 Here, we have investigated the comparative quantitative proteomics analysis of the molecular response of HeLa cells to biocompatible Fe2C@C nanoparticles (NPs) using 16O/18O isotopic labelling of the cell culture.
Here, we have investigated the comparative quantitative proteomics analysis of the molecular response of HeLa cells to biocompatible Fe2C@C nanoparticles (NPs) using 16O/18O isotopic labelling of the cell culture.
Abstract
The effective detection of molecular biomarkers, such as proteins, lipids, carbohydrates, and pathogens, in a living body is a huge challenge in the field of nanomedicine. Here, we have investigated the comparative quantitative proteomics analysis of the molecular response of HeLa cells to biocompatible Fe2C@C nanoparticles (NPs) using 16O/18O isotopic labelling of the cell culture. The relative binding efficiency of proteins to Fe2C@C NPs was calculated. HPLC-ESI-orbit-trap analysis found 51 differentially expressed proteins, out of which 23 were over-expressed and 28 down-regulated. This study showed that Fe2C@C NPs alter the expression of the proteins involved in endocytosis, cell-cycle regulation, and cell membrane protrusion. Further, the quantification and validation of the mass spectrometry (MS) results was successfully confirmed by western blot analysis of cytochrome C. The change in the expression of proteins can be useful for early stage disease diagnoses and the development of tailored therapeutic strategies. This study is the first large-scale characterization of low abundance proteins on Fe2C@C NPs, providing the biochemical basis for the assessment of the suitability of magnetic NPs as biomedical markers and emerging functional probes.
1. Introduction
In recent years, a large number of reports have focused on the potential use of magnetic nanoparticles (NPs) as effective tools for biomedical applications, for example, disease diagnosis using magnetic nanomaterials for magnetic resonance imaging (MRI) and targeted therapy using hyperthermia, which can be combined with theranostic approaches.1,2 However, very limited information is available regarding the response of cells to magnetic NPs at the protein level. The induction of magnetic NPs within a cell provides an exciting area of study, such as the evaluation of changing protein structure and functions.3–7 The alteration of the protein structure and function may provide a useful molecular explanation of the physiological responses to magnetic NPs. In this regard, the HeLa cell line was selected to screen the unique proteins in response to biocompatible magnetic NPs. These cell lines are used to obtain crucial information for understanding cell–NP interactions, before moving to in vivo analysis. This information, in the form of proteomics as biomarkers for various diseases, such as cancer, has been discovered using magnetic NPs.8 Mass spectrophotometry (MS) based quantitative proteomics analysis has proven to be valuable in cancer research, but the complicated process of tumor genesis may involve complex protein networks.9 However, pre-existing proteomics data focused on specific groups of proteins, which may be present at relatively low levels. These proteins are considered to be biomarkers and can play imperative roles in cell–cell communication, growth, and many other phenomena.8–11 Moreover, this information might represent a useful source of biomarkers and potential targets to treat diseases. Various proteomics approaches have been used to discover biomarkers in blood/plasma, body fluids and tissues, exosomes and conditioned media from cultured or primary cells.12–14 The MS proteomics analysis of proteins that adhere to super paramagnetic iron oxide (SPIO) NPs might be a new way for the detection of the key proteins involved in the early diagnosis of diseases. It is obvious that characterizing the protein absorption of the Fe2C@C NPs has not been quantitatively documented previously. To fill this knowledge gap, there is need of a comprehensive study to investigate the biochemical properties of the proteins around the Fe2C@C NPs using a proteomics approach. There are many proteomics strategies that are being widely used for the comparative study of cells and tissues using techniques such as isotope coded affinity tags (ICATs), isobaric tags for relative and absolute quantification (ITRAQ), isotopically labeled amino acids in cell cultures and the enzymatic incorporation of 18O atoms into the C-terminal side of cleaved peptides.15,16
Here, we have employed a 16O/18O-labelling technique due to the small sample size requirement to study two different groups of in vitro culture. This technique not only allows the identification of the proteins having interaction with the NPs, but also measures the enrichment level of each protein at the level of organelles and functional basis in response to this material. This study will provide useful information regarding the impact of NPs on cellular physiology during the intake of these NPs not only for drug delivery but help in the safe diagnosis of several diseases, especially cancer.
2. Experimental methods
2.1. Synthesis of the Fe2C@C NPs and their cell viability
The synthesis of the Fe2C@C NPs and their surface coating and cell viability procedures are provided in the Materials and method section in the ESI.†
2.2. Protein sample preparation
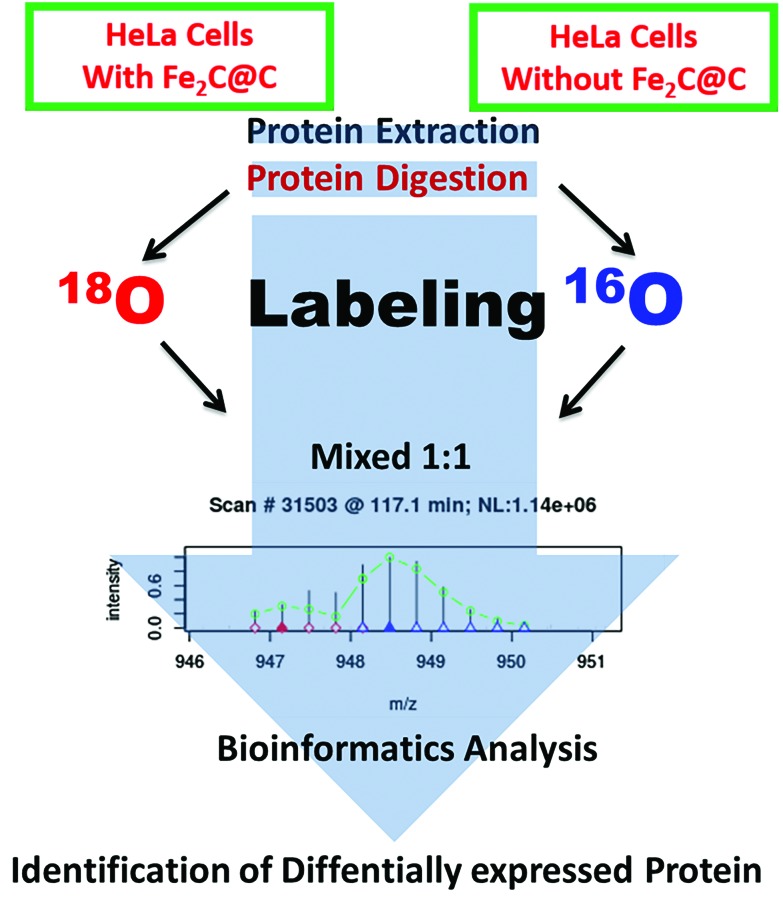
The HeLa cells were harvested and the proteins were extracted. Afterwards, the control and experimentally grouped proteins were digested with trypsin, labelled with isotopes, and quantified using HPLC-ESI-orbit (Fig. 1). The HeLa cells were uprooted after 24 h of incubation with the Fe2C NPs and suspended in 0.5 mL of isoelectric focusing buffer containing 40 mM Tris, 5 M urea, 2 M thiourea, 4% CHAPS, 10 mM 1,4-dithioerythritol, 1.0 mM EDTA (3–10), and a mixture of protease inhibitors (Roche diagnostics, 1 tablet per 10 mL of buffer suspension). The cell suspensions were sonicated for ∼30 s and centrifuged at 53 000 rpm for 50 min at 4 °C. The protein content in the supernatant was determined using Bradford reagent (BioRad) to make the concentration of protein in each sample up to 100 μg. At the end, both the control and experimental samples contained 500 μg of protein.
Fig. 1. A schematic illustration of the harvesting cell, the proteins were extracted, digested with trypsin, isotopically labelled, and quantified using HPLC-ESI-orbit.
2.3. Protein digestion
The protein samples were digested to obtain the peptides. Initially, denaturation was done using 60 μL of 8 M urea solution and then the peptides were protected to reduce the disulphide bridges by the addition of 6 μL of 100 mM of DTT solution, and finally the samples were lyophilized and kept at 37 °C for 4 h. Later on, 30 μL of 100 mM iodoacetamide (IAA) solution was added to the samples to alkylate them while they were kept under darkness for 1 h. Finally, 600 μL of 50 mM NH4CO3 (pH 8.2) and 24 μL of trypsin (0.5 mg mL–1) were added to digest the samples and the reaction was continued for 20 h at 37 °C.
2.4. Trypsin-catalyzed 16O/18O labelling
Trypsin-catalyzed 16O/18O labelling was done according to the literature.17 After the residual trypsin activity was quenched through boiling and then sudden cooling, aliquots of the peptides (100 μg of each) were taken from the control and experimental samples for lyophilization. Then all the samples were dissolved in a KH2PO4–K2HPO4 buffer with a pH of 5.0 and trypsin (Promega, USA) of 1 : 50 (w/w) was prepared with either 18O-enriched water (97%, Cambridge Isotope Laboratories, USA) or regular 16O-enriched water. The peptide samples were incubated for 24 h at 37 °C, where the controlled peptides were labelled with 16O and the experimental ones were labelled with 18O. After labelling, centrifugation was done for 5 min at 15 000g to collect the supernatant from each sample. The corresponding 16O/18O-labeled samples were pooled, combined, and then lyophilized.
2.5. Cation exchange chromatographic separation
The strong cation exchange chromatographic separation of the peptide mixtures was carried out by an Agilent 300 Å SCX. A column of 6 mm × 250 mm was used with a flow rate of 0.7 mL min–1 of the mobile phase solvents maintained. The mobile phase consists of two solvents (A) 10 mM ammonium format with 25% acetonitrile and a pH of 3.0 and (B) 500 mM ammonium format with 25% acetonitrile and a pH of 6.8. The peptides were separated with a linear gradient from 3 to 40% B over 50 min at 280 nm. The fractions were collected every 2 min and stored at –80 °C. Each SCX fraction was analyzed with a capillary HPLC system coupled to an electrospray-ion trap (ESI-OrbitTrap) mass spectrometer from Thermo Fisher (nanoLC (model: easy nLC1000)-Orbitrap Elite). A 5 mL, 150 mL, i.e. 150 mm, C-18 column (Column Technology Inc., Fremont, CA, USA) was eluted with a linear acetonitrile gradient elution from 100% solvent A (water with 0.1% formic acid) to 60% solvent A + 40% solvent B (acetonitrile with 0.1% formic acid) over 60 min at a flow rate of 1.8 mL min–1, and from 40% to 95% solvent B over 10 min, and finally 100% solvent B for another 10 min. During ESI-TOF and ESI-Trap analysis, the nebulizer pressure was 15 psi. The drying gas flow and temperature were 7 L min–1 and 325 °C, respectively. The mass spectrometer scan range was set between a mass of 100 and 1500.
2.6. Western blot
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed with a Bio-Rad mini protein III apparatus. The samples, with and without exposure of the magnetic nanoparticles, were separated by SDS-PAGE (10% gel). The gels were electro-blotted onto 0.45 mL of a PVDF membrane. The membrane was blocked with 10% non-fat milk in PBS for 1 h at 37 °C and then incubated with antibodies raised against the COX5A binding protein (COX5A) (beta actin, internal standard) and appropriate secondary antibodies. The detection of the immune complex was performed by enhanced chemiluminescence.
2.7. Data analysis and bioinformatics
The Swiss-Prot protein database was explored through the MASCOT (Matrix Science, UK) server (http://www.expasy.ch/sprot/). The parameters utilized in the present work are as follows: no restriction on the protein molecular weight, one missed cleavage, trypsin digestion, and monoisotopic mass. Other parameters for MASCOT were a peptide mass tolerance of 2 Da, fragment mass tolerance of 0.8 Da, and peptide charges of +1, +2, and +3. For Q-TOF, the peptide mass tolerance and fragment mass tolerance values were 20 ppm and 0.05 Da, respectively. The protein matching probabilities were measured using the MASCOT protein scores, with the identification confidence shown by the number of matches and the coverage of the protein sequence by the matching peptides. Only statistically significant MASCOT score results (p < 0.05) were used in the data analysis. For the quantification of the HPLC-ESI-TOF data, the Mascot summary report along with the output files from ESI-TOF were subjected to quantification on an in-house built pep-match quantification tool, which was built specifically for measuring the low to high (L/H) molecular mass ratios. Furthermore, unique peptides were used for quantification. The Q-TOF data were quantified by spectrum mill and mascot distiller inbuilt quantification tools. Significantly, the differentially expressed proteins involved in the response to the magnetic NPs were sorted by keeping to the threshold frequency, which showed an expression significance level of between <0.5 and <1.5.
3. Results
3.1. Biocompatibility of the Fe2C@C NPs to HEk293 and HeLa cells
The transmission electron microscopy (TEM) images shown in Fig. S1† reveal that the particles are well dispersed with good homogeneity while having an average size of 16.5 nm. Surface modification of the NPs was carried out to keep them well-dispersed and biocompatible to obtain a good response. The biocompatibility of the Fe2C@C NPs was tested with the HeLa cells over a 24-hour incubation time. It has been observed that Fe2C@C NPs lack lethality with both kinds of cells and there is no aggregation, as shown in the ESI (Fig. S2†). MS-based shotgun proteomics were employed to investigate the significant differential regulation of the proteins in response to the Fe2C@C NPs. The HeLa cells were exposed to 20 μg mL–1 Fe2C@C NPs for 24 h. After the preparation of a cell lysate, the proteins were digested with trypsin and the peptides were labeled with 16O/18O and quantified by MS using the untreated cells as the experimental control, as illustrated in scheme (Fig. 1).
3.2. Protein identification
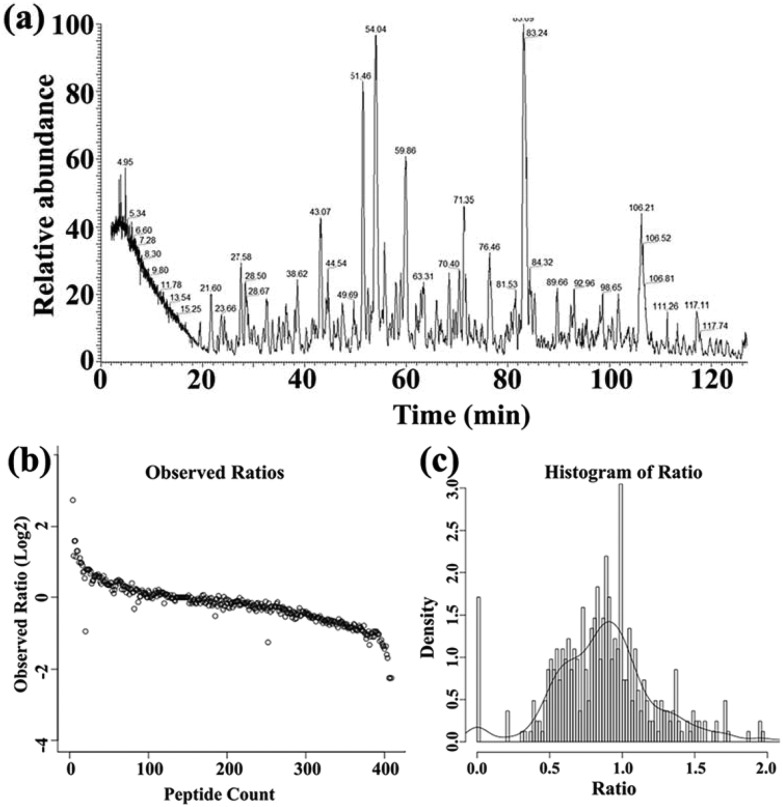
The proteins and peptides of the HeLa cells in response to the biocompatible Fe2C@C NPs were identified on the basis of the retention time and the 16O/18O-labelling techniques after the digestion with trypsin, as shown in Fig. 2a. The 16O/18O-labelling technology confirmed the number of peptides for all the identified proteins, and it was also found that both the peptides have a 1 : 1 ratio, as shown in Fig. 2b. However, this 1 : 1 ratio further confirmed that the density of protein was at a maximum at this ratio, as shown in Fig. 2c. The classification of the overall proteins was carried out using Swiss-Prot accession numbers along with the protein description, symbol of the protein, 16O/18O ratio, molecular weight, total number of identified peptides, location of the protein, and their specific function, as shown in Table S2.† Using the experimental strategy depicted in Fig. 1, 51 proteins were found to be differentially expressed (Table S2†) out of a totally identified 394 proteins (Table S1†) and quantified with p-value 0.01.
Fig. 2. The MS spectrum of protein peptides identified on the basis of 16O/18O-labelling and retention time (a). The total protein observed ratio with peptide count (b) and the total protein ratio with density (c).
3.3. Functional categorization of the identified proteins
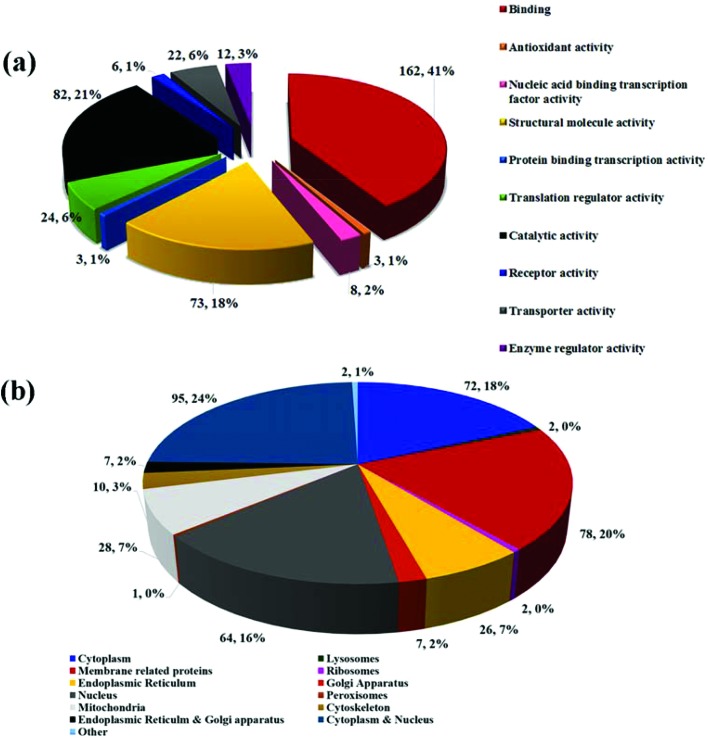
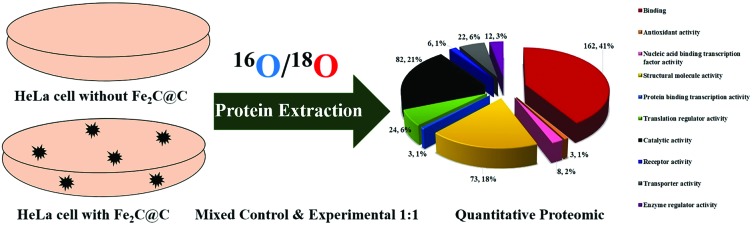
Fig. 3a delineates the documentation of the proteins and shows the level of expression in response to the biocompatible Fe2C@C NPs in the HeLa cell; 162 proteins (41.12%) have binding characteristics, 82 proteins (21.11%) have catalytic activity, 73 proteins (18.11%) are involved in the structural and molecular activity, 24 proteins (6.83%) are found to have translation regular activity, and 22 proteins (6.16%) are identified to have transported activity, while only 8 (2.66%) proteins are nucleic acid binding transcription factor activity proteins and only 3 proteins (1.12%) have enzyme regulator activity, receptor activity, and antioxidant and protein binding transcription activity.
Fig. 3. GO fact analysis of the major biological processes performed by the identified proteins involved in the different cellular processes (a). The classification of the differential expressed protein in response to the Fe2C@C NPs in the HeLa cell on the basis of their location (b).
3.4. Subcellular localization of the identified proteins
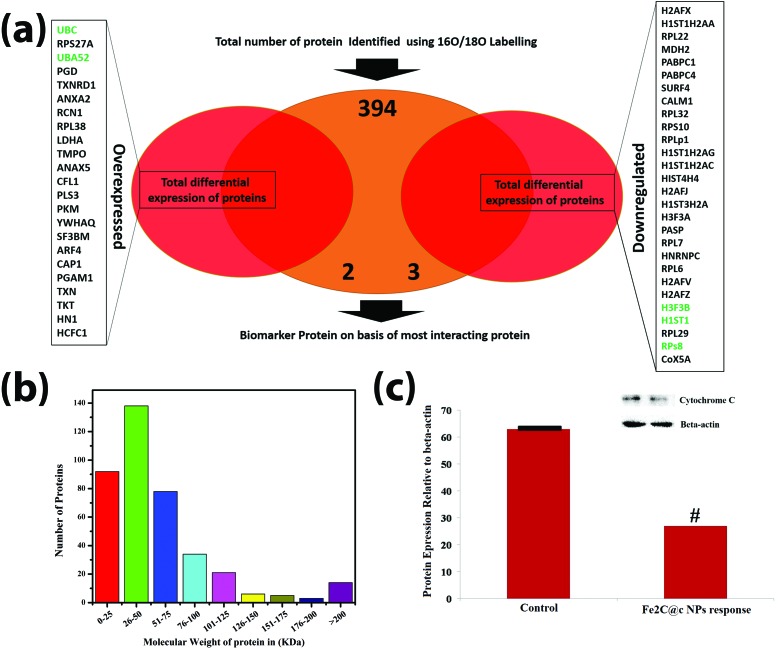
The location of the HeLa cell proteins in response to the Fe2C@C NPs was identified using gene ontology (GO) analysis. Fig. 3b delineates the proteins on the basis of their location; 72 proteins (18.27%) are cytoplasmic, 2 proteins (0.51%) are related to lysosomes, 78 proteins (20%) belong to all types of membrane proteins (mitochondrial membrane, endoplasmic reticulum membrane, Golgi body membrane, ribosomal membrane, and nuclear membrane), 64 (16.24%) proteins are nucleus related, 28 (7.11%) proteins are mitochondrial, 7 (1.68%) proteins are found in both the endoplasmic reticulum and Golgi apparatus, 2 (0.51%) proteins are found in the ribosomes, 10 proteins (2.78%) are specific to the cytoskeleton, 1 protein (0.25%) is from the peroxisomes, 95 (24.11%) proteins are found in both the nucleus and cytoplasm, and a few other proteins (0.50%) are isolated without identification. Moreover, the gene expression in response to the Fe2C@C NPs was summarized via a Venn diagram to identify the key protein involved in the biocompatibility process, as shown in Fig. 4a. However, the relative abundance of the proteins expressed in response to the Fe2C@C NPs in vitro was furthered classified on the basis of the molecular mass, as shown in Fig. 4b. A maximum number of proteins was found with molecular masses in the range of 26–50 kDa, while a minimum number of proteins was found with molecular masses in the range of 176–200 kDa. It was found that UBA and UBc52 were the key proteins out of the 23 proteins that were differentially over-expressed and identified by unique peptide methods. Whereas H3F3B, H1ST1 and RPS8 were the key proteins out of the 28 proteins that were down-regulated. Furthermore, the retention time of the identified proteins followed a sequence of unique peptides. The accession numbers of the proteins and the generic names are given to each of the differentially expressed proteins in Table S1.†
Fig. 4. The analysis of the gene expression via a Venn diagram showing the protein biomarker involved in the biocompatibility process (a). The distributions of the proteins on the basis of their molecular weight using a 16O/18O-labelling technique in HeLa cells (b). The quantification and validation of the cytochrome C protein by western blotting (c). Each experiment was performed in triplicate (n = 3). p < 0.05 compared with beta actin.
3.5. Western blotting
Cytochrome c belongs to the cytochrome family of proteins and is an essential component of the electron transport chain. It is capable of undergoing oxidation and reduction, but does not bind oxygen. In this work, cytochrome c protein was selected for the validation of the mass spectrum because this protein involves processing and transportation (via the endoplasmic reticulum), and the energy-producing centers of the cells (mitochondria). It was observed during western blotting that the cytochrome c protein was highly down-regulated in response to the Fe2C@C NPs, compared to the control, as shown in Fig. 4c.
3.6. Analysis of the protein–protein interactions
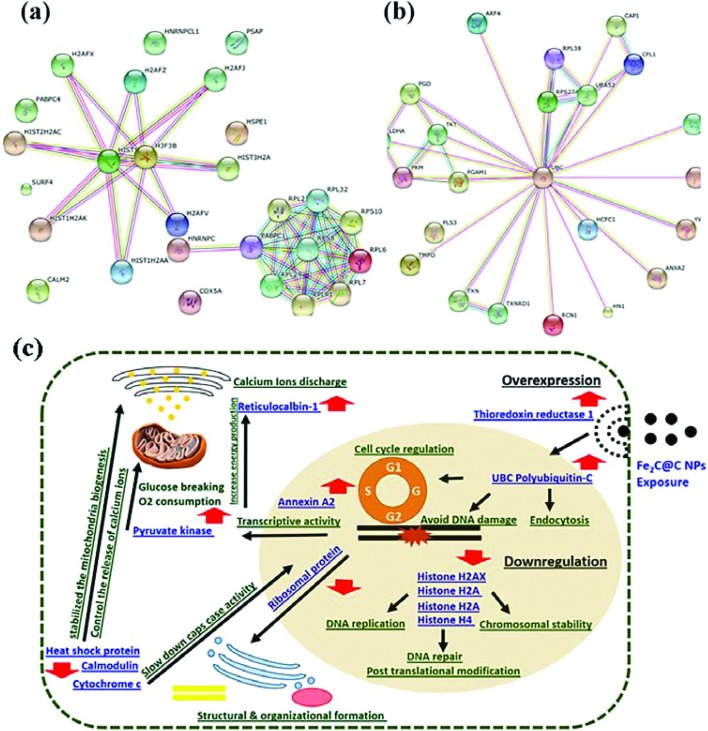
To better understand the molecular mechanisms underlying the cellular responses triggered by the Fe2C@C NPs, we used the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) algorithm to build protein–protein interaction networks (Fig. 5). Fig. 5a represents the protein–protein interactions of the down-regulated proteins, whereas Fig. 5b depicts the interaction networks of the up-regulated ones. Many molecular and cellular processes within a cell are carried out by molecular machines that are built from a large number of protein components organized by their protein–protein interactions. These interactions make up the so-called interactomics of the organism, while abnormal protein–protein interactions are the basis of multiple aggregation-related diseases, such as Creutzfeldt–Jakob and Alzheimer's disease, and may lead to cancer. Previously, many researchers have illustrated the cell response via proteomics analysis and categorised these proteins on the basis of cellular, functional, and molecular analyses.18–20
Fig. 5. The string analysis of the biomarker protein (UBC, H3F3B, and RPS8) showing the integration with the other protein in the HeLa cell exposure to the Fe2C@C NPs (a and b). The schematic diagram of the various protein expressions in response to the biocompatible surface coated Fe2C@C NPs in the HeLa cell (c).
Fig. 5b illustrates all the over-expressed proteins in response to the diversity of polyubiquitin (UBC), which contributes to the regulation of many cellular events. UBC gene transcription is induced during stress and provides extra ubiquitin necessary to remove damaged/unfolded proteins. These proteins have an important role in diverse biological processes, such as innate immunity, DNA repair, and kinase activity. Un-anchored polyubiquitin-C is also a key signaling molecule that connects and coordinates proteasomes and autophagy to eliminate toxic protein aggregates.
4. Discussion
The use of magnetic NPs has become essential in the field of medical sciences. These NPs are used as an efficient tool for drug delivery, and MRI and PET in cells and tissues for early disease diagnosis. The observation of the proteomic changes within the cell in response to the NPs will provide valuable information for their safe use and the detection of various diseases in their early stages. The results have proven the biocompatible nature of the magnetic Fe2C@C NPs within the cell and lead us one step towards their safe usage in the early detection of diseases, such as cancer. The overall expression of the gene in response to the Fe2C@C NPs is shown in Fig. 5c. The results showed that the polyubiquitin (UBC) C, ubiquitin-40S, ribosomal S27a, and ubiquitin-60S ribosomal L40 proteins are over-expressed in response to the Fe2C@C NPs. These proteins are covalently attached to another protein that targets the function of various parts. Previously, it was reported that the polyubiquitin protein is involved in endoplasmic reticulum-associated degradation and in cell-cycle regulation.21 Moreover, these proteins also play a key role in lysosomal degradation, kinase modification, protein degradation via the proteasome, endocytosis, and DNA-damage responses, as well as in the signaling processes leading to the activation of the transcription factor NF-kappa-B.21 Here our results illustrate the comprehensive role of polyubiquitin (UBC) C, especially in the process of endocytosis of the Fe2C@C NPs, avoiding DNA damage, cell signaling, and most importantly the regulation of the cell cycle under the exposure to the Fe2C@C NPs. In thioredoxin reductase 1, the cytoplasm possesses glutaredoxin activity, as well as thioredoxin reductase activity, and induces actin and tubulin polymerization, leading to the formation of cell membrane protrusions. Previous studies reported that this protein regulates the activity of the transcription factors, such as p53, hypoxia-inducible factor, and AP-1.22 The over-expression of this protein in our studies suggested that this protein has a unique and distinct expression pattern in human cells and suggests that thioredoxin reductase 1 can guide actin polymerization in relation to the cell membrane re-structuring. It has been observed that this protein safely induces cell membrane protrusions with the entry of foreign particles, especially magnetic NPs as in the present study, as shown in Fig. 5c.
Annexin A2 is another interesting protein with calcium-regulated membrane-binding properties whose affinity for calcium is greatly enhanced by anionic phospholipids. It has been involved in the heat-stress response and inhibits PCSK9-enhanced LDLR degradation.23 The present study confirmed the transcription process of annexin A2 with up-regulation to promote the activity by induction of Fe2C@C NP interaction within the HeLa cell. Moreover, it will possibly trigger the release of calcium ions to overcome the stress situation in the cell (Fig. 5c). Reticulocalbin-1 regulates the calcium-dependent activities in the endoplasmic reticulum lumen or the post-ER compartment. Here in this study, exposure of the Fe2C@C NPs to the HeLa cells for 24 h enhanced the calcium ion binding activities, which are mostly involved with many energy production processes (Fig. 5c).
The over-expression of the 60S ribosomal protein L38, l-lactate dehydrogenase-a chain, lamina-associated polypeptide 2, and isoform alpha in the present study helps the HeLa cell biocompatibility process, especially concerning the structural organization of the nucleus and the post-mitotic nuclear assembly. All these proteins play an important role in T-cell development and function. Cofilin-1 is a kind of protein that binds to F-actin and exhibits pH-sensitive F-actin depolymerizing activity. It has already been reported that this protein regulates the actin cytoskeleton dynamics with normal progress through mitosis and cytokinesis. Instead of this, Cofilin-1 plays a vital role in the regulation of the cell morphology and cytoskeletal organization.24 The results presented in this study reconfirmed the proteomics data of the present study that Cofilin-1 is over-expressed during the flooding of the biocompatible Fe2C@C NPs. Up-regulation of Plastin-3 helps the actin-bundling protein found in the developmental process. Pyruvate kinase (PK) is a rate-limiting enzyme in glycolysis that is converted to a less active dimer form of PKM2 during oncogenesis. PHD3 knockdown inhibits the PKM2 coactivator function, reduces glucose uptake and lactate production, and increases O2 consumption in cancer cells. Thus, PKM2 participates in a positive feedback loop that promotes HIF-1 transactivation and reprograms glucose metabolism in cancer cells.25 It is concluded, based on the previous reports, that the over-expression of Pyruvate kinase in response to the biocompatible Fe2C@C NPs activates glucose breaking, which is useful for tumor cell proliferation and survival. The up-regulation of many proteins, such as 14-3-3, ADP-ribosylation factor 4, adenylyl cyclase-associated protein 1, transketolas, host cell factor 1 and thioredoxin, during the incubation of Fe2C@C NPs is shown in Fig. 5c. These proteins are involved in protein trafficking, modulating vesicle budding, regulating filament dynamics with morphological processes, the catalysis process including mRNA localization, and the establishment of cell polarity and functions in gene regulation and their opposing effects on the cell cycle.
A group of histone family proteins, such as Histone H2AX, Histone H2A type 1-A, Histone H2A.Z and Histone H4, wrap the nucleosomes and compact DNA into chromatin, limiting the DNA accessibility to the cellular machineries, which require DNA as a template. Previously, it was reported that these proteins require checkpoint-mediated arrest of the cell cycle progression in response to low doses of ionizing radiation and for efficient repair of the DNA double strand breaks (DSBs), specifically when modified by C-terminal phosphorylation.26 In another report, H2A.Z may be involved in the formation of constitutive heterochromatin, which is required for chromosome segregation during cell division. The decrease in the expression of the histome family proteins here in this study confirmed the previous findings about keeping the HeLa cell cycle arrested to cope with the external stimulus in the form of Fe2C@C NPs exposure. Another set of proteins, such as 60S ribosomal protein L22, 60S ribosomal protein L32, 40S ribosomal protein S10, 60S acidic ribosomal protein P1, 60S ribosomal protein L7, 60S ribosomal protein L29, 60S ribosomal protein L6, and 40S ribosomal protein S8, are known as the family of ribosomal proteins located in the cytoplasm. These proteins played a key role in the structural and organizational formation of the cell. The decrease in the level of expression in the HeLa cell under the incubation of the biocompatible surface coated Fe2C@C NPs confirms the adoptability characterization and no correlation was observed between the HeLa cell lethality under the exposure of Fe2C@C NPs. Calmodulin mediates the control of a large number of enzymes, ion channels, aquaporins, and other proteins by Ca2+. Among the enzymes to be stimulated by the calmodulin-Ca2+ complex are a number of protein kinases and phosphatases. Together with CCP110 and centrin, these are involved in a genetic pathway that regulates the centrosome cycle and progression through cytokinesis.27 The deceasing level of calmodulin influenced the level of calcium ions within the cell and confirmed our investigation with the previously reported results.
The 10 kDa heat shock protein, which is mitochondrial, found in the Eukaryotic CPN10 homolog, and essential for mitochondrial protein biogenesis, together with CPN60, binds to CPN60 in the presence of Mg-ATP and suppresses the ATPase activity of the latter. The low level of down-regulation helps to stabilize the mitochondria biogenesis system by specially inducing the cell in a different situation. The cytochrome c oxidase subunit 5A, which is mitochondrial, is a heme A-containing chain of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport. It is also the pathway that controls cell death and DNA damaging. In previous reports, stress was applied in the form of microgravity and the down-regulation of cytochrome C has been shown, which ultimately suppresses the caspase activation and slows down the process of cell death.28 The decrease in the level of cytochrome c expression in this study, as shown in Fig. 5c, supports the previous investigation reporting that the cell has a self-defense system to alternate the level of expression in the gene to cope with environmental stimuli.
5. Conclusion
In summary, our study is an attempt to quantify proteins and highlight the specific molecular mechanisms of purified proteins with their pathways affected by the foreign magnetic Fe2C@C NPs in the HeLa cells. This study helps to broaden future use of magnetic Fe2C@C NPs for biomedical applications in at least two aspects. Firstly, proteins with a high affinity to bind with Fe2C@C NPs are potential targets after these NPs enter the cell. The conformation and functions of these proteins may be affected, resulting in disruptions to the cellular structure and functioning. Secondly, the identification of Fe2C@C NP-binding proteins may have implications on the future applications of Fe2C@C NPs. The binding of these proteins to the Fe2C@C NPs could complicate the efficacy of the Fe2C@C NPs as bio-sensing probes or delivery tools for therapeutic cargos. These results will offer future direction for the surface modification and blocking procedures that will minimize these non-specific protein interactions. All of these results provide novel insights into the nanoproteome pathways affected by Fe2C@C NPs, which could prove to be useful in designing future pharmacological interventions to counteract the harmful effects of magnetic NPs.
Conflicts of interest
The authors declared no conflicts of interest.
Supplementary Material
Acknowledgments
This work was supported in part by the Research Fellowship for International Young Scientists of the National Natural Science Foundation of China (51550110502, 51450110437), National Natural Science Foundation of China (NSFC) (81421004, 51590882, 51125001, 51172005), Doctoral Program (20090001120010), NSFC/RGC Joint Research Scheme (51361165201), and Opening Project of Beijing National Laboratory for Molecular Science. The authors also would like to acknowledge the mass spectrometry facility of the National Center for Protein Sciences at Peking University and Dr Wen Zhou for assistance with LC-MS analysis.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c7tx00248c
References
- (a) Jing Y. Nano Res. 2012;5:679–694. [Google Scholar]; (b) Murtaza H., Wenlong Y., Yanmin J., Xin C., Yun W., Yulin D., Nasir M., Yanglong H. Nano Res. 2017;14:1912–1923. [Google Scholar]
- Hao R. Nanoscale. 2013;5:11954–11963. doi: 10.1039/c3nr04157c. [DOI] [PubMed] [Google Scholar]
- Walczyk D. J. Am. Chem. Soc. 2010;132:5761–5768. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- Mu Q. J. Phys. Chem. 2008;112:3300–3307. [Google Scholar]
- Francois M. B., Yun W. L., Douglas L., Anguis I. L. Mol. Cell. Proteomics. 2009;9:457–470. [Google Scholar]
- Ghazala M., Katsumi S., Zahed H., Setsuko K. J. Proteomics. 2015;122:100–108. [Google Scholar]
- Ghazala M., Katsumi S., Zahed H., Setsuko K. J. Proteomics. 2015;128:280–287. [Google Scholar]
- Tenzer S., Docter D., Rosfa S., Wlodarski A., Kuharev J., Rekik A. ACS Nano. 2011;5(9):7155–7157. doi: 10.1021/nn201950e. [DOI] [PubMed] [Google Scholar]
- Nader Rifai M. A., Gillette S. A. Nat. Biotechnol. 2006;24:971–973. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- Mermelekas G., Zoidakis J. Expert Rev. Mol. Diagn. 2014;14:549–543. doi: 10.1586/14737159.2014.917965. [DOI] [PubMed] [Google Scholar]
- Simberg D., Park J. H., Karmali P. P., Zhang W. M., Merkulov S., McCrae K. Biomaterials. 2009;30:3926–3923. doi: 10.1016/j.biomaterials.2009.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis J. E., Gromov P., Cabezón T., Moreira J. M., Ambartsumian N., Sandelin K., Rank F., Gromova I. Mol. Cell. Proteomics. 2004;3:327–324. doi: 10.1074/mcp.M400009-MCP200. [DOI] [PubMed] [Google Scholar]
- Raimondo F., Morosi L., Chinello C., Magni F., Pitto M. Proteomics. 2011;11:709–720. doi: 10.1002/pmic.201000422. [DOI] [PubMed] [Google Scholar]
- Dowling P., Clynes M. Proteomics. 2011;11:794–794. doi: 10.1002/pmic.201000530. [DOI] [PubMed] [Google Scholar]
- Ross P., Huang Y., Marchese J. N., Williamson B. Mol. Cell. Proteomics. 2004;3:1154–1159. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Zhou J. Y., Dann G. P., Liew C. W., Smith R. D., Kulkarni R. N. Mol. Cell. Proteomics. 2002;5:376–376. [Google Scholar]
- Javed I., Wang L., Murtaza H., Yuan J. L., Kaleem U. Proteomics. 2014;14:262–263. [Google Scholar]
- Javed I., Wang L., Kaleem U., Murtaza H., Guo L. Proteomics. 2013;13:2455–2468. [Google Scholar]
- Javed I., Wang L., Murtaza H., Kefu L., Umer A. Proteomics. 2014;14:1424–1433. [Google Scholar]
- Yongqian Z., Hongbin W., Chengjun L., Lu W., Yulin D. Astrobiology. 2013;3:143–150. [Google Scholar]
- Kristariyanto Y. A., Abdul R. S. A., Campbell D. G., Morrice N. A., Johnson C. Mol. Cell. 2015;58:83–84. doi: 10.1016/j.molcel.2015.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammeyer P., Damdimopoulos A. E., Nordman T., Jiménez A., Miranda-Vizuete A., Arnér E. S. J. Biol. Chem. 2008;283:2814–2811. doi: 10.1074/jbc.M708939200. [DOI] [PubMed] [Google Scholar]
- Seidah N. G., Poirier S., Denis M., Parker R., Miao B. PLoS One. 2012;7:e41865. doi: 10.1371/journal.pone.0041865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohla A., Birkenfeld J., Bokoch G. M. Nat. Cell Biol. 2005;1:21–29. doi: 10.1038/ncb1201. [DOI] [PubMed] [Google Scholar]
- Luo W., Hu H., Chang R., Zhong J., Knabel M. Cell. 2011;145:732–734. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffler K., Schulz K., Hirmer A., Wiesner J., Grimm M. RNA. 2010;8:1488–1489. doi: 10.1261/rna.2146910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichow S. L., Clemens D. M., Freites J. A., Németh-Cahalan K. L., Heyden M. Nat. Struct. Mol. Biol. 2013;9:1085–1092. doi: 10.1038/nsmb.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed I., Wang L., Murtaza H., Yu J. L., Ullah K. Proteomics. 2014;14:262–263. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.