΔuhpC lux L. pneumophila growth in broth containing 1/16 normal amount of cysteine (0.21 mM) in the presence of no stimulation (no stim), 1.0 mM glucose, 1.0 mM G6P, and 10.0 μg/ml chloramphenicol (Chlor) as measured by light emission (RLU). *, P < 0.05; ***, P < 0.001; ns, not significant (two-way ANOVA with Bonferroni posttest to control for multiple comparisons). (c) CFU of LP02 ΔflaA lux and LP02 ΔflaA ΔuhpC lux L. pneumophila at 0 h and after 48 h in broth in the presence of no stimulation (no stim), 0.25 mM 2DG, 0.25 mM 2DGP, or 0.25 mM G6P. ***, P < 0.001; ns, not significant (two-tailed Student t test). Data represent at least two independent experiments. Error bars reflect the standard deviations from three technical replicates.
FIG 6.
Complementation of ΔuhpC L. pneumophila with wild-type uhpC confers sensitivity to 2DG-phosphate in broth. (a) Replication of LP02 ΔflaA lux complemented with pJB908 or pJB908 expressing wild-type (LP02) uhpC in broth in the presence of no stimulation (no stim), 0.25 mM 2DGP, and 1.0 mM 2DGP as measured by light emission (RLU). (b) Replication of LP02 ΔflaA ΔuhpC lux complemented with pJB908 or pJB908 expressing wild-type (LP02) uhpC in broth in the presence of no stimulation (no stim), 0.25 mM 2DGP, and 1.0 mM 2DGP as measured by light emission (RLU). ***, P < 0.001, ns, not significant (two-way ANOVA with Bonferroni posttest to control for multiple comparisons in panels a and b). (c) CFU of LP02 ΔflaA lux and LP02 ΔflaA ΔuhpC lux L. pneumophila complemented with pJB908 or pJB908 expressing wild-type (LP02) uhpC after 48 h in broth in the presence of no stimulation (no stim) and 0.25 mM 2DG. **, P < 0.01; ns, not significant (two-tailed Student t test). Data represent at least two independent experiments. Error bars reflect the standard deviations from three technical replicates.
DISCUSSION
There is an increasing appreciation for how changes in macrophage metabolism are critical for the immune response against pathogens, particularly intracellular bacterial pathogens (7, 31). Increased glycolysis is part of a characteristic metabolic shift in macrophages in response to bacterial pattern recognition signaling. This metabolic change is thought to allow macrophages to produce the high levels of inflammatory mediators necessary for initiation of an immune response. We observed that BMMs upregulate glycolysis in response to infection with L. pneumophila, a response we characterized to be largely dependent on intact macrophage pattern recognition signaling. Elevated glycolysis is associated with proinflammatory activities in a variety of immune cells, and increased glycolysis has been associated with restriction of intracellular bacterial replication in macrophages (29, 32). Because L. pneumophila is able to replicate in glycolytic macrophages, we initially hypothesized that L. pneumophila was taking advantage of macrophage glycolysis during intracellular replication. This hypothesis was supported by our result showing that the glycolysis inhibitor 2DG blocked L. pneumophila replication in BMMs, an effect that has been reported in other studies (22–25). However, other experiments in which we inhibited host macrophage glycolysis had no effect on intracellular L. pneumophila replication and demonstrated that the inhibitory effect of 2DG on intracellular L. pneumophila was unlikely via induction of a shift in host cell metabolism. Ultimately, we observed that L. pneumophila hexose-phosphate transporter UhpC was required for bacterial sensitivity to 2DG during infection of BMMs, defining a potential mechanism for the sensitivity of L. pneumophila to 2DG during intracellular growth.
We propose that in BMMs mammalian hexokinase phosphorylates 2DG to produce 2DGP, which is taken up via L. pneumophila UhpC. Once inside the bacterium, 2DGP interferes with L. pneumophila growth through an as-yet-undetermined mechanism. The lack of effect of 2DG on L. pneumophila replication in broth may be explained by the inability of certain bacteria to phosphorylate 2DG (33). Similarly, 2DG phosphorylation may not occur in A. castellanii. This revised model of how 2DG restricts L. pneumophila replication in BMMs is supported by our result that 2DGP potently inhibits the growth of ΔflaA but not ΔflaA ΔuhpC L. pneumophila in broth.
We did not observe defects in ΔuhpC L. pneumophila replication in BMMs, broth, or in Acanthamoeba castellanii, indicating that this gene is not required for L. pneumophila growth in normal laboratory conditions. Of particular note, ΔuhpC L. pneumophila replicates without a growth defect in BMMs treated with high levels of 2DG. This result indicates that in contrast to our initial hypothesis, increased macrophage glycolysis is not required to support intracellular L. pneumophila growth. Increased macrophage glycolysis may be part of a coordinated multicellular immune response in response to L. pneumophila infection in vivo. Accordingly, the identification of 2DG-resistant L. pneumophila may provide a useful tool for the study of metabolic responses to infection in more complex experimental models.
During intracellular and extracellular growth, L. pneumophila is thought to depend on amino acids as a carbon source (34, 35). While the L. pneumophila genome is predicted to contain intact glucose metabolism pathways and the bacteria have been shown to incorporate carbon from glucose into its central carbon metabolism, the addition of glucose does not boost L. pneumophila growth in broth and cannot replace amino acids as a carbon source (35–37). The importance of L. pneumophila glucose metabolism during intracellular replication remains obscure, since a range of defects have been reported in L. pneumophila harboring mutations in glucose metabolism pathways (37, 38). L. pneumophila uhpC is a putative hexose-phosphate transporter gene based on sequence similarity to characterized transporters in other species of bacteria, including Chlamydia, Salmonella, and Escherichia coli (39, 40). In Listeria monocytogenes, the hexose-phosphate transporter gene hpt is a virulence (PrfA)-regulated gene, and hpt mutant bacteria display impaired intracellular growth and a virulence defect during mouse infection (41). The prevalence of hexose-phosphate transporter genes across species of pathogenic bacteria indicates that hexose-phosphates may be an advantageous nutrient source during intracellular replication. Presumably, L. pneumophila UhpC facilitates the uptake of phosphorylated hexose sugars, similar to UhpC-like proteins in other bacteria. However, we did not observe increased L. pneumophila growth in broth supplemented with glucose-6-phosphate. Inhibition of L. pneumophila replication by 2DGP suggests that poisoning L. pneumophila glucose metabolic pathways does have a deleterious effect on bacterial growth. Alternatively, 2DGP may have off-target toxic effects on L. pneumophila. Future metabolite profiling of wild-type and UhpC-deficient L. pneumophila may reveal a role for UhpC in terms of facilitating uptake of carbon and/or phosphate-containing molecules, which may allow L. pneumophila to access a variety of metabolites that could be critical for persistence or replication in specific environments or host species encountered in nature.
MATERIALS AND METHODS
Ethics statement.
We conducted this study in accordance with guidelines contained in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (42) and under a protocol approved by the Animal Care and Use Committee at the University of California, Berkeley (protocol AUP-2014-09-6665).
Cell culture.
We used BMMs from WT C57BL/6J mice for all experiments unless otherwise indicated. We derived macrophages from the bone marrow of C57BL/6J mice (Jackson Laboratory) or Myd88−/− Nod1−/− Nod2−/− mice on the C57BL/6J background (26) in RPMI medium supplemented with 10.0% fetal bovine serum, 2.0 mM l-glutamine, and 100 μM streptomycin (all from Life Technologies), as well as 5.0% supernatant from 3T3 cells expressing macrophage colony-stimulating factor (generated in-house). BMMs from LysMCre+/+ and LysMCre+/+Atg5fl/fl mice in a C57BL/6J background were provided by the lab of Daniel Portnoy at the University of California, Berkeley.
Bacterial strains and culture.
LP01 is a streptomycin-resistant derivative of a clinical isolate of Philadelphia-1 strain L. pneumophila (43). LP02 is derived from LP01 L. pneumophila and is a thymidine auxotroph (43). ΔflaA, ΔdotA, and lux strains of L. pneumophila have been described previously (16, 27, 44). We cultured L. pneumophila in AYE (ACES-buffered yeast extract broth) or on ACES-buffered charcoal-yeast extract (BCYE) agar plates at 37°C. We made in-frame deletion of uhpC (ΔuhpC) using the sucrose-sensitive suicide plasmid pSR47S as described previously (45). We complemented ΔuhpC strains with wild-type uhpC cloned from LP02 genomic DNA and expressed from the plasmid pJB908, which encodes thymidine synthetase for selection. We list sequences of primers used to generate deletion and complementation constructs in Table 1. To measure growth of L. pneumophila in AYE broth, we diluted bacteria to an optical density at 600 nm (OD600) of 1.0 with or without 2-deoxyglucose (2DG; Abcam) at the indicated concentrations and measured bacterial growth by measuring the OD600 or diluting bacteria to enumerate CFU on BYCE agar plates. To measure lux strain L. pneumophila growth in broth, we diluted bacteria to an estimated 5.0 × 104 L. pneumophila cells/well in opaque white TC-treated 96-well microtiter plates (Corning) in AYE broth in the presence of glucose (Sigma), glucose-6-phosphate (G6P; Santa Cruz Biotechnology), 2DG, 2DG-6-phosphate (2DGP; Santa Cruz Biotechnology), or chloramphenicol (Sigma) as indicated. We measured bacterial growth by detection of luminescence at λ470 using a Spectramax L luminometer (Bio-Rad) or by dilution on BYCE agar plates for enumeration of CFU. For experiments using heat-killed bacteria, we incubated L. pneumophila in phosphate-buffered saline (PBS) at 100°C for 15 min to kill bacteria before performing infections. We verified death of bacteria by incubation on BYCE agar plates.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′–3′)a |
|---|---|
| uhpC deletion construct in pSR47S | |
| uhpC forward | TAAGCAGGATCCCGTATGCCGATAGCTTGAAACG (BamHI) |
| uhpC internal reverse | CTAAGCAAATTCGGATTTGCCTGAAGGTTTTAGAAAACTCAATAATGCCAT |
| uhpC internal forward | ATGGCATTATTGAGTTTTCTAAAACCTTCAGGCAAATCCGAATTTGCTTAG |
| uhpC reverse | TGCTTAGCGGCCGCATCCAGGTGCCTTGGTATGTCC (NotI) |
| uhpC complementation construct in pJB908 | |
| uhpC comp forward | TAAGCAGGATCCTTTTCATGATCATGGACAATC (BamHI) |
| uhpC comp reverse | ATGTAGTCGACGAAGCCAATCACTAAGCAAAT (SalI) |
Restriction enzymes are indicated in parentheses; restriction sites are indicated in boldface.
Infection of BMMs.
For measurement of intracellular L. pneumophila growth by luminescence, we plated 1.0 × 105 BMMs/well in opaque white TC-treated 96-well microtiter plates and infected the samples with L. pneumophila at a multiplicity of infection (MOI) of 0.01 or 0.05 as indicated. For measurement L. pneumophila growth by CFU, we plated 5.0 × 105 BMMs/well in TC-treated 24-well plates (Corning) and infected with L. pneumophila at an MOI of 0.01. At 1 h postinfection by centrifugation at 287 × g, we replaced the media of infected BMMs with media containing 2DG, sodium fluoride (NaF, Sigma), or sodium oxamate (NaO; Sigma) at the indicated concentrations. We maintained 2DG or other drugs in the infection media for the duration of the experiment. At the indicated times after infection, we measured bacterial growth by detection of luminescence or by dilution of infected cultures on BYCE agar plates for the enumeration of CFU. To measure lactate and glucose in the supernatant of BMMs infected with L. pneumophila, we plated 5.0 × 105 BMMs/well in 1.0 ml/well in TC-treated 24-well plates (Fig. 1a, d, and e) or 2.0 × 106 BMMs/well in 2.0 ml/well in TC-treated 6-well plates (Fig. 1b and c) and infected them with L. pneumophila as described above. At 1 h postinfection, we replaced the media of infected BMMs with media with or without 2DG at the indicated concentrations. We measured lactate and glucose in the supernatant of BMMs cultures using assay kits (Sigma) according to the manufacturer's instructions. For altered-sugar experiments, we performed infections using glucose-free RPMI (Life Technologies). We added 11.11 mM galactose (Sigma) as indicated.
Serial enrichment of L. pneumophila mutants.
We plated 1.0 × 106 BMMs/well in 6-well TC-treated plates (Corning) for a total of 12 wells. We infected each well with LP02 ΔflaA lux L. pneumophila at an MOI of 0.05 as described above. At 1 h postinfection, we replaced the media of infected BMMs in 11 wells with medium containing 0.2 mM 2DG and one well with medium containing no 2DG. At 96 to 120 h postinfection, we removed the culture supernatant and lysed the remaining BMMs in sterile H2O. We combined L. pneumophila from BMM lysis with L. pneumophila collected from the culture supernatant for reinfection of fresh BMMs. We diluted L. pneumophila for reinfection based on the luminescence signal measured from recovered bacteria with the aim of reinfecting at an MOI between 0.01 and 0.05. We repeated the infection procedure as described above at least three times in two independent experiments. At each reinfection step, a fraction of L. pneumophila recovered from each well were tested in a luminescence growth assay using 1.0 × 105 BMMs/well in opaque white TC-treated 96-well microtiter plates with or without 2DG. After the third round of reinfection, when we observed robust growth of L. pneumophila in 2DG-treated BMMs from several independent source wells versus the control well, we isolated individual clones of L. pneumophila from each of the 12 wells by dilution on BYCE agar plates. After testing of these clonal populations of L. pneumophila for the ability to replicate in 2DG-treated BMMs, we extracted genomic DNA using a blood and tissue DNA extraction kit (Qiagen) and submitted the DNA for library preparation and paired-end sequencing on a MiSeq instrument at the QB3 functional genomics facility at UC Berkeley (http://qb3.berkeley.edu/fgl/). We analyzed the sequencing data for variations compared to the Philadelphia-1 L. pneumophila reference genome (GenBank accession no. CP013742.1) using CLC Genomics Workbench software (Qiagen).
Infection of Acanthamoeba castellanii.
We maintained A. castellanii strain Neff (ATCC 30010) in PYG broth (2.0% proteose-peptone [BD Biosciences], 0.1% yeast extract [BD Biosciences], 0.1 M glucose, 4.0 mM MgSO4 [Fisher Scientific], 0.4 M CaCl2 (Fisher Scientific) 0.1% sodium citrate (Sigma), 0.05 mM Fe(NH4)2(SO4)2 · 6H2O [Sigma], 2.5 mM NaH2PO4 [Fisher Scientific], 2.5 mM K2HPO4 [Fisher Scientific], and 100 μM streptomycin [Life Technologies]) at room temperature. For infection, we plated 1.0 × 105 A. castellanii cells/well in opaque white TC-treated 96-well microtiter plates in PBS (Life Technologies). We diluted L. pneumophila to the desired MOI in PBS, followed by centrifugation at 287 × g. At 1 h postinfection, we replaced the media of infected A. castellanii with PBS containing 2DG at the indicated concentrations and maintained infected cultures at 37°C. At the indicated times after infection, we measured the growth of L. pneumophila by the detection of luminescence.
ACKNOWLEDGMENTS
R.E.V. is supported by an Investigator Award from the Howard Hughes Medical Institute and by National Institutes of Health grants AI063302 and AI075039.
We acknowledge stimulating discussions with Sarah Stanley, Jonathan Braverman, Greg Barton, Daniel Portnoy, members of the Vance, Stanley, Barton, and Portnoy Labs, and members of the P01 Intracellular Pathogens and Innate Immunity research group.
We have no conflicts of interest with regard to the results presented in this study.
REFERENCES
- 1.Rodriguez-Prados JC, Traves PG, Cuenca J, Rico D, Aragones J, Martin-Sanz P, Cascante M, Bosca L. 2010. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol 185:605–614. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- 2.Krawczyk CM, Holowka T, Sun J, Blagih J, Amiel E, DeBerardinis RJ, Cross JR, Jung E, Thompson CB, Jones RG, Pearce EJ. 2010. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 115:4742–4749. doi: 10.1182/blood-2009-10-249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Neill LA, Pearce EJ. 2016. Immunometabolism governs dendritic cell and macrophage function. J Exp Med 213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly B, O'Neill LA. 2015. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res 25:771–784. doi: 10.1038/cr.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, O'Neill LA. 2013. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jha AK, Huang SC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, Pearce EJ, Driggers EM, Artyomov MN. 2015. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Price JV, Vance RE. 2014. The macrophage paradox. Immunity 41:685–693. doi: 10.1016/j.immuni.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 8.Czyz DM, Willett JW, Crosson S. 2017. Brucella abortus induces a Warburg shift in host metabolism that is linked to enhanced intracellular survival of the pathogen. J Bacteriol 199:e00227-. doi: 10.1128/JB.00227-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah-Simpson S, Lentini G, Dumoulin PC, Burleigh BA. 2017. Modulation of host central carbon metabolism and in situ glucose uptake by intracellular Trypanosoma cruzi amastigotes. PLoS Pathog 13:e1006747. doi: 10.1371/journal.ppat.1006747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, Sharrar RG, Harris J, Mallison GF, Martin SM, McDade JE, Shepard CC, Brachman PS. 1977. Legionnaires' disease: description of an epidemic of pneumonia. N Engl J Med 297:1189–1197. doi: 10.1056/NEJM197712012972201. [DOI] [PubMed] [Google Scholar]
- 11.Horwitz MA, Silverstein SC. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J Clin Invest 66:441–450. doi: 10.1172/JCI109874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nash TW, Libby DM, Horwitz MA. 1984. Interaction between the legionnaires' disease bacterium (Legionella pneumophila) and human alveolar macrophages: influence of antibody, lymphokines, and hydrocortisone. J Clin Invest 74:771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubber A, Roy CR. 2010. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 14.Isberg RR, O'Connor TJ, Heidtman M. 2009. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molofsky AB, Byrne BG, Whitfield NN, Madigan CA, Fuse ET, Tateda K, Swanson MS. 2006. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med 203:1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. 2006. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog 2:e18. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lightfield KL, Persson J, Brubaker SW, Witte CE, von Moltke J, Dunipace EA, Henry T, Sun YH, Cado D, Dietrich WF, Monack DM, Tsolis RM, Vance RE. 2008. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol 9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zamboni DS, Kobayashi KS, Kohlsdorf T, Ogura Y, Long EM, Vance RE, Kuida K, Mariathasan S, Dixit VM, Flavell RA, Dietrich WF, Roy CR. 2006. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol 7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- 19.Amer A, Franchi L, Kanneganti TD, Body-Malapel M, Ozoren N, Brady G, Meshinchi S, Jagirdar R, Gewirtz A, Akira S, Nunez G. 2006. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem 281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 20.Wright EK, Goodart SA, Growney JD, Hadinoto V, Endrizzi MG, Long EM, Sadigh K, Abney AL, Bernstein-Hanley I, Dietrich WF. 2003. Naip5 affects host susceptibility to the intracellular pathogen Legionella pneumophila. Curr Biol 13:27–36. doi: 10.1016/S0960-9822(02)01359-3. [DOI] [PubMed] [Google Scholar]
- 21.Diez E, Lee SH, Gauthier S, Yaraghi Z, Tremblay M, Vidal S, Gros P. 2003. Birc1e is the gene within the Lgn1 locus associated with resistance to Legionella pneumophila. Nat Genet 33:55–60. doi: 10.1038/ng1065. [DOI] [PubMed] [Google Scholar]
- 22.Escoll P, Song OR, Viana F, Steiner B, Lagache T, Olivo-Marin JC, Impens F, Brodin P, Hilbi H, Buchrieser C. 2017. Legionella pneumophila modulates mitochondrial dynamics to trigger metabolic repurposing of infected macrophages. Cell Host Microbe 22:302–316. doi: 10.1016/j.chom.2017.07.020. [DOI] [PubMed] [Google Scholar]
- 23.Ogawa M, Yoshida S, Mizuguchi Y. 1994. 2-Deoxy-d-glucose inhibits intracellular multiplication and promotes intracellular killing of Legionella pneumophila in A/J mouse macrophages. Infect Immun 62:266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldoni P, Cattani L, Carrara S, Pastoris MC, Sinibaldi L, Orsi N. 1998. Multiplication of Legionella pneumophila in HeLa cells in the presence of cytoskeleton and metabolic inhibitors. Microbiol Immunol 42:271–279. doi: 10.1111/j.1348-0421.1998.tb02283.x. [DOI] [PubMed] [Google Scholar]
- 25.Matsuda F, Fujii J, Yoshida S. 2009. Autophagy induced by 2-deoxy-d-glucose suppresses intracellular multiplication of Legionella pneumophila in A/J mouse macrophages. Autophagy 5:484–493. doi: 10.4161/auto.5.4.7760. [DOI] [PubMed] [Google Scholar]
- 26.Fontana MF, Shin S, Vance RE. 2012. Activation of host mitogen-activated protein kinases by secreted Legionella pneumophila effectors that inhibit host protein translation. Infect Immun 80:3570–3575. doi: 10.1128/IAI.00557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coers J, Vance RE, Fontana MF, Dietrich WF. 2007. Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signaling pathways. Cell Microbiol 9:2344–2357. doi: 10.1111/j.1462-5822.2007.00963.x. [DOI] [PubMed] [Google Scholar]
- 28.Yang Z, Klionsky DJ. 2010. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braverman J, Sogi KM, Benjamin D, Nomura DK, Stanley SA. 2016. HIF-1α is an essential mediator of IFN-γ-dependent immunity to Mycobacterium tuberculosis. J Immunol 197:1287–1297. doi: 10.4049/jimmunol.1600266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sexton JA, Pinkner JS, Roth R, Heuser JE, Hultgren SJ, Vogel JP. 2004. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J Bacteriol 186:1658–1666. doi: 10.1128/JB.186.6.1658-1666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisenreich W, Heesemann J, Rudel T, Goebel W. 2013. Metabolic host responses to infection by intracellular bacterial pathogens. Front Cell Infect Microbiol 3:24. doi: 10.3389/fcimb.2013.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Neill LA, Kishton RJ, Rathmell J. 2016. A guide to immunometabolism for immunologists. Nat Rev Immunol 16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romano AH, Trifone JD, Brustolon M. 1979. Distribution of the phosphoenolpyruvate:glucose phosphotransferase system in fermentative bacteria. J Bacteriol 139:93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fonseca MV, Swanson MS. 2014. Nutrient salvaging and metabolism by the intracellular pathogen Legionella pneumophila. Front Cell Infect Microbiol 4:12. doi: 10.3389/fcimb.2014.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tesh MJ, Miller RD. 1981. Amino acid requirements for Legionella pneumophila growth. J Clin Microbiol 13:865–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, Asamani G, Hill K, Nuara J, Feder M, Rineer J, Greenberg JJ, Steshenko V, Park SH, Zhao B, Teplitskaya E, Edwards JR, Pampou S, Georghiou A, Chou IC, Iannuccilli W, Ulz ME, Kim DH, Geringer-Sameth A, Goldsberry C, Morozov P, Fischer SG, Segal G, Qu X, Rzhetsky A, Zhang P, Cayanis E, De Jong PJ, Ju J, Kalachikov S, Shuman HA, Russo JJ. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- 37.Eylert E, Herrmann V, Jules M, Gillmaier N, Lautner M, Buchrieser C, Eisenreich W, Heuner K. 2010. Isotopologue profiling of Legionella pneumophila: role of serine and glucose as carbon substrates. J Biol Chem 285:22232–22243. doi: 10.1074/jbc.M110.128678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harada E, Iida K, Shiota S, Nakayama H, Yoshida S. 2010. Glucose metabolism in Legionella pneumophila: dependence on the Entner-Doudoroff pathway and connection with intracellular bacterial growth. J Bacteriol 192:2892–2899. doi: 10.1128/JB.01535-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwoppe C, Winkler HH, Neuhaus HE. 2002. Properties of the glucose-6-phosphate transporter from Chlamydia pneumoniae (HPTcp) and the glucose-6-phosphate sensor from Escherichia coli (UhpC). J Bacteriol 184:2108–2115. doi: 10.1128/JB.184.8.2108-2115.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Island MD, Wei BY, Kadner RJ. 1992. Structure and function of the uhp genes for the sugar phosphate transport system in Escherichia coli and Salmonella Typhimurium. J Bacteriol 174:2754–2762. doi: 10.1128/jb.174.9.2754-2762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chico-Calero I, Suarez M, Gonzalez-Zorn B, Scortti M, Slaghuis J, Goebel W, Vazquez-Boland JA, European Listeria Genome Consortium . 2002. Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc Natl Acad Sci U S A 99:431–436. doi: 10.1073/pnas.012363899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.National Academies Press. 2011. Guide for the care and use of laboratory animals. National Academies Press, Washington, DC: http://grants.nih.gov/grants/olaw/Guide-for-the-Care-and-use-of-laboratory-animals.pdf. [Google Scholar]
- 43.Berger KH, Isberg RR. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol 7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 44.Berger KH, Merriam JJ, Isberg RR. 1994. Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotA gene. Mol Microbiol 14:809–822. doi: 10.1111/j.1365-2958.1994.tb01317.x. [DOI] [PubMed] [Google Scholar]
- 45.Shen X, Banga S, Liu Y, Xu L, Gao P, Shamovsky I, Nudler E, Luo ZQ. 2009. Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol 11:911–926. doi: 10.1111/j.1462-5822.2009.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]