Abstract
Transcription factors and chromatin remodeling proteins control the transcriptional variability for ESC lineage commitment. During ESC differentiation, chromatin modifiers are recruited to the regulatory regions by transcription factors, thereby activating the lineage-specific genes or silencing the transcription of active ESC genes. However, the underlying mechanisms that link transcription factors to exit from pluripotency are yet to be identified. In this study, we show that the Ctbp2-interacting zinc finger proteins, Zfp217 and Zfp516, function as linkers for the chromatin regulators during ESC differentiation. CRISPR-Cas9-mediated knock-outs of both Zfp217 and Zfp516 in ESCs prevent the exit from pluripotency. Both zinc finger proteins regulate the Ctbp2-mediated recruitment of the NuRD complex and polycomb repressive complex 2 (PRC2) to active ESC genes, subsequently switching the H3K27ac to H3K27me3 during ESC differentiation for active gene silencing. We therefore suggest that some zinc finger proteins orchestrate to control the concise epigenetic states on active ESC genes during differentiation, resulting in natural lineage commitment.
INTRODUCTION
Transcription factors and chromatin regulators are important in establishing the epigenetic changes during embryonic stem cell (ESC) development, and in defining cell identity (1,2). Transcription factors bind to the promoters and enhancer regions, and recruit chromatin modifiers for activation or repression of cell specific gene expressions (3). During the ESC differentiation, the promoter and the enhancer regions of the pluripotency-associated genes require to be shut down at the proper time (4,5). Transcription factors that initiate differentiation and lineage specification (6,7) mediate this accurate regulation. A combination of transcription factors are further able to artificially reprogram the fully differentiated cells into the induced pluripotent stem (iPS) cells (2,8). Thus, it is essential to define the transcriptional and the chromatin-based regulatory mechanisms involved in stem cell development and reprogramming.
Ctbp2 (C-terminal binding protein 2), a transcriptional corepressor, displays early embryonic lethality in Ctbp2-null mice (embryonic day 10.5), indicating that Ctbp2 is essential for the normal development (9). A recent study identified the function of Ctbp2 in exit from pluripotency which regulates β-catenin level and its accessibility to the active ESC gene regions (10). Also, Ctbp2 controls the epigenetic states of H3K27 in active ESC genes mediated by the nucleosome remodeling and deacetylation (NuRD) complex and the polycomb repressive complex 2 (PRC2) during stem cell differentiation (11). The components of the NuRD complex, namely Mbd3, Mi2β and Lsd1, are essential for a proper induction of stem cell differentiation and lineage commitment (4,12,13). PRC2 is a transcriptional repressor complex having a methyltransferase activity on H3K27 and is required for gene silencing in ES cells (14). Thus, it is necessary to search for transcription factors that fine-tune these epigenetic regulators on active ESC genes.
In order to seek functional transcription factors that aid in the exit from pluripotency, assessment by shRNA screening against Ctbp2-interacting zinc finger proteins lead to identification of the zinc finger protein 217 (Zfp217) and zinc finger protein 516 (Zfp516). ZNF217 is an oncogenic protein frequently amplified in human tumors (15–17). Its mRNA expression levels are also increased in tumors which induce metastasis (18,19). Notably, ZNF217 overexpression promotes the epithelial-mesenchymal transition (EMT) process which is associated with the early lineage commitment step in ESCs (20) by TGF-β activation, significant reduction of epithelial markers, and upregulation of EMT drivers such as SNAIl1/2, TWIST1/2 and ZEB1/2 (19,21). In addition, ZNF217 is also a known transcriptional repressor and a component of the histone deacetylase (HDAC) and the CoREST complex and is found to develop complexes with CTBP, lysine-specific demethylase 1A (LSD1), and enhancer of zeste homolog 2 (EZH2) (22–25). Another zinc finger protein, Zfp516 depletion causes embryonic lethality and has recently been identified as a cold-inducible transcription factor promoting the BAT program by Lsd1-mediated activation (26,27). Zfp516 contains PXDLS and RRT motifs which are known as Ctbp interacting sites (16). Also, in human breast cancer cells, ZNF516 transcriptionally repress EGFR related genes and cancer proliferation by associating with CTBP, LSD1, and the CoREST complex (28). Taken together, these studies raise the possibility that Zfp217 and Zfp516 might modulate the chromatin modifiers for appropriate ESC differentiation.
In this study, we found that knock-out of the Ctbp2-interacting zinc finger proteins, Zfp217 and Zfp516 in ESCs had an effect on the deregulation of exit from pluripotency during ESC differentiation. This underlies the mechanism that both zinc finger proteins associate to the NuRD-mediated H3K27 deacetylation and PRC2-mediated H3K27 tri-methylation, to active the ESC gene silencing for natural ESC development. This indicates that zinc finger proteins regulate the proper exit from pluripotency by targeting the chromatin regulators to site-specific, temporal, and spatial transcriptional regulation in an organized process.
MATERIALS AND METHODS
Cell culture
Embryonic stem cells were cultured and passaged on 0.1% gelatinized (Sigma-Aldrich, St. Louis, MO, USA) dishes, as reported previously (29). E14 mouse ESCs were cultured in DMEM (Hyclone, Logan, Utah) or KNOCK-OUT™ DMEM (Gibco, Grand Island, NY, USA) supplemented with 15% FBS (Gibco, Grand Island, NY, USA), 2 mM l-glutamine, 55 μM β-mercaptoethanol, 1% (v/v) non-essential amino acid, 100 U/ml penicillin and 100 μg/ml streptomycin (all from Gibco, Grand Island, New York) and 500 U/ml ESGRO LIF (Millipore, Germany). ZHBTc4 cells were kindly provided by Hitoshi Niwa (RIKEN, Kobe, Japan).
ESC differentiation was induced by LIF withdrawal from ESC medium in monolayer cultures. For embryonic body (EB) formation, ESCs were cultured in low-attachment dishes that contained ESC medium without LIF. Oct4 depletion of ZHBTc4 ESCs was performed as described (29).
Self-renewal assay
Self-renewal assay (colony-forming assay) was done as described (30). Briefly, ESCs were plated at a density of 600 cells/well in a six-well plate. After incubation for 5 to 6 days with or without LIF, colonies were stained for alkaline phosphatase and grouped by differentiation status.
Genome editing with CRISPR/Cas9
The sgRNAs targeting exon 2 of Zfp217 and exon 2 of Zfp516 were designed using the CRISPR Design Tool (http://crispr.mit.edu/) (31,32). sgRNA oligomers were annealed and inserted into the pSpCas9n(BB)-2A-GFP Addgene vectors (32) and transiently co-transfected into the E14 ESCs using Lipofectamine® 2000 (Invitrogen, Carlsbad, California). ESCs expressing GFP were sorted through fluorescence-activated cell sorting (FACS) at 24 h post-transfection. Sorted ESCs were grown for 5 days at single-cell density for clonal cell line expansion and isolation. Each clone was isolated and screened by western blot. The verified clones were further confirmed by genomic DNA PCR and sequencing.
Lentiviral shRNA-mediated knock-down
pLKO.1 lentiviral vectors for shRNAs (Sigma-Aldrich, St. Louis, MO, USA) were purchased for knock-down experiments. 293FT cell was used for lentivirus production by co-transfection of 0.5 μg each of pMD2.G, pMDLg/pRRE, pRSV-rev, and pLKO.1-shRNA using Lipofectamine 2000 (Invitrogen, Carlsbad, California). Forty eight hours after transfection, the virus-containing medium was collected and filtered through 0.45 μm filters. Polybrene (8 μg/ml) was added just before target cell infection and infection was performed for 6 h. Post infection, puromycin selection (2 μg/ml) was performed for a minimum of 2 days.
Plasmids constructs
Full-length mouse Zfp217, Zfp516, Hic2, Zbtb8b, Wiz, Zfp518b, Zscan10 and Zscan4f cDNAs were obtained from E14 ESC cDNA by PCR and further cloned into the pCAG-Flag vector. Human CTBP2 cDNA was cloned into the pcDNA3.1-Myc vector (11). pCAG-Flag-Zfp217 NL680AS, RRT746AAA and NL680AS/RRT746AAA mutant constructs were generated by site-directed mutagenesis.
Generation of stable ESC lines
Zfp217 wild-type (WT) and Zfp217 NL680AS/RRT746AAA mutant (MUT) were cloned into the pCAG-Flag-vector. Stably expressed Flag-tagged Zfp217 WT and Zfp217 MUT rescued ESCs were generated by introducing the pCAG-Flag-Zfp217 WT and pCAG-Flag-Zfp217 MUT into the Zfp217 knock-out ESCs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Forty eight hours after transfection, cells were selected by culturing with 20 μg/μl blastocidin containing medium for 7 days to collect stably integrated ESC cell lines.
Immunoprecipitation and Western blot
Using the wild-type ESCs, the Flag-tagged CTBP2 expressing ESCs, the Flag-tagged Zfp217 WT rescued ESCs and the Flag-tagged Zfp217 MUT rescued ESCs, immunoprecipitation and western blot were done as previously described (29). Anti-Zfp217, anti-Zfp516, anti-Zfp518b, anti-Zbtb8b, anti-Hic2, anti-Wiz, anti-Zscan10, anti-Zscan4f antibodies were made by GenScript (Piscataway, New Jersey). Anti-Oct4 (sc-5279) and anti-Znf217 (sc-55351) were acquired from Santa Cruz Biotechnology (Dallas, TX, USA); anti-Nanog (ab14959), anti-Hdac1 (ab7028), anti-Mi-2b (ab72418), anti-Lsd1 (ab17721), anti-H3 (ab1792), anti-H3K4me1 (ab8895), anti-H3K4me3 (ab8580), and anti-H3K27ac (ab4729) were purchased from Abcam (UK); anti-Ctbp1 (612042), anti-Ctbp2 (612044), and anti-Ezh2 (612667) were acquired from BD Transduction Laboratories (San Jose, CA, USA); anti-Esrrb (H6707) and anti-Sox2 (MAB2018) were obtained from R&D System (Minneapolis, MN, USA); anti-Suz12 (3737S), anti-Bax (2772), anti-Caspase-3 (9662S), and anti-Cleaved Caspase-3 (9661S) were from Cell Signaling Technology (Danvers, Massachusetts); anti-CoREST (07-455) and anti-H3K27me3 (07-449) was obtained from Millipore (Germany); anti-HA tag and anti-Myc tag were acquired from Covance (Princeton, NJ, USA); anti-Flag tag and anti-β-Actin were purchased from (Sigma-Aldrich, St. Louis, MO, USA).
Immunofluorescence and confocal microscopy
Pluripotent state of ES cells were grown on coverslips for 2 days in LIF-containing media and ES cells were induced to differentiate on coverslips by LIF withdrawal for 5 days. Cells were fixed in 4% paraformaldehyde and permeabilized in 0.25% Triton X-100 in PBS for 30 min at room temperature (RT). Next, the cells were blocked with 2% BSA in PBS for an hour at RT and were incubated overnight with each indicated primary antibodies at 4°C. Antibody dilutions were 1:500 for anti-Oct4 (Santa Cruz Biotechnology, Dallas, TX, sc-5279), 1:500 for anti-Nanog (Abcam, UK, ab14959), 1:500 for anti-Flag (Sigma-Aldrich, St. Louis, MO, F1804), and 1:100 for anti-Zfp217 (this study). Secondary antibodies used in immunostaining were Alexa Fluor 488 and 568 (Invitrogen, Carlsbad, CA) and the nucleus was stained with Hoechst 33342 and DAPI. Confocal micro-images were obtained by a confocal laser scanning microscope (Olympus, Japan, Confocal-FV1000).
Real-time qPCR
Total RNA was extracted from mESCs with TRIzol reagent (Invitrogen, Carlsbad, California). RNAs were extracted and synthesized into cDNAs, according to the manufacturer's protocol (AMV Reverse Transcriptase, Takara, Japan). Quantitative real time-PCR for cDNAs was performed using the SYBR premix Ex Taq (Takara, Japan) on the CFX Connect Real-time PCR Detection System (Bio-Rad, Hercules, California), and normalized to 18s rRNA. For quantitating ChIP results, SYBR® Green qPCR mix (Thermofisher-scientific, Waltham, MA, Finnzymes, F-410) and SYBR premix Ex Taq (Takara, Japan) were used and normalized to 1% input chromatin. Sequences of the primers for real-time PCR are listed in Supplementary Table S1.
Apoptosis assay
Apoptosis assay was performed using FITC Annexin V Apoptosis Detection Kit (BD Pharmingen™, Franklin Lakes, New Jersey) and detected with BD-FACS Canto with following manufacturer's instructions.
Chromatin Immunoprecipitation (ChIP) assay
Chromatin immunoprecipitation (ChIP) assays were performed as described (29). Briefly, mES cells cross-linked with 1% formaldehyde were lysed and sonicated. Lysates were diluted 10-fold in IP buffer and incubated overnight at 4°C with appropriate amount of antibodies. Next day, the chromatin samples were incubated with protein A/G Plus agarose for 4 h at 4°C, washed and further eluted. The eluted chromatin samples were then reverse cross-linked overnight at 65°C. DNA was precipitated with ethanol and stored at −20°C till further use. The sequences of the ChIP-primers for real-time PCR are listed in Supplementary Table S1.
ChIP-seq data analysis
Public ChIP-seq data sets and GEO accession numbers are described in Supplementary Table S2. Reads after sequencing were mapped against the mouse genome (NCBI build 37/mm9) using Bowtie2 v.2.2.6 (33) with the default parameters. The BAM formatted outputs were sorted by genomic coordinates (samtools sort) and the reliable reads based on the mapping score were used in subsequent processes (34). MACS v.1.4.0 (35) was used to locate the binding regions of transcription factors, chromatin regulators, and histone modifications. HOMER (36) was used for analyzing co-bound regions of multiple factors. For visualization of histone modification and chromatin modifier profiles in co-enriched regions of Zfp217 and Ctbp2, ±50 kb peak spanning regions were binned into 200 bp windows and read densities normalizations were generated using HOMER. Cluster v3.0 (37) was then applied to cluster read densities into three groups. Heatmaps were created by Java TreeView (http://jtreeview.sourceforge.net). Co-bound regions of Zfp217 and Ctbp2 were annotated into promoters, exons, introns, intergenic regions and other features according to the RefSeq transcripts by HOMER (36). To annotate chromatin states of the co-bound regions, we applied ChromHMM v. 1.0.6 (38) to segment the E14 chromatin and then used BEDOPS v2.2.0 (39) to intersect with the co-bound regions. Gene ontology (GO) annotations, de novo motif, and known motif discovery algorithms were performed using HOMER.
RNA-seq data analysis
Reads of each sample were aligned to the mouse genome (NCBI build 37/mm9) using STAR 2.4.0.1 (40) with the default settings. HOMER tools were used to quantify FPKM values and normalize genes defined from RefSeq transcripts. Unsupervised hierarchical clustering analysis of gene expression values were done by Cluster 3.0 (37) and the results were visualized with Java TreeView (http://jtreeview.sourceforge.net).
RESULTS
Ctbp2 interacts with zinc finger proteins in mouse ESCs
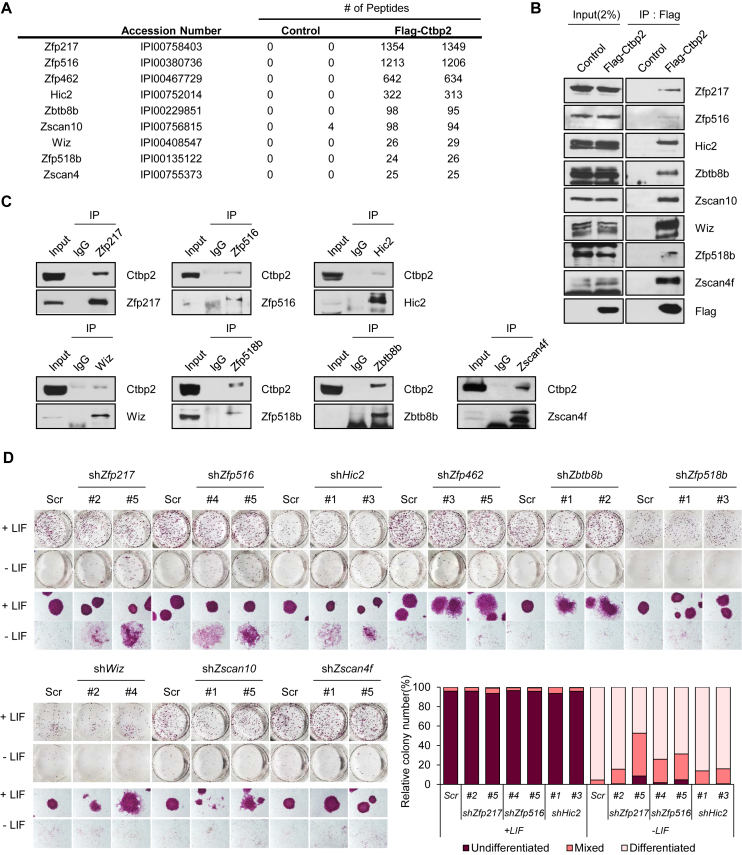
C-terminal binding protein (Ctbp) has been known to associate with repressive chromatin modifiers and repress certain transcription factors related with development and tumorigenesis (41). In addition, Ctbp2 primes the active ESC genes and participates in the exit from ESC pluripotency (11). However, Ctbp2 is supposed to control the exit from pluripotency through specific binding to certain transcription factors, because Ctbp2 basically has no ability to directly bind to the DNA sequences. Thus, we reassessed the Ctbp2 interactome (11) to find Ctbp2-interacting transcription factors. We found that many types of zinc finger proteins are remarkably associated with Ctbp2 and decided to extract zinc finger proteins that recorded the highest binding results (Figure 1A). Interaction of Ctbp2 with zinc finger proteins were verified by immunoprecipitation using stably expressing Flag-CTBP2 ESCs and transiently transfected 293T cells (Figure 1B and Supplementary Figure S1A). Flag-CTBP2-expressed stable ES cell lines showed an undifferentiated stem cell state similar to wild type ESCs (11). We then generated rabbit polyclonal antibodies against each of the selected zinc finger proteins, Zfp516, Zfp217, Zfp518b, Hic2, Wiz, Zscan4f, Zscan10 and Zbtb8b, and confirmed their specificity by western blot (Supplementary Figure S1B). The zinc finger proteins also endogenously interacted with Ctbp2 in ESCs using each of the antibodies (Figure 1C).
Figure 1.
Ctbp2 interacts with zinc finger proteins in mouse ESCs. (A) A list of Ctbp2-interacting zinc finger proteins with reasonable peptide counts were extracted from the Ctbp2 binding complex in ESCs (11). (B) Stably incorporated Flag-tagged CTBP2 in ESCs were used for immunoprecipitation with Flag antibodies, and endogenous Zfp217, Zfp516, Hic2, Zbtb8b, Zscan10, Wiz, Zfp518b and Zscan4f were detected with the indicated antibodies. (C) Immunoprecipitation with Zfp217, Zfp516, Hic2, Zbtb8b, Wiz, Zfp518b, and Zscan4f antibodies in E14 ESCs, endogenous Ctbp2 was detected with indicated antibodies. (D) Self-renewal assay and alkaline phosphatase (AP) staining in wild-type and each of Zfp217, Zfp516, Hic2, Zfp462, Zbtb8b, Zscan10, Wiz, Zfp518b and Zscan4f-knock-down ESCs. Delayed differentiation phenotype was shown in Zfp217, Zfp516 and Hic2-knock-down ESCs.
Next, the expressions of Ctbp2-binding zinc finger proteins during ESC differentiation were evaluated. Publically available RNA-sequencing data were analyzed (42) to cluster zinc finger proteins by expression patterns. Zinc finger proteins were clustered into three large groups: increment, decrement, and no change in expression patterns (Supplementary Figure S2A). The group containing Zfp217, Hic2, Zfp462, Zscan10, Zbtb8b, and Zfp518b showed decreased expression pattern upon ESC differentiation, similar to that of Ctbp2. Expression patterns were further confirmed by differentiating ESCs into embryonic bodies, and by withdrawing leukemia inhibitory factor (LIF) (Supplementary Figures S2B–D). To analyze whether these zinc finger proteins are regulated directly by Oct4, a master regulator in stem cell pluripotency, we used genetically modified ZHBTc4 cells to deplete the Oct4 expression; this resulted in decreased levels of Zfp217, Zfp518b, and Zscan10 (Supplementary Figures S2E and F).
To screen zinc finger proteins possessing the delayed differentiation phenotype similar to that of Ctbp2, we introduced small hairpin RNAs (shRNAs) into ESCs (Supplementary Figure S3A) and triggered stem cell differentiation. Delayed differentiation phenotype was shown in Zfp217, Zfp516 and Hic2 knock-down ESCs upon LIF withdrawal by displaying positive alkaline phosphatase (AP) colonies (Figure 1D). Moreover, shZfp217, shZfp516 and shHic2 ES cells were induced to differentiate by withdrawing LIF from the culture media for 5 days and stained with specific antibodies to Oct4 and Nanog. Though the expression levels of Oct4 and Nanog were significantly diminished in differentiated wild-type ESCs, certain expression levels of Oct4 and Nanog were still sustained in shZfp217, shZfp516, and shHic2 ESCs (Supplementary Figure S3B). Of three possible candidates, we primarily focused Zfp217, since it is regulated by Oct4, its expression decreases upon stem cell differentiation, and its knock-down shows delayed differentiation.
Zfp217 depletion retards ESC differentiation
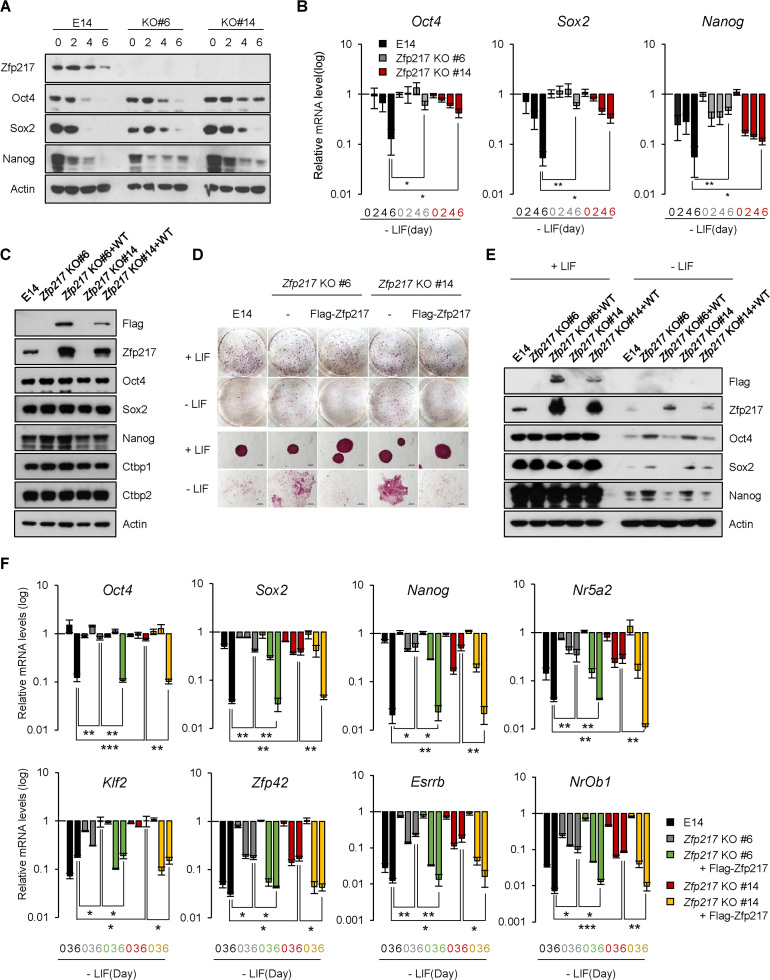
When generating shZfp217 ESCs, knock-down of Zfp217 triggered apoptotic cell death immediately after puromycin selection (Supplementary Figure S4A), while stabilized shZfp217 ESCs no longer induced apoptosis (Supplementary Figure S4B). Thus, to further define the role of Zfp217 in ESC identity, we designed guide RNAs (gRNAs) targeting exon 2 of the Zfp217 gene and cloned into the Cas9 D10A nickase mutant plasmid (pSpCas9n(BB)-2A-GFP) (Supplementary Figure S5B) (32,43,44). ESCs were co-transfected with plasmids encoding Cas9 nickase mutant and Zfp217 gRNAs, following which the cells expressing GFP signals were sorted by fluorescence-activated cell sorting (FACS). Each colony was grown to verify CRISPR-Cas9-mediated knock-out by western blot. We found that Zfp217 was completely depleted in clone number 6 and 14, and these clones were further confirmed to have flanked target sequences which lead to a premature stop codon (Supplementary Figures S5C and D). Zfp217 knock-out (KO) ESCs maintained their stem cell morphology as well as AP positive colonies; also, pluripotency-related ESC genes remained unaffected (Supplementary Figures S5E and F). This phenotype was also shown in shRNA-mediated Zfp217-knock-down (KD) ESC lines (Supplementary Figures S6A and S6C). Although Zfp217-knock-down exhibited severe retardation in cell growth (Supplementary Figure S6B), Zfp217 KO ESCs exhibited only about 20% reduction in growth rate (Supplementary Figure S5G) and comparable cell cycle profile (Supplementary Figure S5H). Moreover,
Zfp217 KO ESCs clone 6 and 14 were maintained in long-term cultures until passage 5, 10, 15 and 20. All Zfp217 KO ESCs were cultured till passage 20 and had no significant morphological changes (Supplementary Figure S5I) and showed positive AP stained colonies (Supplementary Figure S5J). Also, pluripotency-related ESC genes remained unaffected (Supplementary Figure S5K and S5L). Constant expressions of Bax and Caspase-3 and negative expression of cleaved Caspase-3 depict that Zfp217 KO ESCs maintain self-renewal without apoptosis (Supplementary Figure S5K). Thus, by the facts that immediate knock-down of Zfp217 lead some population to apoptosis and that Zfp217 KO ESCs were able to be generated and maintained in long-term passages, in overall, Zfp217 might not seem to have a profound impact on stem cell maintenance.
In another experiment, the Zfp217 KO ESCs were induced to differentiate by LIF removal at 2-day intervals. We observed that the pluripotency related genes, namely Oct4, Sox2 and Nanog, were downregulated in wild-type ESCs after differentiation, but were sustained in Zfp217 KO ESCs (Figure 2A and B). Also, Zfp217 KD ESCs generated a higher portion of AP-positive colonies relative to knock-down efficiency in differentiation conditions (Figure 1D) and exhibited comparable levels of pluripotency gene expression as the undifferentiated state (Supplementary Figures S6D and S6E). Zfp217 KD cells were then cultured under differentiating conditions for a maximum of 30 days, to determine the function of Zfp217 in pluripotency exit. The Zfp217 KD ESCs in differentiation medium remained AP-positive and maintained substantial mRNA levels of pluripotency genes (Supplementary Figures S6F and G). To verify the functional role of Zfp217 in ESCs, the Zfp217 KO ESC clone 6 and 14 were rescued with exogenous Zfp217, which displayed similar expression levels of pluripotency genes as the wild-type ESCs (Figure 2C). Upon differentiation cue, ectopic expression of Flag–Zfp217 restored the Zfp217 KO ESCs back to the differentiation process and exit from pluripotency, similar to that seen in wild-type ESCs (Figure 2D–F). This indicates that Zfp217 is involved in regulating the proper pluripotency-exit during ESC differentiation.
Figure 2.
Loss of Zfp217 retards differentiation of embryonic stem cells. (A and B) The protein and mRNA levels of Zfp217 and pluripotency-associated genes in wild-type and Zfp217 KO ESCs (clone #6 and #14) upon LIF withdrawal for indicated days. Expression was detected by indicated antibodies; Actin was used as a control. (n = 3) Presented as means ± SEM (*P ≤ 0.05, **P ≤ 0.01). (C) Zfp217 expression was rescued by Flag-tagged Zfp217 overexpression in Zfp217 KO ESCs (Zfp217-rescued ESCs). Western blot analysis of Zfp217 KO and Zfp217-rescued ESCs, using Flag and Zfp217 antibodies. Expression was detected by indicated antibodies; Actin was used as a control. (D) Self-renewal assay and alkaline phosphatase (AP) staining of Zfp217 KO and Zfp217-rescued ESCs in undifferentiated and differentiated conditions. Knock-out of Zfp217 leads to incomplete exit from pluripotency during ESC differentiation; this phenotype was rescued upon Zfp217 introduction. (E and F) The protein and mRNA levels of Zfp217, and pluripotency-associated genes in wild-type, Zfp217 KO, and Zfp217-rescued ESCs upon LIF withdrawal for indicated days. Expression was detected by indicated antibodies; Actin was used as a control (n = 3) Presented as means ± SEM (* P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001)
Zfp217 and Ctbp2 are co-localized at actively transcribed regions in ESCs
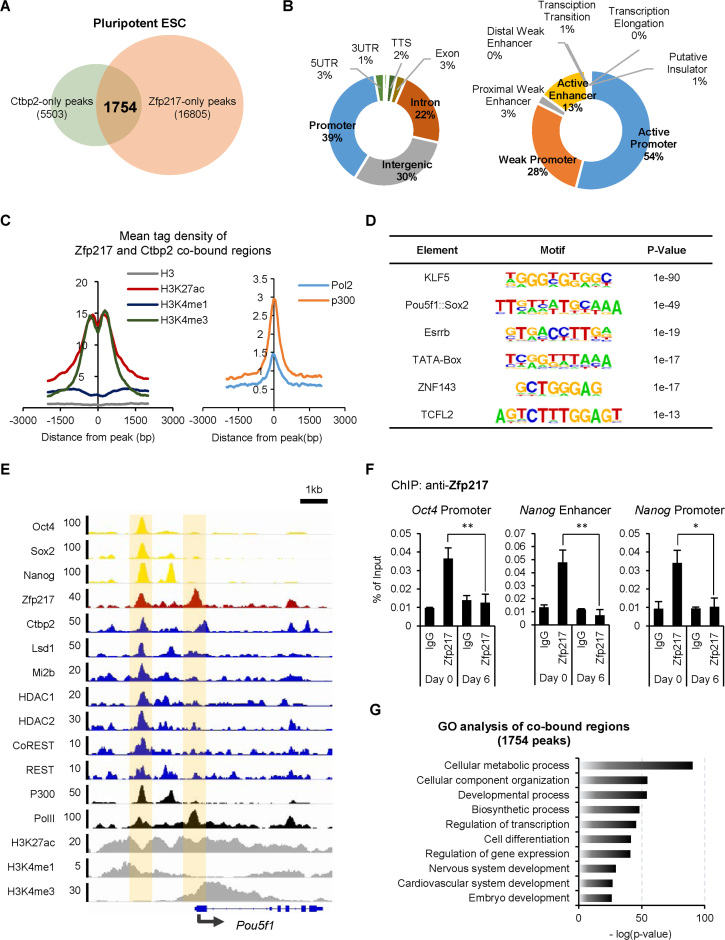
Publically available chromatin immunoprecipitation (ChIP) sequencing datasets were processed to determine whether Ctbp2 and its binding partner Zfp217 share common targets in the state of ESCs. We identified 1754 possible co-bound peaks of Ctbp2 and Zfp217 (Figure 3A) which were mostly enriched in promoter, intergenic and intron regions (Figure 3B). The co-bound peaks of Ctbp2 and Zfp217 were further comprehensively analyzed by chromatin states using ChromHMM (38). We observed that the co-bound regions were usually occupied at the promoter regions and some at the active enhancer regions (Figure 3B). Mean tag densities of active marks, where H3K27ac and p300 represent active enhancer regions and H3K4me3 and Pol2 represent active promoter regions, were centered on the co-bound peaks of Zfp217 and Ctbp2, indicating that Zfp217 and Ctbp2 are mostly co-localized at actively transcribed regions in ESCs (Figure 3C). Motifs of factors related to pluripotency in stem cells were significantly enriched in co-bound peaks of Zfp217 and Ctbp2 (Figure 3D). These results were further verified through Zfp217 and Ctbp2 co-localization at the Oct4 loci, along with the active transcription factors Oct4, Sox2 and Nanog (Figure 3E). Furthermore, localization of Zfp217 at the regions of Oct4 and Nanog were also confirmed by ChIP assays using the Zfp217 specific antibody (Figure 3F). By gene ontology analysis, the co-bound regions were found to be mostly associated with the developmental processes (Figure 3G), suggesting that both Zfp217 and Ctbp2 are involved in proper regulation of stem cell development.
Figure 3.
Zfp217 and Ctbp2 co-localize at actively transcribed ESC gene regions at undifferentiated ESCs. (A) A diagram of co-occupancy regions of Zfp217 and Ctbp2 using published ChIP-seq data (11,66). 1754 peaks were identified to be co-occupied by Zfp217 and Ctbp2. Annotation of the co-bound regions of Zfp217 and Ctbp2 is displayed in Supplementary Table S3. (B) The proportion of transcript coordinates (left pie chart) and chromatin states (right pie chart) of co-enriched regions of Ctbp2 and Zfp217. Co-occupied regions of Zfp217 and Ctbp2 (87%) are predominantly active enhancers and promoters. (C) The mean tag density of H3K27ac, H3K4me1, H3K4me3, RNA polymerase 2 (Pol2), and p300 on co-occupied regions. All active histone markers representing promoter and enhancer regions were enriched in co-occupied peaks. (D) Using motif discovery algorithms, pluripotency-associated gene motifs were predicted with high scores within the co-bound peaks of Zfp217 and Ctbp2. (E) Integrated genomics viewer (IGV) represents Ctbp2, Zfp217, Oct4, Sox2, Nanog, repressive chromatin regulators, and active histone markers in Oct4 gene loci in ESCs. (F) ChIP-qPCR analyses of Zfp217 at active ESC regions in undifferentiated and differentiated ESCs (day 6 of LIF withdrawal). The enrichment of Zfp217 was observed in undifferentiated ESCs, but disappeared upon ESC differentiation (n = 3) Presented as means ± SEM (*P ≤ 0.05, **P ≤ 0.01). (G) The functional gene ontology (GO) analysis of co-occupied regions of Zfp217 and Ctbp2. Enriched regions were mostly associated with the developmental process.
Zfp217 associates with repressive chromatin modifiers on active ESC genes
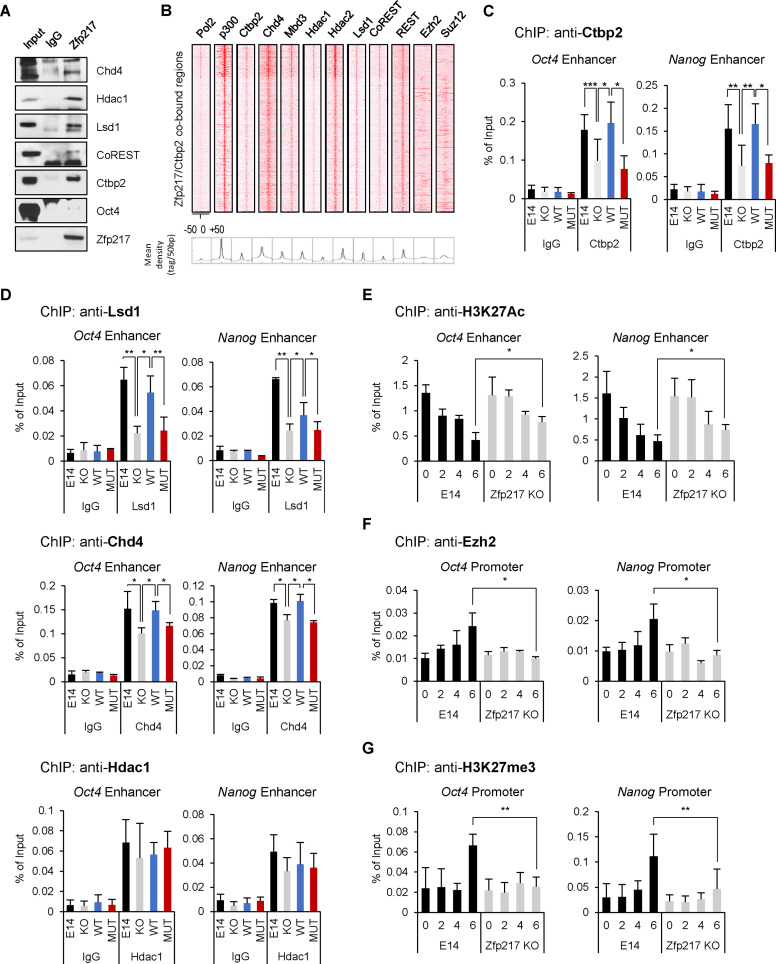
To find the functional association between Zfp217 and Ctbp2 during ESC differentiation, we focused on the chromatin regulators that interact with Zfp217 in ESCs. In the previous report, ZNF217 is known to interact with JARID1B/PLU-1, G9a, LSD1, CoREST, CTBP1 and EZH2 in MCF7 cells, resulting in transcriptional repression (24). Thus, in this study, we attempted to associate Zfp217 endogenously with chromatin regulators in ESCs. We found that the main components of the NuRD complex, the HDAC complex, and the CoREST complex bind to Zfp217 in ESCs (Figure 4A). Next, to examine the function of Zfp217 on genome regulation, we aligned the ChIP-seq peaks of Zfp217 and Ctbp2 with that of histone modifiers. The co-bound regions of Zfp217 and Ctbp2 aligned well with Chd4, Mbd3, Hdac1, Hdac2, CoREST, REST and p300, where the active enhancer mark is also located (Figure 4B). The components of the NuRD complex, namely Chd4, Mbd3 and Hdac1, as well as Lsd1, are all essential for epigenetic silencing of pluripotency genes during ESC differentiation (4,5,13,45,46). The association of Zfp217 with these chromatin repressive regulators on active enhancer regions provides a clue that Zfp217 regulates the exit from pluripotency in ESCs.
Figure 4.
Zfp217 associates with repressive chromatin modifiers on active ESC genes. (A) E14 ESC lysates were immunoprecipitated with Zfp217 antibodies; endogenous Chd4, Hdac1, Lsd1, CoREST, Ctbp2 and Oct4 were detected with the indicated antibodies. (B) The co-occupied regions of Zfp217 and Ctbp2 (1754 peaks) were aligned with the components of the NuRD complex and the CoREST complex in ESCs. (C and D) ChIP-qPCR analysis of Ctbp2, Lsd1, Chd4 and Hdac1 on active ESC gene regions in wild-type, Zfp217 KO, Zfp217 WT-rescued, and Zfp217 MUT-rescued ESCs. (n = 3) Presented as means ± SEM (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). (E–G) ChIP-qPCR analysis of H3K27ac, Ezh2, and H3K27me3 on active ESC gene regions in wild-type and Zfp217 KO ESCs during ESC differentiation by LIF withdrawal for indicated days (n = 3). Presented as means ± SEM (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001)
To clarify the association of Ctbp2 with Zfp217 and chromatin regulators on actively transcribed ESC genes, we generated a Ctbp2-binding defective Zfp217 mutant (NL680AA/RRT746AAA) (23) (Supplementary Figure S7A). The abolished interaction between Ctbp2 and Zfp217 mutants were validated by co-immunoprecipitation assays (Supplementary Figure S7B). Flag-Zfp217 NL680AA/RRT746AAA mutant (MUT) was then introduced to Zfp217 KO ESCs and it was confirmed that pluripotency genes were unaffected as compared to the wild-type (Supplementary Figure S7C). Through the self-renewal assay upon LIF withdrawal, Zfp217 MUT ESCs formed AP-positive colonies, while Zfp217 WT ESCs got fully differentiated (Supplementary Figure S7D).
On initiation of differentiation signal, components of the NuRD complex instantly silence the pluripotency related genes for proper lineage commitment (5). We therefore examined whether the interaction between Zfp217 and Ctbp2 affects the recruitment of the NuRD complex to activate ESC gene regions in ESCs. We observed that Zfp217 KO efficiently blocked the positioning of Ctbp2 on the enhancers or promoters of active ESC genes (Figure 4C and Supplementary Figure S8A) and the recruitment of the NuRD complex components, Lsd1 and Chd4 (Figure 4D and Supplementary Figure S8B). However, the recruitment of Hdac1 was not altered by Zfp217 knock-out in ESCs (Figure 4D and Supplementary Figure S8B). Ctbp2-binding defective Zfp217 mutant significantly reduced the recruitment of the NuRD complexes as well as Ctbp2 to the regions of active ESC genes in Zfp217 MUT-rescued ESCs (Figure 4C–D and Supplementary Figures S8A–B), indicating that the interaction between Ctbp2 and Zfp217 is important for recruiting the NuRD complex to active ESC genes.
Upon ESC differentiation, the NuRD complex deacetylates the acetylated H3K27 for gene suppression (5). Therefore, we investigated the effect of Zfp217 on H3K27 deacetylation on active ESC genes during differentiation, by comparing the wild-type ESC with Zfp217 KO ESCs. We found that upon differentiation, the H3K27ac levels on the active ESC genes were gradually decreased in wild-type ESCs. However, the decrease in H3K27ac level was relatively slow in Zfp217 KO ESCs compared to that of wild-type ESCs (Figure 4E and Supplementary Figure S8C).
The NuRD complex-associated H3K27 deacetylation on active ESC genes results in the PRC2-mediated H3K27 methylation during ESC differentiation (11). Thus, we further examined whether Zfp217 affects the recruitment of PRC2 for active ESC gene silencing. Enrichment of Ezh2, a catalytic component of PRC2, on active ESC genes were examined during ESC differentiation in wild-type and Zfp217 KO ESCs. While Ezh2 was recruited to the active ESC genes in the wild-type ESCs on differentiation day 6, Ezh2 enrichment was not detected in Zfp217 KO ESCs (Figure 4F and Supplementary Figure S8D). Consequently, upon differentiation, we detected lower H3K27me3 levels in Zfp217 KO ESCs than in wild-type ESCs (Figure 4G and Supplementary Figure S8E), suggesting that Zfp217 primes active ESC genes to quickly associate with the NuRD and the PRC2 for H3K27me3-mediated repression of active ESC genes during ESC differentiation.
Ctbp2-associated zinc finger proteins control over ESC differentiation
We next speculated if other Ctbp2-interacting zinc finger proteins are also subjected to H3K27-mediated active ESC gene repression and if multiple zinc finger proteins cooperate in the ESC differentiation process. For this study, we selected Zfp516 which showed retarded differentiation phenotype upon shRNA-mediated knock-down in ESCs (Figure 1D). We cloned gRNAs targeting exon 2 of the Zfp516 gene into the Cas9 nickase mutant plasmid (pSpCas9n(BB)-2A-GFP) (Supplementary Figure S9A), transfected the plasmid into E14 cells and Zfp217 KO ESCs, and sorted by FACS. Sorted CRISPR-Cas9-mediated Zfp516 knock-out ESC colonies were verified by western blot and were further sequenced (Supplementary Figures S9B and S9C). Zfp516 knock-out (KO) ESCs and Zfp217 and Zfp516 double knock-out (DKO) ESCs displayed similar expression levels of pluripotency-related ESC genes (Supplementary Figure S9D). Also, Zfp516 KO and DKO ESCs were significantly ineffective on stem cell proliferation (Supplementary Figure S9E).
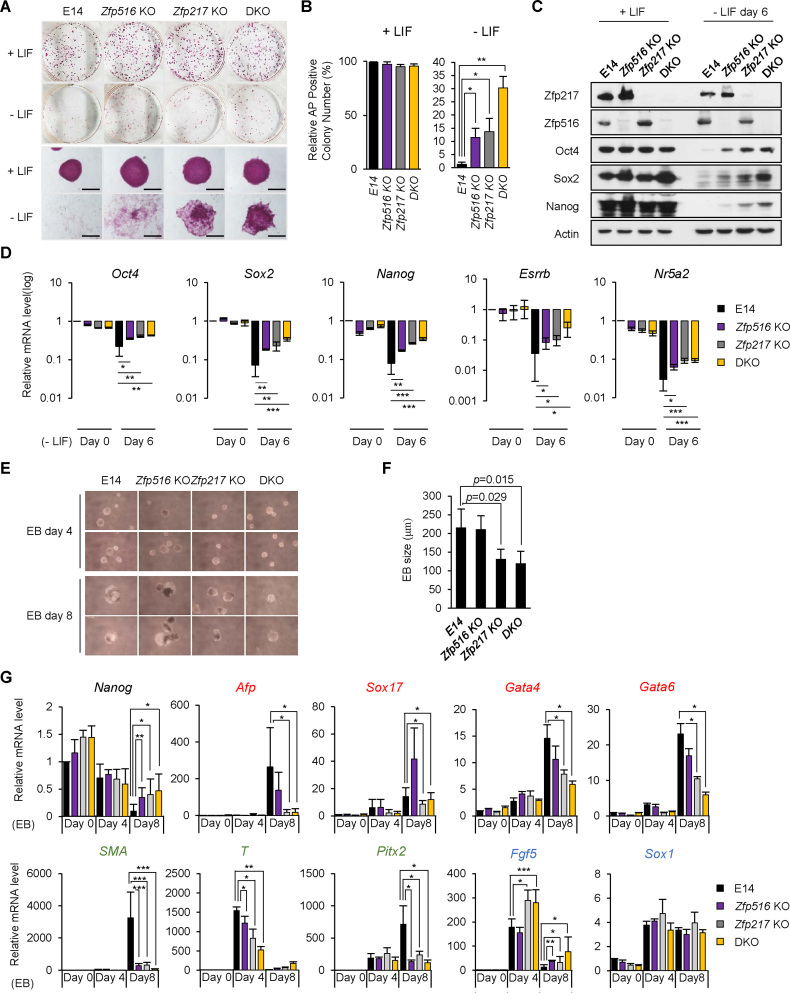
To investigate the knock-out effect of both Zfp217 and Zfp516, we differentiated all three types of knock-out ESCs and performed the self-renewal assays. All of Zfp516 KO, Zfp217 KO, and DKO ESCs maintained stem cell morphology and AP positive colonies in undifferentiated conditions. However, under differentiation conditions, more undifferentiated colonies were sustained in Zfp516 KO, Zfp217 KO, and DKO ESCs than in wild-type ESCs (Figure 5A and B). We then induced differentiation of KO and DKO ESCs by removing LIF from the culture medium and measured the expression levels of pluripotency-associated genes. Zfp516 KO, Zfp217 KO and DKO ESCs all displayed minor decrease in pluripotency-related gene levels than the wild-type ESCs (Figure 5C and D), indicating that both Zfp217 and Zfp516 contribute to exit from pluripotency.
Figure 5.
Zfp217 and Zfp516 double knock-out ESCs fail to differentiate properly. (A and B) Self-renewal assay and alkaline phosphatase (AP) staining of wild-type, Zfp516 KO, Zfp217 KO and Zfp217 and Zfp516 double knock-out (DKO) ESCs in undifferentiated and differentiated conditions. Double knock-out of Zfp217 and Zfp516 leads to incomplete exit from pluripotency during ESC differentiation by sustaining more positive AP colonies than wild-type ESCs (n = 3). Presented as means ± SEM. (C and D) The protein and mRNA levels of Zfp217, Zfp516 and pluripotency-associated genes in wild-type, Zfp516 KO, Zfp217 KO and DKO ESCs upon ESC differentiation by LIF withdrawal for 6 days. Expression was detected by indicated antibodies; Actin was used as a control (n = 3). Presented as means ± SEM (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). (E and F) Embryoid body (EB) formation of wild-type, Zfp516 KO, Zfp217 KO and DKO ESCs. Diameter of EBs were measured for size determination. Representative cells are shown at 40 × magnification from EB cultures (n = 8). Presented as means ± SEM. (G) After forming embryonic bodies (EB) (4 and 8 days) of wild-type, Zfp516 KO, Zfp217 KO and DKO ESCs, the mRNA levels of Nanog and the three germ layer markers [mesoderm (Sma, T, Pitx2) (labeled in green); endoderm (Afp, Sox17,Gata4, Gata6) (labeled in red); ectoderm (Sox1, Fgf5) (labeled in blue)] were analyzed by qRT-PCR (n = 3). Presented as means ± SEM (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
To determine the function of Ctbp2-interacting zinc finger proteins in germ layer formation, we measured and compared the expression levels of germ layer-related genes during embryoid body (EB) formation in wild-type, Zfp516 KO, Zfp217 KO and DKO ESCs. Zfp217 KO and DKO ESCs formed smaller EBs compared to wild-type ESCs, while Zfp516 KO ESCs showed comparable size to wild-type (Figure 5E and F). A representative of the three lineage-specific genes was induced during wild-type ESC differentiation, but the endoderm and mesoderm genes were barely induced in Zfp217 KO ESCs. Moreover, mesoderm gene induction was diminished in Zfp516 KO ESCs. However, ectoderm-related genes were comparably induced in both Zfp516 and Zfp217 KO ESCs (Figure 5G). These results suggest that Zfp217 and Zfp516 probably play their own roles in appropriate lineage commitment.
Moreover, we tempted to compare their pluripotency ability between Ctbp2 KO and DKO ESCs. Ctbp2 KO and DKO ESCs were differentiated for 5 days and were stained with alkaline phosphatase (Supplementary Figure S10A). Both Ctbp2 KO and DKO ESCs displayed more positive AP stained colonies than wild-type. Ctbp2 KO ESC showed higher rate of defect in differentiation than the DKO ESCs. (Supplementary Figure S10B). Next, Ctbp2 KO and DKO ESCs were induced to differentiate by 6 days of LIF removal at 2-day intervals. Protein levels of Oct4, Sox2, and Nanog in Ctbp2 KO ESCs sustained longer than those of DKO ESCs (Supplementary Figure S10C). To further compare their defect in differentiation potency between them, the effect on each lineage-specific genes was assessed during embryoid body (EB) formation of Ctbp2 KO and DKO ESCs. The expression levels of endoderm and mesoderm genes were poorly induced in both Ctbp2 KO and DKO ESCs compared to the wild-type ESCs (Supplementary Figure S10D). These results suggest that Ctbp2 KO and DKO ESCs have similar control over lineage specification but the loss of Ctbp2 in ESCs has more significant effect on regulating exit from pluripotency than either Zfp217 or Zfp516.
Zfp217 and Zfp516 facilitate Ctbp2-mediated repression on active ESC genes during differentiation
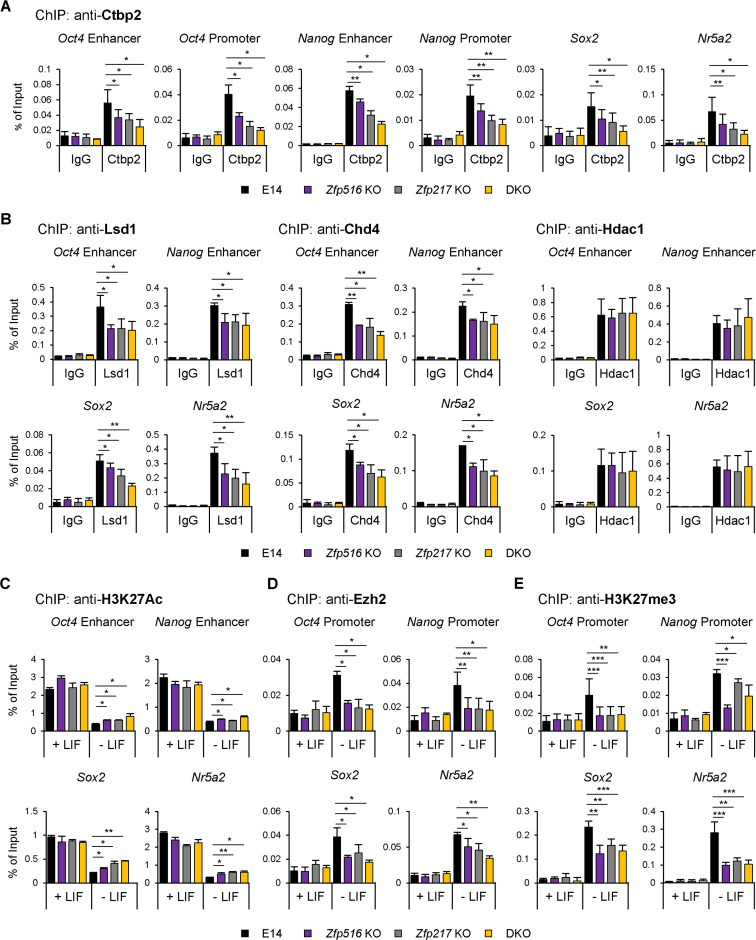
We further examined the knock-out effect of Zfp516 and Zfp217 on the recruitment of Ctbp2 to active gene regions. Enrichment of Ctbp2 on the regions of active ESC genes was diminished in Zfp217 KO, Zfp516 KO, and DKO ESCs compared to the wild-type ESCs (Figure 6A). Moreover, we identified the repressive process of active ESC genes during differentiation by Zfp217 and Zfp516-mediated chromatin regulators. The occupancies of Chd4 and Lsd1 on active ESC gene regions were more decreased in each of Zfp516 KO, Zfp217 KO and DKO ESCs than wild-type, but the enrichment of Hdac1 in all three KO ESCs was comparable to that of the wild-type ESCs (Figure 6B and Supplementary Figure S11A). As expected, reduced occupancy of Chd4 and Lsd1 in Zfp516 KO, Zfp217 KO and DKO ESCs resulted in less reduction of H3K27ac levels on the active ESC gene regions upon differentiation (Figure 6C). Remaining level of H3K27ac in DKO ESCs affected the recruitment of Ezh2 on the regions of active ESC genes (Figure 6D), resulting in minor induction of H3K27me3 levels in Zfp516 KO, Zfp217 KO, and DKO ESCs compared to the wild-type ESCs (Figure 6E). Protein levels of the components of NuRD complex, the HDAC complex, and the PRC2 remained unaffected in the Zfp516 KO, Zfp217 KO and DKO ESCs, indicating that changes in H3K27ac and H3K27me3 levels were regulated without the expression changes in these enzymes (Supplementary Figure S11B). Also, global levels of histones were unaffected by Zfp516 and Zfp217 depletion in undifferentiated conditions (Supplementary Figure S11C). However, when DKO ESCs were induced to differentiate, the protein levels of Chd4, Lsd1, Hdac1 and Ezh2 were sustained while those of wild-type were decreased (Supplementary Figure S11D), implying that repressive chromatin regulators are also affected by the delayed differentiation of DKO ESCs. Taken together, these results represent that the Ctbp2-associated zinc finger proteins, Zfp217 and Zfp516, regulate the epigenetic states of H3K27 on active ESC genes during differentiation, ultimately leading to the proper exit from pluripotency.
Figure 6.
Zfp217 and Zfp516 facilitate Ctbp2-mediated repression on active ESC genes during differentiation. (A) ChIP-qPCR analysis of Ctbp2 on active ESC gene regions in wild-type, Zfp516 KO, Zfp217 KO and Zfp217 and Zfp516 double knock-out (DKO) ESCs (n = 3). Presented as means ± SEM. Zfp217 KO, Zfp516 KO ESCs and DKO ESCs showed lower enrichment of Ctbp2 than wild-type ESCs (n = 3). Presented as means ± SEM (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). (B) ChIP-qPCR analysis of Lsd1, Chd4, and Hdac1 on active ESC gene regions in wild-type, Zfp516 KO, Zfp217 KO and Zfp217 and Zfp516 double knock-out (DKO) ESCs (n = 3). Presented as means ± SEM. Zfp217 KO, Zfp516 KO ESCs and DKO ESCs showed lower enrichment of Chd4 and Lsd1 than wild-type ESCs, but not Hdac1 (n = 3). Presented as means ± SEM (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). (C–E) ChIP-qPCR analysis of H3K27ac, Ezh2 and H3K27me3 on active ESC gene regions in wild-type, Zfp516 KO, Zfp217 KO and DKO ESCs in undifferentiated and differentiated (LIF withdrawal for 6 days) conditions (n = 3). Presented as means ± SEM (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
DISCUSSION
The epigenetic status of embryonic stem cells (ESCs) differs remarkably from the fully differentiated somatic cells (47). This epigenetic difference is due to the differential binding of transcription factors and various chromatin remodeling complexes during ESC differentiation (47,48). Transcription factors directly bind to gene regulatory regions and modulate chromatin modifiers to activate or repress cell-type specific genes on onset of ESC differentiation, thereby determining the cell identity (6,12,49). However, it needs to be unraveled how and which transcription factors contribute to exit from pluripotency and commit to specific lineages.
Ctbp2, a transcriptional corepressor and a regulator during exit from pluripotency, binds to transcription factors through a Pro-X-Asp-Leu-Ser (PXDLS) motif, and recruits epigenetic remodelers (23). A variety of transcription factors interact directly or indirectly with CTBP (26,41,50–55), of which some are related to stem cell maintenance or development. For example, Zscan10, also known as Zfp206, maintains pluripotency of ESCs by regulating ES-specific transcripts (56). Zscan4 requires genome stability and development potency in ESCs (57,58). Moreover, Zfp462 maintains pluripotency of P19 cells (59,60). Although these contributions of Ctbp-interacting factors in stem cells were verified, the detailed mechanisms of association of Ctbp2 and its interacting zinc finger proteins in ESC development remain uninvestigated.
In this study, we demonstrate that Ctbp2-associated zinc finger proteins cooperate for the delicate control of exit from pluripotency by regulating the epigenetic states of H3K27 on active ESC gene regions. One of the Ctbp2-binding zinc finger proteins, Zfp217 (Figure 1) is known as an oncogenic protein associated in various cancers (16,17,61). ZNF217 negatively regulates E-cadherin, an epithelial marker, and induces mesenchymal makers for the epithelial-mesenchymal transition (EMT) process (19,62). EMT occurs during normal embryonic development and is important for lineage determination. For example, an EMT induce marker, Snai1 is in control of epiblast stem cell exit and mesoderm lineage commitment (20). In turn, reprogramming fibroblasts to induced pluripotent stem (iPS) cells resembles the MET program (63).
ZNF217 is identified to form a complex with repressive histone modifiers, JARID1B/PLU-1, G9a, LSD1, CoREST, CTBP1 and EZH2, and regulate transcriptional repression (22–24). Also, Zfp217 was confirmed to interact with Lsd1, Chd4, CoREST and Ctbp2 in undifferentiated ES cell state (Figure 4). These histone modifiers are mostly involved in regulation of ESC differentiation. Lsd1 and the NuRD complex regulate enhancer silencing during ESC differentiation (4,5), and Ezh2, a component of PRC2, catalyzes the H3K27 tri-methylation to stably silence the ESC genes (64,65). We observed that Zfp217 co-occupies with Ctbp2 at the active promoter and enhancer regions in ESCs (Figure 3). Zfp217 recruits the components of the NuRD complex through Ctbp2 and induces the deacetylation of H3K27 at active ESC genes; this transcription inhibition triggers the tri-methylation of H3K27 by the recruitment of PRC2 during ESC differentiation (Figure 4). This supports an idea that Zfp217 initiates the suppression of active ESC genes leading to PRC2 complex-mediated complete silencing.
CRISPR-Cas9 mediated Zfp217 knock-out in ESCs displayed similar stem cell characteristics to ESCs lacking Lsd1 or Mbd3. ESCs lacking Lsd1 showed impaired differentiation of ESCs without affecting self-renewal or pluripotency (4) and loss of Mbd3 disrupted the NuRD complex which led to incomplete ESC differentiation but had no significant effect on ESC self-renewal (45). Zfp217 knock-out ESCs also exhibited features of defect in morphological changes, silencing of active ESC genes, and upregulation of developmental markers upon ESC differentiation, supporting that Zfp217 is also essential for embryonic development. On the other hand, Zfp217 was reported to sustain undifferentiated state of ESCs and support its pluripotent state by epigenetically regulating N6-methyladenosine (m6A) deposition on ESC transcripts, indicating that Zfp217 functions as an activator to pluripotency genes (66). The different conclusions in phenotype are probably assumed to the experimental conditions such as the timing of experiments followed by Zfp217 deletion. After viral infection, knock-down of Zfp217 led to apoptotic cell death immediately after selection, however, once Zfp217-knock-down cells were stabilized, apoptosis no longer occurred. Thus, abrupt loss of Zfp217 possibly destabilizes some cell population and leads to cell death. Despite the notable differences, Zfp217 is a possible driver for reprogramming since chromatin remodeling factors involved in stem cell development, such as Mbd3, also facilitate the reprogramming process (66,67). These findings support that Zfp217 adjusts the balance between LIF-mediated transcriptional activation and NuRD-mediated repression in undifferentiated ESCs.
Moreover, not only Zfp217 but also Zfp516 contributed to exit from pluripotency by H3K27me-mediated repressive mechanisms (Figure 6). Also a strong binding protein to Ctbp2, Zfp516 affected the exit from pluripotency upon stem cell differentiation (Figure 5). On the other hand, Zfp516 increased its expression as development progresses, which is a different pattern from Zfp217 (Figure 1), and also each of Zfp217 and Zfp516 affected different lineage-related genes during ESC differentiation. Zfp217 KO ESCs failed to induce endoderm and mesoderm genes and Zfp516 KO ESCs barely induced mesoderm genes (Figure 5). Even though only Zfp217 but Zfp516 had an effect on endoderm gene induction, Zfp217/Zfp516 double KO embryoid bodies also failed to induce the endoderm genes. This gives a clue that Zfp217 influences on lineage determination temporally and spatially apart from Zfp516 and thus both zinc finger proteins cooperatively participate in the developmental process.
Based on the previous finding that Ctbp2 primes the active ESC genes even in undifferentiated ESCs (11), Zfp217 and Zfp516, along with Ctbp2, are likely to participate in priming the active ESC genes. Besides their priming role in active ESC regions, Ctbp2-interacting zinc finger proteins are also likely to be applied to Ctbp2-mediated lineage-specific gene repression. Moreover, we expect that there might be other transcription factors which carry functional redundancy since Ctbp2 KO carried more significant effect in stem cell differentiation than Zfp217/Zfp516 double knock-out ESCs (Supplementary Figure S10). On the basis that Ctbp2 associates with various zinc finger proteins (Figure 1), other new zinc finger proteins, besides Zfp217 and Zfp516, might bring Ctbp2 to the regulatory regions of the pluripotency genes. Further detailed investigations are in need to identify other new zinc finger proteins and whether these multiple zinc finger proteins act separately or simultaneously during ESC differentiation process.
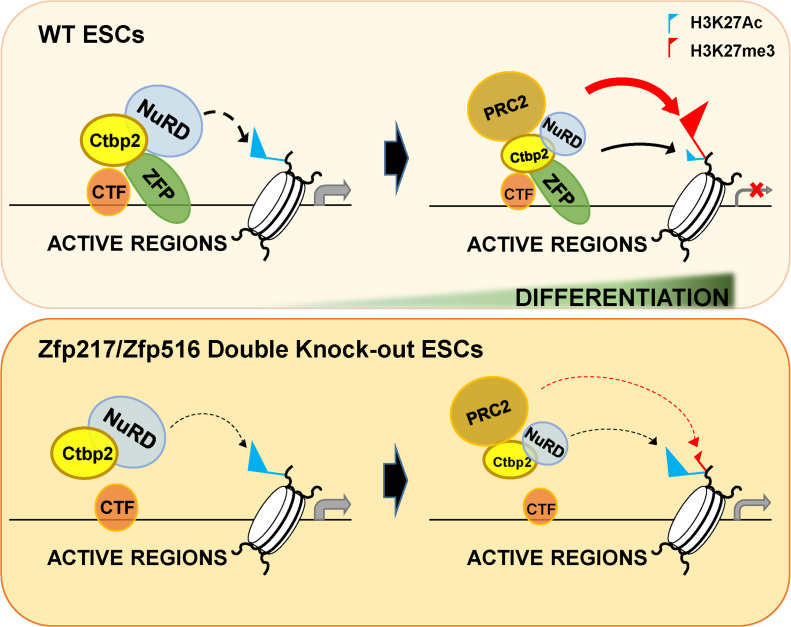
This study reveals that Zfp217 and Zfp516 directly interact with Ctbp2 and trigger active ESC gene silencing by modulating the NuRD complex and PRC2 (Figure 7). We propose that Zfp217 and Zfp516 aptly support chromatin regulators to fine-tune epigenetic changes during cell transitions. Also, the combinatorial effect of multiple zinc finger proteins on stem cell development provides us an insight that certain combinations of transcription factors cooperatively orchestrate the full process of embryonic development.
Figure 7.
A diagram illustrating the contribution of Ctbp2-interacting zinc finger proteins in exit from pluripotency. Ctbp2-interacting zinc finger proteins, such as Zfp217 and Zfp516, bind to the active ESC gene regions and recruit the NuRD complex to prime for H3K27 deacetylation upon ESC differentiation. (The black and thick dotted line shows priming state of the NuRD complex on active ESC regions.) Further differentiation allows the zinc finger proteins to recruit the PRC2 for H3K27 tri-methylation on the active ESC genes, and to silence transcription (shown by the red line). Conversely, when such zinc finger proteins are depleted, the NuRD complex-mediated H3K27 deacetylation is weakened (shown by the thin and black dotted line) and the active gene transcription is sustained. Thus, remaining H3K27ac levels and active transcription affects the PRC2-mediated active ESC gene silencing and result in improper ESC differentiation. (The weakened effect by the PRC2 on active ESC genes is presented with the red dotted line.)
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Yoichi Shinkai (Kyoto University, Japan) for the Wiz expression construct.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea grant, funded by the Korean government (MSIP) [2012R1A3A2048767 to H.-D.Y.]; Education and Research Encouragement Fund of Seoul National University Hospital. Funding for open access charge: We pay the publication charges by myself or grants.
Conflict of interest statement. None declared.
REFERENCES
- 1. Young R.A. Control of the embryonic stem cell state. Cell. 2011; 144:940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Takahashi K., Yamanaka S.. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006; 126:663–676. [DOI] [PubMed] [Google Scholar]
- 3. Li B., Carey M., Workman J.L.. The role of chromatin during transcription. Cell. 2007; 128:707–719. [DOI] [PubMed] [Google Scholar]
- 4. Whyte W.A., Bilodeau S., Orlando D.A., Hoke H.A., Frampton G.M., Foster C.T., Cowley S.M., Young R.A.. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012; 482:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reynolds N., Latos P., Hynes-Allen A., Loos R., Leaford D., O'Shaughnessy A., Mosaku O., Signolet J., Brennecke P., Kalkan T. et al. . NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012; 10:583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamamizu K., Piao Y., Sharov A.A., Zsiros V., Yu H., Nakazawa K., Schlessinger D., Ko M.S.. Identification of transcription factors for lineage-specific ESC differentiation. Stem Cell Rep. 2013; 1:545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Panman L., Andersson E., Alekseenko Z., Hedlund E., Kee N., Mong J., Uhde C.W., Deng Q., Sandberg R., Stanton L.W. et al. . Transcription factor-induced lineage selection of stem-cell-derived neural progenitor cells. Cell Stem Cell. 2011; 8:663–675. [DOI] [PubMed] [Google Scholar]
- 8. Feng B., Jiang J., Kraus P., Ng J.H., Heng J.C., Chan Y.S., Yaw L.P., Zhang W., Loh Y.H., Han J. et al. . Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat. Cell Biol. 2009; 11:197–203. [DOI] [PubMed] [Google Scholar]
- 9. Hildebrand J.D., Soriano P.. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell Biol. 2002; 22:5296–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim T.W., Kwak S., Shin J., Kang B.H., Lee S.E., Suh M.Y., Kim J.H., Hwang I.Y., Lee J.H., Choi J. et al. . Ctbp2-mediated beta-catenin regulation is required for exit from pluripotency. Exp. Mol. Med. 2017; 49:e385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim T.W., Kang B.H., Jang H., Kwak S., Shin J., Kim H., Lee S.E., Lee S.M., Lee J.H., Kim J.H. et al. . Ctbp2 modulates NuRD-Mediated deacetylation of H3K27 and facilitates PRC2-Mediated H3K27me3 in active embryonic stem cell genes during exit from pluripotency. Stem Cells. 2015; 33:2442–2455. [DOI] [PubMed] [Google Scholar]
- 12. Reynolds N., Salmon-Divon M., Dvinge H., Hynes-Allen A., Balasooriya G., Leaford D., Behrens A., Bertone P., Hendrich B.. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 2012; 31:593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaji K., Nichols J., Hendrich B.. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007; 134:1123–1132. [DOI] [PubMed] [Google Scholar]
- 14. Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K. et al. . Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006; 125:301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rahman M.T., Nakayama K., Rahman M., Nakayama N., Ishikawa M., Katagiri A., Iida K., Nakayama S., Otsuki Y., Shih Ie M. et al. . Prognostic and therapeutic impact of the chromosome 20q13.2 ZNF217 locus amplification in ovarian clear cell carcinoma. Cancer. 2012; 118:2846–2857. [DOI] [PubMed] [Google Scholar]
- 16. Quinlan K.G., Verger A., Yaswen P., Crossley M.. Amplification of zinc finger gene 217 (ZNF217) and cancer: when good fingers go bad. Biochim. Biophys. Acta. 2007; 1775:333–340. [DOI] [PubMed] [Google Scholar]
- 17. Rooney P.H., Boonsong A., McFadyen M.C., McLeod H.L., Cassidy J., Curran S., Murray G.I.. The candidate oncogene ZNF217 is frequently amplified in colon cancer. J. Pathol. 2004; 204:282–288. [DOI] [PubMed] [Google Scholar]
- 18. Collins C., Rommens J.M., Kowbel D., Godfrey T., Tanner M., Hwang S.I., Polikoff D., Nonet G., Cochran J., Myambo K. et al. . Positional cloning of ZNF217 and NABC1: genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:8703–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vendrell J.A., Thollet A., Nguyen N.T., Ghayad S.E., Vinot S., Bieche I., Grisard E., Josserand V., Coll J.L., Roux P. et al. . ZNF217 is a marker of poor prognosis in breast cancer that drives epithelial-mesenchymal transition and invasion. Cancer Res. 2012; 72:3593–3606. [DOI] [PubMed] [Google Scholar]
- 20. Lin Y., Li X.Y., Willis A.L., Liu C., Chen G., Weiss S.J.. Snail1-dependent control of embryonic stem cell pluripotency and lineage commitment. Nat. Commun. 2014; 5:3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cohen P.A., Donini C.F., Nguyen N.T., Lincet H., Vendrell J.A.. The dark side of ZNF217, a key regulator of tumorigenesis with powerful biomarker value. Oncotarget. 2015; 6:41566–41581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cowger J.J., Zhao Q., Isovic M., Torchia J.. Biochemical characterization of the zinc-finger protein 217 transcriptional repressor complex: identification of a ZNF217 consensus recognition sequence. Oncogene. 2007; 26:3378–3386. [DOI] [PubMed] [Google Scholar]
- 23. Quinlan K.G., Nardini M., Verger A., Francescato P., Yaswen P., Corda D., Bolognesi M., Crossley M.. Specific recognition of ZNF217 and other zinc finger proteins at a surface groove of C-terminal binding proteins. Mol. Cell Biol. 2006; 26:8159–8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Banck M.S., Li S., Nishio H., Wang C., Beutler A.S., Walsh M.J.. The ZNF217 oncogene is a candidate organizer of repressive histone modifiers. Epigenetics. 2009; 4:100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thillainadesan G., Chitilian J.M., Isovic M., Ablack J.N., Mymryk J.S., Tini M., Torchia J.. TGF-beta-dependent active demethylation and expression of the p15ink4b tumor suppressor are impaired by the ZNF217/CoREST complex. Mol. Cell. 2012; 46:636–649. [DOI] [PubMed] [Google Scholar]
- 26. Dempersmier J., Sambeat A., Gulyaeva O., Paul S.M., Hudak C.S., Raposo H.F., Kwan H.Y., Kang C., Wong R.H., Sul H.S.. Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat. Mol. Cell. 2015; 57:235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sambeat A., Gulyaeva O., Dempersmier J., Tharp K.M., Stahl A., Paul S.M., Sul H.S.. LSD1 interacts with Zfp516 to promote UCP1 transcription and brown fat program. Cell Rep. 2016; 15:2536–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li L., Liu X., He L., Yang J., Pei F., Li W., Liu S., Chen Z., Xie G., Xu B. et al. . ZNF516 suppresses EGFR by targeting the CtBP/LSD1/CoREST complex to chromatin. Nat. Commun. 2017; 8:691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jang H., Kim T.W., Yoon S., Choi S.Y., Kang T.W., Kim S.Y., Kwon Y.W., Cho E.J., Youn H.D.. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell. 2012; 11:62–74. [DOI] [PubMed] [Google Scholar]
- 30. Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A.. Nanog safeguards pluripotency and mediates germline development. Nature. 2007; 450:1230–1234. [DOI] [PubMed] [Google Scholar]
- 31. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. et al. . DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013; 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Hoon M.J., Imoto S., Nolan J., Miyano S.. Open source clustering software. Bioinformatics. 2004; 20:1453–1454. [DOI] [PubMed] [Google Scholar]
- 38. Ernst J., Kellis M.. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012; 9:215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neph S., Kuehn M.S., Reynolds A.P., Haugen E., Thurman R.E., Johnson A.K., Rynes E., Maurano M.T., Vierstra J., Thomas S. et al. . BEDOPS: high-performance genomic feature operations. Bioinformatics. 2012; 28:1919–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell. 2002; 9:213–224. [DOI] [PubMed] [Google Scholar]
- 42. Teif V.B., Vainshtein Y., Caudron-Herger M., Mallm J.P., Marth C., Hofer T., Rippe K.. Genome-wide nucleosome positioning during embryonic stem cell development. Nat. Struct. Mol. Biol. 2012; 19:1185–1192. [DOI] [PubMed] [Google Scholar]
- 43. Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y. et al. . Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013; 154:1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L., Wang L., Hodgkins A., Iyer V., Huang X. et al. . Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods. 2014; 11:399–402. [DOI] [PubMed] [Google Scholar]
- 45. Kaji K., Caballero I.M., MacLeod R., Nichols J., Wilson V.A., Hendrich B.. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 2006; 8:285–292. [DOI] [PubMed] [Google Scholar]
- 46. Zhu D., Fang J., Li Y., Zhang J.. Mbd3, a component of NuRD/Mi-2 complex, helps maintain pluripotency of mouse embryonic stem cells by repressing trophectoderm differentiation. PLoS One. 2009; 4:e7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hawkins R.D., Hon G.C., Lee L.K., Ngo Q., Lister R., Pelizzola M., Edsall L.E., Kuan S., Luu Y., Klugman S. et al. . Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010; 6:479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gifford C.A., Ziller M.J., Gu H., Trapnell C., Donaghey J., Tsankov A., Shalek A.K., Kelley D.R., Shishkin A.A., Issner R. et al. . Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013; 153:1149–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tsankov A.M., Gu H., Akopian V., Ziller M.J., Donaghey J., Amit I., Gnirke A., Meissner A.. Transcription factor binding dynamics during human ES cell differentiation. Nature. 2015; 518:344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boxer L.D., Barajas B., Tao S., Zhang J., Khavari P.A.. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes Dev. 2014; 28:2013–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu B., Shats I., Angus S.P., Gatza M.L., Nevins J.R.. Interaction of E2F7 transcription factor with E2F1 and C-terminal-binding protein (CtBP) provides a mechanism for E2F7-dependent transcription repression. J. Biol. Chem. 2013; 288:24581–24589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Basu A., Atchison M.L.. CtBP levels control intergenic transcripts, PHO/YY1 DNA binding, and PcG recruitment to DNA. J. Cell Biochem. 2010; 110:62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kajimura S., Seale P., Tomaru T., Erdjument-Bromage H., Cooper M.P., Ruas J.L., Chin S., Tempst P., Lazar M.A., Spiegelman B.M.. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008; 22:1397–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bian C., Chen Q., Yu X.. The zinc finger proteins ZNF644 and WIZ regulate the G9a/GLP complex for gene repression. Elife. 2015; 4:e05606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ueda J., Tachibana M., Ikura T., Shinkai Y.. Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J. Biol. Chem. 2006; 281:20120–20128. [DOI] [PubMed] [Google Scholar]
- 56. Zhang W., Walker E., Tamplin O.J., Rossant J., Stanford W.L., Hughes T.R.. Zfp206 regulates ES cell gene expression and differentiation. Nucleic Acids Res. 2006; 34:4780–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Amano T., Hirata T., Falco G., Monti M., Sharova L.V., Amano M., Sheer S., Hoang H.G., Piao Y., Stagg C.A. et al. . Zscan4 restores the developmental potency of embryonic stem cells. Nat. Commun. 2013; 4:1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zalzman M., Falco G., Sharova L.V., Nishiyama A., Thomas M., Lee S.L., Stagg C.A., Hoang H.G., Yang H.T., Indig F.E. et al. . Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010; 464:858–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Masse J., Laurent A., Nicol B., Guerrier D., Pellerin I., Deschamps S.. Involvement of ZFPIP/Zfp462 in chromatin integrity and survival of P19 pluripotent cells. Exp. Cell Res. 2010; 316:1190–1201. [DOI] [PubMed] [Google Scholar]
- 60. Masse J., Piquet-Pellorce C., Viet J., Guerrier D., Pellerin I., Deschamps S.. ZFPIP/Zfp462 is involved in P19 cell pluripotency and in their neuronal fate. Exp. Cell Res. 2011; 317:1922–1934. [DOI] [PubMed] [Google Scholar]
- 61. Rahman M.T., Nakayama K., Rahman M., Katagiri H., Katagiri A., Ishibashi T., Ishikawa M., Iida K., Nakayama N., Otsuki Y. et al. . Gene amplification of ZNF217 located at chr20q13.2 is associated with lymph node metastasis in ovarian clear cell carcinoma. Anticancer Res. 2012; 32:3091–3095. [PubMed] [Google Scholar]
- 62. Littlepage L.E., Adler A.S., Kouros-Mehr H., Huang G., Chou J., Krig S.R., Griffith O.L., Korkola J.E., Qu K., Lawson D.A. et al. . The transcription factor ZNF217 is a prognostic biomarker and therapeutic target during breast cancer progression. Cancer Discov. 2012; 2:638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. et al. . A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010; 7:51–63. [DOI] [PubMed] [Google Scholar]
- 64. Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al. . A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006; 125:315–326. [DOI] [PubMed] [Google Scholar]
- 65. Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K. et al. . Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006; 441:349–353. [DOI] [PubMed] [Google Scholar]
- 66. Aguilo F., Zhang F., Sancho A., Fidalgo M., Di Cecilia S., Vashisht A., Lee D.F., Chen C.H., Rengasamy M., Andino B. et al. . Coordination of m(6)A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell. 2015; 17:689–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. dos Santos R.L., Tosti L., Radzisheuskaya A., Caballero I.M., Kaji K., Hendrich B., Silva J.C.. MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell. 2014; 15:102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.