GABAA receptors are important regulators of neuronal inhibition. Butler et al. identify de novo variants in GABRA2 and GABRA5, encoding α2 and α5 receptor subunits, respectively, as causes of paediatric epilepsy and developmental delay.
Keywords: GABRA2, GABRA5, GABRB3, epileptic encephalopathy, GABAA receptor
Abstract
GABAA receptors are ligand-gated anion channels that are important regulators of neuronal inhibition. Mutations in several genes encoding receptor subunits have been identified in patients with various types of epilepsy, ranging from mild febrile seizures to severe epileptic encephalopathy. Using whole-genome sequencing, we identified a novel de novo missense variant in GABRA5 (c.880G > C, p.V294L) in a patient with severe early-onset epilepsy and developmental delay. Targeted resequencing of 279 additional epilepsy patients identified 19 rare variants from nine GABAA receptor genes, including a novel de novo missense variant in GABRA2 (c.875C > A, p.T292K) and a recurrent missense variant in GABRB3 (c.902C > T, p.P301L). Patients with the GABRA2 and GABRB3 variants also presented with severe epilepsy and developmental delay. We evaluated the effects of the GABRA5, GABRA2 and GABRB3 missense variants on receptor function using whole-cell patch-clamp recordings from human embryonic kidney 293T cells expressing appropriate α, β and γ subunits. The GABRA5 p.V294L variant produced receptors that were 10-times more sensitive to GABA but had reduced maximal GABA-evoked current due to increased receptor desensitization. The GABRA2 p.T292K variant reduced channel expression and produced mutant channels that were tonically open, even in the absence of GABA. Receptors containing the GABRB3 p.P301L variant were less sensitive to GABA and produced less GABA-evoked current. These results provide the first functional evidence that de novo variants in the GABRA5 and GABRA2 genes contribute to early-onset epilepsy and developmental delay, and demonstrate that epilepsy can result from reduced neuronal inhibition via a wide range of alterations in GABAA receptor function.
Introduction
GABA is the major inhibitory neurotransmitter in the brain. GABAA receptors are heteropentameric, ligand-gated anion channels that are activated by GABA to mediate both phasic synaptic transmission and tonic extrasynaptic inhibition in the brain. Functional GABAA receptors are typically formed from 3 of 19 different gene products encoded by GABRA1–GABRA6 (α1–6), GABRB1–GABRB3 (β1–3), GABRG1–GABRG3 (γ1–3), GABRR1–GABRR3 (ρ1–3), GABRD (δ), GABRE (ɛ), GABRP (π), and GABRQ (θ), with most synaptic receptors consisting of two α, two β, and one γ subunit (Olsen and Sieghart, 2009).
Considering the role of GABAA receptors in neuronal inhibition, the genes encoding these receptors (GABRs) are of obvious interest in disorders of altered neuronal excitability such as epilepsy. Pathogenic variants in these genes have been identified in patients with various types of epilepsy. GABRA1 is associated with hereditary generalized epilepsies, like juvenile myoclonic epilepsy, as well as sporadic epileptic encephalopathies, such as Dravet and Ohtahara syndromes (Cossette et al., 2002; Carvill et al., 2014; Kodera et al., 2016). GABRG2 mutations were found in patients with generalized epilepsy with febrile seizures plus (GEFS+), absence epilepsy, and epileptic encephalopathy (Baulac et al., 2001; Wallace et al., 2001; Kang and Macdonald, 2016; Shen et al., 2017). Mutations in GABRB3 have recently emerged as a cause of febrile seizures, GEFS+, focal epilepsy, myoclonic-atonic epilepsy, and early-onset epileptic encephalopathy (Allen et al., 2013; Janve et al., 2016; Møller et al., 2017). There have also been a few reports of epilepsy-associated variants in GABRA3, GABRB1, GABRB2, and GABRD (Srivastava et al., 2014; Okamoto et al., 2015; Janve et al., 2016; Ishii et al., 2017; Niturad et al., 2017). A single de novo missense variant was recently reported in GABRA2 in an individual with epileptic encephalopathy; however, the functional consequence of the variant was not investigated (Orenstein et al., 2018). Additionally, Hernandez et al. (2016) recently identified variants in several GABRs that may modify epilepsy susceptibility.
Here we provide the first functional evidence that de novo variants in the GABRA2 and GABRA5 genes—encoding the α2 and α5 GABAA receptor subunits, respectively—contribute to early-onset epilepsy and developmental delay. We also present functional characterization of a recurrent missense variant identified in GABRB3.
Materials and methods
Next-generation sequencing and Sanger confirmation
Whole-genome sequencing was performed on DNA extracted from whole blood from Patient 1 and his parents at 30× coverage using the Illumina HiSeq X Ten platform with 150-bp paired-end reads. Reads were mapped to the human reference genome using PEMapper, and variants were called using PECaller (Johnston et al., 2017). Variants were annotated using SeqAnt 2.0 (Shetty et al., 2010). De novo variants identified in the proband were confirmed with Sanger sequencing.
Targeted resequencing of DNA samples from 279 epilepsy patients was performed using a custom in-solution hybridization probe library (IDT or SureSelect, Agilent Technologies) to capture the coding exons of ∼4800 genes associated with human disease. The following GABRs were present in the sequencing library: GABRA1, GABRA2, GABRA6, GABRB2, GABRB3, GABRG1, GABRG2, GABRD, and GABRR2. Direct sequencing of the amplified captured regions was performed using next-generation sequencing (2 × 100 base pairs, paired-end reads) on an Illumina HiSeq 2500 in rapid run mode. The individual DNA sequence reads were aligned to the published human genome reference, and variants were called using NextGENe® (SoftGenetics, State College, PA). Variants were called within the coding exons and ±10 bases of flanking intronic sequence. Variants were filtered for call quality and frequency in the population database gnomAD. Peripheral blood or saliva samples were obtained from family members to test variant inheritance after written consent was obtained. The GABRA2 variant (c.875C > A, p.T292K) was determined to be de novo by Sanger sequencing of Patient 2 and her parents. Parental samples were unavailable for Patient 3. This study was approved by the Institutional Review Boards of Emory University and Heidelberg University Hospital.
GABAA receptor cDNAs and mutagenesis
The human GABAA receptor α1 (NM_000806), α2 (NM_000807), α5 (NM_000810), β3 (NM_000814), γ2s (NM_000816), and rat β2 (NM_012957) subunit cDNAs were each cloned into the pcDNA3.1+ expression vector containing a cytomegalovirus promoter. The rat β2 cDNA (NM_012957) was humanized with a single N323S substitution to match the human β2 peptide sequence (NP_000804). Site-directed mutagenesis (QuikChange Lightning, Agilent Technologies) was performed to introduce the following variants: α2(T292K), α5(V294L), and β3(P301L) into their respective clones. Numbering of variants was based on the full-length protein sequences, which include the signal peptides. Sanger sequencing was used to confirm each variant and to ensure the absence of unwanted substitutions.
Cell culture and transfection
Human embryonic kidney 293T (HEK293T) cells (CRL-3216, ATCC®) were maintained at 37°C in 5% CO2 in Eagle’s minimum essential medium supplemented with 5% foetal bovine serum, 40 μM l-glutamine, 100 U/ml penicillin, and 0.1 mM streptomycin. Cells were grown on poly-d-lysine-coated glass coverslips (No.2, VWR) and transfected with X-tremeGENE™ (Roche Diagnostics) with the GABAA receptor cDNAs at a 1:1:1 ratio to express αxβxγ2s receptors (0.5 μg per cDNA) and 0.5 μg green fluorescent protein (GFP). GFP was used as an expression marker for transfection efficiency. Patch-clamp experiments were performed on cells at 24–72 h post-transfection. All experiments were performed at 22°C using cells from at least two transfections and across multiple days to control for cell health and transfection efficiency. All reagents were purchased from Sigma unless otherwise noted.
Whole-cell patch-clamp recordings
Whole-cell patch-clamp remains the gold standard for recording the activation of ligand-gated ion channels, and for the testing of disease variants in epilepsy, and many other neurological and psychiatric disorders (Yuan et al., 2015). No other single technique can generate data that establish the presence of functional receptors on the cell surface and simultaneously define how a variant alters the activation of the receptor on a millisecond timescale. When combined with the recombinant expression of a homogeneous population of variant receptors, the data generated provide the most accurate assessment of variant impact on the amplitude and time course of disease-associated changes in membrane excitability.
Whole-cell patch-clamp recording was performed on HEK293T cells expressing αxβxγ2s GABAA receptors and GFP, similar to methods previously described (Williams et al., 2010). Patch pipettes were fabricated from thin-walled borosilicate glass (TW150F-4, World Precision Instruments, Inc.) using a horizontal puller (P-97, Sutter Instruments, Inc.) to give a resistance of 2–8 MΩ when filled with intracellular solution (120 mM KCl, 2 mM MgCl2, 10 mM EGTA, and 10 mM HEPES, adjusted to pH 7.2 with NaOH, 315 mOsm). Extracellular solution contained 161 mM NaCl, 3 mM KCl, 1 mM MgCl2, 1.5 mM CaCl2, 10 mM HEPES, and 6 mM d-glucose, adjusted to pH 7.4 with NaOH (320–330 mOsm). A rapid solution changer (RSC-160, BioLogics Science Instruments) connected to a 10-channel infusion pump (KD Scientific Inc.) was used to deliver GABA and picrotoxin solutions. The rapid solution changer was controlled by protocols written in pClamp 9 (Molecular Devices, LLC). Whole-cell currents were recorded at −60 mV, filtered at 100 Hz, and sampled at 200 Hz with a MultiClamp 700B amplifier and DigiData 1322A digitizer (Molecular Devices, LLC).
GABA concentration-response assays were performed by exposing each whole-cell patch to increasing concentrations of GABA (0.3, 1, 3, 10, 30, 100, 300 and 1000 μM) for 2 s, with an 8-s washout between concentrations. Recordings were baseline corrected and analysed in MATLAB (MathWorks, Inc.). Peak currents (I) were measured from GABA exposures and fitted using least-of-squares non-linear regression analysis based on the Hill equation: I = Imax × [A]nH / (EC50nH + [A]nH), where I is current peak amplitude, Imax is maximum current amplitude, EC50 is the GABA concentration producing the half-maximal response, A is agonist concentration, and nH is the Hill coefficient. GABA concentration-response assays were individually fitted to the Hill equation for each whole-cell recording. The maximum peak current, EC50, and Hill coefficient were estimated based on averaged values for each mutant receptor and are shown as mean ± standard error of the mean (SEM). The EC50, also known as apparent-affinity, is a compound measure of the binding affinity and gating efficacy of GABA for the receptor (Colquhoun, 1998).
The degree of baseline leak current for cells expressing α2- and α2(T292K)-containing receptors was calculated using whole-cell recordings from GABA concentration-response assays. The first 41 points (0.2 s) of whole-cell baseline current in extracellular solution was averaged for each patch to give a measurement of baseline leak. This was performed for all eight concentrations in the GABA concentration assay, and the values averaged for each cell.
Desensitization was measured for α5- and α5(V294L)-containing receptors from the whole-cell recordings of GABA concentration-response assays. Whole-cell analysis of desensitization was performed using previously described methods (Moody et al., 2017). Briefly, desensitization was measured from 2-s GABA exposures as follows: (Ipeak − Iend) / Ipeak × 100, where Ipeak was the amplitude of the total peak current response and Iend was the amplitude of the peak current response at the end of the agonist exposure (at 2 s). For each cell, desensitization was measured for the eight GABA concentration responses. The log-linear concentration-desensitization relationship was fitted by linear regression. The slope of this function describes the extent of desensitization as GABA concentration increased.
Picrotoxin assay
Picrotoxin (Sigma) was dissolved in 0.1% DMSO and diluted in extracellular solutions to final concentrations of 1, 10, and 100 μM. Picrotoxin solutions were applied in increasing concentrations to patched cells for 3 s, with an 8-s washout between each concentration. Recordings were baseline corrected and peak current amplitude was measured at each picrotoxin concentration.
Biotinylation assay and western blotting
Cell surface biotinylation was performed on transfected HEK293T cells as previously described (Thompson et al., 2012). Total and surface fractions were separated by SDS-PAGE (4–15% gel) and transferred to a PVDF membrane (Bio-Rad). Membranes were blocked with 5% non-fat milk, then incubated with anti-α2 (1:500; 822-GA2CL Phosphosolutions) or anti-α5 (1:500; N415/24 NeuroMab) and Na+/K+-ATPase (1:10 000; ab76020 Abcam) primary antibodies. Membranes were stripped and incubated with monoclonal mouse anti-β-actin (1:2000; A00702 GenScript) primary antibody to verify that the biotinylating reagent did not cross the cell membrane. Signal intensities were quantified using Image Lab™ software (Bio-Rad). GABAA receptor expression levels were normalized to Na+/K+-ATPase.
Structural modelling and lollipop diagrams
A 3D model of the assembled GABAA receptor was generated using PyMOL software and is based on the crystal structure of the human GABAA β3 homopentamer (PDB: 4COF) presented by Miller and Aricescu (2014).
Lollipop diagrams were generated using the freely available Lollipops software (Jay and Brouwer, 2016). After downloading the source code, the software was run directly from the command line interface using the following UniProt accession numbers to draw the protein domains: P31644 (GABRA5), P47869 (GABRA2), and P28472 (GABRB3). Synonymous and non-synonymous variants were downloaded from the gnomAD browser for each gene. The resulting diagrams for the synonymous and non-synonymous variants were merged into a single image for easier visualization.
Statistical analysis
Individual parameters from the whole-cell patch-clamp recordings and western blot experiments were compared using unpaired two-way t-tests (α = 0.05). A two-way unpaired t-test (α = 0.05) with Welch’s correction was used to evaluate group differences in baseline leak current. A linear regression analysis was used to evaluate differences in receptor desensitization as GABA concentration increased. For the picrotoxin assay, a two-way repeated measures ANOVA (α = 0.05) with a Sidak post hoc test for multiple comparisons was performed to compare levels of picrotoxin block across groups. Statistical analyses were carried out using Prism 7.0 (GraphPad Software, Inc.). All data are presented as mean ± SEM. Statistical differences are indicated in the figures and tables using the following symbols: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001.
Data availability
The authors are willing to provide the raw data related to this manuscript upon request.
Results
GABAA receptor variants detected from individuals with epilepsy
Using trio-based whole-genome sequencing, we identified the novel de novo GABRA5 variant c.880G > C (p.V294L) in a proband (Patient 1) with severe epilepsy and developmental delay. To identify additional disease-associated variants in GABRs, we next examined available sequence data from 279 clinically referred epilepsy patients screened at EGL Genetics using a targeted sequencing panel of approximately 4800 genes. Most of the patients referred for genetic testing were children (average age 7 years) with severe paediatric-onset epilepsy. Nine GABRs were included in the sequencing panel: GABRA1, GABRA2, GABRA6, GABRB2, GABRB3, GABRG1, GABRG2, GABRD, and GABRR2. GABRA5 was not included in the sequencing panel. Variants identified in these nine genes were filtered to remove those seen at a frequency >1% in the gnomAD database (Table 1). There were 19 unique heterozygous missense variants identified from 20 individuals after filtering. Three of the 19 variants were absent from the gnomAD database, which includes whole-exome and genome data from ∼138 600 individuals and excludes individuals with severe paediatric diseases. Although the frameshifting GABRR2 variant c.57_67delCCTCACAGATG was absent from the gnomAD database, at least 31 other loss-of-function GABRR2 variants are listed in the database, suggesting that GABRR2 is tolerant of heterozygous loss-of-function variation. We selected the remaining two variants, GABRA2 c.875C > A (p.T292K) and GABRB3 c.902C > T (p.P301L) for further investigation (Table 1 and Fig. 1). The clinical features of the individuals carrying the α5(V294L), α2(T292K), and β3(P301L) variants are summarized in Table 2.
Table 1.
GABR variants identified from screening of 279 individuals with epilepsy
| Gene | Varianta | mRNA reference | Protein change | Domain | gnomADb | PolyPhen-2c | SIFTc |
|---|---|---|---|---|---|---|---|
| GABRA1 | 5:161281174 C/T | NM_000806.5 | p.P29S | NT | 10 | Benign | Tolerated |
| GABRA2 | 4:46264127 G/T | NM_000807.2 | p.T292K | M2 | 0 | Probably damaging | Deleterious |
| GABRA2 | 4:46252550 A/T | NM_000807.2 | p.N377K | CL | 7 | Benign | Tolerated |
| GABRA6 | 5:161113962 A/G | NM_000811.2 | p.T60A | NT | 30 | Probably damaging | Deleterious |
| GABRA6 | 5:161115980 G/A | NM_000811.2 | p.R84H | NT | 17 | Probably damaging | Tolerated |
| GABRA6 | 5:161119035 G/A | NM_000811.2 | p.W305X | M3 | 3 | - | - |
| GABRA6 | 5:161128559 T/C | NM_000811.2 | p.I381T | CL | 36 | Benign | Tolerated |
| GABRB2 | 5:160753448 C/T | NM_021911.2 | p.R373Q | CL | 17 | Benign | Tolerated |
| GABRB3 | 15:26806257 G/A | NM_000814.5 | p.P301L | M2-M3 | 0 | Probably damaging | Deleterious |
| GABRD | 1:1961528 G/C | NM_000815.4 | p.G389A | CL | 5 | Benign | Tolerated |
| GABRG1 | 4:46060315 T/C | NM_173536.3 | p.I279V | M1 | 4 | Benign | Tolerated |
| GABRG2 | 5:161530954 G/A | NM_000816.3 | p.D231N | NT | 4 | Probably damaging | Deleterious |
| GABRR2 | 6:90024892 ACATCTGTGAGG/A | NM_002043.3 | p.L20Pfs*44 | SP | 0 | - | - |
| GABRR2 | 6:89978866 C/T | NM_002043.3 | p.D126N | NT | 219 | Probably damaging | Deleterious |
| GABRR2 | 6:89977789 A/C | NM_002043.3 | p.M180R | NT | 95 | Probably damaging | Deleterious |
| GABRR2 | 6:89975361 C/T | NM_002043.3 | p.R287H | M1-M2 | 138 | Possibly damaging | Tolerated |
| GABRR2 | 6:89974256 C/T | NM_002043.3 | p.V321I | M2-M3 | 176 | Benign | Tolerated |
| GABRR2 | 6:89974162 T/C | NM_002043.3 | p.Q377R | CL | 951 | Benign | Tolerated |
| GABRR2 | 6:89967489 C/T | NM_002043.3 | p.R433H | CL | 975 | Benign | Tolerated |
aGenomic positions are relative to the GRCh37/hg19 human genome assembly.
bNumber of times the allele was observed in the gnomAD database of ∼277 200 alleles.
cPolyPhen-2 and SIFT prediction algorithms only score missense variants.
Genes in bold highlight variants that were functionally evaluated. CL = cytoplasmic loop between M3 and M4 transmembrane domains; M1-M2 = linker between M1 and M2 transmembrane domains; M2-M3 = linker between M2 and M3 transmembrane domains; NT = N-terminal; SP = signal peptide.
Figure 1.
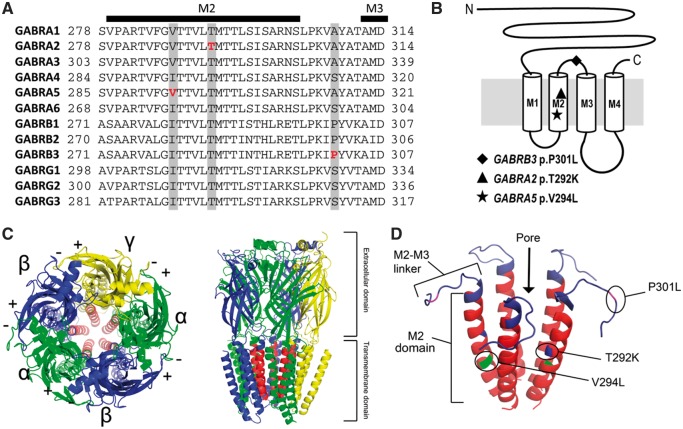
Location of epilepsy variants in the GABAA receptor subunits. (A) Alignment of human α, β, and γ subunits. The location of the α5(V294L), α2(T292K), and β3(P301L) variants are highlighted in grey and the specific amino acids affected are bolded in red. Secondary structures (M2 and M3 transmembrane domains) are also shown above the alignment. The following protein sequences were used to make the alignment: GABRA1, NP_000797; GABRA2, NP_000798; GABRA3, NP_000799; GABRA4, NP_000800; GABRA5, NP_000801; GABRA6, NP_000802; GABRB1, NP_000803; GABRB2, NP_000804; GABRB3, NP_000805; GABRG1, NP_775807; GABRG2, NP_000807; GABRG3, NP_150092. (B) Schematic representation of a single GABAA receptor subunit with the approximate locations of the three missense variants shown. (C) Top-down and side view of an assembled GABAA receptor containing two α (green), two β (blue), and one γ (yellow) subunit. The M2 domain of each subunit has been coloured red to show the location of the channel pore. (D) 3D representation of the five M2 domains of the assembled GABAA receptor pore with the three patient variants indicated.
Table 2.
Genetic and clinical details of epilepsy patients with GABAA receptor variants
| Patient 1 | Patient 2 | Patient 3 | |
|---|---|---|---|
| Age, sex | 2 years, male | 11 years, female | 6 years, male |
| Detection method | WGS | Gene panel | Gene panel |
| Genomic positiona | Chr15:27188364 | Chr4:46264127 | Chr15:26806257 |
| Gene | GABRA5 | GABRA2 | GABRB3 |
| Nucleotide change | c.880G > C (NM_000810.3) | c.875C > A (NM_000807.2) | c.902C > T (NM_000814.5) |
| Protein change | p.V294L | p.T292K | p.P301L |
| Inheritance | De novo | De novo | Unknown |
| Seizures | Myoclonic, tonic, tonic-clonic seizures, oral automatisms, migrating partial seizures | Clustered partial seizures, tonic, tonic-clonic, myoclonic seizures, epileptic spasms | Intractable seizures |
| Onset | 4 months | 6 weeks | NA |
| Development | Secondary microcephaly, delayed milestones, non-verbal | Developmentally delayed, microcephaly, nonverbal, profound intellectual disability | Developmentally delayed |
| Motor development | Truncal hypotonia, tetraspasticity | Central hypotonia, spasticity, non-ambulatory, cerebral palsy | NA |
| EEG | Epileptiform discharges predominantly located within temporal and posterior parts of the brain | Slow and disorganized background, multifocal epileptiform discharges | NA |
| MRI | Hypomyelination | Hypomyelination | NA |
| Other | Autistic features | Cortical visual impairment | Behavioural/psychiatric abnormality |
aGenomic positions are relative to the GRCh37/hg19 human genome assembly.
NA = not available; WGS = whole-genome sequencing.
GABRA5 c.880G > C (p.V294L)
Patient 1 is the second child of unrelated parents with no family history of epilepsy. At 4 months of age, he developed seizures during sleep consisting of myoclonic and tonic seizures, oral automatisms, coughing, tonic-clonic generalized seizures, and migrating partial seizures. Seizure frequency increased from one seizure per week to clusters of up to 100 seizures/day within 6 months. Cognitive and motor development slowed severely at the time of seizure onset. The patient developed secondary microcephaly. At the age of 24 months, Patient 1 shows muscular hypotonia, tetraspasticity, and autistic behaviour. EEG analysis revealed epileptiform discharges, predominantly within the temporal and posterior parts of the brain. MRI showed hypomyelination. Seizures were unresponsive to treatment with phenobarbital, pyridoxine, folinic acid, pyridoxal phosphate, valproate, lacosamide, clonazepam, levetiracetam (alone), and levetiracetam and topiramate in combination; however, the patient became seizure-free at 14 months of age on a combination of zonisamide, levetiracetam, and oxcarbazepine.
Previous metabolic analyses and gene panel testing for epileptic encephalopathies (CeGAT Tubingen) were negative. Through whole-genome sequencing of Patient 1 and his parents, we identified the de novo GABRA5 c.880G > C (p.V294L) variant. Another de novo variant in the MIA2 gene (c.1001_1004delACAA) was also detected but was considered unlikely to contribute to the patient’s phenotype since multiple loss-of-function variants in this gene were observed in the gnomAD database. No other candidate variants were identified by whole-genome sequencing. The GABRA5 p.V294L variant is located in the pore-forming M2 transmembrane domain of the receptor and the affected valine is conserved across the benzodiazepine-sensitive alpha subunits (α1–3, α5, Fig. 1). The V294L substitution was absent from gnomAD and occurs in a region of the receptor where very few missense variants are seen in population controls (Fig. 2A). Finally, GABRA5 p.V294L was predicted to be damaging by PolyPhen-2 and SIFT algorithms.
Figure 2.
Distribution of variants from the gnomAD database across the three GABAA receptor proteins. In each lollipop diagram, the ligand binding domain is shown in green, the transmembrane domain, which includes all four transmembrane segments, is shown in red, and the unstructured regions are shown in grey. The numbers beneath each figure refer to the amino acid number. Missense (blue) and synonymous (grey) variants observed in the gnomAD database are shown. Red lollipops represent the candidate variants identified in this study. (A) Lollipop diagram for α5 (encoded by GABRA5) showing the location of the p.V294L variant in the transmembrane region of the receptor. Protein domain annotation is based on UniProt accession P31644. (B) Lollipop diagram for the α2 receptor showing the location of the p.T292K variant. Based on UniProt accession P47869. (C) Lollipop diagram of the β3 receptor showing the location of p.P301L (red) as well as the locations of previously reported pathogenic GABRB3 variants (gold). Based on UniProt accession P28472.
α5(V294L)β2γ2s receptors are more sensitive to GABA but exhibit increased desensitization
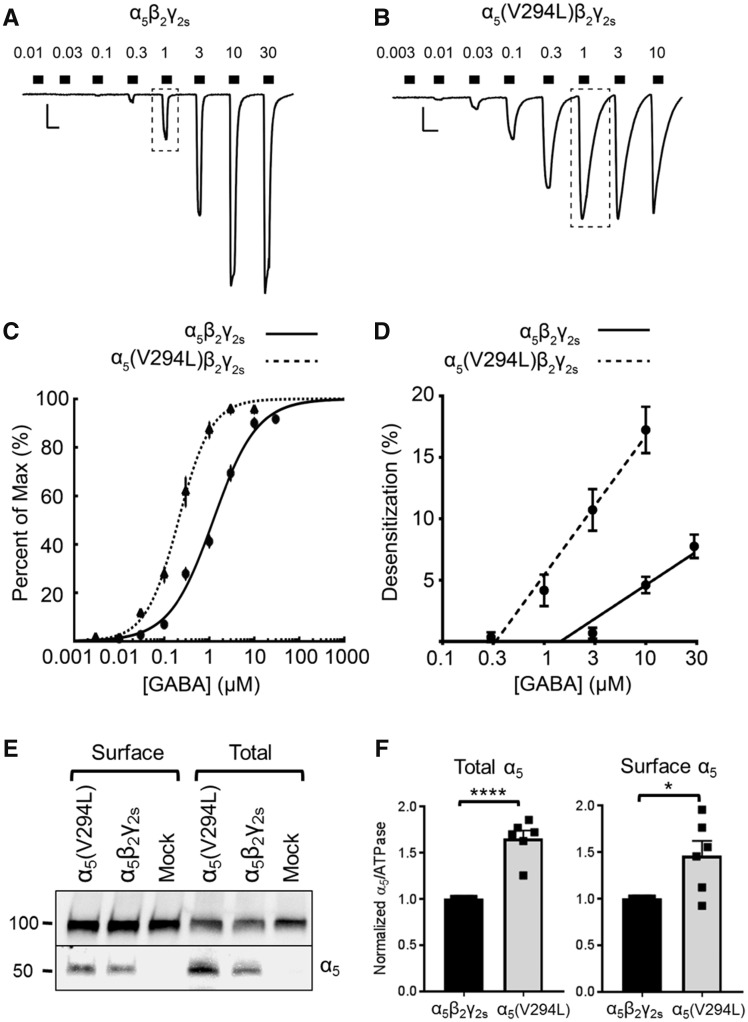
When the α5(V294L) variant was co-expressed with β2 and γ2s subunits in HEK293T cells, the GABA concentration-response relationship was altered relative to channels expressing wild-type α5. First, the maximum GABA-evoked current produced by mutant receptors was significantly lower than that of wild-type receptors [wild-type α5: −4165 ± 314 pA, n = 18 cells; α5(V294L): −2717 ± 324 pA, n = 22 cells, P = 0.0024; Fig. 3A, B and Table 3]. Second, there was a leftward shift in the GABA concentration-response curve relative to wild-type α5β2γ2s receptors (Fig. 3C). This shift is exemplified by the EC50 of α5(V294L)β2γ2s receptors being ∼1/10 that of wild-type receptors [wild-type α5: 2.041 ± 0.314 μM; α5(V294L): 0.238 ± 0.028 μM, P = 0.0024]. Third, the Hill coefficient, a measure of the cooperativity of GABA binding, was significantly higher for mutant receptors [wild-type α5: 1.120 ± 0.061; α5(V294L): 1.562 ± 0.071, P < 0.0001]. These changes are consistent with mutant α5(V294L)β2γ2s receptors having increased GABA apparent-affinity, causing them to approach maximal activation around 1 μM GABA instead of 10 μM GABA like wild-type receptors (Fig. 3A–C).
Figure 3.
Increased GABA apparent-affinity and increased desensitization of α5(V294L)β2γ2s receptors. (A and B) Example traces of GABA concentration-response assays for (A) α5β2γ2s receptors (0.01–30 μM) and (B) α5(V294L)β2γ2s receptors (0.003–10 μM) expressed in HEK293T cells. Scale bars: horizontal = 5 s, vertical = 500 pA. Dotted boxes highlight the 1 μM GABA response, which is larger and more desensitized in mutant receptors. (C) GABA concentration-response curves for α5β2γ2s (solid line, n = 18 cells) and α5(V294L)β2γ2s (dotted line, n = 22 cells) receptors. Points are mean ± SEM and error bars are not shown where bars are smaller than points. Lines are a representative fit based on the average GABA concentration responses. (D) Average desensitization of peak currents from α5β2γ2s (solid line) and α5(V294L)β2γ2s receptors (dotted line). Linear regressions to calculate desensitization are: y = 5.508x + 0.909 (α5β2γ2s) and y = 9.584x – 6.277 [α5(V294L)], where y is the per cent of desensitization, and x is the log[GABA] in micromolar. Points represent mean ± SEM. (E) Total and cell surface protein lysates were analysed by SDS-PAGE and blotted by anti-α5 and anti-ATPase antibodies. Experiments were performed in triplicate on protein samples from two separate transfections. A representative western blot is shown. (F) Band intensities of α5 protein were normalized to the ATPase signal. Bars represent mean ± SEM. An unpaired t-test was used to determine significance. *P ≤ 0.05, ****P < 0.0001.
Table 3.
Electrophysiological recordings from whole-cell patch-clamping of wild-type and mutant GABAA receptors
| Gene | Genotype | Maximum current (pA) | Hill coefficient | EC50 (μM) | n |
|---|---|---|---|---|---|
| GABRA5 | WT | −4165 ± 314 | 1.120 ± 0.061 | 2.041 ± 0.314 | 18 |
| V294L | −2717 ± 324** | 1.562 ± 0.071**** | 0.238 ± 0.028** | 22 | |
| GABRA2 | WT | −3340 ± 392 | 1.344 ± 0.069 | 5.97 ± 1.16 | 9 |
| T292K | − | − | − | 9 | |
| GABRB3 | WT | −1742 ± 157.1 | 1.235 ± 0.046 | 120.0 ± 14.4 | 21 |
| P301L | −540.2 ± 43.02**** | 1.474 ± 0.050*** | 298.1 ± 16.5**** | 20 |
Lack of GABA-evoked currents prevented Hill fit of α2(T292K)β2γ2s receptor data. Mean ± SEM values from n number of cells. WT = wild-type. **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001, unpaired two-way t-test.
One possible explanation for the reduced maximum GABA-evoked currents observed for α5(V294L)β2γ2s receptors is increased receptor desensitization. Since the mutant receptors are more sensitive to GABA, they are more likely to desensitize at lower GABA concentrations. To quantify this, we measured the degree of desensitization occurring over the 2-s GABA exposure during whole-cell GABA concentration-response assays. In whole-cell recordings, desensitization was seen as a decrease in current amplitude in the continued presence of GABA. The relationship between GABA concentration and the degree of desensitization could be described by the lines: y = 5.508x + 0.909 (wild-type) and y = 9.584x − 6.277 [α5(V294L)], where y is the per cent of desensitization, and x is the log[GABA] concentration in micromoles (Fig. 3D). The results show that mutant receptors desensitized more than wild-type receptors as GABA concentration increased [F(1,7) = 15.03, P = 0.0061]. Furthermore, the α5(V294L) variant was not associated with a reduction in the total or cell surface protein expression. In fact, total and cell surface levels of α5(V294L)-containing receptors were increased compared to wild-type (Fig. 3E and F). Increased expression of mutant receptors has been reported previously and could indicate altered subunit stoichiometry compared to wild-type receptors (Janve et al., 2016). Overall, our results show that α5(V294L)β2γ2s receptors are more sensitive to GABA but are also more likely to desensitize, reducing the receptor’s capacity to pass GABAergic chloride current.
GABRA2 c.875C > A (p.T292K)
Patient 2 is the second child of unrelated healthy parents with no family history of epilepsy. Seizures began at 6 weeks of age and included clustered focal seizures and infantile spasms. Clustered focal seizures were accompanied by eye dilation, alternating laughing and crying, breath holding, and behavioural arrest. The patient has also experienced tonic, tonic-clonic, and myoclonic seizures. EEG analysis at 2 years of age showed slow and disorganized background with multifocal epileptiform discharges. MRI performed at age nine showed hypomyelination. Now, at age 11, Patient 2 exhibits microcephaly, cerebral palsy with severe central hypotonia and asymmetric lower extremity spasticity, and cortical visual impairment. She is non-ambulatory and non-verbal and has profound intellectual disability.
Her seizures failed to be controlled by treatment with oxcarbazepine, levetiracetam, phenobarbital, lamotrigine, topiramate, vigabatrin, adrenocorticotropic hormone, pyridoxal phosphate, vitamin E, ketogenic diet, or vagal nerve stimulation. She is currently treated with a combination of valproic acid, phenobarbital, and clobazam but still experiences seizures.
Patient 2 was previously screened using a 110-gene epilepsy panel; however, the results were inconclusive. Using the larger next-generation sequencing library containing 4800 genes, we identified the novel GABRA2 c.875C > A (p.T292K) variant. Sanger sequencing of the parents confirmed that GABRA2 p.T292K was de novo in Patient 2. Thr292 is located in the M2 domain and is conserved across all human α, β, and γ GABAA subunits (Fig. 1A and B). T292K is predicted to be damaging by PolyPhen-2 and SIFT programs (Table 1). It is absent from the gnomAD database and there are few missense variants reported in close proximity to this position (Fig. 2B).
α2(T292K)β2γ2s receptors are tonically open and unresponsive to GABA
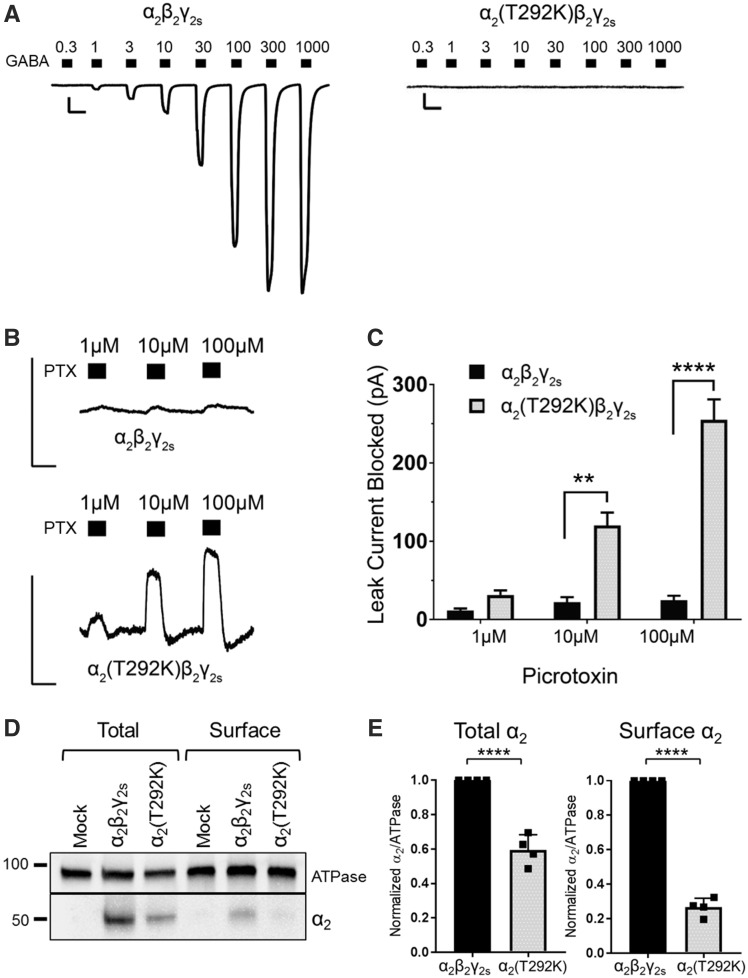
When the α2(T292K) variant was co-expressed with β2 and γ2s subunits, it produced dysfunctional GABAA receptors that did not generate GABA-evoked whole-cell currents within the GABA concentration range of 0.3–1000 μM. This range normally evokes up to several nanoamps of current, as seen with wild-type α2β2γ2s receptors (Fig. 4A and Table 3). For α2(T292K)β2γ2s receptors, the average current responses to 300, 1000, and 3000 μM GABA were: −22.32 ± 7.11 pA, −23.57 ± 76.27 pA, and −7.33 ± 1.82 pA, respectively (n = 9 cells). As a result, no Hill parameters could be estimated from the mutant data.
Figure 4.
α2(T292K)β2γ2s receptors are predominantly open and produce leak current that can be blocked by picrotoxin. (A) Example leak-subtracted trace of GABA concentration-response assays (0.3–1000 μM) for α2β2γ2s and α2(T292K)β2γ2s receptors expressed in HEK293T cells. Traces are aligned for easier visualization, although the mutant example starts at a greater baseline leak current. Scale bars: horizontal = 5 s, vertical = 500 pA. (B) Picrotoxin (PTX) blocked tonic leak current of α2(T292K)β2γ2s receptors in the absence of GABA, while wild-type α2β2γ2s receptors showed little block [wild-type α2: n = 4 cells; α2(T292K): n = 10 cells]. Scale bars: horizontal = 5 s, vertical = 300 pA. (C) Quantification of the leak current (pA) suppressed by picrotoxin. Picrotoxin block was significantly larger for mutant receptors at concentrations 10 μM (P = 0.0017) and 100 μM (P < 0.0001) (two-way repeated-measures ANOVA, Sidak post hoc test). Bars represent mean ± SEM. **P ≤ 0.01, ****P < 0.0001, respectively. (D) Total and cell surface protein lysates were blotted with anti-α2 and anti-ATPase antibodies. Experiments were performed in duplicate on protein samples from two separate transfections. A representative western blot is shown. (E) Band intensities of α2 protein were normalized to the ATPase signal. Bars represent mean ± SEM. An unpaired t-test was used to determine significance. ****P < 0.0001.
We noted that the basal leak current of cells expressing α2(T292K)β2γ2s receptors was twice as large as those expressing wild-type receptors [t(24.45) = 3.37, P < 0.05]. Basal leak current refers to the baseline current that passes into cells in the absence of GABA, in part due to spontaneous channel openings. This observation, in combination with the location of the variant in the pore-forming M2 domain of the receptor, led us to hypothesize that the mutant channels might be trapped in an open state. We used the GABAA receptor antagonist picrotoxin, a known open-channel blocker, to test this hypothesis. Mutant α2(T292K)β2γ2s receptors showed increased suppression of the basal leak current when exposed to increasing concentrations of picrotoxin in the absence of GABA (Fig. 4B). This suppression is due to picrotoxin blocking the tonic GABAA receptor-mediated leak current and is reflected in the membrane current moving closer to zero during picrotoxin exposure. Mutant receptors had significantly larger suppression of leak current in the presence of picrotoxin relative to wild-type receptors at both 10 μM (P = 0.0017) and 100 μM (P < 0.0001) concentrations (Fig. 4B and C). Given that GABAA receptors are generally closed in the absence of GABA, the observation of a leak current that could be blocked by picrotoxin indicates that the mutant α2(T292K)β2γ2s channel is likely trapped in an open state.
Based on the magnitude of leak current suppressed by picrotoxin, we hypothesized that the α2(T292K) variant may also reduce expression of α2-containing receptors. To test this, we performed a cell-surface biotinylation assay and measured total and cell surface expression of wild-type and α2(T292K)-containing receptors (Fig. 4D). There was a significant reduction in the total amount of mutant receptor (∼60% of wild-type levels, P < 0.0001) and a further reduction in the amount of mutant protein at the cell surface (∼27% of wild-type levels, P < 0.0001, Fig. 4E). These results confirm that mutant α2(T292K)β2γ2s receptors are expressed at the cell surface, albeit at lower levels compared to wild-type receptors.
GABRB3 c.902C > T (p.P301L)
Patient 3 is a 6-year-old male with intractable seizures, developmental delay, and an unspecified psychiatric abnormality. He was referred for genetic testing at age five, but a detailed clinical history and parental samples were unavailable.
Patient 3 was previously screened using a 110-gene epilepsy panel but the results were inconclusive. We identified the GABRB3 c.902C > T (p.P301L) variant using the larger sequencing library. Pro301 is located in the extracellular loop between the M2 and M3 transmembrane domains and is conserved across the human beta subunits (Fig. 1A and B). GABRB3 p.P301L is absent from gnomAD and is predicted to be damaging by PolyPhen-2 and SIFT programs (Table 1). Additionally, it is proximal to several reported pathogenic GABRB3 variants (Fig. 2C). Recently, Møller et al. (2017) reported the same GABRB3 variant as de novo in an individual with focal epilepsy, supporting the pathogenicity of this variant.
α1β3(P301L)γ2s receptors are less sensitive to GABA and produce less GABA-evoked current
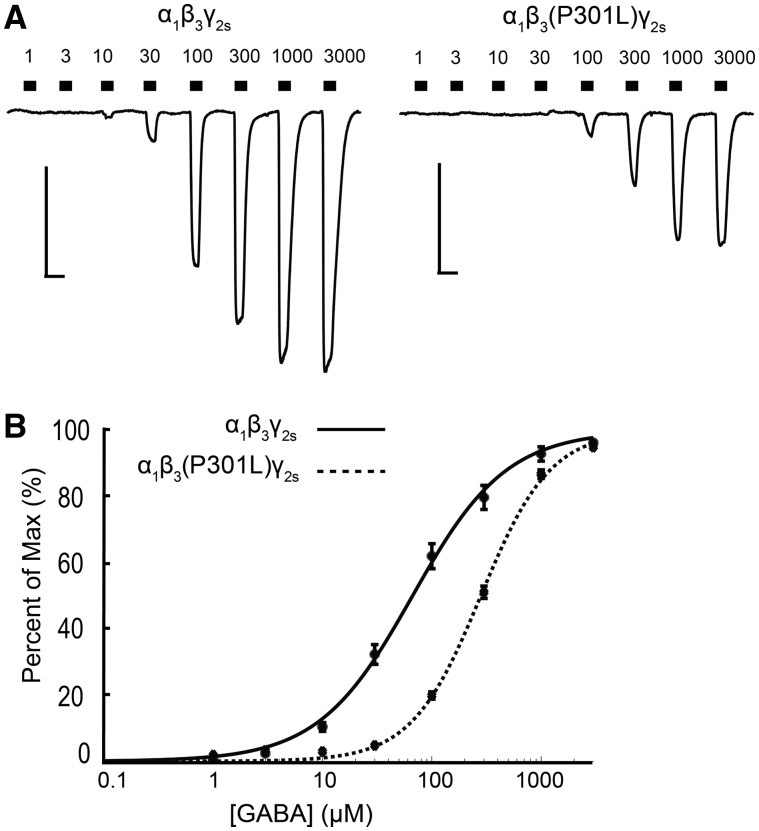
The maximum GABA-evoked current produced by α1β3(P301L)γ2s receptors was significantly lower than for wild-type receptors [wild-type β3: −1742.0 ± 157.1 pA; β3(P301L): −540.2 ± 43.0 pA, P < 0.0001; Fig. 5A and Table 3]. Additionally, expression of α1β3(P301L)γ2s receptors shifted the GABA concentration-response curve rightwards relative to wild-type receptors (Fig. 5B), resulting in a GABA EC50 that was significantly higher for mutant receptors than for wild-type receptors [wild-type β3: 120 ± 14.37 μM, n = 21 cells; β3(P301L): 298.10 ± 16.51 μM, n = 20 cells, P < 0.0001]. These results suggest that β3(P301L)-containing receptors have a reduced capacity for passing current and are less likely to be activated to the same degree as wild-type α1β3γ2s receptors in response to GABAergic synaptic events.
Figure 5.
α1β3(P301L)γ2s receptors are less sensitive to GABA activation. (A) Example trace of GABA concentration-response assays (1–3000 μM) for α1β3γ2s and α1β3(P301L)γ2s receptors expressed in HEK293T cells. Scale bars: horizontal = 5 s, vertical = 500 pA. (B) GABA concentration-response curves for α1β3γ2s (solid line, n = 21 cells) and α1β3(P301L)γ2s (dotted line, n = 20 cells) receptors. Points are mean ± SEM and error bars are not shown where bars are smaller than points. The drawn line is a representative fit based on the average GABA concentration responses.
Discussion
GABAA receptors are important regulators of neuronal inhibition, and mutations in GABRs have been associated with various types of epilepsy. While pathogenic variants in GABRB3 have previously been identified in individuals with mild and severe forms of epilepsy (Janve et al., 2016; Møller et al., 2017), the first potentially pathogenic GABRA2 variant was only recently reported (Orenstein et al., 2018). Additionally, Hernandez et al. (2016) identified three GABRA5 missense variants (p.V204I, p.W280R, and p.P453L) that resulted in gating defects and/or reduced current production from individuals with epilepsy. However, without familial segregation information, they were unable to determine pathogenicity and concluded that these variants might act as modifiers of epilepsy susceptibility. In this study, we describe three individuals with epilepsy and developmental delay who were found to carry heterozygous missense variants in GABRA5, GABRA2, and GABRB3, and present the effects of these variants on channel function.
GABRA5 is highly expressed in the hippocampus and contributes to extrasynaptic tonic inhibition in the brain (Caraiscos et al., 2004). The GABRA5 gene is located on chromosome 15, along with GABRB3 and GABRG3. This genomic region has been implicated in the neurological features of Prader-Willi syndrome, Angelman syndrome, and autism spectrum disorder; three disorders that often include seizures (Hogart et al., 2007; Fatemi et al., 2010). Brain slices from mice lacking Gabra5 exhibit increased excitability of hippocampal pyramidal neurons (Bonin et al., 2007), and reduced Gabra5 mRNA and protein expression has been observed in experimental rat models of pilocarpine-induced temporal lobe epilepsy (Fritschy et al., 1999; Houser and Esclapez, 2003).
We identified the novel de novo GABRA5 variant (p.V294L) in Patient 1, who presented with severe epilepsy and delayed motor and cognitive development. Interestingly, a de novo valine-to-leucine change was observed at the homologous position of GABRA1 (p.V287L) in a patient with early-onset epileptic encephalopathy (Kodera et al., 2016). Although the group who identified the GABRA1 p.V287L variant did not examine the effects of the variant on channel function (Kodera et al., 2016), this observation suggests that substitutions at this position may be associated with severe epilepsy phenotypes.
Whole-cell patch-clamp recordings of α5(V294L)β2γ2s receptors revealed a 10-fold increase in the GABA apparent-affinity, but with a concurrent reduction in maximal current. The observed properties of α5(V294L)β2γ2s channels are consistent with previous mutagenesis of the homologous position in the α2 subunit (denoted as V260W), where mutant receptors became 10 times more sensitive to GABA but had decreased maximal responses (Ueno et al., 2000). Although the α5(V294L)β2γ2s receptors are more sensitive to GABA, this likely causes more channels to accumulate in the desensitized state, removing them from the pool of activatable receptors and thus leading to less neuronal inhibition. During desensitization, the receptors enter a state in which the channel is closed and chloride current is blocked, despite the receptor being activated. In the presence of higher GABA concentrations, the population of activated GABAA receptors increases, and the probability of those receptors entering a desensitized state also increases. Indeed, when we measured the degree of receptor desensitization from the GABA concentration-response assays, we found that α5(V294L)β2γ2s receptors desensitized significantly more than wild-type receptors as GABA concentration increased (Fig. 3D). The impact of this deficit is predicted to be more pronounced for extrasynaptic receptors, such as α5, where the constant presence of GABA would not allow receptors to recover from desensitization.
Given the enhanced sensitivity of α5(V294L)-containing receptors to GABA, the patient harbouring this variant is unlikely to receive therapeutic benefit from traditional positive allosteric modulators targeting GABAA receptors, such as benzodiazepines and barbiturates, as these types of drugs would be expected to further increase the number of desensitized receptors. Patient 1, who had the GABRA5 p.V294L variant, experienced increased seizure frequency while on the barbiturate phenobarbital; however, it was hard to differentiate whether this was the natural course of the patient’s epilepsy or whether this was due to the medication. Treatment with the benzodiazepine clonazepam had no effect on seizure frequency but caused sedation. Interestingly, Patient 1 achieved seizure freedom on a combination of zonisamide, levetiracetam, and oxcarbazepine. These anti-epileptic drugs act predominantly through non-GABAergic mechanisms, such as inhibiting voltage-gated sodium and calcium channels.
GABRA2 is located on chromosome 4 in a gene cluster with GABRB1, GABRA4, and GABRG1. GABRA2 is also highly expressed in the hippocampus, but is localized to the cell soma to mediate synaptic transmission (Tian et al., 2005; Prenosil et al., 2006). In a meta-analysis of 12 genome-wide association studies, an intergenic single nucleotide polymorphism (rs535066) near the 3′ end of GABRA2 was associated with increased risk for epilepsy (International League Against Epilepsy Consortium on Complex Epilepsies, 2014). Hawkins and colleagues (2016) demonstrated that decreased expression of Gabra2 was correlated with greater mortality of heterozygous Scn1a+/− mice that serve as a model of Dravet syndrome. Finally, Gabra2 mRNA expression was observed to be reduced in several brain regions following kainic acid-induced seizures in rats (Drexel et al., 2013).
Patient 2, harbouring the de novo GABRA2 p.T292K variant, presented with intractable epilepsy, profound intellectual disability, and severe cerebral palsy. Orenstein et al. (2018) recently reported a de novo missense variant in GABRA2 (c.1003A > C, p.N335H) in an individual with early-onset epileptic encephalopathy, choreiform movement disorder, and visual impairment. The clinical presentation of their patient is strikingly similar to that of Patient 2, suggesting that pathogenic variants in GABRA2 may be associated with severe disease.
Furthermore, a threonine-to-lysine change was reported at the homologous position of GABRB2 (p.T284K) from a patient with myoclonic encephalopathy who passed away at 17 days of age (Hamdan et al., 2017). Additionally, a threonine-to-isoleucine substitution at the homologous position in GABRA1 (p.T292I) was identified in two unrelated patients with Dravet syndrome and infantile spasms, respectively (de novo in both cases) (Allen et al., 2013; Johannesen et al., 2016). The individual with infantile spasms also shared many features with Patient 2, including poor vision, microcephaly, hypotonia, and cognitive and motor delay (Allen et al., 2013). Therefore, substitution of this highly conserved threonine residue lining the pore of the receptor appears to be associated with severe epilepsy phenotypes.
The α2(T292K) variant occurs at the 10’ position in the M2 domain, which is conserved across the human α, β, and γ subunits. A previous study examined this position of the α2 receptor via tryptophan scanning mutagenesis. The α2(T292W) mutant (numbered as T265W in that study) led to spontaneous channel openings, with no detectable GABA-evoked responses (Ueno et al., 2000). Furthermore, an engineered threonine-to-lysine substitution at the homologous position in the ρ1 subunit (T302K) resulted in channels that exhibited small GABA-evoked currents and high background leak (Wotring and Weiss, 2008). These experiments show that substitution of this highly conserved threonine, especially with bulky, positively charged residues, can alter the gating of the channel.
Functional studies of α2(T292K)β2γ2s receptors revealed channels that did not produce GABA-evoked currents, consistent with previous studies. Although we saw that expression of the mutant receptor was reduced, protein was detectable in the biotinylated surface fraction, indicating that receptors are present at the cell surface. Therefore, reduction in α2(T292K)β2γ2s receptor expression alone cannot completely explain the lack of GABA-evoked currents observed in the GABA concentration-response assays. Cells expressing α2(T292K)β2γ2s receptors also exhibited larger basal leak currents, which could be suppressed with increasing concentrations of the channel blocker picrotoxin. These results are consistent with a receptor that is expressed at the cell surface but unable to transition between open and closed states. A channel that is caught in the open state would be expected to increase the amount of tonic inhibition, which is usually set by a low frequency of spontaneous channel openings in the absence of GABA, or by activation at basal GABA concentrations (<1 µM). Furthermore, this receptor would have reduced ability to respond to temporally-specific GABA stimulation. Consequently, neurons expressing these receptors would pass indiscriminate GABAA receptor-mediated currents, whose polarity would depend on the reversal potential of chloride in the neuron. During early development, the intracellular concentration of chloride is higher, causing chloride ions to move out of the cell when receptors become activated, thereby depolarizing neuronal membranes. Later in development, expression of the KCC2 potassium-chloride co-transporter lowers the intracellular concentration of chloride, allowing GABAA receptors to hyperpolarize cells in response to activation by GABA (Rivera et al., 1999; Hubner et al., 2001). Therefore, α2(T292K) could result in aberrantly high levels of depolarization signals elicited during early brain development when GABAergic synaptic development precedes glutamatergic development (Ben-Ari, 2006). This would likely promote the development of seizures since proper tonic GABAergic currents are important during development (Lee and Maguire, 2014). Also, because GABRA2 expression is highest during early development and declines with age, it is reasonable to expect that the α2(T292K) variant might confer the highest vulnerability to the brain during development (Laurie et al., 1992).
Pathogenic epilepsy variants in GABRB3 were first reported in 2013 by the Epi4K Consortium and the Epilepsy Phenome/Genome Project (Allen et al., 2013). Since then, several GABRB3 variants have been identified in individuals with a spectrum of epilepsies, ranging from mild febrile seizures to severe epileptic encephalopathy (Møller et al., 2017). We identified the GABRB3 p.P301L variant in a patient with intractable epilepsy and developmental delay. Although we were unable to determine variant inheritance, the same variant was recently reported as de novo in a patient with focal epilepsy (Møller et al., 2017). Additionally, GABRB3 p.Y302C was previously identified in three unrelated patients with phenotypes that include focal epilepsy and intractable epileptic encephalopathy (Allen et al., 2013; Møller et al., 2017). The M2-M3 linker, where P301L and Y302C are located, is a highly conserved region that is known to be involved in coupling binding of the agonist to the gating of the channel, a function important for conferring ligand efficacy (O’Shea et al., 2009).
Because the GABRB3 p.P301L substitution was not functionally characterized previously, we performed a GABA concentration-response assay on α1β3(P301L)γ2s receptors. Mutant receptors demonstrated a reduction in GABA apparent-affinity and GABA-evoked current amplitude. This would lead to a receptor that does not respond as strongly to GABA signals as receptors expressing wild-type β3, likely causing reduced neuronal inhibition. The observed reduction in function of β3(P301L) receptors is similar to the reduction seen for receptors containing other GABRB3 epilepsy variants, particularly the adjacent β3(Y302C) variant (Janve et al., 2016; Møller et al., 2017), providing support for the pathogenicity of this variant.
In addition to the three variants that we investigated functionally, we identified 17 other heterozygous GABR variants from individuals with epilepsy (Table 1). The frameshifting GABRR2 c.57_67delCCTCACAGATG variant, which was absent from the gnomAD database, was not considered to be causative of disease since multiple heterozygous loss-of-function GABRR2 variants are listed in the gnomAD database. Similarly, GABRA6 appears to be tolerant of heterozygous loss-of-function variation based on the presence of multiple frameshifting, splice-site, and premature stop variants in gnomAD. Therefore, we excluded the GABRA6 p.W305X variant, seen three times in gnomAD, from further study. The GABRG1 p.I279V variant is also unlikely to be casual since the affected individual also carried a pathogenic frameshifting variant in SCN1A. On the other hand, GABRG2 p.D231N was classified as a variant of uncertain significance. This variant affects the N-terminal ligand-binding domain of the receptor and is predicted to be deleterious by several prediction algorithms; however, it was observed four times in the gnomAD database.
The remaining 13 GABR variants, observed five or more times in gnomAD (Table 1), are unlikely to act as monogenic causes of epilepsy since a recent study showed that dominant pathogenic epilepsy variants were typically absent from or observed only once in the ExAC database, even for milder forms of epilepsy such as GEFS+ and benign familial neonatal-infantile epilepsy (Bennett et al., 2017). However, we cannot exclude the possibility that some of these variants could contribute to the risk for developing epilepsy. Hernandez et al. (2016) evaluated GABR variants identified from individuals with genetic epilepsies and healthy controls and showed that several variants altered channel gating kinetics. Therefore, it is possible that some of the rare variants presented in Table 1 could also contribute to epilepsy susceptibility through complex and polygenic inheritance.
In summary, we present de novo variants in GABRA5 and GABRA2 as contributors to early-onset epilepsy. We provide multiple lines of evidence to support the role of the GABRA5 p.V294L and GABRA2 p.T292K substitutions in disease, including that the variants are de novo, absent from population controls, affect highly conserved and important regions of the GABAA receptor, and alter the function of α5- and α2-containing receptors, respectively. Additionally, we provide functional evidence of pathogenicity for the identified GABRB3 p.P301L variant. This study highlights how different epilepsy-associated variants in GABRs can reduce neuronal inhibition via a range of alterations in GABAA receptor function. The β3(P301L) variant produced functional changes that were consistent with several published pathogenic variants (Janve et al., 2016; Møller et al., 2017), while the α2(T292K) and α5(V294L) variants produced unexpected changes in receptor function that have not been previously reported for human GABAA receptor variants. Further studies are needed to determine whether pathogenic variants in GABRA2 and GABRA5 are associated with distinct epilepsy phenotypes or a spectrum of phenotypes, as observed for GABRA1, GABRG2 and GABRB3.
Acknowledgements
We would like to thank the patients and their families for their participation in this study. We thank Cristina da Silva for assistance with the gene panel data, Caroline Goodroe for her assistance in the patient re-contact process, and Iris Speigel for advice concerning biotinylation and western blotting. We would also like to thank Donald Rainnie for constructive discussions about GABR variants, and Cheryl Strauss for editorial assistance. Whole-genome sequencing was performed through the Emory Integrated Genomics Core, which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities.
Glossary
Abbreviation
- GABR
GABAA receptor gene
Funding
This study was supported by funding from the National Institutes of Health (A.J., R01 NS089719), (K.M.B., 5T32GM008490), (O.A.M., T32NS007480), (O.A.M., T32GM008602), and by funding from Children's Healthcare of Atlanta and the Emory University Research Council to A.E.
References
- Allen AS, Berkovic SF, Cossette P, Delanty N, Dlugos D, Eichler EE et al. . De novo mutations in epileptic encephalopathies. Nature 2013; 501: 217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulac S, Huberfeld G, Gourfinkel-An I, Mitropoulou G, Beranger A, Prud’homme JF et al. . First genetic evidence of GABA(A) receptor dysfunction in epilepsy: a mutation in the gamma2-subunit gene. Nat Genet 2001; 28: 46–8. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y. Seizures beget seizures: the quest for GABA as a key player. Crit Rev Neurobiol 2006; 18: 135–44. [DOI] [PubMed] [Google Scholar]
- Bennett CA, Petrovski S, Oliver KL, Berkovic SF. ExACtly zero or once: a clinically helpful guide to assessing genetic variants in mild epilepsies. Neurol Genet 2017; 3: e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin RP, Martin LJ, MacDonald JF, Orser BA. Alpha5GABAA receptors regulate the intrinsic excitability of mouse hippocampal pyramidal neurons. J Neurophysiol 2007; 98: 2244–54. [DOI] [PubMed] [Google Scholar]
- Caraiscos VB, Elliott EM, You-Ten KE, Cheng VY, Belelli D, Newell JG et al. . Tonic inhibition in mouse hippocampal CA1 pyramidal neurons is mediated by alpha5 subunit-containing gamma-aminobutyric acid type A receptors. Proc Natl Acad Sci USA 2004; 101: 3662–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvill GL, Weckhuysen S, McMahon JM, Hartmann C, Møller RS, Hjalgrim H et al. . GABRA1 and STXBP1: novel genetic causes of Dravet syndrome. Neurology 2014; 82: 1245–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun D. Binding, gating, affinity and efficacy: the interpretation of structure-activity relationships for agonists and of the effects of mutating receptors. Br J Pharmacol 1998; 125: 924–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossette P, Liu L, Brisebois K, Dong H, Lortie A, Vanasse M et al. . Mutation of GABRA1 in an autosomal dominant form of juvenile myoclonic epilepsy. Nat Genet 2002; 31: 184–9. [DOI] [PubMed] [Google Scholar]
- Drexel M, Kirchmair E, Sperk G. Changes in the expression of GABAA receptor subunit mRNAs in parahippocampal areas after kainic acid induced seizures. Front Neural Circuits 2013; 7: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Reutiman TJ, Folsom TD, Rooney RJ, Patel DH, Thuras PD. mRNA and protein levels for GABAAalpha4, alpha5, beta1 and GABABR1 receptors are altered in brains from subjects with autism. J Autism Dev Disord 2010; 40: 743–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritschy JM, Kiener T, Bouilleret V, Loup F. GABAergic neurons and GABA(A)-receptors in temporal lobe epilepsy. Neurochem Int 1999; 34: 435–45. [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Myers CT, Cossette P, Lemay P, Spiegelman D, Laporte AD et al. . High rate of recurrent de novo mutations in developmental and epileptic encephalopathies. Am J Hum Genet 2017; 101: 664–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins NA, Zachwieja NJ, Miller AR, Anderson LL, Kearney JA. Fine mapping of a Dravet syndrome modifier locus on mouse chromosome 5 and candidate gene analysis by RNA-seq. PLoS Genet 2016; 12: e1006398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez CC, Klassen TL, Jackson LG, Gurba K, Hu N, Noebels JL et al. . Deleterious rare variants reveal risk for loss of GABAA receptor function in patients with genetic epilepsy and in the general population. PLoS One 2016; 11: e0162883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogart A, Nagarajan RP, Patzel KA, Yasui DH, Lasalle JM. 15q11‐13 GABAA receptor genes are normally biallelically expressed in brain yet are subject to epigenetic dysregulation in autism-spectrum disorders. Hum Mol Genet 2007; 16: 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser CR, Esclapez M. Downregulation of the alpha5 subunit of the GABA(A) receptor in the pilocarpine model of temporal lobe epilepsy. Hippocampus 2003; 13: 633–45. [DOI] [PubMed] [Google Scholar]
- Hubner CA, Stein V, Hermans-Borgmeyer I, Meyer T, Ballanyi K, Jentsch TJ. Disruption of KCC2 reveals an essential role of K-Cl cotransport already in early synaptic inhibition. Neuron 2001; 30: 515–24. [DOI] [PubMed] [Google Scholar]
- International League Against Epilepsy Consortium on Complex Epilepsies. Genetic determinants of common epilepsies: a meta-analysis of genome-wide association studies. Lancet Neurol 2014; 13: 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii A, Kang JQ, Schornak CC, Hernandez CC, Shen W, Watkins JC et al. . A de novo missense mutation of GABRB2 causes early myoclonic encephalopathy. J Med Genet 2017; 54: 202–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janve VS, Hernandez CC, Verdier KM, Hu N, Macdonald RL. Epileptic encephalopathy de novo GABRB mutations impair GABAA receptor function. Ann Neurol 2016; 79: 806–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay JJ, Brouwer C. Lollipops in the clinic: information dense mutation plots for precision medicine. PLoS One 2016; 11: e0160519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesen K, Marini C, Pfeffer S, Møller RS, Dorn T, Niturad CE et al. . Phenotypic spectrum of GABRA1: from generalized epilepsies to severe epileptic encephalopathies. Neurology 2016; 87: 1140–51. [DOI] [PubMed] [Google Scholar]
- Johnston HR, Chopra P, Wingo TS, Patel V International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome Epstein MP et al. . PEMapper and PECaller provide a simplified approach to whole-genome sequencing. Proc Natl Acad Sci USA 2017; 114: E1923–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JQ, Macdonald RL. Molecular pathogenic basis for GABRG2 mutations associated with a spectrum of epilepsy syndromes, from generalized absence epilepsy to Dravet syndrome. JAMA Neurol 2016; 73: 1009–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodera H, Ohba C, Kato M, Maeda T, Araki K, Tajima D et al. . De novo GABRA1 mutations in Ohtahara and West syndromes. Epilepsia 2016; 57: 566–73. [DOI] [PubMed] [Google Scholar]
- Laurie DJ, Wisden W, Seeburg PH. The distribution of thirteen GABAA receptor subunit mRNAs in the rat brain. III. Embryonic and postnatal development. J Neurosci 1992; 12: 4151–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee V, Maguire J. The impact of tonic GABAA receptor-mediated inhibition on neuronal excitability varies across brain region and cell type. Front Neural Circuits 2014; 8: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller PS, Aricescu AR. Crystal structure of a human GABAA receptor. Nature 2014; 512: 270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller RS, Wuttke TV, Helbig I, Marini C, Johannesen KM, Brilstra EH et al. . Mutations in GABRB3: from febrile seizures to epileptic encephalopathies. Neurology 2017; 88: 483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody OA, Talwar S, Jenkins MA, Freeman AA, Trotti LM, Garcia PS et al. . Rigor, reproducibility, and in vitro cerebrospinal fluid assays: the devil in the details. Ann Neurol 2017; 81: 904–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niturad CE, Lev D, Kalscheuer VM, Charzewska A, Schubert J, Lerman-Sagie T et al. . Rare GABRA3 variants are associated with epileptic seizures, encephalopathy and dysmorphic features. Brain 2017; 140: 2879–94. [DOI] [PubMed] [Google Scholar]
- Okamoto N, Miya F, Tsunoda T, Kato M, Saitoh S, Yamasaki M et al. . Targeted next-generation sequencing in the diagnosis of neurodevelopmental disorders. Clin Genet 2015; 88: 288–92. [DOI] [PubMed] [Google Scholar]
- Olsen RW, Sieghart W. GABA A receptors: subtypes provide diversity of function and pharmacology. Neuropharmacology 2009; 56: 141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein N, Goldberg-Stern H, Straussberg R, Bazak L, Hubshman MW, Kropach N et al. . A de novo GABRA2 missense mutation in severe early-onset epileptic encephalopathy with a choreiform movement disorder. Eur J Paediatr Neurol 2018; 22: 516–24. [DOI] [PubMed] [Google Scholar]
- O’Shea SM, Williams CA, Jenkins A. Inverse effects on gating and modulation caused by a mutation in the M2-M3 Linker of the GABA(A) receptor gamma subunit. Mol Pharmacol 2009; 76: 641–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prenosil GA, Schneider Gasser EM, Rudolph U, Keist R, Fritschy JM, Vogt KE. Specific subtypes of GABAA receptors mediate phasic and tonic forms of inhibition in hippocampal pyramidal neurons. J Neurophysiol 2006; 96: 846–57. [DOI] [PubMed] [Google Scholar]
- Rivera C, Voipio J, Payne JA, Ruusuvuori E, Lahtinen H, Lamsa K et al. . The K+/Cl- co-transporter KCC2 renders GABA hyperpolarizing during neuronal maturation. Nature 1999; 397: 251–5. [DOI] [PubMed] [Google Scholar]
- Shen D, Hernandez CC, Shen W, Hu N, Poduri A, Shiedley B et al. . De novo GABRG2 mutations associated with epileptic encephalopathies. Brain 2017; 140: 49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty AC, Athri P, Mondal K, Horner VL, Steinberg KM, Patel V et al. . SeqAnt: a web service to rapidly identify and annotate DNA sequence variations. BMC Bioinformatics 2010; 11: 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Cohen J, Pevsner J, Aradhya S, McKnight D, Butler E et al. . A novel variant in GABRB2 associated with intellectual disability and epilepsy. Am J Med Genet A 2014; 164A: 2914–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CH, Porter JC, Kahlig KM, Daniels MA, George AL Jr. Nontruncating SCN1A mutations associated with severe myoclonic epilepsy of infancy impair cell surface expression. J Biol Chem 2012; 287: 42001–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Chen HJ, Cross TH, Edenberg HJ. Alternative splicing and promoter use in the human GABRA2 gene. Brain Res Mol Brain Res 2005; 137: 174–83. [DOI] [PubMed] [Google Scholar]
- Ueno S, Lin A, Nikolaeva N, Trudell JR, Mihic SJ, Harris RA et al. . Tryptophan scanning mutagenesis in TM2 of the GABA(A) receptor alpha subunit: effects on channel gating and regulation by ethanol. Br J Pharmacol 2000; 131: 296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RH, Marini C, Petrou S, Harkin LA, Bowser DN, Panchal RG et al. . Mutant GABA(A) receptor gamma2-subunit in childhood absence epilepsy and febrile seizures. Nat Genet 2001; 28: 49–52. [DOI] [PubMed] [Google Scholar]
- Williams CA, Bell SV, Jenkins A. A residue in loop 9 of the beta2-subunit stabilizes the closed state of the GABAA receptor. J Biol Chem 2010; 285: 7281–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotring VE, Weiss DS. Charge scan reveals an extended region at the intracellular end of the GABA receptor pore that can influence ion selectivity. J Gen Physiol 2008; 131: 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Low CM, Moody OA, Jenkins A, Traynelis SF. Ionotropic GABA and glutamate receptor mutations and human neurologic diseases. Mol Pharmacol 2015; 88: 203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors are willing to provide the raw data related to this manuscript upon request.