Abstract
Motivation
Mass spectrometry combined with enrichment strategies for phosphorylated peptides has been successfully employed for two decades to identify sites of phosphorylation. However, unambiguous phosphosite assignment is considered challenging. Given that site-specific phosphorylation events function as different molecular switches, validation of phosphorylation sites is of utmost importance. In our earlier study we developed a method based on simulated phosphopeptide spectral libraries, which enables highly sensitive and accurate phosphosite assignments. To promote more widespread use of this method, we here introduce a software implementation with improved usability and performance.
Results
We present SimPhospho, a fast and user-friendly tool for accurate simulation of phosphopeptide tandem mass spectra. Simulated phosphopeptide spectral libraries are used to validate and supplement database search results, with a goal to improve reliable phosphoproteome identification and reporting. The presented program can be easily used together with the Trans-Proteomic Pipeline and integrated in a phosphoproteomics data analysis workflow.
Availability and implementation
SimPhospho is open source and it is available for Windows, Linux and Mac operating systems. The software and its user’s manual with detailed description of data analysis as well as test data can be found at https://sourceforge.net/projects/simphospho/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Protein phosphorylation is a post-translation modification, which plays a vital role in the regulation of many cellular processes including cell cycle, growth, apoptosis and signal transduction pathways. The leading technology to discover and confirm phosphorylation is tandem mass spectrometry. Due to ongoing developments in enrichment and separation techniques, faster scanning mass spectrometers and data analysis tools, it is now possible to identify tens of thousands of phosphopeptides. Despite its critical importance, phosphopeptide data analysis involves additional unmet challenges compared to analysis of unmodified peptides due to the more complex spectra that are harder to interpret (Zhang et al., 2013). In addition to reliable identification of phosphorylated peptides and proteins, information on the exact phosphorylation sites is essential to understand the interaction and regulation of signaling pathways. Given that specific phosphorylation events function as molecular switches (Vaga et al., 2014), accurate assignment of the precise site of phosphorylation is of utmost importance.
One approach to identify phosphorylation sites involves searching a sequence database followed by analysis using designated localization tools (Beausoleil et al., 2006; Olsen et al., 2006; Taus et al., 2011). Another employs searching a spectral library (Bodenmiller et al., 2008; Hummel et al., 2007; Suni et al., 2015). While it offers better scoring compared to sequence database search scores (Lam et al., 2008; Suni et al., 2015), the downside of this approach has been the lack of readily-available and highly-accurate reference spectral libraries. Recently we reported a method for phosphosite validation that takes advantage of the sensitivity of spectral library searching by overcoming the lack of phosphopeptide libraries (Suni et al., 2015). More specifically, our strategy builds libraries of simulated phosphopeptide spectra based on spectra of unmodified peptides. These simulated phosphopeptide spectra are then used as a reference in the spectral search of observed phosphopeptides. However, the prototype implementation of the simulation method was a command line application, available only for Windows, with no configuration option.
Here, we present further development of our approach through a new tool called SimPhospho. It is the only publically available tool for simulation of higher-energy collisional dissociation (HCD) phosphopeptide spectra. Simulated phosphopeptide spectra by SimPhospho in combination with spectral library searching enable high accuracy and confident phosphosite validation in a comprehensive manner. It follows our previously described workflow but is superior to the prototype version in terms of usability, configuration options and performance. These improvements allow us to optimize various conditions of phosphopeptide simulation, as presented in this study.
2 Implementation
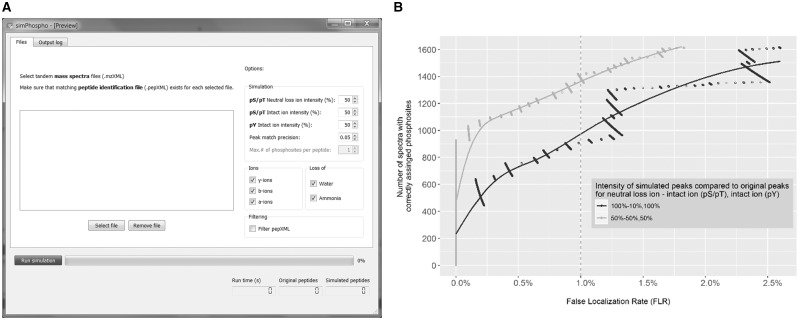
The SimPhospho software (Fig. 1A) was implemented in C++ using components of the Proteowizard project (Kessner et al., 2008) and an XML library (Thomason) and includes a user interface based on the Qt framework (The Qt Company). Two XML files (Keller et al., 2005) serve as an input to SimPhospho: (i) a .pep.xml file that contains the sequence database search results [e.g. Mascot (Matrix Science), X! Tandem (Craig and Beavis, 2004) or COMET (Eng et al., 2013)], validated by PeptideProphet (Keller et al., 2002), and (ii) an .mzXML file that contains mass spectra. First, SimPhospho processes the .pep.xml file. For every peptide identification that contains serine, threonine or tyrosine residues in its sequence, singly phosphorylated peptide isoforms are created and theoretical masses of fragment ions to be phosphorylated are calculated. These masses are then searched in the corresponding spectrum in the .mzXML file and masses and intensities of the found ion peaks are modified for simulating a phosphorylation as described below. The program outputs two files: (i) a .pep.xml file that contains the sequences and modification sites of phosphopeptides, and (ii) an .mzXML file that contains the simulated spectra of the phosphopeptides.
Fig. 1.
(A) Screenshot of SimPhospho displaying the main features of software: simulation options, including ion intensity values, peak match precision, types of ions used for simulation, data filtering switch, as well as output statistics and progress bar. (B) Optimization of intensity values of simulated peaks. To determine the optimal default parameters for SimPhospho, we tested different ion intensity combinations for phosphoric acid neutral loss ions and for intact ions compared to original fragment ion intensities in spectra of nonphosphorylated peptides. We saw the largest number of correctly assigned phosphosites at <1% FLR achieved with a combination of 50 and 50% intensities for intact ions and neutral loss ions for pS and pT, and 50% intensities for intact ions for pY. Data for 100 and 10% (pS, pT) and 100% (pY) are given for reference, as this combination was chosen for the prototype program (Suni et al., 2015). See Supplementary Figure S1 for other intensity combinations
To demonstrate the performance of the SimPhospho program and to determine the optimal default parameters, we tested different ion intensity combinations for simulating phosphopeptide spectra. In the earlier paper we selected 100% of intensity for phosphoric acid neutral loss ions (e.g. y-H3PO4) and 10% for intact ions (e.g. y-ion) compared to original fragment ions for pSer and pThr, and 100% of intensity for intact ions for pTyr. For more details on the simulation rules, refer to (Imanishi et al., 2007; Suni et al., 2015). When testing the simulation criteria using SpectraST 5.0 on an HCD dataset of synthetic phosphopeptides with known phosphosites (Suni et al., 2015), we now observed that the largest number of correctly assigned phosphosites at 1% false localization rate (FLR) was achieved with a combination of 50% intensities (Fig. 1B). Other tested combinations are shown in Supplementary Figure S1.
Users can select which proteins, peptides or scans are used for simulation by adding a .filter text file that lists protein names, peptide sequences or scan numbers of interest. For instance, filtering by scan numbers can be useful when applying identification score cutoffs to the input data. In addition to activating a filter, the other options users can specify are ion types used for simulation (a-, b-, y-ions, ammonia and water losses) and intensities of simulated peaks for intact ions and phosphoric acid neutral loss ions (Fig. 1A). The run time is 50 times faster than when using our prototype program (Suni et al., 2015): simulating 13 000 phosphopeptides from 4000 peptides takes under one minute on a workstation equipped with an Intel Core i5 CPU, 2.30 GHz, 16 GB RAM, Windows 7, 64-bit.
3 Results
SimPhospho is a fast and easy to use tool for simulation of phosphopeptide tandem mass spectra. The program output files can be used directly to build a spectral library using SpectraST (Lam et al., 2008) as either stand-alone version or through Trans-Proteomic Pipeline (TPP) (Keller et al., 2005). This simulated reference spectral library of phosphosites is suitable for phosphopeptide identification, and for validation of phosphopeptides and phosphosites identified by sequence database search programs. Simulated spectral libraries can be searched by stand-alone SpectraST, in TPP, or in Proteome Discoverer (Thermo Fisher Scientific) using SpectraST node. Visualization of the simulated spectra as well as the search results is possible via TPP viewer or in Proteome Discoverer.
The updated version of the software has the following advantages compared to the prototype program: (i) graphical user interface in addition to command line options, (ii) faster data processing, (iii) improved simulation parameters, (iv) optional simulation features, (v) possibility to select a subset of input spectra or peptides to be used for simulation and (vi) cross-platform support. We anticipate that these improvements will facilitate further adoption of this phosphosite validation method, especially in the large scale studies, ultimately leading to fewer false-positive results in the public domain. SimPhospho is available for Windows, Linux and Mac operating systems at https://sourceforge.net/projects/simphospho/. The next version of the software is expected to support simulation of spectra of multiply phosphorylated peptides.
Supplementary Material
Acknowledgement
We thank the Turku Proteomics Facility for their support.
Funding
This work was supported by the Japan Society for the Promotion of Science KAKENHI [JP16K08206 to S.Y.I.]; Academy of Finland [128712 to G.L.C., 304995 and 310561 to L.L.E.]; and Nordforsk [070325 to G.L.C.].
Conflict of Interest: none declared.
References
- Beausoleil S.A. et al. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol., 24, 1285–1292. [DOI] [PubMed] [Google Scholar]
- Bodenmiller B. et al. (2008) PhosphoPep—a database of protein phosphorylation sites in model organisms. Nat. Biotechnol., 26, 1339–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R., Beavis R.C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics, 20, 1466–1467. [DOI] [PubMed] [Google Scholar]
- Eng J.K. et al. (2013) Comet: an open-source MS/MS sequence database search tool. Proteomics, 13, 22–24. [DOI] [PubMed] [Google Scholar]
- Hummel J. et al. (2007) ProMEX: a mass spectral reference database for proteins and protein phosphorylation sites. BMC Bioinformatics, 8, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi S.Y. et al. (2007) Reference-facilitated phosphoproteomics: fast and reliable phosphopeptide validation by microLC-ESI-Q-TOF MS/MS. Mol. Cell. Proteomics, 6, 1380–1391. [DOI] [PubMed] [Google Scholar]
- Keller A. et al. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol., 1, E1. 2005 0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A. et al. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem., 74, 5383–5392. [DOI] [PubMed] [Google Scholar]
- Kessner D. et al. (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics, 24, 2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H. et al. (2008) Building consensus spectral libraries for peptide identification in proteomics. Nat. Methods, 5, 873–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J.V. et al. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell, 127, 635–648. [DOI] [PubMed] [Google Scholar]
- Suni V. et al. (2015) Confident site localization using a simulated phosphopeptide spectral library. J. Proteome Res., 14, 2348–2359. [DOI] [PubMed] [Google Scholar]
- Taus T. et al. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res., 10, 5354–5362. [DOI] [PubMed] [Google Scholar]
- Thomason L. TinyXML. http://sourceforge.net/projects/tinyxml/.
- Vaga S. et al. (2014) Phosphoproteomic analyses reveal novel cross-modulation mechanisms between two signaling pathways in yeast. Mol. Syst. Biol., 10, 767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. (2013) Protein analysis by shotgun/bottom-up proteomics. Chem. Rev., 113, 2343–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.