Abstract
Introduction
Circular RNAs (circRNAs) function as efficient microRNA sponges with gene-regulatory potential and are promising cancer biomarkers. In this study, we used the Arraystar Human circRNA Array to construct a genome-wide circRNA profile of esophageal squamous cell cancer (ESCC) and breast cancer (BC).
Patients and methods
Expression levels between cancer lesions and adjacent normal-appearing tissues were compared. We observed 469 upregulated circRNAs and 275 downregulated circRNAs in ESCC. Hsa_circRNA_103670 was upregulated 20.3-fold, while hsa_circRNA_030162 was downregulated 12.1-fold. For BC, 715 circRNAs were upregulated, and 440 circRNAs were downregulated. Hsa_circRNA_005230 was upregulated 12.2-fold, while hsa_circRNA_406225 was downregulated 12.4-fold.
Results
When we set the criteria as fold change in expression ≥2 between cancer and adjacent normal-appearing tissue with a P-value <0.01, there were 22 common circRNAs (11 upregulated and 11 downregulated) in relation to both ESCC and BC. Gene ontology and the Kyoto encyclopedia of genes and genomes analyses showed that these circRNAs were involved in the tumorigenesis of human cancers.
Conclusion
Our study revealed that circRNAs are promising candidates as valuable biomarkers for ESCC and BC, although relevant research is still in its infancy and the functional role of specific circRNAs in tumorigenesis is just starting to be elucidated.
Keywords: circRNA, noncoding RNA, esophageal squamous cell carcinoma, breast cancer, biomarker
Introduction
Cancer is the leading cause of death in developed countries, and its prevalence is increasing in developing countries as well, which imposes a heavy burden to society at large. According to a GLOBOCAN report, there were an estimated 14.1 million new cancer cases and 8.2 million cancer-related deaths in 2012 worldwide.1,2 Esophageal cancer ranks as the eighth most common cancer worldwide and ranks sixth in cancer-causing deaths. Histologically, the majority of esophageal cancers are divided into squamous cell carcinoma and adenocarcinoma. The high-risk areas include northern Iran, southern Russia, central Asian countries, and northern China, where esophageal squamous cell cancer (ESCC) accounts for 90% of all esophageal cancer cases.3 The prognosis of ESCC is poor, and patients have only 15%–25% of 5-year survival rates after diagnosis.4–6
The poor prognosis and rising incidence of esophageal cancer have highlighted the need for improved detection and prediction methods that are essential prior to treatment.7 Current clinically approved surveillance practices highly depend on expensive, invasive, and sampling-error-prone endoscopic procedures.8 Therefore, there is a great demand to establish reliable biomarkers that could identify patients at higher risks of neoplastic progression who would hence greatly benefit from further monitoring and/or intervention. Emerging molecular tools promise to extend the diagnostic research of the endoscopist and open doors to population screening for ESCC.9 An increasing evidence has shown that noncoding RNAs, such as microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs), play an important role in the development and progression of multiple human cancers and could be used as prognostic factors and therapeutic targets for esophageal cancer.10
CircRNAs are a class of noncoding RNA molecules that lack 5′3′ ends and a poly A tail, covalently forming closed continuous loops.11,12 In general, circRNAs are stable molecules, and some have functioned as efficient miRNA sponges with gene-regulatory ability.13 CircRNAs, with their distinctive characteristics, have superior potential to serve as novel markers for human diseases. The list of endogenous circRNAs involved in cancer continues to grow; however, the functional relevance of the majority of endogenous circRNAs has yet to be discovered.13
In one of our previous studies, we have provided evidence that circRNAs are differentially expressed in breast cancer (BC) and play an important role in carcinogenesis because they participate in cancer-related pathways and sequester miRNAs.14 However, whether circRNAs are sensitive and specific biomarkers of ESCC and BC remains largely unknown. In this study, we used an Arraystar human circRNA array (Arraystar Inc., Rockvile, MD, USA) to construct a genome-wide circRNA profile of ESCC compared with altered expression in BC, with the aim of exploring the potential functions of these circRNAs as diagnostic biomarkers.
Patients and methods
Ethics statement
This study was approved by the Institutional Review Board of Nanjing Medical University, China. Written informed consent was obtained from all participants included in the study.
Patients
Patients with ESCC were enrolled from the First People’s Hospital of Yancheng in October 2016. Tissues in the cancer lesion and adjacent normal-appearing esophagus were collected from patients who underwent surgical resection and met the following criteria: 1) a pathologic diagnosis of ESCC; 2) no previous history of cancer; 3) HIV negative; and 4) no history of radiotherapy or chemotherapy before specimen collection. The recruitment process of patients with BC was detailed in a previously published paper.14 Tissue samples were placed in RNA storage solution (Shanghai Biotechnology Corporation, Shanghai, China) and stored at −80°C until use. Tumor stages were determined according to the tumor-node-metastasis (TNM) staging criteria.15
CircRNA microarray
The RNA was isolated with an RNeasy mini kit (Qiagen, Hilden, Germany) and analyzed using an 8*15K Arraystar human circRNA microarray V2 (Catalog No: AS-CR-H-V2.0). Sample preparation and array hybridization followed the manufacturer’s protocol.
Bioinformatics and data analysis
We performed gene ontology (GO) analysis (http://www.geneontology.org)16 to construct meaningful annotation of genes in any organism covering domains of biological processes, cellular components, and molecular functions. The log10(P-value) denotes the significance of GO term enrichment correlated to the genes producing differentially expressed circRNAs. Kyoto encyclopedia of genes and genomes (KEGG) analysis (http://www.genome.jp/kegg)17 was carried out to confirm the pathway clusters covering molecular interaction and reaction networks in genes producing differentially expressed circRNAs.
Statistical analysis
The fold change in circRNA expression was calculated by comparing expression levels between cancer lesions and control tissues. Student’s t-test was used to estimate the significance of the difference between the two groups. CircRNAs with fold change ≥2 and P<0.05 were considered to be statistically significant. We used the filter criteria of fold change ≥2 and P<0.01 to screen for common differentially expressed circRNAs shared by both ESCC and BC. The false discovery rate was used to adjust for P-values in microarray analysis. Agilent feature extraction software (version 11.0.1.1, Agilent, Santa Clara, CA, USA) was used to analyze the acquired array images. R software version 3.3.1 (https://www.r-project.org/)18 was used to perform quantile normalization and for GO and KEGG analysis.
Results
Differentially expressed circRNAs
Seven paired ESCC samples and four paired BC samples were collected for the Arraystar human circRNA array. The characteristics of the patients are shown in Tables S1 and S2. When we set the criteria as fold change ≥2.0 and P<0.05 between cancer lesions and adjacent normal-appearing tissues, there were 744 differentially expressed circRNAs in ESCC and 1155 differentially expressed circRNAs in BC. For ESCC, 469 circRNAs were upregulated, and 275 circRNAs were downregulated. The top 10 upregulated and top 10 downregulated circRNAs for ESCC are listed in Table 1. Hsa_circRNA_103670 was upregulated 20.3-fold, while hsa_circRNA_030162 was downregulated 12.1-fold. For BC, 715 circRNAs were upregulated, and 440 circRNAs were downregulated. The top 10 upregulated and top 10 downregulated circRNAs of BC are listed in Table 2. Hsa_circRNA_005230 was upregulated 12.2-fold, while hsa_circRNA_406225 was downregulated 12.4-fold.
Table 1.
The top 10 upregulated and top 10 downregulated circRNAs for ESCC
| Upregulated circRNAs | P-value | Fold change | Downregulated circRNAs | P-value | Fold change |
|---|---|---|---|---|---|
| hsa_circRNA_103670 | 0.021 | 20.321 | hsa_circRNA_030162 | 0.014 | 12.071 |
| hsa_circRNA_004390 | 0.004 | 7.870 | hsa_circRNA_001729 | <0.001 | 9.382 |
| hsa_circRNA_104172 | 0.004 | 7.686 | hsa_circRNA_087212 | 0.044 | 9.087 |
| hsa_circRNA_404013 | 0.014 | 7.522 | hsa_circRNA_037767 | 0.015 | 8.543 |
| hsa_circRNA_001937 | 0.015 | 6.517 | hsa_circRNA_000367 | <0.001 | 8.223 |
| hsa_circRNA_100872 | 0.006 | 6.255 | hsa_circRNA_102459 | 0.012 | 7.413 |
| hsa_circRNA_102854 | <0.001 | 5.763 | hsa_circRNA_001640 | 0.001 | 7.021 |
| hsa_circRNA_406826 | 0.004 | 5.552 | hsa_circRNA_020068 | 0.024 | 6.912 |
| hsa_circRNA_013058 | 0.004 | 5.249 | hsa_circRNA_004183 | <0.001 | 6.724 |
| hsa_circRNA_004662 | <0.001 | 5.127 | hsa_circRNA_400850 | 0.001 | 6.529 |
Abbreviations: circRNA, circular RNA; ESCC, esophageal squamous cell cancer.
Table 2.
The top 10 upregulated and top 10 downregulated circRNAs for BC
| Upregulated circRNAs | P-value | Fold change | Downregulated circRNAs | P-value | Fold change |
|---|---|---|---|---|---|
| hsa_circRNA_005230 | 0.045 | 12.198 | hsa_circRNA_406225 | 0.032 | 12.406 |
| hsa_circRNA_103254 | 0.006 | 8.155 | hsa_circRNA_404724 | 0.021 | 12.154 |
| hsa_circRNA_103552 | 0.006 | 7.201 | hsa_circRNA_406697 | 0.008 | 9.130 |
| hsa_circRNA_101981 | 0.003 | 6.762 | hsa_circRNA_002873 | 0.013 | 8.908 |
| hsa_circRNA_063763 | 0.005 | 6.725 | hsa_circRNA_008842 | 0.043 | 8.860 |
| hsa_circRNA_103253 | 0.006 | 6.467 | hsa_circRNA_047418 | 0.018 | 7.477 |
| hsa_circRNA_103902 | 0.010 | 5.929 | hsa_circRNA_401033 | 0.034 | 7.371 |
| hsa_circRNA_002908 | 0.003 | 5.895 | hsa_circRNA_001288 | 0.022 | 7.321 |
| hsa_circRNA_102736 | 0.012 | 5.723 | hsa_circRNA_104858 | 0.025 | 6.891 |
| hsa_circRNA_104327 | 0.020 | 5.682 | hsa_circRNA_001838 | 0.037 | 6.875 |
Abbreviations: circRNA, circular RNA; BC, breast cancer.
Figure 1 illustrates the hierarchical clustering analysis showing the distinct circRNA expression profiling in ESCC (Figure 1A) and BC (Figure 1B). Figure 2 shows the scatter plots demonstrating the heterogeneity of circRNA expression of ESCC cancer lesions (Figure 2A) and BC cancer lesions (Figure 2B) with their adjacent normal-appearing tissues. The expression of the circRNAs above the top reference line and below the bottom reference line changed by >2-fold. Volcano plots were used to visualize the significantly differentially expressed circRNAs for ESCC (Figure 3A) and BC (Figure 3B).
Figure 1.
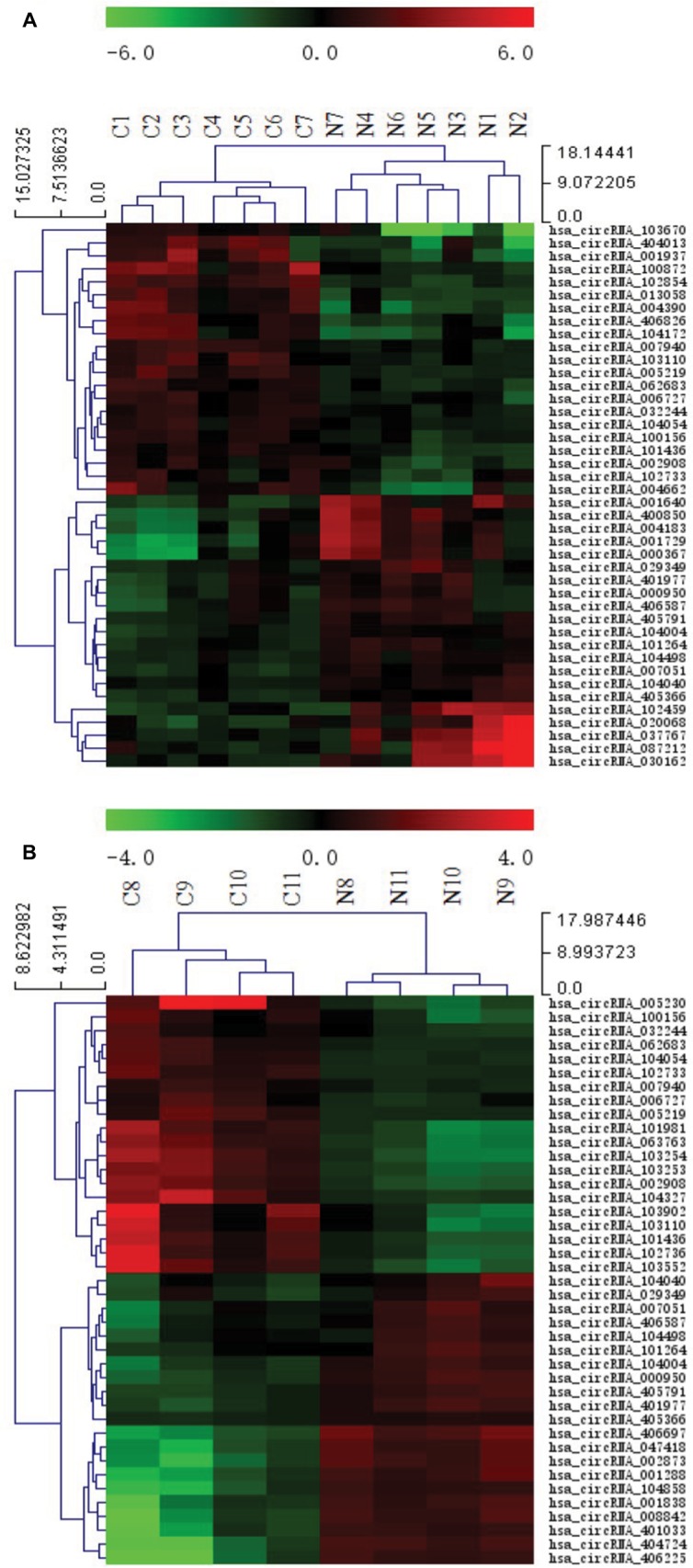
Heat map showing the expression profiles of circRNAs in ESCC (A) and BC (B).
Notes: The expression values are represented by the color scale. The intensity increases from green (relatively lower expression) to red (relatively higher expression). Each column represents one tissue sample, and each row represents a single circRNA. C1–C7 represent seven ESCC specimens collected from ESCC patients. N1–N7 represent paired adjacent normal-appearing tissues. C8–C11 represent four BC specimens collected from BC patients. N8–N11 represent paired adjacent normal-appearing tissues.
Abbreviations: ESCC, esophageal squamous cell cancer; BC, breast cancer.
Figure 2.
Scatter plots demonstrating the heterogeneity of ESCC lesions (A) and BC lesions (B) with their adjacent normal-appearing tissues.
Notes: The values of the X and Y axes represent the averaged normalized signal values of the group (log2-scaled). The green line stands for 2-fold changes. The expression of the circRNAs above the top green line and below the bottom green line indicate changes by >2-fold between the two groups of samples.
Abbreviations: ESCC, esophageal squamous cell cancer; BC, breast cancer.
Figure 3.
Volcano plots visualizing differential expression of ESCC lesions (A) and BC lesions (B) with adjacent normal tissues.
Notes: The vertical lines correspond to 2.0-fold up- and downregulation (log2 ratio), and the horizontal line represents a P-value of 0.05. The red points in the plot represent the differentially expressed circRNAs with statistical significance (fold change >2 and P<0.05), the gray points represent the remaining circRNAs (fold change <2 or P>0.05).
Abbreviations: ESCC, esophageal squamous cell cancer; BC, breast cancer.
When we set the criteria as the fold change in expression ≥2 between cancer and adjacent normal-appearing tissue and P<0.01, there were 22 common circRNAs in relation to both ESCC and BC. Among them, 11 were upregulated and 11 were downregulated (Table 3).
Table 3.
Common circRNAs in relation to both ESCC and BC
| CircRNAs | ESCC
|
BC
|
||
|---|---|---|---|---|
| P-value | Fold change | P-value | Fold change | |
| Upregulated | ||||
| hsa_circRNA_002908 | 0.008 | 2.447 | 0.003 | 5.895 |
| hsa_circRNA_005219 | 0.001 | 3.556 | 0.009 | 2.945 |
| hsa_circRNA_006727 | 0.009 | 2.51 | 0.005 | 2.301 |
| hsa_circRNA_007940 | 0.001 | 2.688 | 0.009 | 2.068 |
| hsa_circRNA_032244 | 0.002 | 2.029 | 0.004 | 2.073 |
| hsa_circRNA_062683 | 0.005 | 2.741 | 0.006 | 2.568 |
| hsa_circRNA_100156 | 0.005 | 2.26 | 0.001 | 3.209 |
| hsa_circRNA_101436 | 0.009 | 2.42 | 0.008 | 5.084 |
| hsa_circRNA_102733 | 0.001 | 2.193 | 0.008 | 2.930 |
| hsa_circRNA_103110 | 0.006 | 2.987 | 0.005 | 5.189 |
| hsa_circRNA_104054 | 0.007 | 2.019 | 0.008 | 2.887 |
| Downregulated | ||||
| hsa_circRNA_000950 | 0.008 | 2.095 | 0.001 | 3.847 |
| hsa_circRNA_007051 | 0.001 | 2.592 | 0.005 | 2.555 |
| hsa_circRNA_029349 | 0.001 | 2.409 | 0.009 | 2.158 |
| hsa_circRNA_101264 | <0.001 | 2.245 | 0.006 | 2.313 |
| hsa_circRNA_104004 | 0.004 | 2.154 | 0.002 | 3.455 |
| hsa_circRNA_104040 | 0.004 | 2.024 | 0.005 | 2.473 |
| hsa_circRNA_104498 | <0.001 | 2.041 | <0.001 | 2.028 |
| hsa_circRNA_401977 | 0.005 | 2.074 | 0.006 | 3.263 |
| hsa_circRNA_405366 | 0.008 | 2.463 | 0.002 | 2.019 |
| hsa_circRNA_405791 | 0.003 | 2.216 | 0.007 | 3.039 |
| hsa_circRNA_406587 | 0.001 | 2.347 | 0.007 | 2.564 |
Abbreviations: circRNA, circular RNA; ESCC, esophageal squamous cell cancer; BC, breast cancer.
GO enrichment and KEGG analysis
We further used GO analysis to explore the roles of these dysregulated circRNAs. The top 10 upregulated circRNAs in ESCC were related to positive regulation of cellular processes, biological processes, and macromolecule metabolic processes (Figure S1A), whereas the top 10 downregulated circRNAs in ESCC were related to positive regulation of cellular metabolic processes, RNA metabolic processes, and cellular processes (Figure S1B). In BC, the top 10 upregulated circRNAs were associated with negative regulation of cellular processes, biological processes, and macromolecule metabolic processes (Figure S1C), while the top 10 downregulated circRNAs were associated with the regulation of macromolecule metabolic processes, metabolic processes, and primary metabolic processes (Figure S1D). We further performed GO analysis on 22 dysregulated circRNAs shared in both ESCC and BC. Our data revealed that 11 shared upregulated circRNAs were involved in the regulation of signaling, cellular processes, and biological processes (Figure S1E), while 11 shared downregulated circRNAs were involved in the regulation of signaling and developmental processes (Figure S1F).
In KEGG pathway analysis, for the top 10 upregulated circRNAs in ESCC, HTLV-I infection, cancer, and cGMP-PKG signaling were the top three pathways (Figure S2A). For the top 10 downregulated circRNAs in ESCC, Wnt signaling, ubiquitin-mediated proteolysis, and HTLV-I infection were the top three pathways (Figure S2B). Moreover, the top three KEGG pathways for the top 10 upregulated circRNAs in BC were MAPK signaling, neurotrophin signaling, and ubiquitin-mediated proteolysis (Figure S2C), and the top three KEGG pathways for the top 10 downregulated circRNAs in BC were MAPK signaling, synaptic vesicle cycle, and hippo signaling pathway-multiple species (Figure S2D). For the 11 upregulated circRNAs shared in ESCC and BC, the three most enriched pathways in the KEGG analysis were endocytosis, thyroid hormone signaling pathway, and proteoglycans in cancer (Figure S2E). The three most enriched pathways for the 11 downregulated circRNAs shared in ESCC and BC were proteoglycans in cancer, endocytosis, and pathways in cancer (Figure S2F).
Discussion
CircRNAs are RNA molecules in which a covalent linkage typically contains the exon sequences and a splice between an upstream 3′ splice site and a downstream 5′ splice site.19,20 CircRNAs have been hypothesized to function as miRNA sponges to offset the impact of miRNAs. In recent years, more roles of circRNAs such as sequestering proteins or regulating transcription have been discovered.21 Studies have shown that abnormal circRNAs are involved in tumorigenesis and disease progression.22–25 However, whether they are general cancer biomarkers or cancer specific biomarkers is not clear. To distinguish the differentially expressed circRNAs between ESCC and BC, we performed a comparative study.
For ESCC, hsa_circRNA_103670 was the most upregulated circRNA and aligned with CNOT6L (CCR4-NOT transcription complex subunit 6 like, Gene ID: 246175), which is a main deadenylase complex regulating gene expression in eukaryotes. The most downregulated was hsa_circRNA_030162. This circRNA derives from the gene TPT1 (tumor protein translationally controlled 1, Gene ID: 7178), also called translationally controlled tumor protein (TCTP),26,27 which encodes a cell growth-associated protein and plays an important role in the development of various organisms. TPT1 has recently been identified as related to human skin squamous cell carcinoma and is targeted by miRNA-216b-5p in pancreatic cancer.28,29
For BC, the top upregulated circRNA was hsa_circRNA_005230. It was spliced from the gene DNM3OS (DNM3 opposite strand/antisense RNA, Gene ID: 10062831), which produces a noncoding RNA (ncRNA) that is involved in the formation of miR-199a2 and miR-214.30 These miRNAs have been reported to be downregulated in hepatocellular cancer.31 The top downregulated circRNA was hsa_circRNA_406225. It aligned with the gene TAMM41 (TAM41 mitochondrial translocator assembly and maintenance homolog, Gene ID: 132001), which is a homolog of mitochondrial translocator assembly and maintenance protein.32,33
We observed 22 dysregulated circRNAs shared by both ESCC and BC. Among them, hsa_circRNA_002908 (hsa_ circ_0002908) was found to be upregulated in peripheral blood mononuclear cells of tuberculosis patients compared with those of paired healthy controls and may be an alternative biomarker for pulmonary tuberculosis.34 Hsa_circRNA_101436 (hsa_circ_0000567) can inhibit colorectal tumor growth and was upregulated in gefitinib-acquired resistant non-small-cell lung cancer cells.35,36 Hsa_circRNA_103110 (hsa_circ_0004771) has been reported to be significantly expressed in BC14 and papillary thyroid carcinoma.37 Hsa_circRNA_000950 (hsa_circ_0001525) and hsa_circRNA_104040 (hsa_circ_0075410) were listed in the top 10 downregulated circRNAs from a microarray on cutaneous squamous cell carcinoma.38 The expression of hsa_circRNA_401977 (hsa_circ_0000567) was found to be downregulated in early stage lung adenocarcinoma tissues and cell lines.39 These findings provided evidence of the role of circRNAs in tumorigenesis.
KEGG analysis indicated that differentially expressed circRNAs were associated with multiple cancers. Many pathways are related to cancer-related functions. For example, the Wnt pathway has vital roles in the prognosis of non-small-cell lung cancer and may have future clinical value.40 The Hippo signaling pathway, consisting of the critical downstream effectors YAP/TAZ, contributes to the development of cancer by resulting in an overgrowth phenotype.41 The MAPK signaling pathway is an essential process for CD97 to promote gastric cancer cell proliferation and invasion.42
Our study has several limitations. First, the sample size is too small to make any reasonable conclusion. These results might not even be educational for further studies by other research groups. Differentially expressed circRNAs discovered in this study need further validation. Second, the paired t-test used in this study to compare circRNA expression between the groups may not have been appropriate for the microarray experiment. Other robust hyperparameter estimations need to be used to protect against hypervariable genes and improve the power to detect differential expression.43,44 Third, the study on the role of circRNAs in human cancers is still in its infancy, and a common standard for reporting and naming circRNAs is lacking. Fourth, we used tissue samples to detect circRNA. More easily acquired and noninvasive clinical samples, such as blood, urine, or saliva, should be explored as sources of biomarkers in future research.
Conclusion
In conclusion, circRNAs are promising candidates as valuable biomarkers for ESCC and BC, although circRNA research is still in its infancy, and the functional role of circRNAs in tumorigenesis is just starting to be elucidated.
Supplementary materials
GO enrichment analysis for differentially expressed genes.
Notes: (A) GO annotation of the top 10 upregulated circRNAs in ESCC; (B) GO annotation of the top 10 downregulated circRNAs in ESCC; (C) GO annotation of the top 10 upregulated circRNAs in BC; (D) GO annotation of the top 10 downregulated circRNAs in BC; (E) GO annotation of 11 shared upregulated circRNAs in ESCC and BC; (F) GO annotation of 11 shared downregulated circRNAs in ESCC and BC.
Abbreviations: circRNA, circular RNA; ESCC, esophageal squamous cell cancer; BC, breast cancer.
Kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis of differentially expressed genes.
Notes: (A) Pathways corresponding to the top 10 upregulated circRNAs in ESCC; (B) Pathways corresponding to the top 10 downregulated circRNAs in ESCC; (C) Pathways corresponding to the top 10 upregulated circRNAs in BC; (D) Pathways corresponding to the top 10 downregulated circRNAs in BC; (E) Pathways corresponding to 11 shared upregulated circRNAs in ESCC and BC; (F) Pathways corresponding to 11 shared downregulated circRNAs in ESCC and BC.
Abbreviations: circRNA, circular RNA; ESCC, esophageal squamous cell cancer; BC, breast cancer.
Table S1.
General information of ESCC patients for microarray test
| Patient no. | Age | Ethnicity | Gender | TNM stage | Histologic differentiation | Tumor location | Histological type |
|---|---|---|---|---|---|---|---|
| 1 | 62 | Chinese Han | Male | T3N1M0 | Moderate-poorly | Middle-lower | Squamous cell cancer |
| 2 | 66 | Chinese Han | Female | T2N0M0 | Well-moderately | Middle | Squamous cell cancer |
| 3 | 66 | Chinese Han | Female | T3N0M0 | Well-moderately | Lower | Squamous cell cancer |
| 4 | 76 | Chinese Han | Male | T2N0M0 | Well-moderately | Middle-lower | Squamous cell cancer |
| 5 | 77 | Chinese Han | Female | T3N0M0 | Well-moderately | Upper | Squamous cell cancer |
| 6 | 69 | Chinese Han | Male | T3N0M0 | Well | Lower | Squamous cell cancer |
| 7 | 75 | Chinese Han | Female | T3N0M0 | Well-moderately | Middle | Squamous cell cancer |
Abbreviation: ESCC, esophageal squamous cell cancer.
Table S2.
General information of BC patients for microarray test
| Patient no. | Age | Ethnic | Menopausal | TNM stage | ER | PR | HER2 | Histological type |
|---|---|---|---|---|---|---|---|---|
| 1 | 46 | Chinese Han | Pre- | T2N0M0 | Positive | Positive | Negative | Invasive ductal cancer |
| 2 | 62 | Chinese Han | Post- | T2N0M0 | Positive | Positive | Positive | Invasive ductal cancer |
| 3 | 41 | Chinese Han | Pre- | T2N2M0 | Positive | Negative | Negative | Invasive ductal cancer |
| 4 | 74 | Chinese Han | Post- | T1N0M0 | Negative | Negative | Positive | Invasive ductal cancer |
Abbreviation: BC, breast cancer.
Acknowledgments
The present study was supported by the National Natural Science Foundation of China (81673249), the Social Development Project in Jiangsu Province (BE2015694), the Scientific Research Innovation Project for Graduate Students in Jiangsu Province (KYCX17_1293), and the Priority Academic Program Development of Jiangsu Higher Education Institutions. The funding agencies had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author contributions
PS and JMW conceived, initiated, and led the study. PS, JS, JMW, and HX analyzed the data with input from all the authors. PS and JMW prepared the manuscript. All authors contributed to data analysis, drafted and revised the paper, and agreed to be accountable for all aspects of the work. All authors reviewed and approved the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Brown JC, Winters-Stone K, Lee A, Schmitz KH. Cancer, physical activity, and exercise. Compr Physiol. 2012;2(4):2775–2809. doi: 10.1002/cphy.c120005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Recio-Boiles A, Babiker HM. Cancer, esophageal. Statpearls; Treasure Island, FL: 2018. [Google Scholar]
- 4.Lao-Sirieix P, Fitzgerald RC. Screening for oesophageal cancer. Nat Rev Clin Oncol. 2012;9(5):278–287. doi: 10.1038/nrclinonc.2012.35. [DOI] [PubMed] [Google Scholar]
- 5.Ohashi S, Miyamoto S, Kikuchi O, Goto T, Amanuma Y, Muto M. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology. 2015;149(7):1700–1715. doi: 10.1053/j.gastro.2015.08.054. [DOI] [PubMed] [Google Scholar]
- 6.Wang B, Song H, Jiang H, Fu Y, Ding X, Zhou C. Early diagnostic potential of APC hypermethylation in esophageal cancer. Cancer Manag Res. 2018;10:181–198. doi: 10.2147/CMAR.S148677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang FL, Yu SJ. Esophageal cancer: risk factors, genetic association, and treatment. Asian J Surg. 2018;41(3):210–215. doi: 10.1016/j.asjsur.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Pusung M, Zeki S, Fitzgerald R. Genomics of esophageal cancer and biomarkers for early detection. Adv Exp Med Biol. 2016;908:237–263. doi: 10.1007/978-3-319-41388-4_12. [DOI] [PubMed] [Google Scholar]
- 9.Anderson BW, Ahlquist DA. Molecular detection of gastrointestinal neoplasia: innovations in early detection and screening. Gastroenterol Clin North Am. 2016;45(3):529–542. doi: 10.1016/j.gtc.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Hou X, Wen J, Ren Z, Zhang G. Non-coding RNAs: new biomarkers and therapeutic targets for esophageal cancer. Oncotarget. 2017;8(26):43571–43578. doi: 10.18632/oncotarget.16721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Liang W, Zhang P, et al. Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Cancer Res. 2017;36(1):152. doi: 10.1186/s13046-017-0624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu L, Sun J, Shi P, et al. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget. 2017;8(27):44096–44107. doi: 10.18632/oncotarget.17307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 16.Gene Ontology Consortium [Accessed June 21, 2018]. Available from: http://www.geneontology.org/
- 17.Kanehisa Laboratories [Accessed June 21, 2018]. Available from: http://www.genome.jp/kegg.
- 18.R Foundation for Statistical Computing [Accessed June 21, 2018]. Available from: https://www.r-project.org/
- 19.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32(5):309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17(11):679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cortes-Lopez M, Miura P. Emerging functions of circular RNAs. Yale J Biol Med. 2016;89(4):527–537. [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 23.He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144. doi: 10.1016/j.canlet.2017.03.027. [DOI] [PubMed] [Google Scholar]
- 24.Xia W, Qiu M, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J, Zhang WW, Peng F, Sun JY, He ZY, Wu SG. Downregulation of hsa_circ_0011946 suppresses the migration and invasion of the breast cancer cell line MCF-7 by targeting RFC3. Cancer Manag Res. 2018;10:535–544. doi: 10.2147/CMAR.S155923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amson R, Pece S, Marine JC, Di Fiore PP, Telerman A. TPT1/TCTP-regulated pathways in phenotypic reprogramming. Trends Cell Biol. 2013;23(1):37–46. doi: 10.1016/j.tcb.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Chen W, Wang H, Tao S, et al. Tumor protein translationally controlled 1 is a p53 target gene that promotes cell survival. Cell Cycle. 2013;12(14):2321–2328. doi: 10.4161/cc.25404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu D, Guo Z, Min W, et al. Upregulation of TCTP expression in human skin squamous cell carcinoma increases tumor cell viability through anti-apoptotic action of the protein. Exp Ther Med. 2012;3(3):437–442. doi: 10.3892/etm.2011.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You Y, Tan J, Gong Y, et al. MicroRNA-216b-5p Functions as a tumor-suppressive RNA by targeting TPT1 in pancreatic cancer cells. J Cancer. 2017;8(14):2854–2865. doi: 10.7150/jca.18931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Zhou Y, Loberg A, Tahara SM, Malik P, Kalra VK. Activated transcription factor 3 in association with histone deacetylase 6 negatively regulates microRNA 199a2 transcription by chromatin remodeling and reduces endothelin-1 expression. Mol Cell Biol. 2016;36(22):2838–2854. doi: 10.1128/MCB.00345-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duan Q, Wang X, Gong W, et al. ER stress negatively modulates the expression of the miR-199a/214 cluster to regulates tumor survival and progression in human hepatocellular cancer. PLoS One. 2012;7(2):e31518. doi: 10.1371/journal.pone.0031518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hicks C, Sitthi-Amorn J, Douglas J, et al. Molecular analysis of central nervous system disease spectrum in childhood acute lymphoblastic leukemia. Clin Med Insights Oncol. 2016;10:5–15. doi: 10.4137/CMO.S18180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kariminejad A, Nafissi S, Nilipoor Y, et al. Intellectual disability, muscle weakness and characteristic face in three siblings: a newly described recessive syndrome mapping to 3p24.3-p25.3. Am J Med Genet A. 2015;167A(11):2508–2515. doi: 10.1002/ajmg.a.37248. [DOI] [PubMed] [Google Scholar]
- 34.Qian Z, Liu H, Li M, et al. Potential diagnostic power of blood circular RNA expression in active pulmonary tuberculosis. EBioMedicine. 2018;27:18–26. doi: 10.1016/j.ebiom.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Li X, Lu L, He L, Hu H, Xu Z. Circular RNA hsa_circ_0000567 can be used as a promising diagnostic biomarker for human colorectal cancer. J Clin Lab Anal. 2018 Jan 15; doi: 10.1002/jcla.22379. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai Y, Huang Y, Huang J, Wen C, Zhou H, Wu L. Differential expression of circular RNAs in gefitinib-acquired resistant non-small cell lung cancer cells. TUMOR. 2017;37(11):1128–1135. [Google Scholar]
- 37.Peng N, Shi L, Zhang Q, Hu Y, Wang N, Ye H. Microarray profiling of circular RNAs in human papillary thyroid carcinoma. PLoS One. 2017;12(3):e0170287. doi: 10.1371/journal.pone.0170287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sand M, Bechara FG, Gambichler T, et al. Circular RNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci. 2016;83(3):210–218. doi: 10.1016/j.jdermsci.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 39.Zhao J, Li L, Wang Q, Han H, Zhan Q, Xu M. CircRNA Expression profile in early-stage lung adenocarcinoma patients. Cell Physiol Biochem. 2017;44(6):2138–2146. doi: 10.1159/000485953. [DOI] [PubMed] [Google Scholar]
- 40.Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst. 2014;106(1):djt356. doi: 10.1093/jnci/djt356. [DOI] [PubMed] [Google Scholar]
- 41.Mo JS, Park HW, Guan KL. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15(6):642–656. doi: 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li C, Liu DR, Li GG, et al. CD97 promotes gastric cancer cell proliferation and invasion through exosome-mediated MAPK signaling pathway. World J Gastroenterol. 2015;21(20):6215–6228. doi: 10.3748/wjg.v21.i20.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat. 2016;10(2):946–963. doi: 10.1214/16-AOAS920. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GO enrichment analysis for differentially expressed genes.
Notes: (A) GO annotation of the top 10 upregulated circRNAs in ESCC; (B) GO annotation of the top 10 downregulated circRNAs in ESCC; (C) GO annotation of the top 10 upregulated circRNAs in BC; (D) GO annotation of the top 10 downregulated circRNAs in BC; (E) GO annotation of 11 shared upregulated circRNAs in ESCC and BC; (F) GO annotation of 11 shared downregulated circRNAs in ESCC and BC.
Abbreviations: circRNA, circular RNA; ESCC, esophageal squamous cell cancer; BC, breast cancer.
Kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis of differentially expressed genes.
Notes: (A) Pathways corresponding to the top 10 upregulated circRNAs in ESCC; (B) Pathways corresponding to the top 10 downregulated circRNAs in ESCC; (C) Pathways corresponding to the top 10 upregulated circRNAs in BC; (D) Pathways corresponding to the top 10 downregulated circRNAs in BC; (E) Pathways corresponding to 11 shared upregulated circRNAs in ESCC and BC; (F) Pathways corresponding to 11 shared downregulated circRNAs in ESCC and BC.
Abbreviations: circRNA, circular RNA; ESCC, esophageal squamous cell cancer; BC, breast cancer.
Table S1.
General information of ESCC patients for microarray test
| Patient no. | Age | Ethnicity | Gender | TNM stage | Histologic differentiation | Tumor location | Histological type |
|---|---|---|---|---|---|---|---|
| 1 | 62 | Chinese Han | Male | T3N1M0 | Moderate-poorly | Middle-lower | Squamous cell cancer |
| 2 | 66 | Chinese Han | Female | T2N0M0 | Well-moderately | Middle | Squamous cell cancer |
| 3 | 66 | Chinese Han | Female | T3N0M0 | Well-moderately | Lower | Squamous cell cancer |
| 4 | 76 | Chinese Han | Male | T2N0M0 | Well-moderately | Middle-lower | Squamous cell cancer |
| 5 | 77 | Chinese Han | Female | T3N0M0 | Well-moderately | Upper | Squamous cell cancer |
| 6 | 69 | Chinese Han | Male | T3N0M0 | Well | Lower | Squamous cell cancer |
| 7 | 75 | Chinese Han | Female | T3N0M0 | Well-moderately | Middle | Squamous cell cancer |
Abbreviation: ESCC, esophageal squamous cell cancer.
Table S2.
General information of BC patients for microarray test
| Patient no. | Age | Ethnic | Menopausal | TNM stage | ER | PR | HER2 | Histological type |
|---|---|---|---|---|---|---|---|---|
| 1 | 46 | Chinese Han | Pre- | T2N0M0 | Positive | Positive | Negative | Invasive ductal cancer |
| 2 | 62 | Chinese Han | Post- | T2N0M0 | Positive | Positive | Positive | Invasive ductal cancer |
| 3 | 41 | Chinese Han | Pre- | T2N2M0 | Positive | Negative | Negative | Invasive ductal cancer |
| 4 | 74 | Chinese Han | Post- | T1N0M0 | Negative | Negative | Positive | Invasive ductal cancer |
Abbreviation: BC, breast cancer.




