Abstract
Background
CMY-2 is the most prevalent pAmpC β-lactamase, but the chromosomal blaCMY-2 gene transfer via horizontal transmission has been seldom reported. This study aimed to describe an ISEcp1-mediated transposition of a chromosomal blaCMY-2 gene from Escherichia coli into a small endogenous ColE1-like plasmid, resulting in elevated resistance to extended-spectrum cephalosporins.
Methods
Three ESCs-resistant ST641 E. coli strains EC6413, EC4103 and EC5106 harbored the blaCMY-2 gene. S1-PFGE, I-ceu I-PFGE, Southern blotting and electroporation experiments were performed to investigate the location and transferability of blaCMY-2. The genetic context and gene expression of blaCMY-2 in the original isolates and the corresponding electroporants were explored by PCR mapping, primer walking strategy and RT-qPCR.
Results
The blaCMY-2-containing region (ISEcp1-blaCMY-2-∆blc-∆yggR-∆tnp1-orf7-orf8-orf9-∆tnp2-∆hsdR) was transposed into endogenous ColE1-like plasmid pSC137 in the process of electroporation at very low frequencies (10–8–10–9). The transpositions resulted in novel larger blaCMY-2-harboring ColE1-like plasmids with size of 14,845 bp, enabling increase in MICs of 2 to 8-fold for cefotaxime, ceftiofur, and ceftazidime in recipient strains over their respective original counterparts. Transcriptional level analysis revealed that the increased blaCMY-2 expression was correlated with elevated MIC values of cephalosporins. The blaCMY-2 transposition unit was identical to that in a clinical isolate E. coli TN44889 from France isolated in 2004.
Conclusions
Our results firstly demonstrated that ISEcp1 mediated a transposition of chromosome-borne blaCMY-2 into an endogenous ColE1-like plasmid by electroporation. Amplification of the blaCMY-2 gene facilitates the strain adaptation to a changed environment with an elevated antibiotic pressure.
Keywords: blaCMY-2, chromosome-borne, ColE1-like plasmid, ISEcp1-mediated transposition, extended-spectrum cephalosporin
Introduction
Third- and fourth-generation extended-spectrum cephalosporins (ESCs) are used to treat both intestinal and extraintestinal Escherichia coli infections in human and veterinary medicine.1,2 However, resistance rates due to extended spectrum β-lactamases and plasmid-mediated AmpC (pAmpC) β-lactamases are increasing.3 Moreover, pAmpC β-lactamases are active against cephamycins, especially cefoxitin, and are not inactivated by β-lactam/β-lactamase inhibitor combinations such as amoxicillin/clavulanic acid combination.4
CMY-2 is the most prevalent pAmpC β-lactamase and has been reported in E. coli worldwide.5 This is largely due to the spread of IncA/C and IncI1 plasmids among E. coli from humans, animals, and even environmental sources.6–8 In addition to IncA/C and IncI1 plasmids, insert sequence, ISEcp1, also plays an important role in spread of blaCMY-2.9–11 ISEcp1 seems to mobilize the adjacent resistance genes through transposition by using a weakly related downstream sequence in combination with left inverted repeat (IRL).12 The plasmid-borne blaCMY-2 most likely originated from the Citrobacter freundii chromosome by ISEcp1-mediated transposition.9,11
Research on antimicrobial resistance plasmids has been mainly focused on large plasmids and the role that small plasmids play in resistance gene transfer is not clear. ColE1-like plasmids are small with sizes ranging from ~2 to ~10 kb and have their replication driven only by host-encoded proteins.13,14 Rather surprisingly, prior to 2006, ColE1-like plasmids and other small plasmids were seldom implicated in the spread of antibiotic resistance.15 However, during the last decade, ColE1-like plasmids were identified which disseminated resistance genes for β-lactams (blaCMY, blaCTX-M, blaIMP-8, blaOXA-181, blaOXA-232, blaKPC-2, blaGES-5 and blaBEL-1), quinolones(qnrB, qnrS, and aac-(6′)-Ib-cr) as well as kanamycin (aph(3′)-I).13,16–23 ColE1 plasmids are not self-transmissible but can be mobilized by a helper plasmid.24 Furthermore, like many other non-conjugative plasmids, ColE1 plasmids are multiple copy plasmids in E. coli.25 The high copy number could maintain their segregational stability in the absence of any active and specific segregation mechanism.26 Therefore, once the resistance genes are acquired by ColE1-like plasmids, their mobility and high copy number may accelerate dissemination of these genes.
Numerous studies have indicated a greater probability of the spread of plasmid-borne blaCMY-2 genes. However, the chromosomal blaCMY-2 gene transfer through horizontal transmission has been seldom documented, except for an SXT/R391-like integrative conjugative element, which was implicated in the spread of chromosomal blaCMY-2 in Proteus mirabilis.27 In our previous study, we found that the blaCMY-2 gene from three ESC-resistant ST641 E. coli strains could not be transferred by conjugation, indicating an alternative gene location of blaCMY-2 in these strains.28 To determine the location of blaCMY-2 gene and its transferability in these E. coli strains, we conducted a series of experiments, including electroporation, gene location, plasmids analysis, and the genetic contexts of blaCMY-2, and confirmed that the chromosomal locations of the blaCMY-2 genes could transfer into an endogenous ColE1-like plasmid through an ISEcp1-mediated transposition.
Materials and methods
Bacterial strains
Three ESC-resistant E. coli strains EC6413, EC4103, and EC5106 were isolated from rectal swab samples from sows on a large farrowing farm in Southern China in August 2011 as previously reported.28 They were identified as clonal ST641 E. coli strains, but only strains EC6413 and EC4103 belonged to the same XbaI-pulsed-field gel electrophoresis (PFGE)-type (>90% similarity).
Gene location and transfer of blaCMY-2
We used S1-PFGE and I-Ceu I-PFGE to determine the genomic locations of the blaCMY-2 genes. Briefly, plasmid analysis was carried out in the three original isolates by DNA linearization with S1 nuclease (Takara, Dalian, China) followed by PFGE.29 Total DNA was also digested with I-Ceu I (NEB, Ipswich, MA, USA) followed by PFGE.30 Southern blotting was carried out on both S1-PFGE and I-Ceu I-PFGE gels with digoxigenin-labeled probes (Roche Diagnostics GmbH, Mannheim, Germany) specific for blaCMY-2 gene and blaCMY-2/23S rDNA gene, respectively.
Plasmid DNA from the original strains was extracted by Qiagen Prep Plasmid Midi Kit (Qiagen NV, Venlo, the Netherlands) and electroporated into electrocompetent E. coli DH5α (TaKaRa, Dalian, China) and E. coli DH10B (stored in our laboratory) by using a Gene Pulser apparatus (Bio-Rad Laboratories Inc., Hercules, California, USA). Electroporants were selected on MacConkey agar plates supplemented with cefoxitin (16 µg/mL). Cells harboring blaCMY-2 were confirmed by polymerase chain reaction (PCR) with specific primers as previously described.31 The minimum inhibitory concentrations (MICs) of cefoxitin (FOX), ceftiofur (CIF), cefotaxime (CTX), ceftazidime (CAZ), kanamycin (KAN), amikacin (AMK), florfenicol (FLF), doxycycline (DOX), and ciprofloxacin (CIP) were determined for the electroporants and the original isolates by the agar dilution method. The results were interpreted according to Clinical and Laboratory Standards Institute (CLSI, 2013; 2015) standards. E. coli strain ATCC 25922 was used as the quality control strain. Transfer frequency was calculated as the number of electroporants harboring blaCMY-2 divided by the starting number of E. coli cells used for electroporation. Incompatibility (Inc) groups were assigned by PCR-based replicon typing (PBRT) of the electroporants as previously described.32
Detection of the flanking regions surrounding the blaCMY-2 gene
The genetic contexts of blaCMY-2 in the original isolates and the complete nucleotide sequences of plasmids harboring blaCMY-2 in the electroporants were explored by PCR mapping and a primer walking strategy (Table S1). In addition, one specific set of primers was designed to detect the endogenous ColE1-like plasmid pSC137. The PCR amplification region contained HP4-IS5-RNAII/RNAI. Another five primer pairs were designed to identify the blaCMY-2 loci containing the conserved blaCMY-2 region and DNA segments from plasmid pSC137.
Plasmid analysis and second-round electroporation
To further determine the location of blaCMY-2 and RNAII (involved in the replication of ColE1-like plasmids), plasmid analysis was carried out in the three electroporants with S1-PFGE followed by Southern blot hybridization with the blaCMY-2 and RNAII probes (Table S1) as described above. A second round of electroporation was performed by using plasmids isolated from the first-round electroporants, and transfer frequencies of blaCMY-2 were scored as described above. Electroporants from the second round were also tested for antimicrobial susceptibility.
Relative quantification of the mRNA expression of blaCMY-2
E. coli DH5α electroporants as well as the original isolates were evaluated for the expression of blaCMY-2 gene. Total RNA was extracted from 1 mL of a 24 h culture in Lysogeny broth (LB) without antibiotics grown at 37°C using an RNAprep pure Cell/Bacteria Kit (Tiangen Biotech, Beijing, China). The total RNA was reverse transcribed by using a PrimeScript RT reagent kit (with DNA Eraser) and random hexamers according to the manufacturer’s instructions (TaKaRa). The cDNA samples were used for quantitative real-time PCR (qPCR). Primers used in qPCR are listed in Table S1, and the 16S rRNA gene was used as an internal control for mRNA quantification. Quantification was performed on a Bio-Rad IQ5 instrument (Bio-Rad Laboratories Inc.) by using SYBR Premix Ex Taq TM (TaKaRa) according to the manufacturer’s instructions. The thermal conditions were initial denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 40 s, 63°C for 40 s, and 72°C for 45 s. qPCR assays were performed in duplicate, and each assay sample was tested in triplicate. Product specificity was verified by melting curve analysis by using the software provided with the instrument. 16S rRNA was used to normalize for gene expression levels. Relative quantification was calculated by the 2−∆∆CT method33 (∆∆CT = (CT,Target–CT,Control)electroporant − (CT,Target–CT,Control)original isolate).
Nucleotide sequence accession number
The complete nucleotide sequences of plasmids pSC137, pEC4103, pEC5106, and pEC6413 have been assigned Gen-Bank accession numbers KT074362, KY612498, KY612499, and KY612500, respectively.
Results
Location and transfer of blaCMY-2
In our original experiments, we identified three ST641 E. coli strains that harbored the blaCMY-2 gene, but our attempts at conjugation were unsuccessful.28 Analysis of genomic DNA from these strains using S1-PFGE identified three or four visible plasmid bands in S1-PFGE gels with sizes ranging from <20 to ~140 kb. Southern blot analyses of I-Ceu I and S1-PFGE gels using blaCMY-2 probe revealed hybridization only to the chromosome. None of the endogenous plasmids were hybridized to the blaCMY-2 probe (Figure S1).
Nonetheless, the three ST641 E. coli strains were still able to transfer cefoxitin resistance as well as the blaCMY-2 gene to the recipient strains E. coli DH5α and DH10B, albeit at low frequencies (10–8–10–9). Interestingly, all the electroporants were ESC resistant with MICs equal to 16–32 µg/mL for CTX, CIF, and CAZ. These values represented 2- to 8-fold increase when compared with their respective original strains (Table 1). No other non-β-lactam resistance was transferred in this process, and no replicon was identified by PBRT in any of the electroporants.
Table 1.
Characteristics of the three CMY-2-ST641 E. coli strains and their electroporants
| Strains | MIC (μg/mL) of the donors
|
Gene location of blaCMY-2 in the original isolates | MIC (μg/mL) of the electroporants
|
Size of pColE1-like CMY from electroporants (bp) | Frequency of the first round of electroporation DH5α/DH10B | Frequency of the second round of electroporation DH5α/DH10B | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FOX | CIF | CTX | CAZ | FOX | CIF | CTX | CAZ | |||||
| EC5106 | 128 | 16 | 4 | 16 | Chromosome | 64 | 32 | 16 | 32 | 14,845 | 10–8–10–9 | 10–2–10–3 |
| EC6413 | 64 | 4 | 2 | 8 | Chromosome | 64 | 32 | 16 | 32 | 14,845 | 10–8–10–9 | 10–2–10–3 |
| EC4103 | 64 | 4 | 2 | 8 | Chromosome | 64 | 32 | 16 | 32 | 14,845 | 10–8–10–9 | 10–2–10–3 |
| DH5α/DH10B | – | – | – | – | – | 2 | 0.125 | 0.03 | 0.03 | – | – | – |
Note: Electroporants were challenged for the electroporation by using recipients DH5α or DH10B.
Abbreviations: FOX, cefoxitin; CIF, ceftiofur; CTX, cefotaxime; CAZ, ceftazidime; MIC, minimal inhibitory concentration.
Detection of the flanking regions of the blaCMY-2 gene
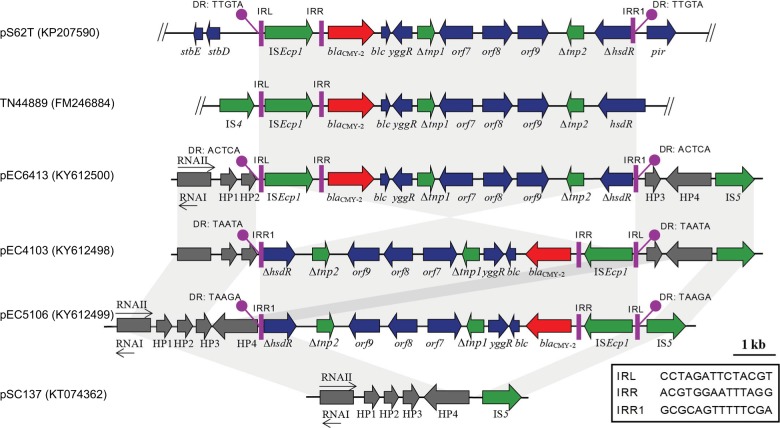
In the three ST641 E. coli isolates, the genetic context of the blaCMY-2 gene was identical to that of a clinical isolate E. coli TN44889 from France in 2004 (Acc. No. FM246884). This region included a blaCMY-2 gene-containing region comprising 11 open reading frames (ORFs) (Figure 1). For the electroporants, we obtained the complete nucleotide sequences of the 14,845 bp circular plasmids (designated pEC6413, pEC4103, and pEC5106) harboring blaCMY-2 (Figures 1 and 2). In each plasmid, the 10,179 bp region containing ISEcp1-blaCMY-2-∆blc-∆yggR-∆tnp1-orf7-orf8-orf9-∆tnp2-∆hsdR was identical to that from the chromosome of each original isolate. “∆tnp1” was 97% identical to the last 144 amino acids of IS200 (Acc. No. 2002282A), and “∆tnp2” was 99% identical to the last 139 amino acids of IS60 Orf2 in Shigella flexneri 2a str. 301 (Acc. No. NP_707701). This region was also identical to that of the IncX4 plasmid pS62T (Acc. No. KP207590) found in our previous study (Figure 1).31 However, each plasmid had a different arrangement of the blaCMY-2-containing region and a complete transposition unit flanked by 5-bp direct repeats (DRs) that bounded this region in pEC6413 (ACTCA), pEC4103 (TAATA), and pEC5106 (TAAGA). One of the putative 5-bp DR was located immediately adjacent to the IRL (CCTAGATTCTACGT) of ISEcp1. The other was located immediately adjacent to the deduced right inverted repeats of ISEcp1 (IRR1: GCGCAGTTTTTCGA).
Figure 1.
Characteristics of the genetic contexts of blaCMY-2. Structural comparison of plasmids pEC6413, pEC4103, and pEC5106 from the electroporants, ColE1-like plasmid pSC137 from the clinical Escherichia coli strain, IncX plasmid pS62T from E. coli strain (Acc. No. KP207590), and the clinical E. coli strain TN44889 (Acc. No. FM246884). Regions of >99% homology are marked by grey shading. The gray rectangles indicate replication-associated genes of ColE1-like plasmids; gray arrows indicate other genes in the original ColE1-like plasmids. The resistance genes, insertion sequences, and other accessory genes are indicated by red, green, and blue arrows, respectively.
Abbreviations: DR, direct repeat sequences generated by ISEcp1-mediated transposition; IRL and IRR1, left and right inverted repeats of ISEcp1, respectively.
Figure 2.
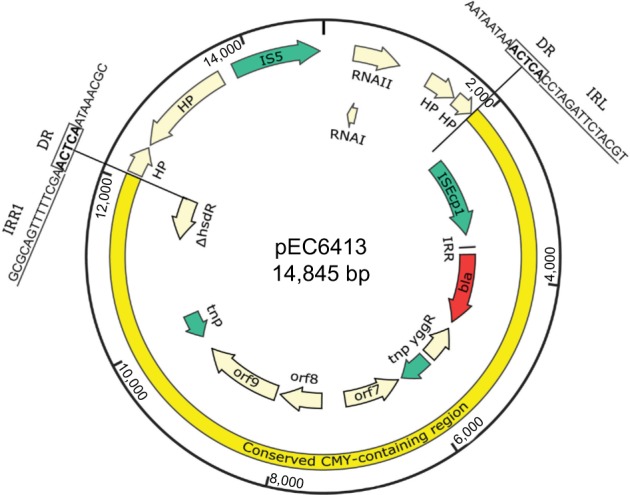
Characteristics of the complete nucleotide sequences of the plasmids pEC6413. The DRs generated by ISEcp1-mediated transposition are highlighted in boldface. The IRL of ISEcp1 and the IRR1 of ISEcp1 are marked by the underlined letters.
Abbreviations: DR, direct repeat sequences generated by ISEcp1-mediated transposition; IRL and IRR1, left and right inverted repeats of ISEcp1, respectively.
Further analysis revealed that this transposition unit had been inserted into a small plasmid pSC137 (Figure 1). Plasmid pSC137 is a 4,661-bp ColE1-like plasmid containing five ORFs. The region from 355 to 899 bp that contained the RNA I/II region was 94% identical to that in the ColE1-like pNPO1 (Acc. No. KF992024). This region is involved in the initiation and control of ColE1-like plasmid replication (Wang et al14). The segments from 120 to 1,077 bp and 686 to 1,796 bp were 91% and 95% identical to that in the ColE1-like plasmids pNPO1 (Acc. No. KF992024; 190-1,170) and pB1022 (Acc. No. JQ319766; 1-1,121), respectively. Furthermore, pSC137 was identical to its counterpart in the CMY-containing ColE1-like plasmids (Figure 2).
PCR confirmed that pSC137-like plasmids existed in the three original strains, while blaCMY-2 loci were absent from pSC137-like plasmids. But the blaCMY-2 regions appeared on pSC137-like plasmids in the electroporants (Table S2). These results demonstrated that the chromosomal blaCMY-2 region could be transferred to the endogenous pSC137-like plasmids, generating larger ColE1-like plasmids containing blaCMY-2 by electroporation.
Plasmid analysis and second-round electroporation
Southern blot analysis was performed on the uncut plasmids from both the original isolates and E. coli DH5α electroporants to determine the location of the blaCMY-2 and RNAII genes. In the original isolates, none of the endogenous plasmids was hybridized with the blaCMY-2 gene which indicated that this gene may be located on the chromosome (Figure S1). On the other hand, a small endogenous plasmid was hybridized with the RNAII probe. In the electroporants, both the blaCMY-2 and RNAII probes were hybridized to the same plasmids, and they were larger than the small endogenous plasmids that were hybridized with the RNAII probe. These data indicated that the chromosomal blaCMY-2 gene had transferred to endogenous plasmid pSC137 that contained the RNAII gene.
Compared to that in the first round of electroporation, the plasmids could be transferred at high frequencies (10–2–10–3) in the second round of electroporation, but the MIC values of FOX, CTX, CIF, and CAZ in these electroporants were the same as that obtained in the first round of electroporation (Table 1).
Relative quantification of the mRNA expression of blaCMY-2
Transfer of blaCMY-2 gene resulted in increased ESC resistance in the electroporants over that of the respective original isolates. This indicated that gene expression of blaCMY-2 was most likely increased. The steady state levels of blaCMY-2 mRNA were elevated 14.420-fold (±1.084), 14.455-fold (±1.309), and 7.980-fold (±0.833) in EC6413T, EC4103T, and EC5106T with respect to the original isolates, respectively.
Discussion
In this study, we describe an ISEcp1-mediated transposition of the chromosomally encoded blaCMY-2 gene into an endogenous ColE1-like plasmid in three ESC-resistant ST641 E. coli strains, which was supported by evidence: 1) the region including 10 ORFs (ISEcp1-blaCMY-2-∆blc-∆yggR-∆tnp1-orf7-orf8-orf9-∆tnp2-∆hsdR) was identical in the ColE1-like plasmids in electroporants and in chromosome of the original isolates; 2) the size of ColE1-like plasmids in electroporants were larger than that in the respective original isolates; 3) a suspected DR exactly emerged neighboring IRL (CCTAGATTCTACGT) and the proposed IRR1 (GCGCAGTTTTTCGA) of ISEcp1 (Figures 1 and 2). Thus, we speculated that the fragment carrying blaCMY-2 could be introduced into the ColE1-like plasmid during the electroporation experiments.
ISEcp1 plays an important role in the mobilization of blaCMY-2 gene, and in general, ISEcp1 located in front of the antimicrobial resistance gene and moves toward its adjacent region by recognizing its own IRL and supposed IRR, resulting in 5-bp DRs.34 In our present study, the proposed transposition fragment comprises a typical ISEcp1-mediated unit, in which ISEcp1 was 116 bp in front of blaCMY-2 and followed by ∆blc-∆yggR-∆tnp1-orf7-orf8-orf9-∆tnp2-∆hsdR. All the transposition fragments shared 100% identity in the three strains, transposed in identical ColE1-like plasmids. However, the transposition unit in pEC5106 and pEC4103 was completely reversed compared with that in pEC6413, and it was inserted in different locations in ColE1-like plasmids (Figure 1). The transposition generated 5-bp different DRs (ACTCA, TAATA, and TAAGA) adjacent to the IRLs and IRR1s of ISEcp1 elements in the three strains. It agreed with previous research where ISEcp1-mediated transposition always resulted in AT-rich DR.34 In addition to blaCMY-2 gene, ISEcp1-mediated transposition was also reported to be related to the spread of blaCTX-M,34,35 blaKPC,36 qnrB-like genes, 37 and rmtC gene.38
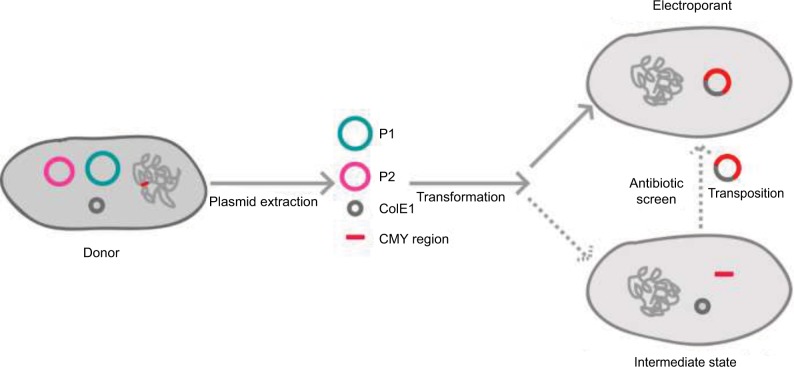
Indeed, ISEcp1-mediated transposition might take place in both the original strain and electroporation. However, PCR detection and Southern blotting confirmed the absence of blaCMY-2 loci in pSC137 in the original strains, and it was also unsuccessful for our several attempts to obtain the transconjugants harbored blaCMY-2 by conjugation (Table S2). Therefore, we speculate that ISEcp1-mediated transposition of blaCMY-2 probably occurred in recipient strains rather than original strains. But it could not be excluded that they occurred in the original strains and could not be detected by Southern blot or PCR. Based on the observed structure and detailed sequence analysis, we propose a most probable model for the route of chromosome-borne blaCMY-2 into an endogenous ColE1-like plasmid. First, the CMY-containing region was dropped from chromosomal DNA in the process of ISEcp1-mediated transposition. Second, both this region and plasmid DNA were acquired through plasmid extraction and electroporation. Subsequently, the CMY region was integrated into the ColE1-like plasmid under cefoxitin selective pressure, generating a novel ColE1-like plasmid carrying blaCMY-2 (Figure 3).
Figure 3.
Schematic representation of the transfer of the chromosome-encoded blaCMY-2 gene into ColE1-like plasmid. On the left side is the donor strain. On the right side is the electroporant involved in the transposition phenomena by electroporation.
In the present study, the efficiency of blaCMY-2 translocation in the second-round was much higher than that in the first-round electroporation, which was consistent with, that ColE1-like plasmid could be transformed into E. coli with high efficiency.39 Interestingly, the transfer of blaCMY-2 into the electrocompetent cells contributed to increasing resistance to ESCs even over that of the respective original isolates (Table 1). Positive correlations between β-lactam MICs and β-lactamase gene expression have been previously shown.40,41 In our study, the relative expression (steady-state mRNA levels) of blaCMY-2 was significantly increased in electroporants compared with the original isolates, which might result from the high copy number of ColE1-like plasmids.42 The chromosome-borne ISEcp1-mediated transposition of blaCMY-2 gene into high copy number ColE1-like plasmids would not only increase the resistance levels against cephalosporins but also greatly improve the potential to spread the blaCMY-2 gene due to the raised transfer efficiency.
Conclusion
This is the first report of ISEcp1-mediated transposition of the chromosome-borne blaCMY-2 gene into a small endogenous plasmid with high copy numbers in E. coli. This may increase the levels of cephalosporins resistance, providing an alternative adaptive survival mechanism for bacteria, especially at high cephalosporin concentrations, and facilitate the spread of blaCMY-2 gene in E. coli strains.
Supplementary materials
(A) I-Ceu I-PFGE and Southern blot hybridization with the 23S rDNA and blaCMY-2 probes; (B) S1 nuclease-PFGE Southern blot hybridization with the blaCMY-2 and ColE1-like probe. Line M: H9812 marker; Lines 1–6: EC6413, EC4103, and EC5106, and their corresponding electroporants EC6413T, eC4103T, and EC5106T. Arrows represent the band hybridized with the blaCMY-2 or ColE1-like probe.
Abbreviation: PFGE, pulsed-field gel electrophoresis.
Table S1.
Primers used for screening for genes and PCR mapping
| PCR | Primer | Primer sequence (5′→3′) | Product length (kb) | Target | Reference |
|---|---|---|---|---|---|
| A | AF | GCACTTAGCCACCTATACGGCAG | 0.758 | blaCMY-2 for digoxigenin-labeled probes | 1 |
| AR | GCTTTTCAAGAATGCGCCAGG | ||||
| B | BF | AATGATGGCCAGGCTGTCTCC | – | 23SrRNA for digoxigenin-labeled probes | 2 |
| BR | CCGCCGTCGATATGAACTCTTG | ||||
| C | CF | GAAGGTTCTCAGAGCTGCAAC | 0.3 | RNAII for digoxigenin-labeled probes | This study |
| CR | GCCGCGTTTATCTCATTCCAC | ||||
| D | DF | ATCGTCTTTTACCGCCTGTCC | 3.1 | Screening for the plasmid of pSC137 | This study |
| DR | GTTAGCCCTATCCTGCATCGT | ||||
| E | EF | GTCATCGCTGGGAAATCGAAC | 3.8 | Junction between the 1S4 and yggR | 3 |
| ER | GCATAACGTCTCGGATCTACACC | ||||
| F | FF | AACTTGACGCCGAAGCCTA | 4.8 | Junction between the blaCMY-2 and orf9 | 3 |
| FR | TACGCCTGCAAAATATCACCA | ||||
| G | GF | TTTGTACTGCCAACGTATCCAA | 2.2 | Junction between the orf9 and ΔhsdR | 3 |
| GR | AAAGAACGGGAAATTGCCAAC | ||||
| H | HF | CCATAACAGCGGAATGACACC | 3.0 | Junction between RNAII and blaCMY-2 in the plasmid of pEC6413 | This study |
| HR | CAGAGCGCAGCATAACGAT | ||||
| I | IF | ACCGGGTTTTCATCCACGA | 1.12 | Junction between ΔhsdR and HP3 in the plasmid of pEC6413 | This study |
| IR | GTGCGTCTGACCAATATCCAC | ||||
| J | JF | CCATAACAGCGGAATGACACC | 3.2/4.8 | Junction between RNAII and ΔhsdR in the plasmids of pEC4103 or pEC5106 | This study |
| JR | AGGGCTTTACCTGTCAGCTC | ||||
| K | KF | CAGAGCGCAGCATAACGAT | 2.3/5.3 | Junction between blaCMY-2 and HP3 in the plasmid of pEC4103 or pEC5106 | This study |
| KR | CGTAATCCGTTGCCAGAGCC | ||||
| L | LF | TATTGTAGCATCGGTTTCCCA | 5.4 | Junction between the ISEcp1and ΔhsdR including pSC137 | This study |
| LR | GCGCGAACATACATATCCAGT | ||||
| M | MF | GCTGCTGACAGCCTCTTT | 0.197 | qCMY2 for quantitative real-time PCR assays | 4 |
| MR | GCGTGACTGGGTGGTTAT | ||||
| N | NF | GGCCGCAAGGTTAAAACTCAAATG | 0.243 | 16S rRNA for quantitative real-time PCR assays | 5 |
| NR | AACCGCTGGCAACAAAGGATAAGG |
Notes: F, forward primer; R, reverse primer. “L” represents reverse PCR primers and its 4 amplification region containing ISEcp1-pSC137-ΔhsdR.
Abbreviation: PCR, polymerase chain reaction.
Table S2.
PCR-typing blaCMY-2 gene-containing loci transferred into endogenous plasmid pSC137
| PCR typing | Target | Size (kb) | Electroporants | Wild strains | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| EC5106T | EC6413T | EC4103T | EC5106 | EC6413 | EC4103 | |||
| H | Junction between RNAII and blaCMY-2 | 3.0 | + | − | − | − | − | − |
| I | Junction between ΔhsdR and HP3 | 1.12 | + | − | − | − | − | − |
| J | Junction between RNAII and ΔhsdR | 3.2/4.8 | − | + | + | − | − | − |
| K | Junction between blaCMY-2 and HP3 | 2.3/5.3 | − | + | + | − | − | − |
| L | Region containing ISEcp1-pSC137-ΔhsdR | 5.4 | + | + | + | − | − | − |
Note: (+) positive and (–) negative.
Abbreviation: PCR, polymerase chain reaction.
References
- 1.Kiiru J, Kariuki S, Goddeeris BM, Butaye P. Analysis of β-lactamase phenotypes and carriage of selected β-lactamase genes among Escherichia coli strains obtained from Kenyan patients during an 18-year period. BMC Microbiol. 2012;12:155. doi: 10.1186/1471-2180-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harada S, Ishii Y, Saga T, Tateda K, Yamaguchi K. Chromosomally encoded blaCMY-2 located on a novel SXT/R391-related integrating conjugative element in a Proteus mirabilis clinical isolate. Antimicrob Agents Chemother. 2010;54(9):3545–3550. doi: 10.1128/AAC.00111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang LX, Sun J, Li L, et al. Dissemination of the chromosomally encoded CMY-2 cephalosporinase gene in Escherichia coli isolated from animals. Int J Antimicrob Agents. 2015;46(2):209–213. doi: 10.1016/j.ijantimicag.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Lee K, Kusumoto M, Sekizuka T, et al. Extensive amplification of GI-VII-6, a multidrug resistance genomic island of Salmonella enterica serovar Typhimurium, increases resistance to extended-spectrum cephalosporins. Front Microbiol. 2015;6:78. doi: 10.3389/fmicb.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karczmarczyk M, Martins M, Quinn T, Leonard N, Fanning S. Mechanisms of fluoroquinolone resistance in Escherichia coli isolates from food-producing animals. Appl Environ Microbiol. 2011;77(20):7113–7120. doi: 10.1128/AEM.00600-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Acknowledgments
This work was supported by the Programs of Changjiang Scholars and Innovative Research Team in University of Ministry of Education of China (IRT13063), the National Key Research and Development Program (2016YFD0501300), Pearl River S&T Nova Program of Guangzhou (Grant no. 201610010036), the National Natural Science Fund of China (Grant no. 31402247), the National Natural Science Fund of China (Grant no. 31402186), and the Natural Science Foundation of Shandong Province (Grant no. ZR2014CP021).
Footnotes
Author contributions
J Sun, Y-H Liu, and P-X Liao designed the experiments and provided reagents and supplies. L-X Fang performed the experiments, analyzed the data, and wrote the manuscript. J Sun, L Li and X-P Li analyzed the data and revised the manuscript. X-P Li, M-Y Chen, C-Y Wu, and L-L Li performed the experiments. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Collignon P, Powers JH, Chiller TM, Aidara-Kane A, Aarestrup FM. Clin Infect Dis. 1. Vol. 49. World Health Organization; 2009. ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies for the use of antimicrobials in food production animals; pp. 132–141. [DOI] [PubMed] [Google Scholar]
- 2.Collignon PC, Conly JM, Andremont A, et al. Clin Infect Dis. 8. Vol. 63. World Health Organization; 2016. ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies to control antimicrobial resistance from food animal production; pp. 1087–1093. [DOI] [PubMed] [Google Scholar]
- 3.Seiffert SN, Hilty M, Perreten V, Endimiani A. Extended-spectrum cephalosporin-resistant Gram-negative organisms in livestock: an emerging problem for human health? Drug Resist Updat. 2013;16(1–2):22–45. doi: 10.1016/j.drup.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Medeiros AA. Evolution and dissemination of beta-lactamases accelerated by generations of beta-lactam antibiotics. Clin Infect Dis. 1997;24(Suppl 1):S19–S45. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 5.Philippon A, Arlet G, Jacoby GA. Plasmid-determined AmpC-type-lactamases. Antimicrob Agents Chemother. 2002;46(1):1–11. doi: 10.1128/AAC.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan JJ, Hong CY, Ko WC, et al. Dissemination of blaCMY-2 among Escherichia coli isolates from food animals, retail ground meats, and humans in Southern Taiwan. Antimicrob Agents Chemother. 2004;48(4):1353–1356. doi: 10.1128/AAC.48.4.1353-1356.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin LC, Weir EK, Poppe C, Reid-Smith RJ, Boerlin P. Characterization of blaCMY-2 plasmids in Salmonella and Escherichia coli isolates from food animals in Canada. Appl Environ Microbiol. 2012;78(4):1285–1287. doi: 10.1128/AEM.06498-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamang MD, Nam HM, Jang GC, et al. Molecular characterization of extended-spectrum-beta-lactamase-producing and plasmid-mediated AmpC beta-lactamase-producing Escherichia coli isolated from stray dogs in South Korea. Antimicrob Agents Chemother. 2012;56(5):2705–2712. doi: 10.1128/AAC.05598-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verdet C, Gautier V, Chachaty E, et al. Genetic context of plasmid-carried blaCMY-2-like genes in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53(9):4002–4006. doi: 10.1128/AAC.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mata C, Miró E, Alvarado A, et al. Plasmid typing and genetic context of AmpC β-lactamases in Enterobacteriaceae lacking inducible chromosomal ampC genes: findings from a Spanish hospital 1999–2007. J Antimicrob Chemother. 2012;67(1):115–122. doi: 10.1093/jac/dkr412. [DOI] [PubMed] [Google Scholar]
- 11.L-H Su, H-L Chen, J-H Chia, et al. Distribution of a transposon-like element carrying blaCMY-2 among Salmonella and other Enterobacteriaceae. J Antimicrob Chemother. 2006;57(3):424–429. doi: 10.1093/jac/dki478. [DOI] [PubMed] [Google Scholar]
- 12.Partridge SR. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev. 2011;35(5):820–855. doi: 10.1111/j.1574-6976.2011.00277.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen CY, Strobaugh TP, Jr, Frye JG. Characterization of small ColE1-like plasmids conferring kanamycin resistance in Salmonella enterica subsp enterica serovars Typhimurium and Newport. Plasmid. 2010;63(3):150–154. doi: 10.1016/j.plasmid.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Yuan Z, Hengge UR. Processing of plasmid DNA with ColE1-like replication origin. Plasmid. 2004;51(3):149–161. doi: 10.1016/j.plasmid.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Rychlík I, Gregorova D, Hradecka H. Distribution and function of plasmids in Salmonella enterica. Vet Microbiol. 2006;112(1):1–10. doi: 10.1016/j.vetmic.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 16.Papagiannitsis CC, Dolejska M, Izdebski R, et al. Characterization of pKP-M1144, a novel ColE1-like plasmid encoding IMP-8, GES-5, and BEL-1 beta-lactamases, from a Klebsiella pneumoniae sequence type 252 isolate. Antimicrob Agents Chemother. 2015;59(8):5065–5068. doi: 10.1128/AAC.00937-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Toro M, Rodriguez I, Rojo-Bezares B, et al. pMdT1, a small ColE1-like plasmid mobilizing a new variant of the aac(6′)-Ib-cr gene in Salmonella enterica serovar Typhimurium. J Antimicrob Chemother. 2013;68(6):1277–1280. doi: 10.1093/jac/dkt001. [DOI] [PubMed] [Google Scholar]
- 18.Cao V, Lambert T, Courvalin P. ColE1-like plasmid pIP843 of Klebsiella pneumoniae encoding extended-spectrum beta-lactamase CTX-M-17. Antimicrob Agents Chemother. 2002;46(5):1212–1217. doi: 10.1128/AAC.46.5.1212-1217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zioga A, Whichard JM, Kotsakis SD, Tzouvelekis LS, Tzelepi E, Miriagou V. CMY-31 and CMY-36 cephalosporinases encoded by ColE1-like plasmids. Antimicrob Agents Chemother. 2009;53(3):1256–1259. doi: 10.1128/AAC.01284-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu SW, Dornbusch K, Kronvall G, Norgren M. Characterization and nucleotide sequence of a Klebsiella oxytoca cryptic plasmid encoding a CMY-type beta-lactamase: confirmation that the plasmid-mediated cephamycinase originated from the Citrobacter freundii AmpC betalactamase. Antimicrob Agents Chemother. 1999;43(6):1350–1357. doi: 10.1128/aac.43.6.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sidjabat HE, Kennedy K, Silvey A, Collignon P, Paterson DL. Emergence of bla(OXA-181)-carrying ColE plasmid in Klebsiella pneumoniae in Australia. Int J Antimicrob Agents. 2013;41(3):294–296. doi: 10.1016/j.ijantimicag.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Potron A, Rondinaud E, Poirel L, et al. Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D beta-lactamase from Enterobacteriaceae. Int J Antimicrob Agents. 2013;41(4):325–329. doi: 10.1016/j.ijantimicag.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Porres-Osante N, Azcona-Gutierrez JM, Rojo-Bezares B, Undabeitia E, Torres C, Saenz Y. Emergence of a multiresistant KPC-3 and VIM-1 carbapenemase-producing Escherichia coli strain in Spain. J Antimicrob Chemother. 2014;69(7):1792–1795. doi: 10.1093/jac/dku055. [DOI] [PubMed] [Google Scholar]
- 24.Keenleyside WJ, Whitfield C. Lateral transfer of rfb genes: a mobilizable ColE1-type plasmid carries the rfbO:54 (O:54 antigen biosynthesis) gene cluster from Salmonella enterica serovar Borreze. J Bacteriol. 1995;177(18):5247–5253. doi: 10.1128/jb.177.18.5247-5253.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan PT, Ohmori H, Tomizawa J-I, Lebowitz J. Nucleotide sequence and gene organization of ColE1 DNA. J Biol Chem. 1985;260(15):8925–8935. [PubMed] [Google Scholar]
- 26.Stepanek V, Valesova R, Kyslik P. Cryptic plasmid pRK2 from Escherichia coli W: sequence analysis and segregational stability. Plasmid. 2005;54(1):86–91. doi: 10.1016/j.plasmid.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Mata C, Navarro F, Miro E, Walsh TR, Mirelis B, Toleman M. Prevalence of SXT/R391-like integrative and conjugative elements carrying blaCMY-2 in Proteus mirabilis. J Antimicrob Chemother. 2011;66(10):2266–2270. doi: 10.1093/jac/dkr286. [DOI] [PubMed] [Google Scholar]
- 28.Deng H, Si HB, Zeng SY, et al. Prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in a farrowing farm: ST1121 clone harboring IncHI2 plasmid contributes to the dissemination of blaCMY-2. Front Microbiol. 2015(6):1210. doi: 10.3389/fmicb.2015.01210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barton BM, Harding GP, Zuccarelli AJ. A general method for detecting and sizing large plasmids. Anal Biochem. 1995;226(2):235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 30.Liu SL, Hessel A, Sanderson KE. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc Natl Acad Sci U S A. 1993;90(14):6874–6878. doi: 10.1073/pnas.90.14.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang LX, Sun J, Li L, et al. Dissemination of the chromosomally encoded CMY-2 cephalosporinase gene in Escherichia coli isolated from animals. Int J Antimicrob Agents. 2015;46(2):209–213. doi: 10.1016/j.ijantimicag.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Poirel L, Lartigue M-F, Decousser J-W, Nordmann AP. ISEcp1B-mediated transposition of blaCTX-M in. Escherichia coli. Antimicrob Agents Chemother. 2005;49(1):447–550. doi: 10.1128/AAC.49.1.447-450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun J, Li XP, Yang RS, et al. Complete nucleotide sequence of an IncI2 plasmid coharboring blaCTX-M-55 and mcr-1. Antimicrob Agents Chemother. 2016;60(8):5014–5017. doi: 10.1128/AAC.00774-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez T, Vazquez GJ, Aquino EE, Martinez I, Robledo IE. ISEcp1- mediated transposition of blaKPC into the chromosome of a clinical isolate of Acinetobacter baumannii from Puerto Rico. J Med Microbiol. 2014;63(Pt 12):1644–1648. doi: 10.1099/jmm.0.080721-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cattoir V, Nordmann P, Silva-Sanchez J, Espinal P, Poirel L. ISEcp1- mediated transposition of qnrB-like gene in Escherichia coli. Antimicrob Agents Chemother. 2008;52(8):2929–2932. doi: 10.1128/AAC.00349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wachino J, Yamane K, Kimura K, et al. Mode of transposition and expression of 16S rRNA methyltransferase gene rmtC accompanied by ISEcp1. Antimicrob Agents Chemother. 2006;50(9):3212–3215. doi: 10.1128/AAC.00550-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milewska K, Wegrzyn G, Szalewska-Palasz A. Transformation of Shewanella baltica with ColE1-like and P1 plasmids and their maintenance during bacterial growth in cultures. Plasmid. 2015;81:42–49. doi: 10.1016/j.plasmid.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Roth AL, Kurpiel PM, Lister PD, Hanson ND. bla(KPC) RNA expression correlates with two transcriptional start sites but not always with gene copy number in four genera of Gram-negative pathogens. Antimicrob Agents Chemother. 2011;55(8):3936–3938. doi: 10.1128/AAC.01509-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reisbig MD, Hossain A, Hanson ND. Factors influencing gene expression and resistance for Gram-negative organisms expressing plasmid-encoded ampC genes of Enterobacter origin. J Antimicrob Chemother. 2003;51(5):1141–1151. doi: 10.1093/jac/dkg204. [DOI] [PubMed] [Google Scholar]
- 42.Lee K, Kusumoto M, Sekizuka T, et al. Extensive amplification of GI-VII-6, a multidrug resistance genomic island of Salmonella enterica serovar Typhimurium, increases resistance to extended-spectrum cephalosporins. Front Microbiol. 2015(6):78. doi: 10.3389/fmicb.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) I-Ceu I-PFGE and Southern blot hybridization with the 23S rDNA and blaCMY-2 probes; (B) S1 nuclease-PFGE Southern blot hybridization with the blaCMY-2 and ColE1-like probe. Line M: H9812 marker; Lines 1–6: EC6413, EC4103, and EC5106, and their corresponding electroporants EC6413T, eC4103T, and EC5106T. Arrows represent the band hybridized with the blaCMY-2 or ColE1-like probe.
Abbreviation: PFGE, pulsed-field gel electrophoresis.
Table S1.
Primers used for screening for genes and PCR mapping
| PCR | Primer | Primer sequence (5′→3′) | Product length (kb) | Target | Reference |
|---|---|---|---|---|---|
| A | AF | GCACTTAGCCACCTATACGGCAG | 0.758 | blaCMY-2 for digoxigenin-labeled probes | 1 |
| AR | GCTTTTCAAGAATGCGCCAGG | ||||
| B | BF | AATGATGGCCAGGCTGTCTCC | – | 23SrRNA for digoxigenin-labeled probes | 2 |
| BR | CCGCCGTCGATATGAACTCTTG | ||||
| C | CF | GAAGGTTCTCAGAGCTGCAAC | 0.3 | RNAII for digoxigenin-labeled probes | This study |
| CR | GCCGCGTTTATCTCATTCCAC | ||||
| D | DF | ATCGTCTTTTACCGCCTGTCC | 3.1 | Screening for the plasmid of pSC137 | This study |
| DR | GTTAGCCCTATCCTGCATCGT | ||||
| E | EF | GTCATCGCTGGGAAATCGAAC | 3.8 | Junction between the 1S4 and yggR | 3 |
| ER | GCATAACGTCTCGGATCTACACC | ||||
| F | FF | AACTTGACGCCGAAGCCTA | 4.8 | Junction between the blaCMY-2 and orf9 | 3 |
| FR | TACGCCTGCAAAATATCACCA | ||||
| G | GF | TTTGTACTGCCAACGTATCCAA | 2.2 | Junction between the orf9 and ΔhsdR | 3 |
| GR | AAAGAACGGGAAATTGCCAAC | ||||
| H | HF | CCATAACAGCGGAATGACACC | 3.0 | Junction between RNAII and blaCMY-2 in the plasmid of pEC6413 | This study |
| HR | CAGAGCGCAGCATAACGAT | ||||
| I | IF | ACCGGGTTTTCATCCACGA | 1.12 | Junction between ΔhsdR and HP3 in the plasmid of pEC6413 | This study |
| IR | GTGCGTCTGACCAATATCCAC | ||||
| J | JF | CCATAACAGCGGAATGACACC | 3.2/4.8 | Junction between RNAII and ΔhsdR in the plasmids of pEC4103 or pEC5106 | This study |
| JR | AGGGCTTTACCTGTCAGCTC | ||||
| K | KF | CAGAGCGCAGCATAACGAT | 2.3/5.3 | Junction between blaCMY-2 and HP3 in the plasmid of pEC4103 or pEC5106 | This study |
| KR | CGTAATCCGTTGCCAGAGCC | ||||
| L | LF | TATTGTAGCATCGGTTTCCCA | 5.4 | Junction between the ISEcp1and ΔhsdR including pSC137 | This study |
| LR | GCGCGAACATACATATCCAGT | ||||
| M | MF | GCTGCTGACAGCCTCTTT | 0.197 | qCMY2 for quantitative real-time PCR assays | 4 |
| MR | GCGTGACTGGGTGGTTAT | ||||
| N | NF | GGCCGCAAGGTTAAAACTCAAATG | 0.243 | 16S rRNA for quantitative real-time PCR assays | 5 |
| NR | AACCGCTGGCAACAAAGGATAAGG |
Notes: F, forward primer; R, reverse primer. “L” represents reverse PCR primers and its 4 amplification region containing ISEcp1-pSC137-ΔhsdR.
Abbreviation: PCR, polymerase chain reaction.
Table S2.
PCR-typing blaCMY-2 gene-containing loci transferred into endogenous plasmid pSC137
| PCR typing | Target | Size (kb) | Electroporants | Wild strains | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| EC5106T | EC6413T | EC4103T | EC5106 | EC6413 | EC4103 | |||
| H | Junction between RNAII and blaCMY-2 | 3.0 | + | − | − | − | − | − |
| I | Junction between ΔhsdR and HP3 | 1.12 | + | − | − | − | − | − |
| J | Junction between RNAII and ΔhsdR | 3.2/4.8 | − | + | + | − | − | − |
| K | Junction between blaCMY-2 and HP3 | 2.3/5.3 | − | + | + | − | − | − |
| L | Region containing ISEcp1-pSC137-ΔhsdR | 5.4 | + | + | + | − | − | − |
Note: (+) positive and (–) negative.
Abbreviation: PCR, polymerase chain reaction.