Abstract
Genetically encoded live-cell reporters measure signaling pathway activity at the cellular level with high temporal resolution, often revealing a high degree of cell-to-cell heterogeneity. By using multiple spectrally distinct reporters within the same cell, signal transmission from one node to another within a signaling pathway can be analyzed to quantify factors such as signaling efficiency and delay. With other reporter configurations, correlation between different signaling pathways can be quantified. Such analyses are useful in establishing the mechanisms and consequences of cell-to-cell heterogeneity and can inform new models of the functional properties of signaling pathways. In this unit, we describe an approach for designing and executing live-cell multiplexed reporter experiments. We also describe approaches for analyzing the resulting time-course data to quantify correlations and trends between the measured parameters at the single-cell level.
Keywords: Live-cell microscopy, signal transduction, fluorescent protein, reporter, biosensor, image informatics
Introduction
Live-cell imaging has become a very useful tool for signal transduction research, and can be essential for making accurate measurements of dynamic signaling activity, particularly in heterogeneous cell populations (Locke and Elowitz, 2009; Purvis and Lahav, 2013). However, measuring any single signaling event in isolation leaves many question unanswered, due to the highly interconnected nature of intracellular signaling networks. Each signaling event measured by a reporter may be influenced by many additional factors, including the activity of other pathways, metabolism, and the cell cycle, which vary from cell to cell at any time. Recording multiple signaling components within the same cell allows the dynamics of related pathways to be directly correlated without the interference of cell-to-cell variance. The techniques in this unit are focused on reliably measuring and determining the relationships among intracellular signaling components.
This unit describes techniques and design principles for the simultaneous dynamic measurement of multiple gene products or enzyme activities in single cells. Applied to multiple parts of a signaling pathway, this approach facilitates the identification and modeling of intracellular signaling and regulatory processes. An increasing array of signaling molecules can now be observed in live cells via fluorescence microscopy. Molecular processes including kinase activity (Komatsu et al., 2011; Regot et al., 2014), transcription (Chubb et al., 2006), translation (Han et al., 2014), and intracellular translocation (Nelson et al., 2002) can be reliably imaged using existing live-cell reporters. Moreover, CRISPR-based knock-in of fluorescent proteins to genomic loci is expected to add to the number and diversity of regulatory events that can be imaged in living cells (Stewart-Ornstein and Lahav, 2016). In each case, live-cell imaging allows the observation of signal dynamics in single cells (Albeck et al., 2013) and the evaluation of intra- vs. extra-cellular noise (Elowitz et al., 2002).
This unit is designed to provide a conceptual overview of the experiment design, and deliver critical details for managing practical implementations. Four basic topics are covered: 1) Construction of suitable cell lines, 2) Long term time-lapse imaging of live cells, 3) Image processing to automate cell segmentation and extract fluorescent reporter values, and 4) Signal analysis of relationships among the reporters.
Strategic Planning
General
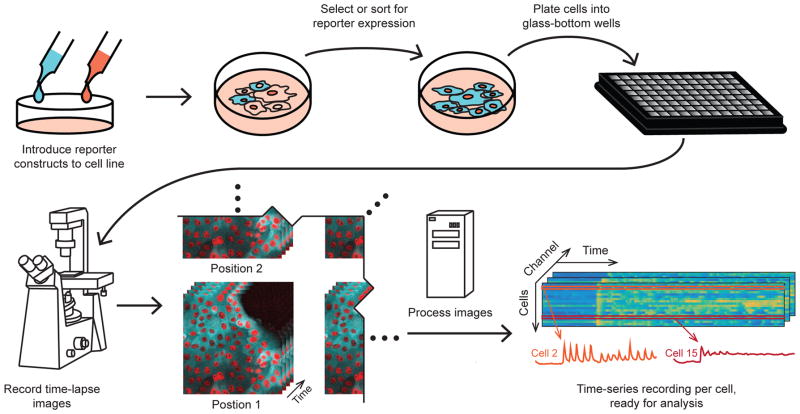
Efficiently executing a multiple reporter live-cell imaging experiment requires coordination of several key operations. Because the construction of cell lines bearing stable fluorescent reporter genes can be quite lengthy, careful planning is critical. In addition to the principles of the experiment, the targets to measure, fluorophores to use, and final analyses should be planned prior to embarking. Illustrated in Figure 1, the complete experiment consists of: 1) Acquiring/producing DNA constructs for reporters, 2) Inserting all constructs into desired cell line(s), 3) Imaging cells under desired treatments, and 4) Processing image data and analyzing time-series data. Review the Time Considerations section following the protocols for suggestions regarding time-consuming steps.
Figure 1.
Schematic example figure showing the construction of a cell line with multiple reporters, live-cell imaging, and data processing.
Selection of reporters and fluorophores
Reporter selection is a balance of choosing targets suitable for the study that are feasible to measure and minimally challenging to implement. Establishing fundamental reporter function can be very application-specific (Komatsu et al., 2011; Slattery and Hahn, 2014) and is beyond the scope here. Instead, this protocol addresses the choice of fluorophores, especially fluorescent proteins, and methods to drive their expression. Five aspects of the fluorescent proteins themselves are emphasized: spectral overlap, chromophore maturation rate, aggregation, photobleaching, and brightness. Each candidate fluorophore should be evaluated against alternatives based on these features.
Basic protocol 1: Paired reporter cell line construction
This protocol addresses the preparation of cell lines expressing multiple fluorescent reporters, including the selection of reporters, transfection and transduction into the desired cell line, selection, and validation of the line. Specific DNA constructs are very application-specific, and the details of their construction are varied, but are well established using a variety of methods and are not addressed here.
Materials
Cell line(s) to receive reporters
Culture medium appropriate for selected cell line(s)
Sterile culture materials: dishes, pipettes, conical tubes
DNA construct(s) for reporters
Transfection reagents, depending on the technique chosen, e.g. Optimem, Fugene, Lipofectamine, PEI (polyethylenimene), Electroporation apparatus, etc.
Selection antibiotics (optional)
Epifluorescence microscope (to view cells and evaluate reporter expression)
Select fluorophores for each reporter
Table 1 enumerates critical fluorescent protein properties and indicates the concerns when considering each. Comparative lists of properties for different fluorescent proteins are available from a variety of sources (such as www.fpvis.org), though none are exhaustive.
Table 1.
Criteria for choosing FPs and reporters.
| Property | Design preference | Complications |
|---|---|---|
| Spectra | Narrow, and distinct, for separability by filters (see Figure 2) | Spectra (excitation or emission) that are particularly broad, have multiple distant peaks, or overlap greatly (for two FPs) can lead to bleed-through noise |
| Quantum yield (QY) | High, for brightness | When low, FP is inefficient and dim, requiring more intense excitation |
| Extinction coefficient | High, for brightness | When low, FP absorbs light poorly, requiring more intense excitation |
| Maturation time | Low, for expression sensors | When high, protein is slow to develop its chromophore, and not suitable for reporting expression levels dynamically |
| Aggregation | Low, for image quality | When high, imagery is corrupted by puncta and activity may be altered by dimer/multimer formation |
| Photobleaching time | Long (slow bleaching), for dynamics | When short (rapid bleaching), dynamic measurements may be corrupted |
| pKa | Low, for selectivity | When near physiological pH (6–7.4), fluorescence can be sensitive to pH changes, esp. in lysosomes |
-
1
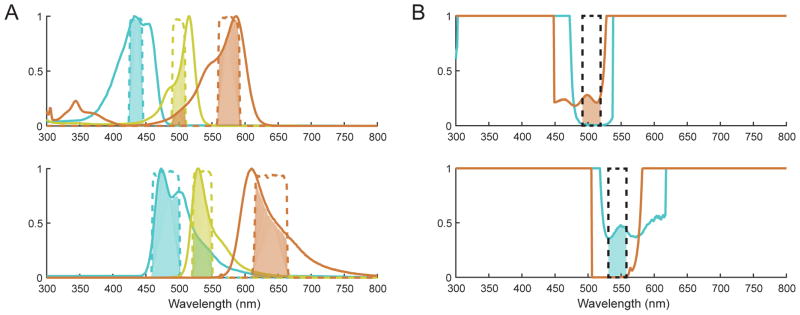
Evaluate fluorophore options for spectral overlap. Review the excitation and emission spectra for candidates and compare with available filters to ensure that each chosen filter will allow less than 1% of the on-target intensity from off-target fluorophores. Minimizing cross-talk becomes increasingly important if reporters are not expressed at the same levels, as higher concentration reporters will affect other filtered images more. Contact filter manufacturers for details and recommendations. See Figure 2 for example spectral comparisons.
Ensure that a nuclear marker is available for image processing, either by a reporter confined to the nucleus or a dedicated nuclear marker. If a nuclear dye is to be added at the time of imaging, ensure that its spectra are evaluated alongside those of the other reporters. Typically, three colors can be imaged with little difficulty, e.g. with cyan, yellow, and red fluorescent proteins (a UV dye, such as DAPI, may be used as well). -
2
For protein expression level reporters, rule out any fluorescent proteins with long maturation times (greater than 1 hour). Notably fast maturing fluorescent proteins include mNeonGreen, mVenus, mCherry, and mKate2.
Availability of maturation time measurements is not consistent. Refer to www.fpvis.org and primary literature regarding the development of particular fluorescent proteins. -
3
Evaluate the tendency of remaining candidates to aggregate, a common issue for fluorescent proteins. While tolerable in some cases, aggregation is generally detrimental and leads to punctate imagery and increased risk of perturbing the function of anything fused to the protein.
Direct data on aggregation are often difficult to find, and decisions rely on more qualitative commentary. It is often necessary to evaluate the trade-off between maturation rate, aggregation and brightness (see step 4). Selections are generally qualitative based on which characteristics are most important for a particular application. -
4
Consider the quantum yield, extinction coefficient and relative photobleaching time of remaining candidates and determine an acceptable compromise between brightness (estimated as quantum yield times extinction coefficient) and bleaching.
Compiled properties may be found at www.fpvis.org, and primary literature typically includes quantification of these properties.
Figure 2.
A. Absorption (upper) and emission (lower) spectra of an example fluorescent protein combination suitable for multiplexed imaging, comprising a CFP (mTurquoise2, in cyan), a YFP (mVenus, in yellow) and a RFP (mCherry, in orange). Appropriate filters are indicated by dashed lines, color-coded according the protein they are intended to measure. The relative contribution of each protein through each filter is shown as the shaded area beneath the filter band. The contributions of RFP to YFP excitation and CFP to YFP emission are allowable because neither contributes significantly to both filters. B. Example filter selection for YFP, by plotting the interference ratio of the other fluorophores (RFP in orange, CFP in cyan). The ratio is defined as the interfering spectrum divided by the intended spectrum, bounded on 0 to 1 for convenient visualization. Suitable filter regions are shown by dashed black lines, with shaded regions indicating the interference contribution. Filters should be placed where the ratio is minimal, and contribution of any interfering fluorophore through both filters should be strictly avoided.
Select reporter gene delivery and co-expression strategy
-
5
Compare co-expression options to determine a suitable compromise between the expected expression conditions and the complexity of the reporter insertion process. Multiple reporters may be included on one plasmid using an internal ribosome entry site (IRES), or a “self-cleaving” 2A peptide. Multiple plasmids or viruses may also be used to introduce reporters.
It is useful to consult experimental comparisons of bicistronic expression systems (Goedhart et al., 2011) and promoter expression output (Qin et al., 2010) when choosing a co-expression strategy. Due to possible interference between promoters, it is not recommended to use multiple promoters on any one plasmid. -
6
Determine the reporter insertion technique to use. See Table 2 for a comparison of insertion techniques, including references for appropriate protocols.
Transient expression can be achieved by simple transfection with the appropriate plasmid DNA (e.g. using polyethylenimine, FuGene, Lipofectimine, electroporation, etc.). However, expression levels will be variable over time and typically fade 5–8 days after transfection as cell division dilutes the plasmid DNA. The useful length of the experiment is thus limited and frequently yields noisy data that are difficult to interpret; transient transfection is therefore not recommended.
Table 2.
Comparison of reporter gene delivery and expression strategies.
| Technique | Description | Add’l Requirements | Pros/Cons |
|---|---|---|---|
| Transfection | Insertion of plasmid DNA only; genomic integration occurs at a low frequency | None | ↓ Transient (X-Y days) |
| Transposon | Use a co-transfected transposase to insert the reporter into genome (Cadinanos and Bradley, 2007; Geurts et al., 2003) | Transposase plasmid | ↑ Stable integration with high efficiency in transfected cells ↓ Limited by transfection frequency of target cell |
| Retrovirus | Use retroviral particles to insert the reporter into the genome (Cepko and Pear, 2001) | Packaging cell line Viral packaging plasmids Filtration devices Polybrene |
↑ Stable integration ↑ High efficiency in a broad range of target cells ↓ Virus handling |
| CRISPR | Introduce a fluorescent protein under the control of a genomic promoter of interest. (Dickinson et al., 2013) | Cas9 Homology plasmid |
↑ Stable integration ↑ Targeted insertion ↓ Low success rate |
Insert reporter constructs into target cells
-
7
Prepare purified plasmid DNA for each reporter, and for each associated genomic insertion method as needed. Modify any reporters via molecular cloning techniques of choice. Similarly, prepare any additional constructs necessary (helper plasmids for transposon expression, genomic targeting, or retroviral packaging). See step 9 for commentary on selection antibiotic resistance genes that may be included.
DNA for transfection into a cell line should be highly purified, for optimal performance. While DNA from a standard alkaline lysis “mini-prep” can be used, it is advantageous to purify by alcohol precipitation or, preferably, by paramagnetic beads. -
8
Introduce reporters to the target cells by transfection, transduction, etc., depending on the technique chosen in steps 5 and 6.
-
9
Select and/or sort cells for adequate stable expression of reporter constructs. Selection can be carried out by inclusion of an appropriate antibiotic resistance gene in the same construct as the reporter. To select for multiple reporters, each construct must bear a different resistance gene, such as puromycin, neomycin, blasticidin, or hygromycin. Alternatively, cells may be sorted by flow cytometry, avoiding the need for selection antibiotics. As multiple antibiotic resistance genes can become cumbersome, sorting is recommended for more than 2 insertions. If the insertion efficiency for each reporter is high enough, it is feasible to transfect/transduce in tandem and sort (or select) for multiple insertions rather than sorting after each.
Validate reporter insertion and function
-
10
Evaluate the insertion of fluorescent reporters initially by viewing cells on an epifluorescence microscope. In particular, look for side-by-side pairs of positive cells suggestive of a successful cell division.
If expression is weak and the insertion efficiency low, it may be quite difficult to visually identify the presence of positive cells even when there are enough to successfully sort. Analysis by flow cytometry provides the best assessment of brightness and frequency. -
11
After selection or sorting, evaluate reporter function via a preliminary experiment (following Basic Protocol 2). Identify two conditions that will induce minimal and maximal reporter response. Plate cells for a live-cell experiment (Basic Protocol 2 and Support Protocol 1), and apply both conditions in the same experiment, preferably in the same well via spike-in of a drug or stimulant. Evaluate the resulting movie for appropriate responses.
For example, with an ERK reporter in a cell line responsive to EGF stimulation, withdraw growth factors for >2 hours before imaging. After beginning imaging, spike in EGF to a saturating concentration, image for enough time to observe the response (1–2 hours). Then spike in an ERK, MEK or EGFR inhibitor to saturating conditions and continue imaging to observe the inhibition (1–2 hours). -
12
If any insertions are targeted to specific sites in the genome, validate the insertion site by sequencing the local region of the genome and/or the appropriate transcript.
-
13
Store frozen aliquots of cells lines with successful reporter integration for future use.
It is often useful to freeze cells after each successful insertion to retain stocks that can be used for future combinations of reporters.If a genetically uniform clonal line is desired, it can be produced at this point (e.g. by limiting dilution).
Basic protocol 2: Time-lapse imaging of living cells
This protocol describes the culture and imaging of cells for a long-term live cell imaging experiment. The process with multiple reporters is nearly identical to that with a single reporter. See (Pargett et al., 2017) for a specific implementation focused on a single reporter in MCF-10A epithelial cells. The goal is to keep the cells viable in an environment accessible to the microscope with high throughput and the ability to add treatments. To achieve this, cell monolayers are produced on a glass-bottom multi-well plate by coating with collagen, laminin, or another appropriate substrate. To provide adequate media for longer experiments (those longer than 24 hours), use larger wells (e.g. a 24-well plate) and spot only a small portion with the substrate. To facilitate robust image processing, it is advantageous to plan for one well to be left without cells; background images will be taken from this well.
Materials
Glass-bottom culture plates for imaging. (24-well: Cellvis P24-1.5H-N, 96-well: Cellvis P96-1.5H-N, square well 96-well: Brooks Life Science Systems MGB096-1-2-LG-L)
Substrate coating solution, e.g. using collagen (ThermoFisher A1048301) or laminin (ThermoFisher 23017015)
Pipetting supplies, typically 20 μl, 1 μl pipettors, 5–300 μl 8- or 12- channel multipipettor (or similar), multichannel aspirating tool, sterile reagent reservoirs.
Imaging medium for cells, preferably without phenol red, riboflavin and folic acid, to reduce background fluorescence (e.g. Fluorobrite from Invitrogen, or custom media formulations that are available from a variety of vendors)
Wide-field epifluorescence microscope equipped with a digital camera, automated stage and capture controls, and an environmental control chamber for samples. E.g. Nikon Eclipse Ti with NIS Elements control software, Andor Zyla 5.5 camera, Lumencor SOLA light source, and OKO Labs stage-top incubator.
Prepare imaging culture plate
-
1
Coat each well of the glass-bottom plate with the substrate solution, using enough solution to cover the desired region. For short-term experiments, simply coat the entire well. For long-term experiments (>24 hours), it is preferable to increase the ratio of media volume to cells by using a square well 96-well plate or a 24-well plate and applying only a 3 μl “spot” of the substrate solution in the center of each well. This strategy creates a region of cells at a density suitable for imaging, while reducing the total number of cells and the depletion of medium factors need for growth. Cover and incubate at least 30 minutes.
The substrate is intended to promote cell adhesion, which is often poor on bare glass, and may also provide substrate-specific interactions that modify cell signaling and phenotype. Choose substrates as desired for the particular cell type and experiment. When using a substrate spot to limit cell adhesion area, sparse adhesion outside of the spot is of no concern. However, if the cells used grow robustly on bare glass, the remaining area may be treated to repel them (e.g. with bovine serum albumin), or non-adhered cells may be carefully washed out after step 6. In larger wells (e.g. 24-well), a larger spot may be used, but 3 μl typically provides more than enough area for multiple frames viewed at 20X magnification. -
2
Aspirate any remaining substrate solution using a Pasteur pipette or aspirating tool. Wash gently with sterile PBS, and aspirate. Avoid touching the plate with pipette tips, as it will mar the coated substrate.
-
3
Obtain a suspension of the desired cells by treating with trypsin and passaging as usual. Count cell concentration by hemocytometer or automated cell counter. Cells will settle; remix suspension prior to use.
-
4
Add the desired number of cells to each well. Determining a suitable cell count may require experimentation. For medium density of MCF-10A cells, 15,000 cells per well is sufficient for 96-well plate. If only a “spot” was coated in step 1, add 5,000 cells in 10 μl media pipetted directly onto the substrate spot to yield near confluence. It is recommended to leave a well empty of cells to provide a convenient background measurement during imaging.
When seeding cells onto a substrate spot, the goal is to cover the entire spot without allowing the cell suspension to contact the wall, as it will wick away from the center. -
5
Add PBS or sterile water to spaces between wells to provide humidity and prevent drying of the cell suspension. Cover the plate and incubate for 2 to 2.5 hours.
-
6
Observe with an inverted microscope to determine if cells have adhered. Elongated cells and small processes indicate adhesion and that more media may be added. If cells still appear spherical, incubate for an additional hour.
-
7
When cells are adhered, gently add media (e.g. 100 μl for 96-well, 500 μl for 24 well). Pipet slowly and avoid contacting the adhered cells to minimize disturbance. Incubate overnight to allow cells to fully attach and acclimate.
If using a substrate spot and the cells in use grow well enough on bare glass to compete for media nutrients, the unadhered cells may be washed off before adding media. Determining the best timing to wash may require experimentation to balance retention of the desired cells on the substrate vs. those over bare glass. -
8
Check cells with an inverted microscope to verify attachment and appearance. When cells appear fully attached and healthy, experimental treatments may be started. Switch to an imaging media, if available, well before imaging (4 hours or more) to avoid observing transient perturbations from the media change.
For optimal fluorescent imaging, it is recommended to use an imaging media lacking or reduced in factors that autofluoresce in the spectrum of interest. Such media can significantly lower background intensity and improve the ability to resolve faint expression and to quantify spectral shifts in reporters (e.g. FRET). Phenol red, riboflavin, folic acid, and serum are often the primary contributors to fluorescence background.
Prepare microscope and imaging procedure
-
9
Prior to desired start of imaging, power on the microscope and environmental controller. Set environment temperature and gas mixture and allow time for the system to stabilize (time required will vary over different hardware).
-
10
When the environmental controller is stable at the set point, transfer the plate to the microscope stage and mount it securely. Any flexibility in the mounting often results in frame shifts during imaging, especially when any treatments are added.
-
11
Start the microscope control software, focus on cells, and set desired acquisition parameters in the software (i.e. filters to use, plate positions to image, acquisition interval, total number of time points, etc.). See Support Protocol 1 for instructions and recommendations specific to NIS Elements control software.
All mechanical movements require time, so it is beneficial to minimize movement distances by choosing plate positions in an order that always moves to an adjacent well and minimizes the total travel. Similarly, selecting filters in the order they are physically positioned limits the movement from one to the next.When choosing excitation intensity and exposure time, there are several factors to consider: 1) maximum image intensities should be well below the saturation point of the camera; 2) the intensities of different channels should be kept similar for later image processing steps; 3) cellular phototoxicity can often be reduced by using lower intensity excitation for longer exposure times.
Perform imaging experiment
-
12
When all imaging parameters are set, begin imaging. It is recommended to review a trial run of all filters at all plate positions to verify that the views are suitable.
-
13
If timed treatments are required, pause imaging after completion of all plate positions, prior to the start of the next time point. Apply treatments (e.g. spike-in of a drug, or washing and replacement with a new media) as quickly as possible while being careful to avoid disturbing or contaminating the plate.
Contacting the plate directly often results in minor image frame shifts even when it is firmly secured. Shifts can be troublesome for image processing, but software implementations of image registration can correct minor cases.A spike solution containing the treatment at 20-fold the final concentration diluted in imaging medium allows for easy pipetting with a minimal (5%) change in the culture volume. Direct addition of solutions with different viscosity than the imaging medium (such as DMSO or glycerol) should be avoided due to the potential for poor mixing. The disturbance of the imaging medium upon treatment can also stimulate responses in signaling pathways due to redistribution of localized signaling molecules or changes in fluorescence readings due to local photobleaching of imaging medium; vehicle-only treatment is an important control -
14
After all imaging is complete, copy the image file(s) to a suitable location for processing, and retain a backup copy in a separate location.
Basic protocol 3: Processing for multi-channel time-series images
This protocol describes the procedure and algorithms for automated processing of time-lapse images to extract single cell data. As computation procedures often vary widely among implementations, we describe here the concepts of the procedure and provide notes regarding specific software implementations using MATLAB. Image processing procedures are complex with multiple layers of subroutines, so this protocol should be reviewed carefully to understand the full process prior to developing or implementing software. A detailed implementation for MATLAB is available in (Pargett et al., 2017). Alternative software solutions that perform the same functions are equally appropriate; for example, CellProfiler provides a publicly available solution with many of the features described here in a graphical user interface (Carpenter et al., 2006).
Materials
Computer with at least 4 GB RAM
Scripting software (e.g. MATLAB)
(Optional) Bio-Formats software (http://www.openmicroscopy.org/site/products/bio-formats)
Particle tracking software, e.g. uTrack (Jaqaman et al., 2008)
Preparation of an image processing pipeline
-
1
Install all necessary software per distributor instructions (e.g. MATLAB, Python, etc.). The Bio-Formats library is useful for reading images from proprietary microscopy files. Particle tracking software (such as uTrack) is used to track cells over time.
-
2
Make images available to the processing computer by mounting on a network drive or copying the image data file(s) locally.
As accessing network storage is often relatively slow, it is preferable to copy the data to the processing computer ahead of time. -
3
Set up access to microscopy files, or extract data to image files (e.g. tiff). If using Bio-Formats, review the documentation (http://www.openmicroscopy.org/site/support/bio-formats5.4/) for methods to access imagery data from different microscopy file types using the chosen scripting software. Microscopy data or image files will be read one frame at a time, each followed by the processing below.
It is convenient to load all color channels for each frame. If these are exported to different tiff files, all channels will need to be loaded at once (e.g. into a Xpixels-by-Ypixels-by-Channels matrix).
Correct images for known biases and background intensity
-
4
(Optional) Correct non-uniform intensity via a recorded image of a uniform field. Scale each pixel intensity by dividing by the relative “intensity” it receives from a uniform field, with the highest intensity pixel valued at 1. With a recorded image of a uniform field, simply divide by the maximum value to give the pixel scalings. It is prudent to filter (i.e. smooth) noise or remove outlier pixels first.
-
5
Identify background intensity in each color channel and subtract from images. To get intensity values, define the well and region to sample for background, load the relevant image(s) if necessary and take the mean value of the selected pixels for each color channel.
Ideally, a background value should be determined for each time point. Retaining and imaging a well with media and treatment compounds, but no cells, provides a convenient reference for background. In this case, it is more efficient to evaluate and store background values prior to processing other frames. Without a designated well, a cell-free region from the current or other wells may be used.
Segment images to estimate nuclear regions
-
6
Working with the channel containing the nuclear marker, reduce noise for segmentation by filtering (i.e. smoothing). A Gaussian filter sized to cover approximate 1/10th of a cell nucleus is typically sufficient.
-
7
(Optional) If the only reliable marker in the cells is a nuclear-excluded cytoplasmic marker, use a gradient magnitude filter to highlight “edges”, i.e. where the image intensity changes quickly, targeting the border between the bright cytoplasm and dark nucleus. Invert the resulting image intensities by subtracting it from its maximum value (this makes the cytoplasm dark and both the nuclei and the background bright, which aligns with the following procedures).
Gradient magnitude filtering may be carried out by applying a Sobel filter in both the X and Y directions and taking the L2 norm of the two filtered images (square root of sum of squares). -
8
Segment nuclei by selecting regions that exceed a threshold value and filtering for shape and size. To be robust to varied expression levels, consider multiple thresholds spanning the range of intensities in the image. Using 20 thresholds linearly spaced between the 5th and 95th percentile intensity values has been successful and robust over a variety of imaging conditions and cell types.
Binary morphological operations and analyses are commonly included in packages focused on image analysis. For example, MATLAB includes the functionregionpropsthat calculates metrics for each region of a binary image (e.g. area, perimeter, etc.) -
9
Remove “salt and pepper” noise in the binary image by morphological dilation and erosion. Both dark and light spots can be removed by first dilating by 1 pixel, eroding by 2 pixels, and re-dilating by 1 pixel. Dilation and erosion sizes may be scaled up to suit the application, e.g. when desired regions (nuclei) are larger.
-
10
Identify each distinct binary feature and select only those matching the feasible size and shape of a cell nucleus. For size, evaluate the total area, and for shape, use the squared ratio of the ideal perimeter of a circle (with the measured area) to the actual perimeter. Store passing features by setting corresponding pixels in a logical mask to TRUE.
A feature found again for the next threshold will be the same size or larger, and will simply overwrite the logical mask.
Create representative masks over the nucleus and cytoplasm of each segmented cell
-
11
Create nuclear masks. The logical mask from segmentation may be used directly for nuclear masks. If appropriate, regions may be eroded to confine masks further within nuclei to ensure robust sampling. Such approaches may be adjusted based on performance for a particular cell type.
-
12
Create cytoplasmic masks by dilating the nuclear mask to extend over a portion of the (presumed) cytoplasm region. From that expanded mask, remove a smaller dilation of the nuclear mask, making a hole over the nuclear region. The size of dilations may be scaled based on the expected nucleus size.
-
13
(Optional) Remove pixels in the cytoplasmic mask that could be associated with more than one nucleus. If using label matrices as masks (each nucleus or cytoplasm is represented by regions of a particular integer value), perform cytoplasm mask construction again with inverted (negative) label values. In grayscale morphological operations, this will give dilations that overlap in the opposite manner. Invert the resulting mask, then set to zero (i.e. background) all pixels with values that differ between the two masks.
-
14
Extract channel data by taking the average over each mask for each color channel. Optionally, to guard against outliers, exclude a maximum and minimum fraction of the intensity values within each mask. Robust performance can be achieved using only the values between the 20th and 80th percentiles, provided masks are large enough (hundreds of pixels). Store values in a matrix alongside X- and Y-coordinate values for the centroids of each nucleus, one row for each cell.
Track cells over time
After all images have been processed via steps 4–14, the cells may be tracked across frames using the X and Y coordinate data stored in step 14.
-
15
Track cells over time using the centroid coordinates and a particle tracking algorithm (or similar software). uTrack is a suitable software for the MATLAB environment, with a usage example available in (Pargett et al., 2017).
-
16
Align track coordinates with segmentation centroid coordinates. The values extracted in step 14 need to be rearranged (within each time point) to provide coherent cell tracks. Find the nearest centroid coordinate to each tracked coordinate, and rearrange the order of cells (e.g. the row entries in a cells-by-time-by-channel data matrix) for each time point to create a dataset where each track is a single row.
Basic protocol 4: Paired signal analysis
This protocol provides a framework and describes a variety of analyses that may be valuable for paired time-series data. Analyses comprise quality control, correlation metrics, and modeling. These procedures are independent and may be included as desired, though initial quality control is highly recommended. All detailed computational procedures are presented for use with MATLAB, but any such scripting software is suitable with some variation in the availability and features of packages. See Anticipated Results for discussion of the outputs from these analyses.
Materials
Computer with at least 4 GB RAM
Scripting software, e.g. MATLAB
Quality control
Preliminary processing to ensure the quality of tracks is critical for the accuracy of all further analyses. Quality control corrects for the errors that arise in automated image processing, as well as the occasional outlier behavior from the biological system.
-
1
Correct for gaps in tracking. Tracking software will typically allow for some gaps in the data (e.g. linking points that appear in frames 3 and 5, but not 4), but many signal analyses require the track to be complete. Choose a maximum gap size that is acceptable, and fill any gaps up to that size by interpolating between the data on either side. If multiple track segments remain that are separated by oversized gaps, keep the longest of these tracks.
It is recommended to choose a maximum gap size according to the expected rate of change for the signals at hand, relative to the imaging interval. If the signal changes rapidly, and especially if it may be pulsatile or oscillatory, a shorter gap size will be necessary to ensure that relevant activity is not missed. Gaps are expected even with effective image processing, as cells may transiently change shape such that they no longer fit the filters, and passing debris in the media may occlude the view of some cells for a short time. -
2
Filter tracks for desired length by completely removing any tracks that are shorter than a desired threshold.
Choose a minimum track length that is suitable for the desired analyses (e.g. based on the duration of the expected activity). When a timed stimulation or drug treatment has been applied early in the experiment, it may be convenient to remove tracks that do not start at the beginning of imaging (the first time point). -
3
Calculate any derived signals, such as normalized values of translocation reporters (ratio of cytoplasmic to nuclear signal) or bi-color reporters like FRET where the values of two color channels are divided. This calculation may be performed at any point up to here, but it can be important to have complete for the following filtering step.
-
4
Filter data for consistency by removing tracks that contain grossly outlier behavior. Examples of outlier behavior include values outside of a reasonable range, and changes that occur too quickly (i.e. sudden shifts in value, evaluated as derivative magnitudes beyond a bound). Bounds need to be determined with care based on the system being measured and imaging interval. To avoid biasing data, it is not recommended at this stage to reject outliers based solely on a statistical basis (e.g. lying far from a normal distribution).
The primary aim on this quality-filtering step is to limit any remaining errors from image processing (e.g. poor segmentation of the nucleus vs. cytoplasm or background, or misaligned tracking due excess motion of nearby cells). Reasonable bounds should be chosen based on the combination of the biochemical process and underlying signal being recorded, and should err on the side of keeping noisy data. For example, both FRET and translocation reporters are automatically normalized with a distinct range of signal value for any given reporter; if typical ratios fall in the range of 0.5 (when inhibited) and 2 (under saturating stimulation), tracks with many values over 4 could safely be excluded. Note that this, and all, filtering steps should be reported in publication.
Direct signal comparisons
Because each cell contains multiple reporters, direct comparisons may be made between the reporters in each cell. These comparisons are essentially forms of correlation, some of which accommodate non-linearity or dynamic lag.
-
5
Compute correlation coefficients for each track. The most common correlation coefficient, Pearson’s rho, specifically evaluates linear relationships, while Spearman’s tau does not diminish for non-linear relationships. Both are typically implemented in basic and/or statistics packages for scripting software.
-
6
Compute mutual information for each track. Mutual information is the most general form of “correlation”, and evaluates how much knowing one signal’s value restricts possible values of the other. It is implemented in some statistical packages for scripting software, though less widely than basic correlation coefficients.
Computation of mutual information relies on binning data to make a histogram, or on summing up nearest neighbors. As such, it is particularly important to have many data points for a robust estimate of mutual information. -
7
Compute cross-correlation for each track. Cross-correlation is equivalent to correlation, but allowing for a dynamic lag between signals. Properly normalized, it represents the linear correlation coefficient evaluated for each possible time lag (i.e. shifting one signal backward in time and calculating correlation, neglecting points that no longer overlap at the ends). Typically, the critical values to record are the peak (maximum) correlation value and the lag at which it occurs. Cross-correlation is typically included in signal processing packages.
Signal response modeling
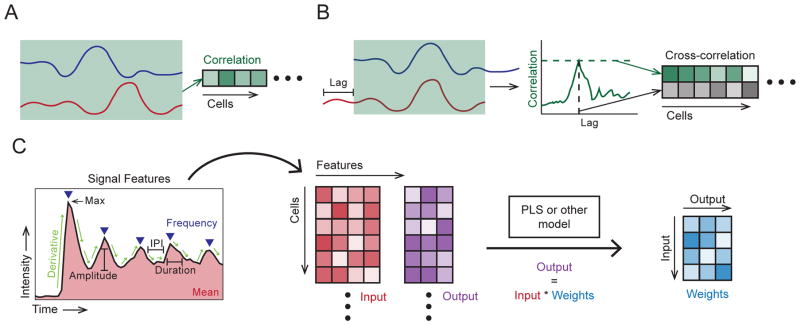
Signal response models may be constructed to estimate the specific relationship between signals across a population of cells. Each pair of input and output signals from individual cells becomes one observation, and the mean relationship is estimated over all observations. The time-series signals are broken down into a smaller number of “characteristics” to comprise an observation. Partial least squares modeling (PLS) is particularly useful as it extracts the most informative features from the covariance structure, allowing many input and output characteristics to be used simultaneously.
-
8
Determine a suitable set of characteristics to describe the single cell tracks. Basic statistics, such as mean value, variance, maximum and minimum values, etc. are likely to be informative. Signals that fluctuate significantly may be informed by mean (and/or max and min) rates of change. Signals with pulsatile or oscillatory behavior may require evaluation of mean (and/or max, min) frequency and amplitude. It is often useful to apply some degree of smoothing when calculating maximum and minimum values, or rates of change, as these are highly sensitive to noise.
There are no hard limitations on what decompositions are applicable. If convenient for the data and further desired analyses, rigorous mathematical expansions may be applied, such as the Fourier Transform or an orthogonal basis decomposition. Such expansions offer a relatively unbiased breakdown of the signal, but are often more difficult to interpret conceptually. -
9
Calculate the characteristic values for every track. Each track will result in an observation vector containing the values for each of the characteristics, for each of the signals recorded.
-
10
Infer models using one or more characteristics of two selected signals as input and output. Any model fitting routine is applicable, and the selection and application of such a scheme is beyond this protocol. However, it is highly recommended to use PLS regression as it can simultaneously consider all of the characteristics in the input and/or the output.
PLS regression and similar procedures are fundamentally linear problems with each of the terms considered being one of the signal characteristics. However, any non-linear combinations of characteristics may be added as additional terms when desired, e.g. x^2 or e^(x*y).
Support Protocol 1: Imaging with NIS Elements
In NIS Elements, open the ND Acquisition panel.
Set path and filename for saving data. Data are automatically saved when experiment is complete.
In the Time tab, select the imaging interval and total imaging duration.
In the Wavelength tab, select the desired fluorescent channels for the experiment.
Navigate to the first well in the imaging sequence and find cells. Turn on “Perfect Focus” and set the appropriate focal plane using the focus wheel.
Open the Zyla Settings panel and set the exposure for each channel to be used in imaging experiment.
Using the joystick, navigate to a suitable area for imaging; try to avoid areas where cells are overcrowded.
In the ND Acquisition panel select the XY tab. Navigate to the first desired ROI using the joystick and select the XY point by checking the box in the XY selection tab.
Continue selecting ROIs in each desired well by navigating to each ROI and selecting the coordinate using the checkbox. Adjust focal plane at each ROI as necessary using Perfect Focus.
To select a background ROI (optional), navigate to an XY position that contains no cells in the field of view. This will be used later to calculate background during image processing.
When all ROIs have been selected return to first ROI and check that focus and exposure are set correctly and that the plate has not shifted.
Start imaging experiment by hitting the “Run Now” button.
To add treatments during imaging, pause imaging using the “Pause” button on the Acquisition Window. Then, remove plate lid and environmental control box, add treatments to each well, minimize contact with plate. Return lid and environmental control box to plate, and resume imaging using the “Resume” button on the Acquisition Window.
Once the time course has completed, the file will be saved in the location specified during imaging set up. Alternatively, terminate the image acquisition using the “Finish” button.
When finished, close NIS Elements and shut down the microscope.
Reagents and Solutions
Collagen coating solution: 20 μg/ml rat tail type I collagen (e.g ThermoFisher A1048301) in 0.1 N acetic acid. Sterilize acetic acid and water by filtration through 0.2 μm filter before adding irradiated collagen; do not filter after adding collagen. Store at 4° C.
Laminin coating solution: 50 mg/mL laminin 111 (ThermoFisher 23017015) in 20 mM sodium acetate (pH 4), containing 1 mM CaCl2. Prepare fresh for each experiment.
Commentary
Background
Live-cell studies of signal transduction have revealed kinetic aspects of signaling that are invisible to both population-based measurements (such as immunoblots) and static single-cell methods (such as immunofluorescence). For example, live-cell reporters of the p53 tumor suppressor protein have revealed that this key regulator of DNA repair is activated in discrete hour-long bursts following certain forms of DNA damage, with cells varying in both the spacing and number of bursts (Batchelor et al., 2011; Lahav et al., 2004). Kinetics observed in single cells are often incompatible with existing models for signaling pathways, prompting significant revisions to long-standing models (Batchelor et al., 2008). However, to move beyond simple description, the relationship of the single-cell kinetics for a given molecule to its upstream and downstream processes must be established, and this task typically requires that the variables of interest be imaged simultaneously in the same cell. In the case of p53, simultaneous imaging of p53 activity and the resolution of DNA damage (upstream) or activation of the target gene p21 (downstream) was necessary to understand the importance of p53 kinetics in encoding responses to different types of DNA damage (Loewer et al., 2013; Purvis et al., 2012). Similar examples can be found in multiplexed single-cell studies of death receptor-induced apoptosis (Albeck et al., 2008), EGFR/Ras/ERK-induced proliferation (Albeck et al., 2013; Shankaran et al., 2009), and NF-kB-mediated gene induction (Nelson et al., 2004).
Single-cell experiments have revealed that “noise” plays a significant role in determining the signaling response of a given pathway to a stimulus, resulting in a distribution of responses for a population of cells responding to a uniform stimulus (Cheong et al., 2011; Raser and O’Shea, 2005). While this variation may result from stochastic effects in the biochemical process of signaling, the observed variation in any one pathway can also result from deterministic but unmeasured cell-to-cell variation in other pathways and cellular processes (Selimkhanov et al., 2014). Multiparameter analysis as presented here can be useful in identifying the sources of such variation by making it possible to quantify the amount of variance in one pathway that can be explained by measured variance in a second (or further additional) pathways. When choosing reporters for multiparameter analysis, it is important to perform a careful literature search to take into account known dependencies (or “crosstalk”) between the pathways of interest and consider how these will influence the quality of the data collected.
Critical Parameters
Throughout the processes of establishing, imaging and analyzing a multiplexed reporter cell line, it is important to double check the status and quality at every opportunity. In cell line construction, this comprises evaluating the cells after each reporter insertion, sorting/selection, and/or cloning step. Verify that cells retain the phenotype and any characteristic behavior of the parental line and that reporter expression levels are suitable for measurement, but not too high (excess expression can both perturb the cell itself and interfere with the measurement of other reporters). Additionally, be certain to perform the experiments described (Basic Protocol 1, step 11) to validate the function of the reporters.
When setting up the microscopy experiment, take care to choose frame positions with minimal obstructions (floating debris, distortions from the substrate, cells overlaying each over, etc.), and carefully double check the focus. Ensure that a background region is available (see Basic Protocol 3, step 5). After starting imaging, it is recommended to observe at least one frame of each position to ensure that it meets expectations. When adding a treatment during imaging (Basic Protocol 2, step 13), be especially cautious not to disturb the plate, and observe the first frames taken after restarting imaging to verify that they align with the previous frames. If the plate was disturbed and a large shift occurred, it is possible to realign it (to some degree) with minimal loss of data by carefully pressing the direction needed and checking the view (select a notable feature in the desired view as a point of reference).
Prior to processing images (Basic Protocol 3), ensure that the parameters are appropriate for the current experiment. Especially after multiple experiments have been processed, previously used parameters can be overlooked and give sub-optimal results. In particular, check that the limits on nucleus size (see step 8) are suitable for the cell type and the magnification that was used. Similarly, an inappropriate background can degrade processing. If the background is to be taken from frames containing cells, carefully ensure that neither cells nor fluorescent materials are present in the background regions. Accurate background removal is particularly important when using ratiometric measures, such as FRET or translocation reporters.
While a well-constructed image processing and quality control pipeline enable high-throughput analysis, it is critical to review the data at different points. Review the raw imagery for microscopy issues, such as out-of-focus frames, misaligned filters, or obstructions in some frames. After processing, visualize tracks in bulk to ensure both that a reasonable number were generated and that overall quality is acceptable. Having many jagged or extremely noisy tracks is suggestive of a processing error. Review tracks again after quality control (Basic Protocol 4, steps 1–4) to ensure that final data are high quality. In such review, it is best to view all individual tracks, as well as averages, to ensure all outliers have been addressed. Individual tracks may be viewed simultaneously by plotting them overlaid, as grossly aberrant tracks will remain apparent. If the data are used for modeling (steps 8–10), it is important to carefully consider how the models are interpreted, as with any modeling endeavor. In particular, it is essential to evaluate for a parsimonious model, i.e. the simplest model that satisfies the data available (Burnham and Anderson, 2003).
Anticipated results
Each imaging experiment in which multiple reporters are measured (Basic Protocol 2) will result in a series of digital images corresponding to each time point, fluorescence channel, and microscope stage position. Software such as ImageJ, MATLAB, or packages from the microscope manufacturer can be used to view each recorded image series. Processing of the raw images using Basic Protocol 3 will result in a dataset of 10s to 1,000s of individual cell “tracks” per stage position, with each track consisting of time-course measurements of all measured reporters for an individual cell. Heatmap visualizations are often helpful in quickly obtaining an overview of the full dataset and identifying major trends within each reporter (Fig. 1). It is also helpful to display a line plot of the mean or median for each of the measured reporters, along with lines corresponding to 25th and 75th percentile values for the population; overlaying line plots of individual cells on top of the mean can also be of use in determining the extent to which individual cells depart from the average population behavior. Visualizing the relationship between reporter measurements within each cell is also critical but can be more tedious, as it is difficult to meaningfully display more than one cell’s multiparameter data on a single 2-dimensional axis, and it is often necessary to adjust the axes limits or normalization to obtain the best visualization for each cell. An appropriate strategy is to combine visual inspection of at least 20 individual cells with the multiparameter analyses described in Basic Protocol 4 for each condition.
The paired signal analyses described in Basic Protocol 4 will result in scalar values that allow for quantitation of the relationship between the reporters analyzed. Correlation analysis, for example, will produce a single value between 0 and 1 for each cell analyzed, with higher numbers indicating increasing similarity between the reporter signals. Cross-correlation analysis will produce two values for each cell, one indicating the lag time between signals and one indicating correlation value (between 0 and 1). Mutual information will also produce a single positive value for each cell, with higher values indicating an increased level of dependence between the reporter signals analyzed. In the case of signal response modeling, each dataset modeled will produce a series of coefficients that indicate the weight with which each considered parameter contributes to the overall response, with negative values indicating an opposing influence and positive values a supporting influence. Interpretation of the biological significance of these values will depend on the particular experimental context. It may be appropriate to compare, for example, the average cross correlation for all cells in one condition in which two reporters are expected to behave similarly (such as when both pathways are induced by the same stimulus) to the same measurement for all cells in a condition in which the reporters are expected to behave differently (such as when one pathway is stimulated and the other is blocked by an inhibitor). Alternatively, it may be useful to compare the calculated values for one pair of reporters to similar measures in a different pair of reporters, such as a control fluorescent proteins that are not expected to vary specifically in response to similar stimuli. In the absence of either different conditions or different reporters that can serve as a point of reference, reporter signals from each cell in the dataset can be “scrambled” such that reporter signals are mismatched; this calculation can be used as a point of comparison to determine whether correlations in the original dataset are meaningful.
Independent replicates of experiments are also critical for establishing reproducibility and statistical significance for comparisons between conditions. It is not recommended to perform a standard t-test between conditions using each cell as an independent measurement, as the large number of cellular measurements collected will result in very low p values and statistically “significant” differences even between replicates of the same condition. It is more appropriate to perform a t-test on measurements of the average correlation (or other metrics) value for each conditions performed as experimental replicates on different days. As always, if more than two conditions are being compared, a t-test alone is inadequate and the family-wise error rate (or false discovery rate) must be considered, e.g. using the Bonferroni correction, Benjamini-Hochberg procedure, or a suitable statistical post-test following ANOVA.
Time Considerations
Several aspects of establishing successful live-cell experiments require long-term time investment. The most time-consuming operations are in producing the necessary DNA constructs and cell lines, which can require weeks to months. Naturally, establishing the computational pipeline for processing image data may also require significant time and troubleshooting, and should be started early. Once in place, image processing should consume only hours to days, depending on processor speeds and availability of parallel computing (via desktop machines or clusters).
Some of the experimental operations can be performed in parallel to avoid unnecessary delays, especially the preparation of different DNA constructs, and the (co-)transfection of cells for reporter insertion, when feasible. However, for cell lines or techniques with low insertion rates (such as targeted genomic insertions via CRISPR), transfection is more efficiently done in series, selecting for positive cells between insertions. Establishing a clonal population of reporter-bearing cells can require an additional month; evaluate the trade-off between the extra time invested and having uniform genotype and reporter expression. In practice, it is productive to begin performing and refining imaging experiments as soon as cells bearing a reporter are available, rather than waiting for the perfect cell line to be complete.
Figure 3.
Examples of paired signal analyses. A. Correlation, computed for each cell, yielding a vector of coefficients. B. Cross-correlation, computed from the correlation of shifted (lagged) traces. For each cell, the peak correlation and corresponding lag are stored, yielding a vector for each. C. Signal response modeling example, using a feature decomposition (left) extracting the: mean value, max value, frequency, amplitudes, derivatives, pulse durations and inter-pulse intervals (IPI). Mean and/or maximum values of the latter four features would be stored (center). Stored features are used to fit model weights (right, e.g. via PLS), so that output values may be estimated by these weights and the input values.
Significance statement.
Live-cell reporters have become an increasingly important tool for the study of intracellular signaling pathways. A reporter can provide data on detailed kinetics and cell-to-cell heterogeneity in signaling that are needed to update and refine models of signal transduction. However, multiple reporters are often essential to link these new views of signaling to other pathways and cellular fate choices. The simultaneous analysis of two or more reporters within the same cell presents a new set of challenges in experimental design, implementation, and data processing. The protocols described here enable multiple signals to be tracked over time and related to each other, providing the data needed to quantify links between signaling pathways in the same cell.
Acknowledgments
Funding for this work was provided by the American Cancer Society (IRG-95-125-13), the National Institute of General Medical Sciences (1R01GM115650), the Department of Defense Neurofibromatosis Research Program (W81XWH-16-1-0085) and the American Association for Cancer Research Stand Up To Cancer (SU2C-AACR-IRG-01-16). Stand Up To Cancer is a program of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. Flow cytometry services were supported by the UC Davis Comprehensive Cancer Center Support Grant (CCSG) awarded by the National Cancer Institute (NCI P30CA093373), and we acknowledge the expert cell sorting assistance of Dr. Bridget McLaughlin and Jonathan Van Dyke.
Footnotes
Conflicts of Interest
The authors have no conflicts of interest to declare.
Internet Resources (annotated)
www.openmicroscopy.org/site/products/bio-formats
Website for Bio-Formats
www.utsouthwestern.edu/labs/danuser/software
Website hosting u-Track software
Website hosting aggregated fluorescent protein properties
Literature Cited
- Albeck JG, Burke JM, Spencer SL, Lauffenburger DA, Sorger PK. Modeling a snap-action, variable-delay switch controlling extrinsic cell death. PLoS Biol. 2008;6:2831–2852. doi: 10.1371/journal.pbio.0060299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albeck JG, Mills GB, Brugge JS. Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol Cell. 2013;49:249–261. doi: 10.1016/j.molcel.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor E, Loewer A, Mock C, Lahav G. Stimulus-dependent dynamics of p53 in single cells. Mol Syst Biol. 2011;7:488. doi: 10.1038/msb.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor E, Mock CS, Bhan I, Loewer A, Lahav G. Recurrent initiation: a mechanism for triggering p53 pulses in response to DNA damage. Mol Cell. 2008;30:277–289. doi: 10.1016/j.molcel.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. Model selection and multimodel inference: a practical information-theoretic approach. Springer Science & Business Media; 2003. [Google Scholar]
- Cadinanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepko C, Pear W. Overview of the retrovirus transduction system. Curr Protoc Mol Biol. 2001;Chapter 9(Unit9):9. doi: 10.1002/0471142727.mb0909s36. [DOI] [PubMed] [Google Scholar]
- Cheong R, Rhee A, Wang CJ, Nemenman I, Levchenko A. Information transduction capacity of noisy biochemical signaling networks. Science. 2011;334:354–358. doi: 10.1126/science.1204553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, Bell JB, Largaespada DA, Hackett PB. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Goedhart J, van Weeren L, Adjobo-Hermans MJ, Elzenaar I, Hink MA, Gadella TW., Jr Quantitative co-expression of proteins at the single cell level--application to a multimeric FRET sensor. PLoS One. 2011;6:e27321. doi: 10.1371/journal.pone.0027321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Jaimovich A, Dey G, Ruggero D, Meyuhas O, Sonenberg N, Meyer T. Parallel measurement of dynamic changes in translation rates in single cells. Nat Methods. 2014;11:86–93. doi: 10.1038/nmeth.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu N, Aoki K, Yamada M, Yukinaga H, Fujita Y, Kamioka Y, Matsuda M. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- Locke JC, Elowitz MB. Using movies to analyse gene circuit dynamics in single cells. Nat Rev Microbiol. 2009;7:383–392. doi: 10.1038/nrmicro2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer A, Karanam K, Mock C, Lahav G. The p53 response in single cells is linearly correlated to the number of DNA breaks without a distinct threshold. BMC Biol. 2013;11:114. doi: 10.1186/1741-7007-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- Nelson G, Paraoan L, Spiller DG, Wilde GJ, Browne MA, Djali PK, Unitt JF, Sullivan E, Floettmann E, White MR. Multi-parameter analysis of the kinetics of NF-kappaB signalling and transcription in single living cells. J Cell Sci. 2002;115:1137–1148. doi: 10.1242/jcs.115.6.1137. [DOI] [PubMed] [Google Scholar]
- Pargett M, Gillies TE, Teragawa CK, Sparta B, Albeck JG. Kinase Signaling Networks. Springer; 2017. Single-Cell Imaging of ERK Signaling Using Fluorescent Biosensors; pp. 35–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin JY, Zhang L, Clift KL, Hulur I, Xiang AP, Ren BZ, Lahn BT. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One. 2010;5:e10611. doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O’Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regot S, Hughey JJ, Bajar BT, Carrasco S, Covert MW. High-sensitivity measurements of multiple kinase activities in live single cells. Cell. 2014;157:1724–1734. doi: 10.1016/j.cell.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selimkhanov J, Taylor B, Yao J, Pilko A, Albeck J, Hoffmann A, Tsimring L, Wollman R. Systems biology. Accurate information transmission through dynamic biochemical signaling networks. Science. 2014;346:1370–1373. doi: 10.1126/science.1254933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaran H, Ippolito DL, Chrisler WB, Resat H, Bollinger N, Opresko LK, Wiley HS. Rapid and sustained nuclear-cytoplasmic ERK oscillations induced by epidermal growth factor. Mol Syst Biol. 2009;5:332. doi: 10.1038/msb.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery SD, Hahn KM. A High-Content Assay for Biosensor Validation and for Examining Stimuli that Affect Biosensor Activity. Curr Protoc Cell Biol. 2014;65:14, 1511–31. doi: 10.1002/0471143030.cb1415s65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Ornstein J, Lahav G. Dynamics of CDKN1A in Single Cells Defined by an Endogenous Fluorescent Tagging Toolkit. Cell Rep. 2016;14:1800–1811. doi: 10.1016/j.celrep.2016.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]