 Recent studies indicated that bisphenol A (BPA) can disrupt spermatogenesis and then cause male infertility.
Recent studies indicated that bisphenol A (BPA) can disrupt spermatogenesis and then cause male infertility.
Abstract
Recent studies indicated that bisphenol A (BPA) can disrupt spermatogenesis and then cause male infertility. The present study revealed that BPA greater than 10–6 M inhibited the proliferation of Leydig TM3 cells via a concentration dependent manner. The proteomic study revealed that 50 proteins were modulated in TM3 cells following exposure to BPA, which was relevant to structure, motility, cell metabolism, protein and nucleotide processing, and cell proliferation. Furthermore, BPA increased the in vitro migration and invasion of Leydig TM3 cells, which might be due to the BPA's modulation of proteins related to cell structure and motility such as actin and heat shock protein (HSP). Silencing of galectin-1, which was up regulated by BPA, significantly abolished the BPA-induced migration of TM3 cells. BPA treatment obviously increased the phosphorylation of ERK1/2 and Akt, while only PD98509 (ERK1/2 inhibitor) significantly attenuated BPA induced up regulation of galectin-1. Furthermore, PD98509 also reversed BPA induced migration of TM3 cells. Our study demonstrated that xenoestrogen BPA at micromolar or greater concentrations can modulate protein profiles, inhibit cell proliferation, and promote the in vitro migration and invasion of Leydig TM3 cells. It provided new insight into the mechanisms responsible for BPA induced male infertility.
1. Introduction
Bisphenol A (BPA), 2,2-bis(4-hydroxyphenyl)propane, is one of the highest volume chemicals produced worldwide.1 It can be easily accumulated in various human tissues such as blood and lipid via food intake or inhalation.2 As a known endocrine disruptor chemical (EDC), multiple studies have indicated that BPA can affect various endocrine related pathways and then cause the origination and development of various diseases such as cancer, obesity, sexual behavior, thyroid function and neurological effects.3 Among these health issues, male infertility caused by BPA is attracting more and more attention. It was demonstrated that BPA can disrupt spermatogenesis and then impair male fertility in animal models.4,5 In vivo studies have documented that prenatal and neonatal exposure of male rats to low doses of BPA cause significant impairments in testicular development and spermatogenesis.6 Furthermore, increasing urine BPA levels were significantly correlated with a decrease of the total count, concentration and vitality of sperm.7,8 However, the exact molecular mechanisms of BPA-induced male infertility were still unclear.
The Leydig cell, located between the seminiferous tubules of the testis, is the major cell type within the interstitium and the principal source for testosterone.9,10 Testosterone secreted by Leydig cells under the stimulus of luteinizing hormone (LH) can not only diffuse into seminiferous tubules and drive spermatogenesis but also inhibit germ cell apoptosis.11 This dependency of the seminiferous epithelium on testosterone illustrates the significance of the Leydig cell in spermatogenesis. Previous studies indicated that estrogen may act in a paracrine fashion in the testis to control Leydig cell development and steroidgenesis.12 Therefore it is reasonable to hypothesize that BPA, an endocrine-disrupting chemical that mimics the hormone estrogen, can modulate the development and function of Leydig cells via the estrogen–estrogen receptor system. Our recent study revealed that nanomolar BPA can significantly stimulate the proliferation of Sertoli cells, which share morphological and functional properties with resident Leydig cells, via activating ERK1/2 through GPR30 and ERα/β.13 However micromolar BPA can inhibit the proliferation of Sertoli cells via elevating the production of reactive oxygen species (ROS).14 Considering that GPR30 and ERα/β have been greatly detected in Leydig cells,15 BPA may modulate the biological effect of Leydig cells via these signal pathways.
There are very limited data about the effects of BPA on the function and proliferation of Leydig cells. Exposure to BPA during pregnancy reduced plasma testosterone at postnatal day 3 in the rat.16 Another study revealed that BPA exposure at less than 50 mg kg–1 day–1 had no effect on the anogenital distance (AGD) in male pups.17 There was no effect of BPA on AGD after a gestational gavage even as high as 50 000 mg kg–1 day–1.18 Therefore, further studies are needed to confirm the role of BPA in the function and proliferation of Leydig cells. The present study revealed that BPA at greater than micromolar concentration significantly inhibited the proliferation of Leydig TM3 cells. The protein profiles of TM3 cells treated with 10–8 M and 10–5 M BPA for 48 h were compared with the control. The results revealed that BPA can promote the in vitro motility of TM3 cells by up regulating galectin-1 (Gal-1). Generally, this study not only found that BPA can suppress the growth and promote migration of TM3 cells, but also provided valuable resources for further study about molecular mechanisms of BPA on spermatogenesis.
2. Materials and methods
2.1. Reagents
All reagents used in two-dimensional electrophoresis (2-DE) were bought from Bio-Rad (Hercules, CA, USA). PD 98059 (PD, ERK1/2 kinase inhibitor) and LY294002 (LY, PI3K/Akt inhibitor) were purchased from Selleck Chemicals (Houston, TX, USA). BPA and other chemicals were purchased from Sigma Chemical Co. (St Louis, MO, USA). Monoclonal antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). The horseradish peroxidase-conjugated secondary antibody was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). All compounds were solubilized in dimethyl sulfoxide (DMSO). A steroid-free medium containing DMSO (0.5% v/v) was used as the control.
2.2. Cell culture
The mouse Leydig cell line TM3 (American Type Culture Collection, CRL-1714) was kindly provided by Prof. Liang Tao (Zhongshan Medical School of Sun Yat-sen University). Cells (passage 12–15) were cultured in phenol red-free Dulbecco's Modified Eagle Medium (DMEM) nutrient mixture F-12 Ham (Sigma-Aldrich, St Louis, MO, USA) containing 5% heat-inactivated fetal bovine serum (FBS, Invitrogen, Carlsbad, CA), 5% heat-inactivated horse serum, and 10 μg ml–1 penicillin–streptomycin at 37 °C under a 5% CO2 atmosphere. Both plastic items used for the experiments and the water used to prepare reagents were pretreated by enhanced sonochemical degradation to reduce any potential background BPA.13
2.3. Cell proliferation assay
The cell proliferation of TM3 exposure to various concentrations of BPA was detected by using the Cell Counting Kit-8 (CCK-8) according to previously described procedures.19 Briefly, cells were seeded in 96-well plates at a cell density of 1 × 104 per well. After treatment with BPA for 24, 48, or 72 h, 10 μl of a CCK-8 solution was added to each well. The plates were incubated for another 2 h, and the absorbance was measured at 450 nm using a microplate reader. The experiments were repeated six times.
2.4. 2-DE, protein visualization and image analysis
The 2-DE, protein visualization and image analysis were conducted according to previously described procedures.20 Control and treated cells were lysed in a lysis buffer for 30 min at 4 °C. Samples were then centrifuged at 12 000 rpm for 30 min at 4 °C. After centrifugation, supernatants were collected, and protein concentrations were determined by the Bio-Rad Dc protein assay. Isoelectric focusing (IEF) was carried out using the Bio-Rad PROTEAN IEF cell (Bio-Rad) and 17 cm Immobiline dry strips with a linear pH gradient of 3–10. Protein samples (1 mg) were loaded during the rehydration step (12 h). IEF was then performed at 20 °C in a stepwise manner: 200 V (1 h), 500 V (1 h), 1 kV (1 h), gradient 8 kV (0.5 h), and finally 8 kV for a total of 48 000 Vhs. Following IEF, the IPG strips were equilibrated for 15 min in equilibration buffer, and then the gel strip was equilibrated once more in the same equilibration buffer containing 2.5% (w/v) iodoacetamide instead of dithiothreitol (DTT). The second dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on 10% acrylamide gels, and electrophoresis was undertaken at 18 °C in Laemmli running buffer (20 mM Tris-HCl, 192 mM glycine, 0.1% SDS) using a PROTEIN II xi Multi-Cell system (Bio-Rad). Separation was carried out at 10 mA per gel for 1 h and 30 mA per gel overnight. After electrophoresis, gels were stained with Coomassie Brilliant Blue (CBB) G-250. To ensure data reliability, sample preparation (both control and treated cells) and 2-DE were performed in triplicate.
2-DE gels were scanned at a 600 dpi resolution with a UMAX Power Look 2100XL scanner (Maxium Technologies, Taipei, China). Image analysis was performed with PDQuest version 7.3 (Bio-Rad, Hercules, CA, USA). The optimized parameters were as follows: saliency 2.0, partial threshold 4, and minimum area 50. The gel images were normalized according to the total quantity in the analysis set. This normalization method provided by PDQuest software divided each spot abundance value by the sum of total spot abundance values to obtain individual relative spot abundances. To study the changes in expression in more detail, patterns for control samples were set as references to be compared qualitatively and quantitatively with those of cells treated with 10–8 M or 10–5 M BPA. Relative comparison of the intensity abundance between control and treated cells (3 replicate samples for each group) was performed using the Student's t test after being checked for normality and homogeneity of variance. Expression intensity larger than 2.0 (p ≤ 0.05) or smaller than 0.5 (p ≤ 0.05) were set as thresholds indicating significant changes.
2.5. In-gel tryptic digestion of proteins and MALDI-TOF-MS/MS analysis
Protein spots were manually excised from CBB-stained gels and transferred to V-bottom 96-well microplates containing 100 μL of 50% acetonitrile/25 mM ammonium bicarbonate solution per well. After being destained for 1 h at 40 °C, gel plugs were dehydrated with 100 μL of 100% acetonitrile for 20 min and then thoroughly dried in a SpeedVac concentrator (Thermo Fisher Scientific, Waltham, MA) for 30 min. The dried gel particles were rehydrated at 4 °C for 45 min with 10 ng sequencing grade modified trypsin (Promega, Madison, WI, USA) dissolved in 25 mM ammonium bicarbonate and then incubated at 37 °C for 12 h. After trypsin digestion, the peptide mixtures were extracted with 8 μL of extraction solution (50% acetonitrile/0.5% trifluoroacetic acid) per well at 37 °C for 1 h. Finally, the extracts were dried under nitrogen gas.
Peptides were eluted with a 0.8 μL matrix solution (α-cyano-4-hydroxy-cinnamic acid in 0.1% trifluoroacetic acid, 50% acetonitrile) before application to the target plate. Samples were allowed to air-dry and analyzed using an Ultraflex II MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). The ultraviolet (UV) laser was operated at a 200 Hz repetition rate with a wavelength of 355 nm. The accelerated voltage was operated at 25 kV. MS spectra were acquired over a mass range of 800–5000 m/z. Peptide Calibration Standard II (Bruker Daltonics) was used to calibrate the mass instrument with internal calibration mode. From the peptide mass fingerprinting (PMF) for each spot the 10 largest peaks, with a signal-to-noise threshold >30, were automatically selected for MS/MS fragmentation in LIFT mode without addition of a collision gas. Flex Analysis software (version 3.3, Bruker Daltonics) was used to perform the spectral processing and peak list generation for both mass (MS) and MS/MS spectra. Processed peak lists were subjected to Mascot (version 2.3, Matrix Science, London, U.K.) via Biotools 3.1 (Bruker Daltonics) and searched using the following parameters: NCBInr database, taxonomy of Homo sapiens (human), trypsin of the digestion enzyme, one missed cleavage site, carbamidomethylation (C) as a fixed modification and oxidation (M) as a variable modification, MS tolerance of 100 ppm, MS/MS tolerance of 0.6 Da, and the significance threshold was set at p < 0.05.
2.6. Western blot analysis
Western blot analysis was performed as previously described.21 Briefly, cells were lysed in cell lysis buffer, and then lysates were cleared by centrifugation and denatured by boiling in Laemmli buffer. Protein concentration was measured using the Bio-Rad protein assay kit. Approximately 50 μg proteins were separated on 10% SDS-PAGE and electrophoretically transferred to nitrocellulose membranes. Following blocking with 5% non-fat milk at room temperature for 2 h, the membranes were washed three times with PBS, incubated with the primary antibody at 1 : 1000 dilution overnight at 4 °C and then incubated with a horseradish peroxidase-conjugated secondary antibody at 1 : 5000 dilution for 2 h at room temperature, and detected with the Western Lightning Chemiluminescent detection reagent (Perkin-Elmer Life Sciences, Wellesley, MA). The results of densitometric analyses of western blots, obtained using ImageJ software, were presented as the relative optical density (%) to the control (GAPDH).
2.7. RNA interference
For RNA interference, TM3 cells were transfected with a 100 pmol siRNA oligomer mixed with the lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) in serum reduced medium according to the manufacturer's instructions. The target sequences for Gal-1 siRNAs (si-Gal-1) were 5′-UGAUGCACACCUCUGCAACACUUCC-3′; si-S100A4 5′-GAG GAA AGA CTA CAG TCC AAG-3′, si-γ-actin 5′-AAG AGA TCG CCG CGC TGG TCA-3′, si-Prof-1 5′-GGA AUU UAG CAU GGA UCU U-3′, negative control siRNA (si-NC) 5′-CAG CUU UGG CUG AGC GUA U-3′. All the siRNA products were obtained from Ambion (Austin, TX, USA).
2.8. Wound healing and transwell migration/invasion assay
For the in vitro wound healing assay, confluent monolayers (60–70%) of TM3 cells were scratched by the use of a 100 μl tip followed by the addition of BPA at different times. The closure of the scratch was analyzed under a microscope and images were captured after incubation for the indicated times (0–72 h). Migration and invasion assays were performed in Boyden chambers according to the previous studies.22 Briefly, migration and invasion assays were performed in Boyden chambers (8 μm pore size, Corning). A Matrigel matrix (20 μg, BD Biosciences) was used for the invasion assay, and uncoated filters were used for the migration assay. TM3 cells treated with or without BPA were added to transwell chambers. 10% charcoal-stripped (CS)-FCS was added to the bottom wells of the chambers to induce cell migration and invasion. After incubation for the indicated times (0–72 h), cells that had migrated and invaded through the membrane were stained with a 0.5% methylrosaniline chloride solution and counted under an upright microscope (5 fields per chamber). Each migration and invasion assay was repeated in three independent experiments.
2.9. Statistical analysis
All values were reported as mean ± SD of three independent experiments unless otherwise specified. Data were analyzed by two-tailed unpaired Student's t-test between two groups and by One-Way ANOVA followed by the Bonferroni test for the multiple comparisons involved. The statistical analyses were performed using SPSS 13.0 for Windows. A p-value of <0.05 was considered to be statistically significant.
3. Results
3.1. Effects of BPA on cell proliferation
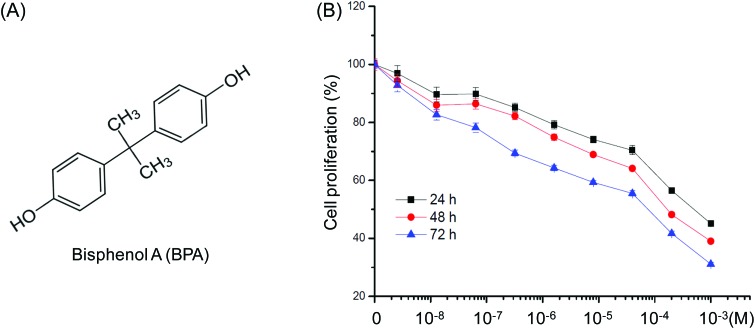
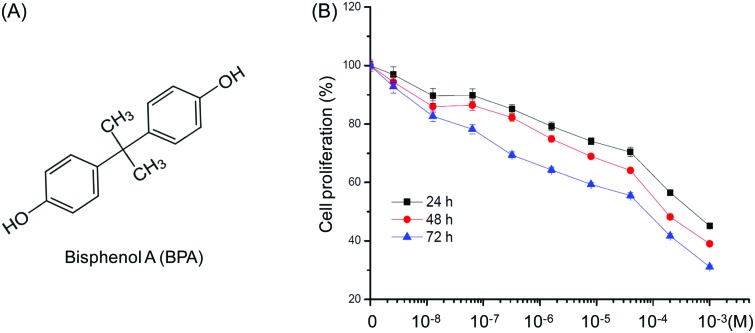
When treated with concentrations of BPA (Fig. 1A) ranging from 10–9 to 10–3 M for 24, 48 and 72 h, we found that BPA greater than 10–6 M obviously inhibited proliferation of TM3 cells after exposure for 48 h. The IC50 values of BPA to TM3 cells were 7.7 × 10–4 M, 2.3 × 10–4 M, and 4.0 × 10–5 M for 24, 48 h, and 72 h, respectively. Therefore, 10–8 M (limited effects) and 10–5 M (significant inhibition effects) were chosen for further proteomic studies.
Fig. 1. Effects of increasing concentrations of BPA on proliferation of Leydig TM3 cells. (A) Chemical structure of BPA. (B) Cells were treated with various concentrations (10–8 to 10–3 M) of BPA for 24, 48, and 72 h, and then cell proliferation was assessed by the CCK8 kit assay. Data were presented as means ± SD of three independent experiments.
3.2. BPA-induced proteome alterations in Leydig TM3 cells
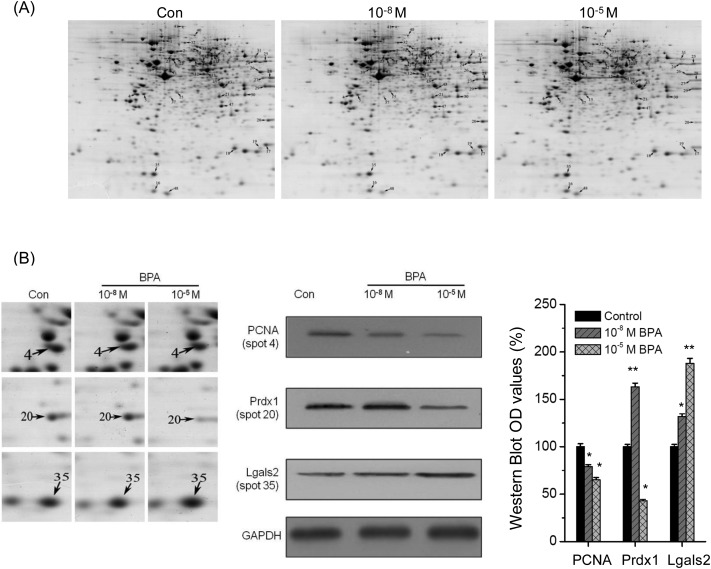
The protein profiles in Leydig TM3 cells treated with BPA were analyzed by MALDI-TOF-MS/MS. The representative two-dimensional gel images of control, 10–8 M and 10–5 M BPA-treated cells are shown in Fig. 2. The 2-DE maps were compared with PDQuest software to identify protein spots that varied among treatments. After exposure to BPA, significant (p < 0.05) differentially expressed protein spots observed in all replicate gels were scored. The fold difference was represented by the ratio of the intensity value of the BPA-treated group to the value of the control group (Table 1). Totally, 51 differently expressed proteins were found as indicated by the spots marked with arrows (Fig. 2A). All these spots were excised from gels for protein identification. And then 50 differentially expressed proteins were successfully identified by MALDI-TOF-MS/MS analysis (Table 1).
Fig. 2. 2-DE analysis and validation of protein expression in TM3 cells treated with BPA. (A) 2-DE analysis of protein expression in TM3 cells exposed to control, 10–8 M and 10–5 M BPA for 48 h, respectively. Proteins indicated by arrows were differentially regulated according to treatment and identified by MS and MS/MS, numbers are correlated with these spot no. listed in Table 1. (B) Validation of differentially expressed proteins. Close up of selective differential expression protein spots in Fig. 2A (left), western blot analysis of the selective differential expression proteins (middle), and the histogram of western blot OD values (right).
Table 1. Identification of differentially expressed proteins in TM4 cells treated with BPA by MALDI-TOF-MS/MS.
| Spot no. a | Protein name | Gene symbol | Accession no. b | MW c (kDa) | pI d | Matched peptides | Protein score e | Coverage f (%) | % change (treated × 100/control)
g
|
|
| Nanomolar | Micromolar | |||||||||
| Cell motility and structure | ||||||||||
| 5 | Annexin A5 | Anxa5 | gi|6753060 | 35 787 | 4.83 | 21 | 545 | 58 | 101 ± 32 | 152 ± 24 |
| 13 | Gamma-actin | actG | gi|809561 | 41 335 | 5.56 | 18 | 455 | 54 | 89.6 ± 15 | 478 ± 59 |
| 15 | Annexin III | ANXA3 | gi|2437840 | 36 520 | 5.33 | 13 | 405 | 43 | 155 ± 31 | 180 ± 21 |
| 16 | Protein S100-A4 | S100a4 | gi|33859624 | 11 942 | 5.23 | 4 | 81 | 16 | 110 ± 13 | 268 ± 39 |
| 17 | Cofilin-1 | Cfl1 | gi|6680924 | 18 776 | 8.22 | 11 | 337 | 61 | 113 ± 9.8 | 30.4 ± 5.2 |
| 19 | Destrin | Dstn | gi|9790219 | 18 852 | 8.14 | 10 | 221 | 46 | 96.8 ± 9.6 | 0.82 ± 0.12 |
| 32 | Gelsolin isoform 2 | Gsn2 | gi|329755239 | 80 997 | 5.52 | 26 | 751 | 37 | 104 ± 12 | 95.7 ± 32 |
| 33 | Ezrin | Ezr | gi|83921618 | 69 478 | 5.83 | 21 | 484 | 32 | 85.8 ± 18 | 51.9 ± 15 |
| 35 | Galectin-1 | Lgals2 | gi|6678682 | 15 198 | 5.32 | 12 | 518 | 61 | 121 ± 31 | 162 ± 27 |
| 37 | Annexin IV | Anx4 | gi|1778313 | 36 252 | 5.43 | 17 | 518 | 41 | 73.4 ± 21 | 134 ± 22 |
| 38 | Protein S100-A11 | S100a11 | gi|21886811 | 11 247 | 5.28 | 4 | 249 | 39 | 101 ± 22 | 230 ± 62 |
| 40 | Vinculin | Vcl | gi|31543942 | 117 215 | 5.77 | 51 | 568 | 47 | 105 ± 25 | 61.6 ± 14 |
| 41 | Collagen alpha-1(I) | Col1a1 | gi|34328108 | 138 974 | 5.65 | 41 | 602 | 48 | 101 ± 31 | 71.6 ± 12 |
| 44 | Translationally controlled tumor protein | Tpt1 | gi|6678437 | 19 564 | 4.76 | 12 | 389 | 44 | 153 ± 45 | 162 ± 28 |
| 50 | Profilin-1 | Pfn1 | gi|6755040 | 15 119 | 8.46 | 12 | 315 | 56 | ||
| Cell metabolism/energy production | ||||||||||
| 12 | Adenosylhomocysteinase | Ahcy | gi|262263372 | 48 170 | 6.08 | 23 | 537 | 40 | 86.4 ± 21 | 185 ± 27 |
| 14 | d-3-Phosphoglycerate dehydrogenase | Phgdh | gi|52353955 | 57 347 | 6.12 | 16 | 599 | 36 | 82.8 ± 22 | 166 ± 12 |
| 18 | Nucleoside diphosphate kinase B | Nme2 | gi|6679078 | 17 466 | 6.97 | 12 | 472 | 78 | 120 ± 29 | 174 ± 17 |
| 21 | S-Formylglutathione hydrolase | Esd | gi|13937355 | 31 870 | 6.7 | 14 | 709 | 67 | 101 ± 18 | 170 ± 5.9 |
| 24 | Aldehyde dehydrogenase II | Aldh3a2 | gi|191804 | 55 131 | 7.89 | 22 | 620 | 45 | 123 ± 71 | 52.2 ± 10 |
| 25 | ATP synthase subunit alpha, mitochondrial precursor | ATPase1 | gi|6680748 | 59 830 | 9.22 | 29 | 704 | 54 | 64.3 ± 21 | 46.6 ± 11 |
| 26 | Phosphoglycerate kinase 1 | Pgk1 | gi|70778976 | 44 921 | 8.02 | 21 | 768 | 56 | 124 ± 12 | 43.7 ± 5.4 |
| 27 | Phosphoserine aminotransferase isoform 1 | PSAT1 | gi|54292132 | 40 732 | 8.15 | 19 | 468 | 45 | 109 ± 12 | 43.7 ± 2.9 |
| 28 | Fructose-bisphosphate aldolase A isoform 2 | ALDOA2 | gi|6671539 | 39 787 | 8.31 | 16 | 620 | 47 | 82.7 ± 31 | 0.05 ± 0.02 |
| 29 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDHS | gi|6679937 | 36 072 | 8.44 | 17 | 376 | 49 | 125 ± 19 | 38.7 ± 5.7 |
| 34 | Glycine-tRNA ligase | Gars | gi|93102417 | 82 624 | 6.24 | 19 | 586 | 30 | 105 ± 11 | 115 ± 16 |
| 36 | ATP synthase subunit d | ATPase4 | gi|21313679 | 18 795 | 5.52 | 5 | 222 | 40 | 111 ± 17 | 77.2 ± 14 |
| 43 | Aldose reductase | Akr1b1 | gi|160707894 | 36 052 | 6.71 | 15 | 796 | 50 | 118 ± 13 | 168 ± 21 |
| Protein processing/folding | ||||||||||
| 3 | Eif4a1 protein, partial | Eif4a1 | gi|71051290 | 46 222 | 5.32 | 22 | 821 | 48 | 162 ± 22 | 270 ± 31 |
| 6 | Hspd1 protein | Hspd1 | gi|76779273 | 59 559 | 8.09 | 27 | 1040 | 51 | 129 ± 21 | 138 ± 16 |
| 8 | T-complex protein 1 subunit gamma | Cct3 | gi|6753320 | 61 162 | 6.28 | 31 | 506 | 52 | 180 ± 22 | 190 ± 12 |
| 9 | WD repeat-containing protein 1 | WDR1 | gi|6755995 | 67 049 | 6.11 | 30 | 853 | 57 | 154 ± 14 | 207 ± 11 |
| 10 | Stress-induced-phosphoprotein 1 | Stip1 | gi|14389431 | 63 170 | 6.4 | 45 | 515 | 52 | 115 ± 10 | 143 ± 17 |
| 22 | Chaperonin containing TCP-1 theta subunit | Cctq | gi|5295992 | 60 044 | 5.44 | 22 | 443 | 37 | 104 ± 12 | 146 ± 19 |
| 23 | Heterogeneous nuclear ribonucleoprotein K | hnRNP K | gi|13384620 | 51 230 | 5.39 | 20 | 489 | 34 | 85.2 ± 5.8 | 192 ± 21 |
| 31 | T-complex protein 1 subunit eta | Cct7 | gi|238814391 | 60 127 | 7.95 | 24 | 555 | 46 | 136 ± 20 | 65.4 ± 4.6 |
| 39 | Protein disulfide isomerase associated 6 | P4hb | gi|60502437 | 49 026 | 5.05 | 14 | 717 | 39 | 73.8 ± 12 | 85.1 ± 8.9 |
| 46 | T-complex protein 1 subunit zeta | Cct6 | gi|6753324 | 58 424 | 6.63 | 26 | 243 | 53 | ||
| 47 | Phosphoglycerate mutase 1 | Pgam1 | gi|114326546 | 28 928 | 6.67 | 20 | 769 | 62 | ||
| DNA/RNA processing | ||||||||||
| 1 | Valosin containing protein, isoform CRA_a | VCP | gi|148670553 | 90 868 | 5.14 | 39 | 713 | 54 | 94.3 ± 11 | 30.4 ± 1.8 |
| 7 | PRP19/PSO4 pre-mRNA processing factor 19 homolog | Prpf19 | gi|148709447 | 52 581 | 6.06 | 18 | 595 | 45 | 119 ± 12 | 274 ± 22 |
| 11 | Bifunctional purine biosynthesis protein PURH | Atic Purh | gi|227908823 | 64 690 | 6.3 | 27 | 681 | 43 | 94.8 ± 8.7 | 260 ± 21 |
| 45 | Poly(rC)-binding protein 1 | cpcG1 | gi|6754994 | 37 987 | 6.66 | 19 | 560 | 59 | 153 ± 9.8 | 162 ± 6.7 |
| 51 | 40S ribosomal protein S12-like | Rps12 | gi|149250091 | 14 904 | 8.13 | 8 | 303 | 43 | ||
| Cell proliferation | ||||||||||
| 2 | Stress-70 protein, mitochondrial | Hspa9 | gi|162461907 | 73 701 | 5.81 | 26 | 574 | 39 | 123 ± 11 | 188 ± 6.9 |
| 4 | Proliferating cell nuclear antigen | PCNA | gi|7242171 | 29 108 | 4.66 | 14 | 602 | 57 | 77.1 ± 8.7 | 68.2 ± 6.8 |
| 49 | Elongation factor 1-beta | Eif1b2 | gi|31980922 | 24 849 | 4.53 | 13 | 511 | 50 | ||
| Oxidative stress | ||||||||||
| 20 | Peroxiredoxin-1 | Prdx1 | gi|6754976 | 22 390 | 8.26 | 17 | 807 | 58 | 145 ± 12 | 35.4 ± 5.4 |
| 42 | Thioredoxin reductase 1 | Txnrd1 | gi|22902393 | 55 101 | 5.95 | 15 | 426 | 35 | 180 ± 14 | 135 ± 13 |
| Signal transduction | ||||||||||
| 30 | G protein beta subunit like | Gpb | gi|475012 | 35 453 | 8.08 | 18 | 565 | 59 | 110 ± 7.9 | 96.1 ± 7.5 |
aSpot numbers correspond with 2-DE gel as shown in Fig. 2.
bAccession number in NCBInr database.
cMW (kDa): molecular mass of predicted protein.
dpI: pI of predicted protein.
eProtein score: In MASCOT, the score for an MS/MS match is based on the absolute probability (P), and the observed match between the experimental data and database sequence is a random event. The reported score is –10log(P). So during a search, if 1.5 × 105 peptides fell with the mass tolerance window the precursor mass, and the significance threshold was chosen to be 0.05, this would translate into a score threshold of 65.
fPercentage of predicated protein sequence covered by matched sequence.
gFold change is expressed as a radio of the vol% between of treated/control cells, and each value represents the mean value ± SD of three independent experiments.
Western-blot analysis was conducted to verify the modulation of identified proteins. The same pattern of expression was observed between the 2-DE analysis and western blot analysis (Fig. 2B). These comparisons confirmed down-regulation of PCNA and up regulation of Lgals2 in cells treated with 10–8 M or 10–5 M BPA, respectively. Furthermore, western blot analysis showed that 10–8 M BPA increased while 10–5 M BPA decreased the protein levels of Prdx1, which was also consistent with the proteomic data. The validation confirmed that the proteomic results are credible.
3.3. Functional categories of identified proteins
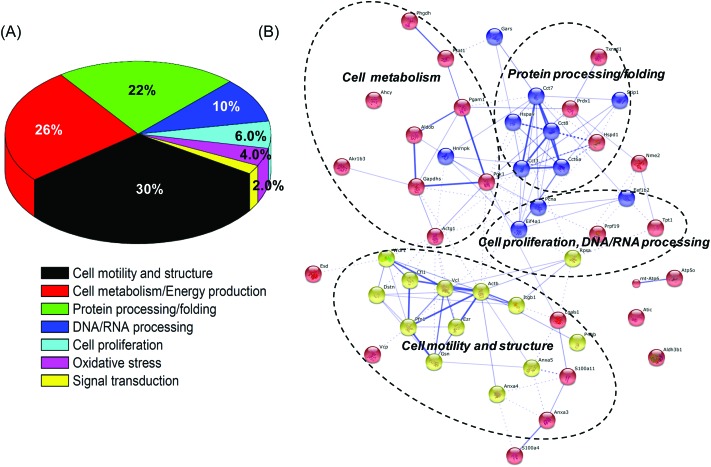
The PANTHER classification system was used to classify the 50 identified proteins.23 These identified proteins were classified into 7 categories according to their functions during biological processes (Fig. 3A). Our results indicated that proteins related to the cell motility and structure, cell metabolism and energy production, protein, DNA, and RNA processing, and cell proliferation accounted for the major proportions among all the identified proteins.
Fig. 3. Functional classification and distribution of all identified proteins. (A) Seven protein groups were categorized based on the putative biological functions of identified proteins and the percentages of each protein group were indicated. (B) The protein–protein interaction network of the identified proteins.
The protein–protein interaction network among the identified proteins was predicted based on the STRING system (string-db.org, version 9.1).24 Notably, four major clusters of interacting proteins were determined by STRING (Fig. 3B), including cell motility and structure, cell proliferation, cell metabolism, and protein processing/folding. This suggested that BPA significantly changed the expression of proteins related to cell structure and metabolism and then influenced the motility and proliferation of Leydig TM3 cells.
3.4. BPA promotes the motility of Leydig TM3 cells
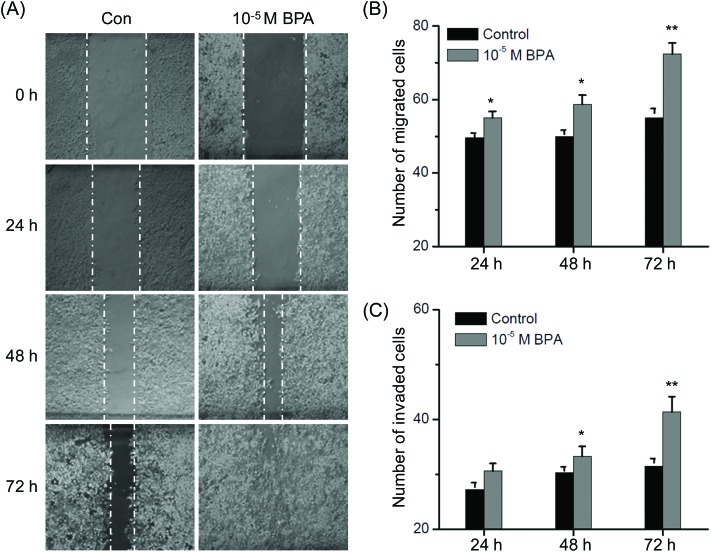
Our recent study indicated that BPA can modulate the colorectal cancer protein profile and promote the metastasis via induction of epithelial to mesenchymal transitions (EMT).20 Results of the proteomic study revealed that proteins related to the cell motility and structure such as annexin and actin were significantly modulated in TM3 cells exposed to BPA. Then the effects of BPA on motility of Leydig TM3 cells were measured by the use of wound healing and trans-well migration/invasion assays. As shown in Fig. 4A, treatment with 10–5 M BPA significantly increased wound closure as compared to the control group for all the indicated times (p < 0.05). Furthermore, the results of transwell revealed that exposure to BPA resulted in a significant increase of migrated (Fig. 4B) and invaded (Fig. 4C) cells for 24, 48 and 72 h. These data revealed that BPA can alter the structure of the Leydig TM3 cells and then promote the in vitro migration and invasion.
Fig. 4. BPA triggered migration and invasion of Leydig TM3 cells. (A) Representative images of the monolayer wound healing assay utilizing the TM3 cell line immediately after scratching 24, 48 and 72 h. Cells were treated with 0.5% DMSO (control) and 10–5 M BPA; TM3 cells were allowed to migrate (B) and invasive (C) transwell chambers for 24, 48, and 72 h in the presence or absence of 10–5 M BPA. Migrated and invaded cells were fixed, stained, and photographed. Data represented the average of three independent experiments. *p < 0.05 compared with control, **p < 0.01 compared with control.
3.5. Galectin-1 is involved in BPA induced migration of Leydig TM3 cells
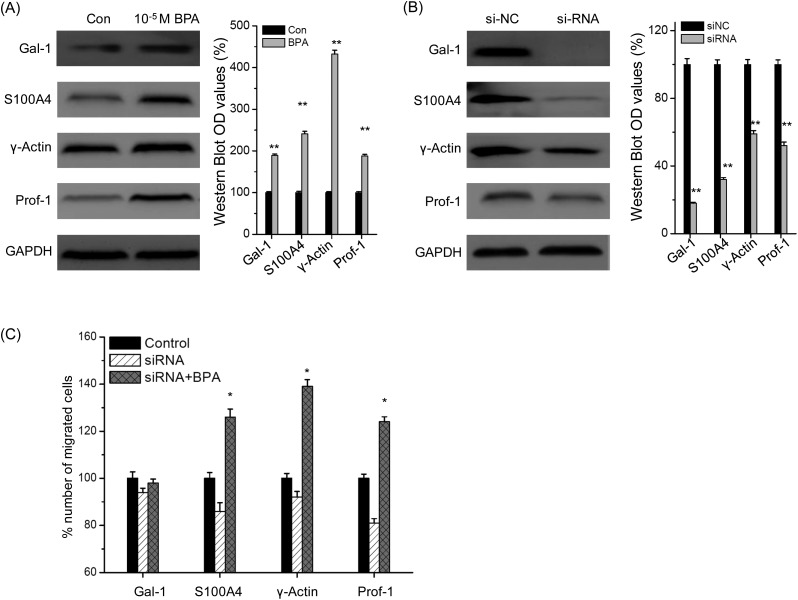
The proteomic results showed that BPA can significantly up regulate the protein levels of γ-actin (4.78-fold), protein S100-A4 (2.68-fold), galectin-1 (1.6-fold), and profilin-1 (1.6-fold), which have been suggested to be significantly correlated with the migration and invasion of cells.25 The up regulation of these proteins was confirmed by western-blot analysis in TM3 cells treated with 10–5 M BPA for 48 h (Fig. 5A).
Fig. 5. Gal-1 mediated BPA-induced migration of Leydig TM3 cells. (A) Cells were treated with 10–5 M BPA for 48 h, and then the protein expression was measured by western blot analysis; (B) TM3 cells transfected with specific si-RNA for Gal-1, A100A4, γ-actin, and prof-1 or negative control si-RNA (si-NC) for 24 h, and then the protein expression was analyzed by western-blot analysis; (C) TM3 cells transfected with specific si-RNA for Gal-1, A100A4, γ-actin, and prof-1 for 24 h and then further treated with 10–5 M BPA for 48 h. Migrated cells were fixed, stained, and photographed by the use of transwell analysis. The % number of migrated cells was calculated to evaluate the effects of siRNA. Data represented the average of three independent experiments. *p < 0.05 compared with control, **p < 0.01 compared with control.
To investigate the roles of these proteins in BPA-induced migration of Leydig cells, TM3 cells were transfected with their corresponding siRNAs, and then migrated cells were measured by transwell. As shown in Fig. 5B, all siRNAs successfully silenced their corresponding proteins in TM3 cells after 24 h. Furthermore, si-Gal-1, but not other siRNAs, successfully abolished the BPA-induced migration of TM3 cells, suggesting that galectin-1 mediated the migration promotion effects of BPA on Leydig cells (Fig. 5C).
3.6. ERK1/2 mediates BPA induced up regulation of galectin-1 and migration of TM3 cells
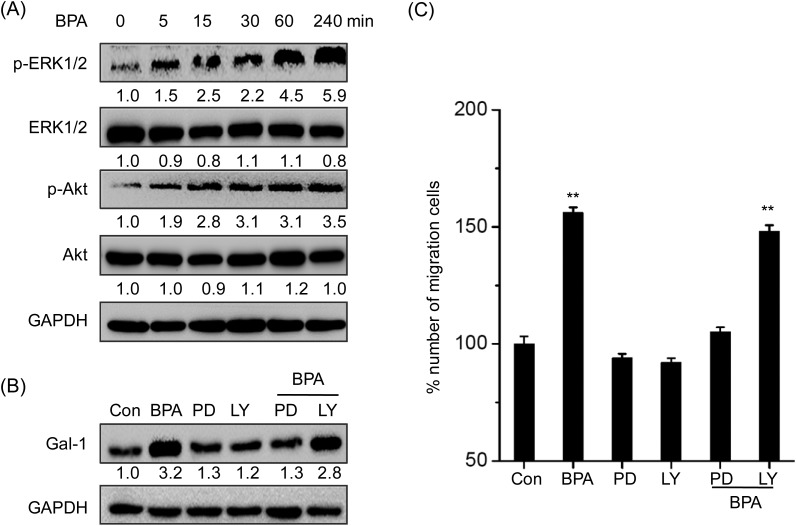
ERK1/2 and PI3K/Akt have been reported as the downstream signals to mediate the biological effect of BPA.13 Therefore the effects of BPA on the phosphorylation of ERK1/2 and PI3K/Akt were checked by western blot analysis. The results showed that BPA treatment markedly increased the phosphorylation of both ERK1/2 and Akt with treatment for 5 min, while it had an obvious effect on the total expression of ERK1/2 or Akt (Fig. 6A). To verify the roles of ERK1/2 and Akt in BPA induced galectin-1 expression, TM3 cells were pretreated with their inhibitors for 90 min. The results showed that ERK1/2 inhibitor PD98509, but not Akt inhibitor LY294002, abolished BPA induced up regulation of Gal-1 (Fig. 6B). Furthermore, only ERK1/2 inhibitor PD98509, but not Akt inhibitor LY294002, significantly attenuated BPA induced migration of TM3 cells (Fig. 6C).
Fig. 6. ERK1/2 mediates BPA induced up regulation of galectin-1 and migration of TM3 cells. (A) Cells were treated with 10–5 M BPA for the indicated time periods, and then the expressions of p-ERK1/2, ERK1/2, p-Akt, and Akt were measured by western blot analysis; (B) TM3 cells were pretreated with 10 μM PD 98059 (PD, ERK1/2 kinase inhibitor) or LY294002 (LY, PI3K/Akt inhibitor) for 90 min and further treated with 10–5 M BPA for 48 h, the expression of Gal-1 was measured by western blot analysis; (C) TM3 cells were pretreated with 10 μM PD 98059 (PD, ERK1/2 kinase inhibitor) or LY294002 (LY, PI3K/Akt inhibitor) for 90 min and further treated with 10–5 M BPA for 48 h, migrated cells were fixed, stained, and photographed by use of transwell analysis. Data represented the average of three independent experiments. **p < 0.01 compared with control.
4. Discussion
4.1. The proteomic modification of Leydig TM3 cells treated with BPA
In the present study, the protein profiles of Leydig TM3 cells changed by BPA were investigated by the use of MALDI-TOF-MS/MS. As for the proteomic study, the doses of 10–8 M and 10–5 M were chosen on the basis of the cytotoxicity test to represent the dose which has limited or significant inhibition effects on cell proliferation, respectively. Totally, 51 differently expressed proteins were found, in which 50 differentially expressed proteins were successfully identified. According to the bioinformatics study of their biological functions, the 50 identified proteins can be classified into 7 categories including cell motility and structure, cell metabolism and energy production, protein, DNA, and RNA processing, and cell proliferation. Considering that the interstitium where Leydig cells reside contributes to the major functions of the testis, testosterone secretion and sperm production, the modification of the above mentioned biological functions of the Leydig TM3 cell might be one of the important reasons responsible for BPA induced male infertility.26 As 30% of the identified proteins are related to the cell motility and structure, and furthermore, the changes of proteins induced by 10–5 M BPA being significantly greater than that of 10–8 M BPA, therefore, the dose of 10–5 M was selected to further investigate the effects of BPA on cell in vitro migration and invasion.
4.2. BPA promotes the in vitro migration and invasion of TM3 cells via up regulating ERK1/2/Gal-1
The proteomic study suggested that 30% of the identified proteins are related to the cell motility and structure. Wound healing and trans-well migration/invasion assays confirmed that BPA can significantly promote the in vitro migration and invasion of TM3 cells. BPA has been reported to influence the migration and cell junction of Sertoli cells.6 The promotion effects of BPA on cell migration have also been observed in breast cancer-associated fibroblasts (CAFs), breast cancer SKBR3 cells,27 colorectal cancer SW480 and HCT-116 cells,20 and neuroblastoma SK-N-SH cells.28 Our results presented here confirmed that BPA can promote the motility of Leydig TM3 cells.
Proteomic studies revealed that BPA can significantly up regulate the motility related proteins such as γ-actin, protein S100A4, Gal-1 and Prof-1. Silencing of Gal-1, but not the others, significantly attenuated the BPA induced migration of TM3 cells. This suggested that Gal-1 mediated the BPA induced migration of Leydig cells. Gal-1 has been greatly detected in Leydig cells29 and considered as a signature of cell invasiveness.30 Gal-1 can increase the motility of glioma cells and re-organize the actin cytoskeleton, conversely, the knockdown of Gal-1 in glioma cells can reduce motility and adhesiveness.31,32 Furthermore, our results suggested that ERK1/2, but not PI3K/Akt, mediates BPA induced migration of TM3 cells, which was consistent with the previous study that showed that BPA can rapidly activate ERK1/2 signals in various cell models.13 In addition, activation of ERK1/2 is reported to participate in the 17β-estradiol induced migration of endometrial cells.33 Together, our results revealed that BPA can promote the migration of Leydig TM3 cells via up regulating ERK1/2/Gal-1.
The proteomic study also identified other proteins and signaling pathways that may be involved in BPA induced migration of TM3 cells. Studies revealed that annexins are well studied receptors for plasminogen, as they convert plasminogen to plasmin after binding and then promote the tumor metastasis.34,35 In TM3 cells, annexin A5, annexin III, and annexin IV were significantly up regulated by BPA treatment, which was similar to the previous study in colorectal cancer SW480 cells.20 The expression of vinculin, which has been suggested to inhibit motility in 3T3 cells,36 was down regulated in TM3 cells treated with BPA. The collagen alpha-1, the most abundant extracellular matrix (ECM) constituent, was also reduced by the treatment of BPA in the present study.
Furthermore, the proteins related to protein processing and folding further supported the promotion effects of BPA on Leydig cells. T-complex protein 1 (TCP-1) and WD repeat containing protein, which can interact with each other, and then modulate the organization of cellular actin and tubulin,37 were both up regulated by BPA. Heterogeneous nuclear ribonucleoprotein K (hnRNP-K), significantly up regulated by 10–5 M BPA, has been reported to promote the tumor metastasis by induction of genes involved in the extracellular matrix, cell movement, and angiogenesis.38 Collectively, the proteomic study and functional experiments revealed that BPA can trigger the in vitro migration of Leydig TM3 cells via Gal-1. Whether other molecules are involved during this process needs further study.
4.3. BPA modulates the proliferation of TM3 cells via multiple signals
Our study revealed that BPA greater than 10–6 M can significantly inhibit cell proliferation. This was also revealed in previous studies that BPA can inhibit the proliferation of Sertoli TM4 cells14 and increase necrosis of human endometrial endothelial cells (HEECs)39 and mice granulose cells.40 Our recent study also revealed that micromolar BPA inhibited cell proliferation while nanomolar BPA had limited effects on SW480 cells.20 The results of the proteomic study on proteins related to energy metabolism, protein, DNA, and RNA processing, oxidative stress and cell proliferation provided important clues to illustrate the mechanisms responsible for inhibition effects of BPA on the proliferation of Leydig TM3 cells. However, Nanjappa et al.26 reported that exposure of male rats to BPA by the gavage of pregnant and lactating Long-Evans dams stimulated Leydig cell division in the prepubertal period and increased Leydig cell numbers, while short-term exposure of prepubertal male rats to BPA at 2.4 μg kg–1 BW from day 21 to day 35 postpartum, but not at higher doses, can decrease serum T levels.41 Combined with the published literature, our data suggested that BPA effects in Leydig cells are affected by the dose, time, pathway and duration of exposure.
Thirty percent of proteins altered by exposure to BPA belong to the “Metabolism/Energy production” group, which was consistent with our previous study that 25% of the identified proteins in Sertoli TM4 cells were related to energy metabolism.13 Regulation of cellular energetic pathways by BPA was evidenced by a low expression of enzymes of the glycolysis (glyceraldehyde-3-phosphate dehydrogenase and fructose-bisphosphate aldolase A isoform 2) and TCA cycle (aldehyde dehydrogenase II, and aldose reductase). Furthermore, proteins related to oxidative phosphorylation such as ATP synthase subunit alpha and ATP synthase subunit d were significantly down regulated by treatment with BPA, particularly for 10–5 M BPA. Considering that activity of ATPase is well documented to associate with cell growth and proliferation,42 our results suggested that BPA might suppress the cell proliferation by modulating the key enzymes involved in cell metabolism and then down regulating the ATP generation.
The proteins related to DNA/RNA processing and proliferation were also significantly modulated by BPA exposure. For example, the down regulation of VCP, a member of AAA family (TPases associated with various cellular activities), has been shown to decrease cell growth and viability of various cancer cells.43 In the present study, 10–5 M BPA significantly down regulated it while 10–8 M BPA had limited effects, which is associated with their corresponding effects on cell proliferation. PCNA, a well-known cell proliferation biomarker, was more down regulated in TM3 cells treated with 10–5 M than 10–8 M BPA. Generally, BPA can modulate the expression of proteins related to energy metabolism, protein, DNA and RNA processing and cell proliferation and then inhibit the in vitro growth of Leydig TM3 cells.
In conclusion, the current study characterized the proliferation inhibition and migration stimulatory effects of BPA on Leydig TM3 cells. With the results of proteomic analysis, it was concluded that BPA can modulate the expression of proteins related to cell structure and motility and trigger the migration. Gal-1 and other unidentified proteins mediated the BPA induced migration of TM3 cells. This study provided new insight to illustrate the mechanisms responsible for BPA induced male infertility and needs further study for the in vivo effects and deeper mechanisms.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (Grant No. 81673454, 81672608, 81472470, 81302317, and 81572270), the Guangdong Natural Science Funds for Distinguished Young Scholars (no. 2014A030306025), the Pearl River S&T Nova Program of Guangzhou (no. 201506010039), the Science & Technology Planning Project of Guangdong Province (2013B060300005), and the Opening Project Program of the State Key Laboratory of Oncology in South China (no. HN2014-09).
References
- Oppeneer S. J., Robien K. Public Health Nutr. 2015;18:1847–1863. doi: 10.1017/S1368980014002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulivo M., Lopez de Alda M., Capri E., Barceló D. Environ. Res. 2016;151:251–264. doi: 10.1016/j.envres.2016.07.011. [DOI] [PubMed] [Google Scholar]
- Rubin B. S. J. Steroid Biochem. Mol. Biol. 2011;127:27–34. doi: 10.1016/j.jsbmb.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Liu C., Duan W., Li R., Xu S., Zhang L., Chen C. Cell Death Dis. 2013;4:e676. doi: 10.1038/cddis.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari D., Vanage G. Reprod. Toxicol. 2013;40:60–68. doi: 10.1016/j.reprotox.2013.05.013. [DOI] [PubMed] [Google Scholar]
- Salian S., Doshi T., Vanage G. Reprod. Toxicol. 2011;31:359–362. doi: 10.1016/j.reprotox.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Lassen T. H., Frederiksen H., Jensen T. K., Petersen J. H., Joensen U. N., Main K. M. Environ. Health Perspect. 2014;122:478–484. doi: 10.1289/ehp.1307309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.-K., Zhou Z., Miao M., He Y., Wang J., Ferber J. Fertil. Steril. 2011;95:625–630. doi: 10.1016/j.fertnstert.2010.09.026. [DOI] [PubMed] [Google Scholar]
- Ge R., Chen G., Hardy M. P. Adv. Exp. Med. Biol. 2008;636:255–269. doi: 10.1007/978-0-387-09597-4_14. [DOI] [PubMed] [Google Scholar]
- Zirkin B. R., Chen H. Biol. Reprod. 2000;63:977–981. doi: 10.1095/biolreprod63.4.977. [DOI] [PubMed] [Google Scholar]
- Singh J., O'Neill C., Handelsman D. J. Endocrinology. 1995;136:5311–5321. doi: 10.1210/endo.136.12.7588276. [DOI] [PubMed] [Google Scholar]
- Abney T. O. Steroids. 1999;64:610–617. doi: 10.1016/s0039-128x(99)00041-0. [DOI] [PubMed] [Google Scholar]
- Ge L.-C., Chen Z.-J., Liu H.-Y., Zhang K.-S., Liu H., Huang H.-B. Toxicol. Lett. 2014;226:81–89. doi: 10.1016/j.toxlet.2014.01.035. [DOI] [PubMed] [Google Scholar]
- Ge L.-C., Chen Z.-J., Liu H., Zhang K.-S., Su Q., Ma X.-Y. Biochim. Biophys. Acta. 2014;1840:2663–2673. doi: 10.1016/j.bbagen.2014.05.018. [DOI] [PubMed] [Google Scholar]
- Chimento A., Casaburi I., Bartucci M., Patrizii M., Dattilo R., Avena P. Cell Death Dis. 2013;4:e747. doi: 10.1038/cddis.2013.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuillier R., Manku G., Wang Y., Culty M. Microsc. Res. Tech. 2009;72:773–786. doi: 10.1002/jemt.20756. [DOI] [PubMed] [Google Scholar]
- Murray T. J., Maffini M. V., Ucci A. A., Sonnenschein C., Soto A. M. Reprod. Toxicol. 2007;23:383–390. doi: 10.1016/j.reprotox.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyl R. W., Myers C. B., Marr M. C., Thomas B. F., Keimowitz A. R., Brine D. R. Toxicol. Sci. 2002;68:121–146. doi: 10.1093/toxsci/68.1.121. [DOI] [PubMed] [Google Scholar]
- Zang Q., Zhang L., Gao N., Huang C. J. Integr. Med. 2016;14:51–59. doi: 10.1016/S2095-4964(16)60238-8. [DOI] [PubMed] [Google Scholar]
- Chen Z.-J., Yang X.-L., Liu H., Wei W., Zhang K.-S., Huang H.-B. Arch. Toxicol. 2015;89:1371–1381. doi: 10.1007/s00204-014-1301-z. [DOI] [PubMed] [Google Scholar]
- Wu Y.-M., Chen Z.-J., Liu H., Wei W.-D., Lu L.-L., Yang X.-L. Oncotarget. 2015;6:25588–25601. doi: 10.18632/oncotarget.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.-J., Wei W., Jiang G.-M., Liu H., Wei W.-D., Yang X. Mol. Oncol. 2016;10:775–788. doi: 10.1016/j.molonc.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Guo N., Kejariwal A., Thomas P. D. Nucleic Acids Res. 2007;35:D247–D252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum M. S. Y., Pasquier E., Po'uha S. T., O'Neill G. M., Chaponnier C., Gunning P. W. FASEB J. 2011;25:4423–4433. doi: 10.1096/fj.11-185447. [DOI] [PubMed] [Google Scholar]
- Nanjappa M. K., Simon L., Akingbemi B. T. Biol. Reprod. 2012;86:135. doi: 10.1095/biolreprod.111.095349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupo M., Pisano A., Lappano R., Santolla M. F., De Francesco E. M., Abonante S. Environ. Health Perspect. 2012;120:1177–1182. doi: 10.1289/ehp.1104526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Zheng J., Xiao X., Zheng S., Dong K., Liu J. Oncol. Rep. 2010;23:129–139. [PubMed] [Google Scholar]
- Biron V. A., Iglesias M. M., Troncoso M. F., Besio-Moreno M., Patrignani Z. J., Pignataro O. P. Glycobiology. 2006;16:810–821. doi: 10.1093/glycob/cwl013. [DOI] [PubMed] [Google Scholar]
- Harvey S., Zhang Y., Landry F., Miller C., Smith J. W. Physiol. Genomics. 2001;5:129–136. doi: 10.1152/physiolgenomics.2001.5.3.129. [DOI] [PubMed] [Google Scholar]
- Camby I., Decaestecker C., Lefranc F., Kaltner H., Gabius H.-J., Kiss R. Biochem. Biophys. Res. Commun. 2005;335:27–35. doi: 10.1016/j.bbrc.2005.07.037. [DOI] [PubMed] [Google Scholar]
- Camby I., Belot N., Lefranc F., Sadeghi N., de Launoit Y., Kaltner H. J. Neuropathol. Exp. Neurol. 2002;61:585–596. doi: 10.1093/jnen/61.7.585. [DOI] [PubMed] [Google Scholar]
- Gentilini D., Busacca M., Di Francesco S., Vignali M., Viganò P., Di Blasio A. M. Mol. Hum. Reprod. 2007;13:317–322. doi: 10.1093/molehr/gam001. [DOI] [PubMed] [Google Scholar]
- Lokman N. A., Ween M. P., Oehler M. K., Ricciardelli C. Cancer Microenviron. 2011;4:199–208. doi: 10.1007/s12307-011-0064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann U., Balabanov S., Giebel J., Teller S., Junker H., Schmoll D. Cancer Lett. 2004;209:111–118. doi: 10.1016/j.canlet.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Rodríguez Fernández J. L., Geiger B., Salomon D., Ben-Ze'ev A. Cell Motil. Cytoskeleton. 1992;22:127–134. doi: 10.1002/cm.970220206. [DOI] [PubMed] [Google Scholar]
- Brackley K. I., Grantham J. Cell Stress Chaperones. 2009;14:23–31. doi: 10.1007/s12192-008-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R., Yu Y., Inoue A., Widodo N., Kaul S. C., Wadhwa R. J. Biol. Chem. 2013;288:15046–15056. doi: 10.1074/jbc.M113.466136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredhult C., Bäcklin B.-M., Olovsson M. Reprod. Toxicol. 2007;23:550–559. doi: 10.1016/j.reprotox.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Xu J., Osuga Y., Yano T., Morita Y., Tang X., Fujiwara T. Biochem. Biophys. Res. Commun. 2002;292:456–462. doi: 10.1006/bbrc.2002.6644. [DOI] [PubMed] [Google Scholar]
- Akingbemi B. T., Sottas C. M., Koulova A. I., Klinefelter G. R., Hardy M. P. Endocrinology. 2004;145:592–603. doi: 10.1210/en.2003-1174. [DOI] [PubMed] [Google Scholar]
- Crouch S. P., Kozlowski R., Slater K. J., Fletcher J. J. Immunol. Methods. 1993;160:81–88. doi: 10.1016/0022-1759(93)90011-u. [DOI] [PubMed] [Google Scholar]
- Vandermoere F., El Yazidi-Belkoura I., Slomianny C., Demont Y., Bidaux G., Adriaenssens E. J. Biol. Chem. 2006;281:14307–14313. doi: 10.1074/jbc.M510003200. [DOI] [PubMed] [Google Scholar]