Abstract
The interaction of the eukaryotic initiation factor 4G (eIF4G) with the cap-binding protein eIF4E initiates cap-dependent translation and is regulated by the 4E-binding proteins (4E-BPs), which compete with eIF4G to repress translation. Metazoan eIF4G and 4E-BPs interact with eIF4E via canonical and non-canonical motifs that bind to the dorsal and lateral surface of eIF4E in a bipartite recognition mode. However, previous studies pointed to mechanistic differences in how fungi and metazoans regulate protein synthesis. We present crystal structures of the yeast eIF4E bound to two yeast 4E-BPs, p20 and Eap1p, as well as crystal structures of a fungal eIF4E–eIF4G complex. We demonstrate that the core principles of molecular recognition of eIF4E are in fact highly conserved among translational activators and repressors in eukaryotes. Finally, we reveal that highly specialized structural motifs do exist and serve to modulate the affinity of protein-protein interactions that regulate cap-dependent translation initiation in fungi.
INTRODUCTION
The assembly of the eukaryotic initiation factor 4F (eIF4F) on the 5′ cap structure of mRNA is essential for cap-dependent translation initiation in eukaryotes. eIF4F is a heterotrimeric complex, consisting of the cap-binding protein eIF4E, the DEAD-box RNA helicase eIF4A and the scaffolding subunit eIF4G. eIF4G plays a central role in complex assembly as it bridges the interaction between eIF4E and eIF4A. Furthermore, eIF4G recruits the 43S preinitiation complex (PIC, comprising the 40S ribosomal subunit, the eIF3 complex, and associated factors) to the mRNA 5′ untranslated region (UTR), which then initiates scanning in search for the initiation codon (1,2).
The assembly of the eIF4F complex is tightly regulated by a large family of translational repressors known as eIF4E-binding proteins (4E-BPs) that compete with eIF4G for binding to eIF4E, thereby inhibiting translation initiation (3,4). Metazoan eIF4G and 4E-BPs bind to the same eIF4E surfaces using a similar bipartite binding region that comprises a canonical and a non-canonical 4E-binding motif connected by a linker region (5–7). The canonical motif, characterized by the YX4LΦ consensus sequence (3,4) (with Y, X, L and Φ representing Tyr, any amino acid, Leu and any hydrophobic residue), mediates binding to a conserved dorsal surface of eIF4E, opposite to the cap-binding pocket (8,9). The non-canonical motif recognizes a conserved eIF4E lateral surface, enhancing the affinity of the interaction (5–7,10–14).
In contrast to the bipartite eIF4E–eIF4G recognition mode observed in the metazoan complexes, previous solution NMR studies of the eIF4E–eIF4G complex from Saccharomyces cerevisae (Sc) revealed a distinct eIF4E recognition mode, whereby eIF4G does not contact the lateral surface of eIF4E (15). Instead, the eIF4G sequences flanking the canonical motif wrap around the poorly conserved N-terminal extension of eIF4E, forming a bracelet-like structure that increases the binding affinity of eIF4G for eIF4E by 400-fold (15).
Budding yeast encodes two 4E-BPs, p20 (or Caf20p) and the eIF4E-associated protein 1 (Eap1p), which have no sequence homology with each other or with metazoan 4E-BPs except for the canonical motif (16–19). These proteins inhibit cap-dependent translation initiation in vitro through competition with eIF4G and have been implicated in different cellular processes through the regulation of translation of specific mRNAs (17,18,20–30).
Although it is well established that p20 and Eap1p act as translational repressors, the molecular details regarding their interaction with eIF4E remain unknown except for the presence of a canonical motif (17,18,29). However, no additional eIF4E-binding elements, such as the non-canonical motifs of metazoan 4E-BPs, have been described for these proteins. Moreover, it is not known whether the yeast 4E-BPs have evolved bracelet-like structures similar to that observed in Sc eIF4G.
In metazoans, the competition mechanism of 4E-BPs is based on molecular mimicry of the bipartite eIF4E–eIF4G interaction. However, since the eIF4E–eIF4G binding modes appear to differ between yeast and metazoans, this raises the question of how do 4E-BPs compete with eIF4G for binding to eIF4E in order to regulate translational initiation in yeast.
To uncover the molecular principles underlying the assembly of eIF4E–eIF4G complexes and competition by 4E-BPs in fungi, we have determined high-resolution crystal structures of the eIF4E–eIF4G complex from Chaetomium thermophilum (Ct)—a thermophilic filamentous fungus. In addition, we also determined crystal structures of the Sc eIF4E in complex with either p20 or Eap1p. Surprisingly, we observed that eIF4G as well as p20 and Eap1p share a conserved bipartite eIF4E-binding mode, as described for the metazoan eIF4G and 4E-BPs, and contact the dorsal and lateral surfaces of eIF4E via canonical and non-canonical motifs, respectively. However, despite this apparent similarity, there are unique structural features in the Ct eIF4E–eIF4G interface that promote complex stability. Finally, we show that yeast 4E-BPs have evolved molecular adaptations to modulate the affinity for this shared binding interface with eIF4E most likely to be able to compete effectively with eIF4G and regulate the initiation of cap-dependent translation.
MATERIALS AND METHODS
DNA constructs
For expression in E. coli, a cDNA fragment encoding Sc eIF4E (residues T25–L213 or V35–L213) was inserted into the NdeI and NheI restriction sites of the pnYC-NpH vector (31) to express proteins with an N-terminal His6-tag cleavable by HRV 3C protease. Codon-optimized synthetic cDNA fragments for expression of Ct eIF4E (full-length), Ct eIF4G (residues A728–P1059), Sc p20 and Sc Eap1p (residues M1–K250) were purchased from GeneArt (Thermo Fisher Scientific). cDNA fragments encoding Ct eIF4E (residues T35–V250, K40–V250 and V45–V250 all carrying the Δ183-202 deletion) were inserted into the NdeI and NheI restriction sites of the pnYC-NpH vector as well as into the NdeI and BamHI restriction sites of the pnYC-CvH vector to express Ct eIF4E with a C-terminal His6 tag cleavable by TEV protease. For the purpose of facilitating crystallization, we used a construct of Ct eIF4E in which we had removed a short internal insertion that is not present in most other eIF4E proteins and is not near the eIF4G-binding surface (Supplementary Figure S1A and B).
cDNA fragments of Sc eIF4G (residues T408–G485) and Ct eIF4G (residues Q954–D1030) were inserted into the NdeI and NheI restriction sites of the pnEA-NpM vector to express the proteins with an N-terminal maltose binding protein (MBP) tag cleavable by HRV 3C protease. cDNA fragments of Sc p20, Sc Eap1p, Sc and Ct eIF4G (Supplementary Table S1) were also inserted into the NdeI and BamHI restriction sites of a modified pnEA-NpM vector additionally encoding a C-terminal fusion of the B1 domain of immunoglobulin-binding protein G (GB1), to produce an MBP-(eIF4G or 4E-BP)-GB1 fusion protein. The GB1 fusion is connected by a GSASG (eIF4G) or GSSRASG (p20 and Eap1p) linker sequence to the eIF4G or 4E-BP fragment. All mutants used in this study were generated by site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene). All constructs used in this study are listed in Supplementary Table S1.
Protein expression and purification
All proteins were expressed in E. coli BL21 Star (DE3) cells (Invitrogen) grown in LB medium overnight at 20°C. To purify the eIF4E–eIF4G and eIF4E–4E-BP complexes used in crystallization, N-terminally His6-tagged Sc eIF4E (residues T25–L213 or V35-L213) or Ct eIF4E (residues T35–V250 Δ183–202) were co-expressed with N-terminally MBP-tagged Sc Eap1p (residues T91–G150), Sc p20 (residues M1–H49) or Ct eIF4G (residues Q954–D1030), respectively. All protein purification steps were performed at 4°C. The cells were lysed by sonication in lysis buffer containing 20 mM Na-phosphate (pH 7.5), 300 mM NaCl and 2 mM dithiothreitol (DTT) supplemented with lysozyme (1 mg/ml), DNase I (5 μg/ml) and a protease inhibitor cocktail (Complete Protease Inhibitor Cocktail, Roche). Except for the Sc eIF4E–p20 complex, all complexes were purified from cleared lysates using amylose resin (New England Biolabs). In the case of the Sc eIF4E–p20 complex, cells were lysed in lysis buffer supplemented with 25 mM imidazole and the cleared cell lysate was applied to a Ni2+-charged HiTrap IMAC HP column (GE Healthcare). The Sc eIF4E–p20 complex was eluted with a gradient of 25–500 mM imidazole. For all protein complexes, MBP- and His6-tags were cleaved overnight with HRV 3C protease while dialyzing into 20 mM HEPES–NaOH (pH 7.5), 100 mM NaCl and 2 mM DTT. The protein complexes were further purified using a HiTrap Heparin HP column (GE Healthcare) and eluted using a gradient from 100 to 500 mM NaCl. As a final step, all complexes were subjected to size exclusion chromatography on a Superdex 75 column (GE Healthcare) equilibrated in 10 mM HEPES–NaOH (pH 7.5), 200 mM NaCl and 2 mM DTT. All proteins were stored at −80°C after flash freezing in liquid nitrogen.
The C-terminally GB1-tagged peptides used in the ITC measurements were purified as described above with the following changes. After cleavage of the MBP tag by HRV 3C protease, the proteins were diluted to obtain a buffer containing 20 mM HEPES–NaOH (pH 7.5), 75 mM NaCl and 2 m DTT and subjected to anion exchange chromatography on a HiTrap Q column (GE Healthcare) to remove the cleaved MBP. The proteins were further purified by size exclusion chromatography on a Superdex 75 column equilibrated in 10 mM Na-phosphate (pH 7.5), 150 mM NaCl and 2 mM DTT, followed by purification over amylose resin to remove any remaining MBP.
The MBP- and GB1-tagged Sc eIF4G, Sc Eap1p, Sc p20 and Ct eIF4G fragments used in pulldown assays were expressed as described above.
The eIF4E proteins used in the ITC measurements and pulldown assays were expressed with an N-terminal His6-tag for Sc eIF4E and a C-terminal His6-tag for Ct eIF4E. The cells were lysed in lysis buffer containing 25 mM imidazole. The cell lysates were applied to Ni2+-charged HiTrap IMAC HP column (GE Healthcare) and eluted with a gradient of 25 to 500 mM imidazole. Cleavage of the Sc eIF4E His6-tag with HRV 3C protease was performed overnight while the proteins were dialyzing in 20 mM HEPES–NaOH (pH 7.5), 300 mM NaCl and 2 mM DTT. After dilution to 20 mM HEPES–NaOH (pH 7.5), 100 mM NaCl and 2 mM DTT, the proteins were further purified on a HiTrap Heparin HP column (GE Healthcare). The Sc eIF4E proteins were subjected to a final purification on a Superdex 75 column (GE Healthcare) in 20 mM Na-phosphate (pH 7.5), 200 mM NaCl and 2 mM DTT.
Note that the removal of the N-terminal His6 and MBP tags by the HRV 3C protease cleavage results in proteins containing a GPHM sequence motif at the N-terminus.
Crystallization
The purified eIF4E–eIF4G complexes were subjected to crystallization trials directly after purification. When indicated, cap analog (m7GpppG; NEB) was added to the purified eIF4E–eIF4G complexes at a 1.05–1.20-fold molar excess prior to crystallization.
Crystals of Ct eIF4E (residues T35–V250 Δ183–202)–Ct eIF4G (residues Q954–D1030) complex at 19 mg/ml [592 μM; in HEPES–NaOH (pH 7.5), 200 mM NaCl and 2 mM DTT]) were obtained after mixing 0.1 μl purified protein complex with 0.1 μl Morpheus 96-well screen condition E9 (Molecular Dimensions). Crystals grew within one day at 20°C using the sitting drop vapor diffusion method. Crystals of Ct eIF4E (residues T35–V250 Δ183–202) in complex with Ct eIF4G (residues Q954–D1030) and m7GpppG were obtained using the sitting drop vapor diffusion method at 20°C. Crystals grew within one day after mixing 0.1 μl of purified protein complex at 20 mg/mL (625 μM protein) and 687 μM cap analog [in 10 mM HEPES–NaOH (pH 7.5), 200 mM NaCl and 2 mM DTT] with 0.1 μl Morpheus 96-well screen condition D9 (Molecular Dimensions).
Crystals of Sc eIF4E (residues V35–L213 carrying the K42A, K168A and K187A substitutions)–Sc Eap1p (residues T91–G150) were obtained using the sitting drop vapor diffusion method at 20°C. The crystals grew within four days after mixing 0.2 μl purified protein complex at 18 mg/ml [in 10 mM HEPES–NaOH (pH 7.5), 200 mM NaCl and 2 mM DTT] with 0.2 μl reservoir solution [0.1 M HEPES–NaOH (pH 7.5), 30% (w/v) Jeffamine ED-2001]. Crystals of Sc eIF4E (residues T25–L213 carrying the K42A, K122A, K168A and K187A substitutions) bound to Sc Eap1p (residues T91–G150) and m7GpppG were obtained using the sitting drop vapor diffusion method at 20°C. Crystals grew within 4 days after mixing 0.2 μl purified protein complex at 18 mg/mL (610 μM protein) and 730 μM cap analog [in 10 mM HEPES–NaOH (pH 7.5), 200 mM NaCl and 1 mM TCEP] with 0.2 μl reservoir solution [0.1 M MIB (sodium malonate:imidazole:boric acid in molar ratio of 2:3:3) buffer (pH 7.0), 21% (w/v) PEG 1500].
Crystals of Sc eIF4E (residues V35–L213 carrying the K42A, K168A and K187A substitutions) in complex with p20 (residues M1–H49) were obtained using sitting drop vapor diffusion at 20°C. Crystals grew within three days after mixing 0.2 μl purified protein complex at 23 mg/ml [850 μM protein in 10 mM HEPES–NaOH (pH 7.5), 200 mM NaCl] with 0.2 μl reservoir solution containing 0.1 M Na-acetate (pH 5.0), 13% (w/v) PEG 6000, 10 mM ZnCl2.
All crystals were briefly transferred into mother liquor supplemented with 18–20% (v/v) glycerol for cryoprotection and then flash cooled in liquid nitrogen.
Data collection
The diffraction data for the Ct eIF4E–eIF4G and Sc eIF4E–Eap1p complexes were collected at 1.0000 Å wavelength at 100 K and on a PILATUS 6M detector (Dectris) at the PXII beamline of the Swiss Light Source (Villigen, Switzerland). The diffraction data for the Sc eIF4E–p20 complex bound to cap analog were recorded at a wavelength of 1.0332 Å at 100 K on a PILATUS 6M detector at the P11 beamline of PETRA III, Deutsches Elektronen-Synchrotron (DESY) (Hamburg, Germany).
Structure determination
Diffraction data were processed using XDS (32) and scaled using AIMLESS as part of the CCP4 package (33–35). The phases were obtained by molecular replacement using PHASER (36). In the case of the Ct eIF4E–eIF4G complex bound to cap analog, the human eIF4E structure [PDB ID: 4TPW (37), chain B] was used as a search model. For the remaining eIF4E complexes, Ct eIF4E of the final model of Ct eIF4E–eIF4G–cap complex was used as a search model. The molecular replacement solutions were used to rebuild the initial models of all complexes using ARP/wARP (38) for the Ct eIF4E–eIF4G complexes, the Sc eIF4E–Eap1p–cap analog complex and the Sc eIF4E–p20–cap analog complex or the PHENIX AutoBuild wizard (39) in the case of the Sc eIF4E–Eap1p complex.
To complete and improve the initial models, iterative cycles of model building were performed using COOT (40). Refinement was performed using PHENIX (41). Friedel reflection pairs were kept separate during the refinement of the Sc eIF4E–p20–cap analog complex as the bound Zn2+ ions contributed a significant anomalous signal. Ligand restraints for the refinement were generated using GRADE (42) and PHENIX ReadySet in the case of the metal ions. In the case of the Ct eIF4E–eIF4G complex and the Sc eIF4E–Eap1p complex bound to cap analog, all atoms other than water were refined with anisotropic individual B-factors. In all the other structures, translation/libration/screw (TLS) parameters were refined for the peptide chains in addition to the individual isotropic B-factors. The unmodified guanosine base of the m7GpppG cap analog used in the crystallization condition was not visible in the electron density map. Therefore, the cap analog was modeled as m7GTP.
The final model of the Ct eIF4E–eIF4G complex comprises one eIF4E–eIF4G complex and 230 water molecules in the asymmetric unit. eIF4E residues S241–A246 are missing in the model. The final model of the Ct eIF4E–eIF4G complex bound to cap analog comprises one eIF4E–eIF4G complex, one m7GTP and 134 water molecules in the asymmetric unit. eIF4E residues G240–R247 are missing in the model.
The final model of the Sc eIF4E–Eap1p complex consists of two eIF4E–Eap1p complexes, one calcium ion (Ca2+) and 218 water molecules in the asymmetric unit. eIF4E residues A202–H206 (chain A) and A202–N203 (chain C) are missing in the model. The final model of the Sc eIF4E–Eap1p complex bound to cap analog comprises two eIF4E–Eap1p complexes, two m7GTP, six glycerol and 517 water molecules in the asymmetric unit. The following residues are missing in the model: eIF4E A31–D34 and S201–P207 (chain A), eIF4E D29–H32 and S201–P207 (chain C), Eap1p S120–E127 and R147–G150 (chain D).
The final model of the Sc eIF4E–p20 complex consists of one eIF4E–p20 complex, two zinc ions (Zn2+) with one at 50% occupancy, three glycerol and 210 water molecules in the asymmetric unit. The following residues are missing in the model: eIF4E W104–D106 and D145–S147 (chain A), and p20 S15–L16 as well as N46–H49 (chain B).
Stereochemical properties for all structures were verified using MOLPROBITY (43), and the structural images were prepared using PyMOL (http://www.pymol.org).
Isothermal titration calorimetry
The ITC experiments were performed using a VP-ITC microcalorimeter (Microcal) at 20°C. All proteins were dialyzed into a buffer containing 20 mM Na-phosphate (pH 7.5) and 150 mM NaCl. A solution of Sc eIF4E (residues T25–L213 or V35–L213; WT or II-AA mutant) 3–11 μM in the calorimetric cell was titrated with tenfold-concentrated solutions of GB1-stabilized fragments of Sc eIF4G (70–100 μM), Sc p20 (100 μM), or Sc Eap1p (30–110 μM). Each titration experiment consisted of a first injection of 2 μl followed by 28 injections of 10 μl at 240 s intervals. At least three independent experiments were performed. The collected data were analyzed using ORIGIN software 7.0 (Microcal). Correction for the heat of dilution and mixing was achieved by subtracting the final baseline, which consisted of small peaks of similar size. To better estimate the heat of dilution and mixing for the titrations of ligands with an affinity of >1 μM, GB1-stabilized fragments were titrated into the buffer whilst the buffer was titrated into the cell containing the corresponding concentration of Sc eIF4E (44). The per-injection averaged sum of both control titrations was subtracted from the eIF4E-ligand thermograms. The first data point was removed from the analysis (45). The thermodynamic parameters were estimated by fitting a single-site binding isotherm yielding the equilibrium association constant (Ka) and enthalpy of binding (ΔH). Representative thermograms are shown in Supplementary Figure S6.
In vitro pulldown assays
Pulldown assays were performed as described previously (5,10). In the pulldown assays shown in Figures 1, 3 and 6, bacterial lysates expressing recombinant eIF4E or purified eIF4E (2 μM; 50 μg) were incubated with Ni-NTA beads for 30 min. The immobilized eIF4E was then incubated for 30 min with bacterial lysates expressing recombinant eIF4G, p20 and Eap1p fragments tagged N-terminally with MBP and C-terminally with GB1. Proteins bound to eIF4E were eluted with imidazole and analyzed by SDS-PAGE followed by Coomassie staining.
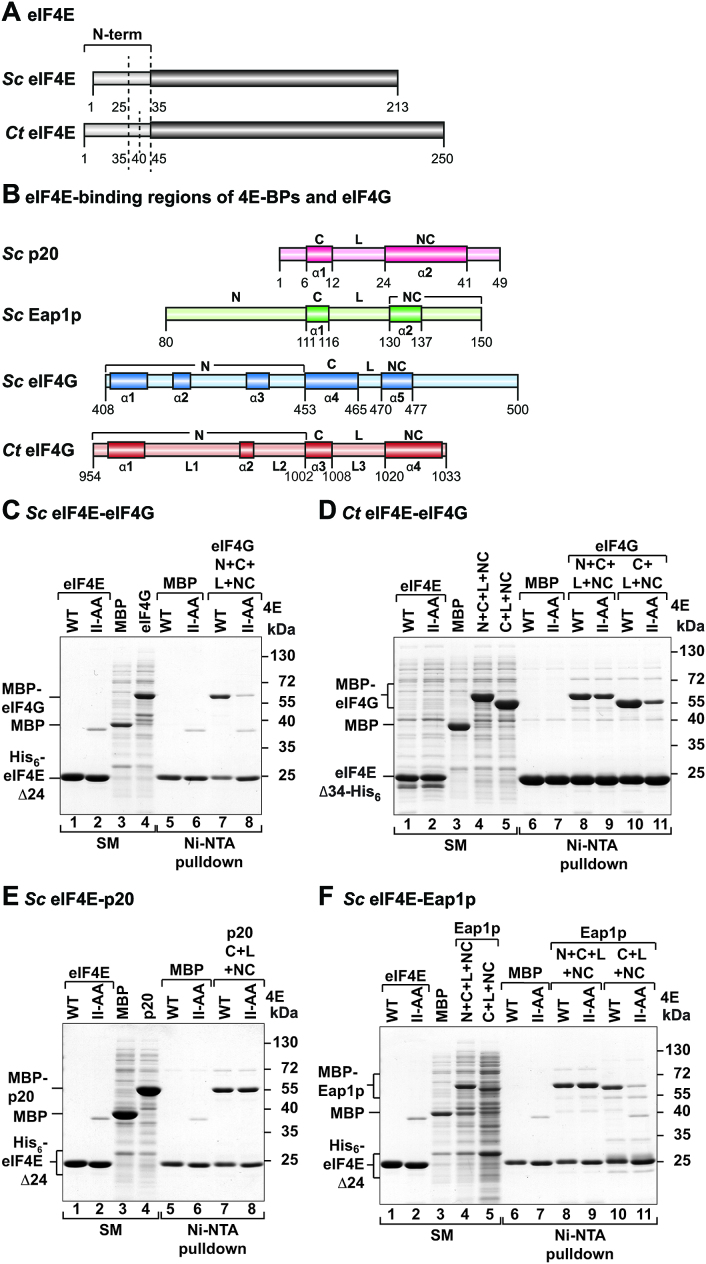
Figure 1.
Sc and Ct eIF4G interact with the lateral surface of eIF4E. (A) Schematic representation of Sc and Ct eIF4E proteins. The eIF4E N-terminal truncations (N-term) used in this study are indicated by dashed lines. The C-terminal conserved folded domain of eIF4E is highlighted in dark gray. (B) Schematic representation of the eIF4E-binding regions of Sc p20, Eap1p, eIF4G and Ct eIF4G proteins. The N-terminal region (N), the canonical (C) and non-canonical (NC) eIF4E-binding motifs, and the connecting linker (L) are indicated. Secondary structural elements are shown below the protein outline. (C, D) The interaction of N-terminally MBP-tagged and C-terminally GB1-tagged eIF4E-binding regions of Sc eIF4G (C) (residues T408–N500, C+L+NC) or Ct eIF4G (D) (residues Q954–S1033, N+C+L+NC or R996–S1033, C+L+NC), with the respective His6-tagged eIF4E protein [wild-type (WT) or lateral surface mutant (II-AA)] were analyzed in vitro using pulldown assays. Sc His6-eIF4E Δ24 or Ct eIF4E Δ34-His6 were used in the pulldown assays. Lanes labeled SM (starting material) show lysates (panel C: 1.5% for the MBP-tagged proteins; panel D: 3% for the Ct eIF4E proteins and 0.5% for MBP-tagged proteins) and purified proteins (panel C: 11% for Sc eIF4E) used in the pulldown assays. Bound fractions (16%) were analyzed by SDS-PAGE and Coomassie blue staining. MBP served as negative control. (E, F) Binding of purified Sc His6-eIF4E Δ24 (wild-type or lateral surface mutant II-AA) to the indicated N-terminally MBP-tagged and C-terminally GB1-tagged fragments of Sc p20 (E) (residues M1–H49, C+L+NC) or Sc Eap1p (F) (residues K80–G150, N+C+L+NC or residues D101–G150, C+L+NC) was analyzed in vitro using Ni-NTA pulldown assays. The starting material (11% for eIF4E proteins and 1.5% or 3% for MBP-tagged proteins in panel E and F, respectively) and bound fractions (16% in panel E and 13% in panel F) were analyzed as in (C, D).
Figure 3.
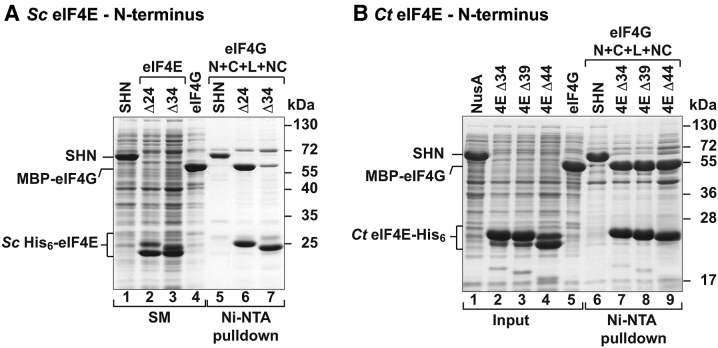
Contribution of the N-terminal extension of fungal eIF4Es to eIF4G binding. (A,B) Ni-NTA pulldown assays were used to probe the binding of N-terminally truncated eIF4E to eIF4G from Sc or Ct. Starting material shows purified proteins (Sc eIF4E) or lysates (all others) (panel A: 1% in lane 1, 2 and 4 and 2% in lane 3; panel B: 0.05% for all) and bound fractions (panel A: 15% for lanes 5, 6 and 30% for lane 7; panel B: 16%) were analyzed by SDS-PAGE and Coomassie blue staining. Strep-His6-NusA (SHN) served as negative control.
Figure 6.
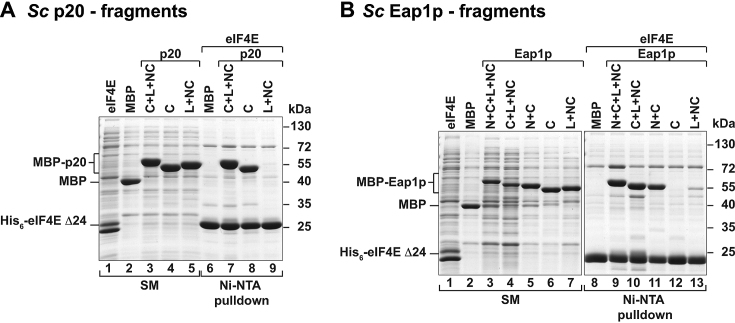
Contribution of individual elements of yeast 4E-BPs to eIF4E binding. (A, B) Ni-NTA pulldown assay showing the binding of Sc His6-eIF4E Δ24 with the indicated MBP-tagged fragments of Sc p20 (panel A) or Sc Eap1p (panel B). The starting material (SM) shows the lysates (panel A: 1% for His6-eIF4E and 1.5% for MBP-tagged proteins; panel B: 1% for His6-eIF4E and 0.5% for MBP-tagged proteins). The bound fractions (16% in panel A and 26% in panel B) were analyzed as described in Figure 1.
Sequence analysis
eIF4E, p20, Eap1p and eIF4G sequences were aligned using MAFFT or MUSCLE as accessible in Jalview (46). Seq2Logo was used to generate the sequence logo for the non-canonical motif of Eap1p (47). The sequence similarity of the eIF4E-binding region of Ct eIF4E (Q954-D1030) and Sc eIF4E (T408–N500) was calculated using the BLOSUM62 matrix as implemented in EMBOSS Water (48).
Small angle X-ray scattering (SAXS)
Small angle X-ray scattering (SAXS) data for the Sc eIF4E–eIF4G–cap complex (Sc eIF4E residues T25–L213 and Sc eIF4G residues T408–G485) was collected on the SWING beamline at SOLEIL synchrotron (Saint-Aubin, France). Cap analog m7GpppG was added in 1.20-fold molar excess to the eIF4E–eIF4G complex. The complex (50 μl at 8 mg/ml) in the presence of the excess of m7GpppG was injected on an online HPLC system (Agilent) equipped with a Superdex 200 Increase 5/150 GL column (GE Healthcare) equilibrated in 10 mM HEPES–NaOH (pH 7.5), 200 mM NaCl and 5 mM DTT (sample flow rate of 0.3 ml min−1). The scattering data were recorded at 1.5 s exposures on Aviex CCD detector at 291 K, at a wavelength of 1.0332 Å and sample-to-detector distance of 1800 mm. Data reduction to absolute units, averaging of frames and subtraction of buffer was performed using FOXTROT (Xenocs, France). Calculation of theoretical SAXS profiles based on atomic models and comparisons with experimental SAXS data were performed using the FoXS server (49) as implemented in CHIMERA (50). Missing loops and terminal residues of the atomic models were modeled using MODELLER (51) version 9.15 interface in CHIMERA. An ensemble of five models per polypeptide chain was used. For the Sc eIF4E–eIF4G–cap model based on the NMR solution structure (PDB ID: 1RF8) (15), the boundaries of the polypeptide chains in the coordinates of the model were adjusted to match those in the reconstituted Sc eIF4E–eIF4G complex (Supplementary Table S1). The homology model of Sc eIF4E–eIF4G–cap complex based on the crystal structure of the Ct eIF4E–eIF4G–cap complex was also generated using MODELLER.
RESULTS
The lateral surface of eIF4E contributes to eIF4G and 4E-BPs binding in fungi
Comparative sequence analysis of the eIF4E proteins across eukaryotes shows that several residues comprising the lateral hydrophobic surface of eIF4E are conserved (Supplementary Figure S1A, B). Furthermore, the eIF4E-binding region of the Sc eIF4G contains a putative non-canonical motif enriched in hydrophobic and aromatic residues (Supplementary Figure S2A, B). Sequence analysis allowed us to identify putative non-canonical motifs in the two known Sc 4E-BPs, p20 and Eap1p (Supplementary Figure S3A–C). This suggests that the lateral surface of eIF4E, as well as the non-canonical motifs of the eIF4E-binding proteins that typically bind to this lateral surface, are under evolutionary selection pressure across eukaryotes. However, the analysis of sequence conservation, which suggests that the fungi eIF4E lateral surface may play a role, possibly by engaging the eIF4G and 4E-BPs non-canonical motifs as was shown in the metazoan complexes (5–7), is in sharp contrast with the solution structure of the Sc eIF4E–eIF4G complex in which the non-canonical region of Sc eIF4G does not contact the lateral surface of eIF4E but forms a part of a bracelet-like structure around the N-terminal extension of eIF4E (15).
To address the question of the functional role of the lateral surface of fungal eIF4E, we initially performed in vitro pulldown assays using a truncated Sc eIF4E that contains the N-terminal extension required for eIF4G binding (eIF4E Δ24; Figure 1A) and an eIF4E (II-AA) mutant in which we had introduced mutations to disrupt binding at the lateral surface (Supplementary Table S1). These eIF4E proteins were tested for binding to protein fragments of Sc eIF4G, p20, and Eap1p that contained both the canonical and putative non-canonical motifs as well as adjacent conserved sequences (Figure 1B; Supplementary Figures S2A, B and S3A, B; Supplementary Table S1). Surprisingly, we observed that the association of Sc eIF4G with Sc eIF4E was substantially weaker with the II-AA mutant (Figure 1C, lane 8 vs. 7), thus suggesting that the lateral surface of eIF4E does contribute to the stability of the Sc eIF4E–eIF4G complex. To address the conservation of the lateral binding to the eIF4E, we performed similar experiments with the eIF4E and eIF4G proteins from the filamentous fungus Ct. However, the equivalent mutations in the lateral surface of Ct eIF4E did not decrease Ct eIF4G binding (Figure 1D; lanes 8 and 9). Rather, binding to the II-AA mutant was only affected when, in addition, the eIF4G sequences N-terminal (N) to the canonical motif, which may form a bracelet-like structure in Ct eIF4G as observed for Sc eIF4G, were also deleted (Figure 1D; lanes 10 and 11). This indicates that the eIF4E lateral surface contributes towards eIF4E interaction with eIF4G in Sc and other fungi, but its contribution may become apparent in some species only when eIF4G binding to eIF4E is impaired by deletion of the eIF4G sequences N-terminal to the canonical motif.
Next, we tested whether the binding of the Sc 4E-BPs, p20 and Eap1p, was similarly sensitive to mutations in the lateral surface of Sc eIF4E. Despite extensive efforts to stabilize full-length p20 in solution following bacterial production, we were unable to prevent nonspecific aggregation and degradation and as both Eap1p and eIF4G are considerably larger than p20 we did not attempt to produce these proteins as intact, full-length polypeptides. Instead, we had used protein fragments of Sc eIF4G, p20 and Eap1p that contained both the canonical and putative non-canonical motifs as well as adjacent conserved sequences for evaluating their binding to the eIF4E proteins (Figure 1B; Supplementary Figures S2A, B and S3A, B; Supplementary Table S1). In contrast to the observations with the eIF4G, the lateral surface mutations did not affect p20 binding to eIF4E under the conditions of the pulldown assay (Figure 1E). This result suggests that the binding mode of p20 to eIF4E is either substantially different from that of eIF4G or that the canonical motif and the non-canonical binding elements contribute differently toward the affinity of the interaction with eIF4E.
The Eap1p fragment (K80–G150) also pulled down the wild-type eIF4E and the II-AA mutant with equal efficiency (Figure 1F, lanes 8 and 9), thus suggesting that Eap1p, like p20, does not form crucial stabilizing contacts to the eIF4E lateral surface. Strikingly, however, the deletion of Eap1p residues N-terminal to the canonical motif rendered this shortened Eap1p fragment (D101–G150) sensitive to mutations in the lateral surface of eIF4E (Figure 1F, lanes 10 and 11). The first important implication of these results is that the lateral surface of the eIF4E participates in interactions with eIF4G and 4E-BPs and, secondly, that the Eap1p sequences N-terminal to the canonical motif (K80–M100, Figure 1B), contribute to the interaction with eIF4E. However, these sequences are not present in the p20 orthologs, because the canonical motif is at the protein N-terminus, suggesting that the interaction modes may have diverged between the two yeast 4E-BPs.
Structure of the fungal eIF4E–eIF4G complex reveals that the bipartite binding mode is conserved in eukaryotes
To gain insight into the architecture and assembly of the eIF4E–eIF4G complex in fungi, we crystallized the Ct eIF4E in complex with the eIF4E-binding region of the Ct eIF4G (residues Q954–D1030). The Ct eIF4E–eIF4G complex was crystallized both in the absence and presence of the m7GpppG cap analog, and the structures were refined to 1.29 Å and 1.50 Å resolution, respectively (Figure 2 and Supplementary Figure S4A; Table 1). The conformation of Ct eIF4G is essentially identical in complex with cap-bound or cap-free eIF4E (Supplementary Figure S4A), consistent with the notion that cap binding does not substantially affect the eIF4E–eIF4G interaction (5,15,52).
Figure 2.
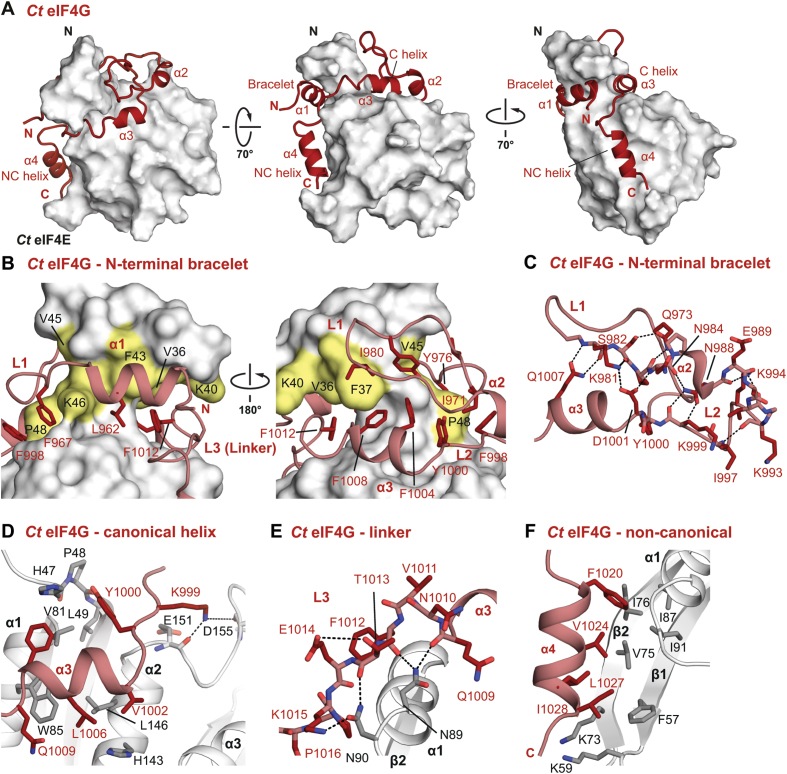
Structure of the Ct eIF4E-eIF4G complex. (A) Cartoon representation of the eIF4E-binding region of Ct eIF4G (dark red) in complex with Ct eIF4E (gray) shown as surface representation in three orientations. Selected secondary structure elements are labeled in black and red for eIF4E and eIF4G, respectively. (B) Close-up views of the molecular bracelet of Ct eIF4G bound to the N-terminus of Ct eIF4E. Selected interface residues are shown as dark red sticks for eIF4G. eIF4E is represented as a surface in two orientations with selected interface residues labelled in yellow. (C) Close-up view of the molecular bracelet of Ct eIF4G. eIF4E was omitted for clarity. Residues involved in intramolecular interactions are shown as sticks and hydrogen bonds as dashed lines. (D) Close-up view of the binding mode of the canonical helix (α3) of Ct eIF4G to the dorsal surface of eIF4E. Residues 34–444E, 954–9904G and L10054G have been omitted for clarity. Selected interface residues are shown as gray sticks for eIF4E and dark red sticks for eIF4G. (E) Close-up view of the linker (L3) region connecting the canonical and non-canonical helices of Ct eIF4G. Dashed lines indicate hydrogen bonds and salt bridges. (F) Close-up view of the Ct eIF4G non-canonical helix bound to the lateral hydrophobic pocket of eIF4E. Selected interface residues are shown as gray sticks for eIF4E and dark red sticks for eIF4G.
Table 1.
Data collection and refinement statistics
| Ct eIF4E-eIF4G complex (cap-bound) | Ct eIF4E-eIF4G complex (cap-free) | Sc eIF4E-Eap1p complex (cap-bound) | Sc eIF4E-Eap1p complex (cap-free) | Sc eIF4E-p20 complex (cap-free) | |
|---|---|---|---|---|---|
| Space group | P21212 | P21212 | C2 | P21212 | C2 |
| Unit Cell | |||||
| Dimensions (Å) | |||||
| a, b, c | 64.0, 75.4, 48.4 | 63.6, 75.2, 48.4 | 91.7, 121.7, 62.4 | 87.6, 95.1, 56.0 | 102.0, 62.8, 44.7 |
| Angles (°) | |||||
| α, β, γ | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 132.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 107.0, 90.0 |
| Data collection | |||||
| Wavelength (Å) | 1.00000 | 1.00000 | 0.99996 | 0.99996 | 1.03320 |
| Resolution (Å) | 48.76–1.50 (1.52–1.50)* | 40.71–1.29 (1.36–1.29) | 46.32–1.35 (1.37–1.35) | 48.26–1.92 (1.97–1.92) | 48.78–1.75 (1.78–1.75) |
| R sym | 0.074 (1.913) | 0.048 (0.713) | 0.045 (1.284) | 0.105 (1.711) | 0.114 (1.056) |
| Mean I/σI | 16.0 (1.2) | 11.5 (1.6) | 16.6 (1.4) | 17.5 (1.6) | 8.8 (1.2) |
| CC (1/2) | 0.999 (0.416) | 0.999 (0.584) | 0.999 (0.643) | 0.999 (0.542) | 0.994 (0.454) |
| Completeness (%) | 99.6 (91.6) | 93.9 (85.3) | 98.1 (92.6) | 100.0 (99.9) | 99.9 (99.9) |
| Multiplicity | 12.7 (10.7) | 3.1 (3.0) | 6.7 (6.6) | 13.2 (12.7) | 4.2 (4.1) |
| Total no. of reflections | 487 466 (18 589) | 172 455 (21 476) | 727 684 (33 379) | 481 553 (30 566) | 114 714 (6189) |
| No. of unique reflections | 38 537 (1742) | 54 870 (7178) | 108 809 (5058) | 36 515 (2406) | 27 294 (1510) |
| Refinement | |||||
| Resolution (Å) | 48.76–1.50 | 40.71–1.29 | 46.32–1.35 | 48.26–1.92 | 42.78–1.75 |
| No. of reflections | 38 394 | 54 840 | 108 796 | 36 466 | 52 161 [27 294]# |
| R work/Rfree | 0.170/0.206 | 0.163/0.201 | 0.138/0.168 | 0.174/0.207 | 0.182/0.217 |
| No. atoms | |||||
| Protein | 2214 | 2239 | 3978 | 3885 | 1798 |
| Cap analog | 33 | - | 66 | - | - |
| Other ligands | - | - | 36 | 1 | 8 |
| Water | 134 | 230 | 514 | 218 | 210 |
| B-factors (Å2) | |||||
| Protein | 38.1 | 23.5 | 28.7 | 45.2 | 27.8 |
| Cap analog | 75.8 | - | 36.1 | - | - |
| Other ligands | - | - | 76.4 | 25.4 | 43.3 |
| Water | 40.7 | 32.8 | 39.7 | 42.2 | 40.2 |
| Ramachandran Plot | |||||
| Favored (%) | 97.0 | 97.8 | 98.7 | 99.1 | 97.6 |
| Disallowed (%) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| RMS Deviation | |||||
| Bond lengths (Å) | 0.022 | 0.008 | 0.012 | 0.010 | 0.020 |
| Bond angles (°) | 1.669 | 0.941 | 1.279 | 0.747 | 1.031 |
*Values shown in parentheses are for the highest-resolution shell.
#Friedel pairs were kept separate during refinement. Value in brackets shows number of non-anomalous reflections.
The overall fold of Ct eIF4E is highly conserved and consists of a strongly curved β-sheet of eight antiparallel strands that adopt a horseshoe-like conformation (9,53). The cap-binding cavity is located on the concave, ventral surface. The convex surface is covered by three long α-helices that form the dorsal binding surface of the protein (Supplementary Figure S4A). The structure of Ct eIF4E bound to cap analog superposes to the Drosophila melanogaster (Dm) and human (Hs) eIF4Es bound to cap analog (5) (Supplementary Figure S4B) with RMSDs of 0.57 Å over 119 Cα atoms and 0.67 Å over 113 Cα atoms, respectively.
The most striking and unanticipated feature in the Ct eIF4E–eIF4G structure is that the eIF4G peptide contacts the lateral surface of eIF4E (Figure 2A) in a mode that is structurally analogous to that observed for diverse 4E-BPs and metazoan eIF4Gs (5–7,54) but is in contrast to the NMR solution structure of the Sc eIF4E–eIF4G complex (15) where the eIF4G is not in contact with the lateral surface of eIF4E (Supplementary Figure S4C).
In addition to the structural elements common to all eIF4E–eIF4G complexes, the Ct eIF4E–eIF4G complex exhibits some striking features that were not observed in the crystal structures of the metazoan complexes. The N-terminal segment of the Ct eIF4G includes two helices (α1 and α2) that, together with the canonical helix α3, wrap around the N-terminus of eIF4E forming a bracelet-like structure (Figure 2A, B; Supplementary Figure S2A, B), analogous to what was observed in the Sc eIF4E–eIF4G complex (Supplementary Figure S4C). Due to the presence of many contacts between the N-terminal regions of eIF4E and eIF4G, the Ct eIF4E–eIF4G interface is thus much more extensive than the metazoan interface (4200 Å2 versus 2900 and 3000 Å2 buried surface area for the Hs and Dm complexes, respectively).
The N-terminal bracelet in eIF4G substantially extends the binding interface with eIF4E
The Ct eIF4G N-terminal bracelet is stabilized in the complex by an extensive network of inter- and intramolecular interactions (Figure 2B, C). The bracelet interior is lined by several aromatic and hydrophobic residues located in the connecting loops L1 (F9674G, Y9764G, I9804G) and L2 (F9984G, Y10004G), helix α1 (L9624G), the canonical helix α3 (F10044G and F10084G) and the linker region (F10124G). Ct eIF4E contributes to the interaction with F434E, V454E and K464E, which face helix α1 and loop L1 of eIF4G, whereas V364E, F374E and K404E contact eIF4G residues located in the canonical helix, loop L1, and the linker region (loop L3). These predominantly hydrophobic interactions are critically positioned to stabilize the whole interface. The residue P484E of the invariant HPL motif of eIF4E forms contacts with aromatic residues of loops L1 and L2, and the canonical motif and together with an extensive hydrogen bond network stabilizes the bracelet on the dorsal surface of eIF4E (Figure 2C).
The binding of the eIF4G canonical helix on the dorsal surface of eIF4E is conserved
The canonical helix α3 of Ct eIF4G interacts with eIF4E through the conserved residues in the consensus sequence in a similar manner as observed in other eIF4G and 4E-BPs in complex with eIF4E (5,6,8,14,15) (Figure 2D). The Tyr residue of the consensus canonical motif (Y10004G) forms a hydrogen bond to the backbone of the invariant HPL motif (H47-P48-L49) of Ct eIF4E and van der Waals interactions with V814E. The hydrophobic residues at the C-terminus of the canonical motif (LΦ – L10054G and L10064G) are in contact with the conserved residues (V814E, W854E and L1464E) on the dorsal surface of eIF4E.
Our previous studies indicated that the canonical motifs of metazoan eIF4G and other 4E-BPs contain Arg/Lys residue at positions 2 and 9 of the canonical motif leading to an extended canonical consensus sequence (6) [YX(R/K)X2LФX2R/K; Figure 2D and Supplementary Figure S2]. In the metazoan complexes, the R/K residues contribute to the interaction with eIF4E, likely by preventing hydrophobic surface patches of eIF4E from being exposed to solvent (5,6,14,54). However, the canonical motif of Ct lacks R/K residues at position 2 and 9. Furthermore, the R/K residues at position 2 of metazoan eIF4G and 4E-BPs form a salt bridge with an acidic residue in eIF4E (6) (Dm D1644E, Hs E1324E), which is conserved in metazoans but absent in fungal eIF4Es (Supplementary Figure S1A, B).
The conformation of eIF4G non-canonical motifs on the lateral surface of eIF4E is preserved in fungal complexes
Adjacent to the canonical helix, the Ct eIF4G linker (N10104G – D10194G) adopts an extended conformation that permits only few intramolecular interactions (one hydrogen bond is formed between T10134G and E10144G; Figure 2E). The linker is anchored to the lateral surface of eIF4E through intermolecular hydrogen bonds via the conserved eIF4E N894E, which interacts with the backbone carbonyls of Q10094G and F10124G and the neighboring N904E residue, which interacts with T10134G and K10154G. These interactions conformationally restrict the linker region and orient it towards the lateral surface of eIF4E.
At the C-terminal end of the linker region, the non-canonical motif of Ct eIF4G binds into a hydrophobic pocket on the lateral surface of eIF4E, which is lined by residues F574E, V754E, I764E, I874E and I914E (Figure 2F). A striking feature of the observed conformation of the Ct eIF4G is the formation of an α-helix (α4) on the lateral surface of the eIF4E, which has been previously observed only in the complex of Dm CUP and invertebrate Mextli proteins bound to eIF4E (14,54). This Ct eIF4G non-canonical helix engages the hydrophobic pocket of eIF4E through the side chains of residues F10204G, V10244G, L10274G and I10284G (Figure 2F).
The eIF4E-binding regions of Ct and Sc eIF4G share a sequence identity and a similarity of 27% and 43%, respectively. Because we were not able to confirm whether the bipartite binding mode observed in the Ct eIF4E–eIF4G complex is representative of the conformation in the Sc complex, which did not crystallize, we employed small angle X-ray scattering (SAXS) to study the conformation of the Sc eIF4E–eIF4G complex in solution. The experimental SAXS profile of the Sc eIF4E–eIF4G complex was in excellent agreement with the scattering profile calculated from a homology model of Sc eIF4E–eIF4G based on the Ct eIF4E–eIF4G crystal structure (χFoxS = 2.0; Supplementary Figure S4D, E). However, the fit to a homology model based on the Sc eIF4E–eIF4G solution NMR structure (15) was convincingly worse (χFoxS = 5.6; Supplementary Figure S4D, F). Thus, the SAXS data indicate that, in solution, the Sc eIF4E–eIF4G complex likely adopts a conformation very similar to that observed in the crystal structure of the Ct eIF4E–eIF4G complex. This is consistent with the results of the pulldown assays, which demonstrate that the lateral surface of eIF4E does contribute to the stability of the Sc eIF4E–eIF4G complex. We therefore propose that the Ct complex may be considered representative of ascomycetous fungi, which include Sc.
The N-terminal extension of eIF4E contributes to the stability of the complex with eIF4G in yeast
Previous reports highlighted the importance of the N-terminal extension of Sc eIF4E for the interaction with eIF4G, whereby deleting the first 35 N-terminal eIF4E residues reduced the binding affinity for the eIF4G by two orders of magnitude (15). Accordingly, we have also observed a pronounced decrease in binding of the Sc eIF4G to an equivalent deletion mutant of the Sc eIF4E (Δ34) compared to the Sc eIF4E Δ24 deletion mutant, which still includes the residues that stabilize the eIF4G bracelet (Figure 3A). In contrast, however, we observed that the equivalent deletions in the Ct eIF4E did not detectably reduce its interaction with the Ct eIF4G (Figure 3B). This observation implies that the Ct eIF4G may be less sensitive to the deletion of the N-terminal residues in the eIF4E under the conditions of the pulldown assay. One explanation for this is that other binding elements form interactions of sufficiently high affinity to compensate for the loss of the N-terminal interface. It is tempting to speculate that such apparent redundancy in stabilizing interfaces may be physiologically relevant to efficiently support translation in thermophilic organisms.
Structures of Sc 4E-BPs bound to eIF4E reveal differences in binding mode
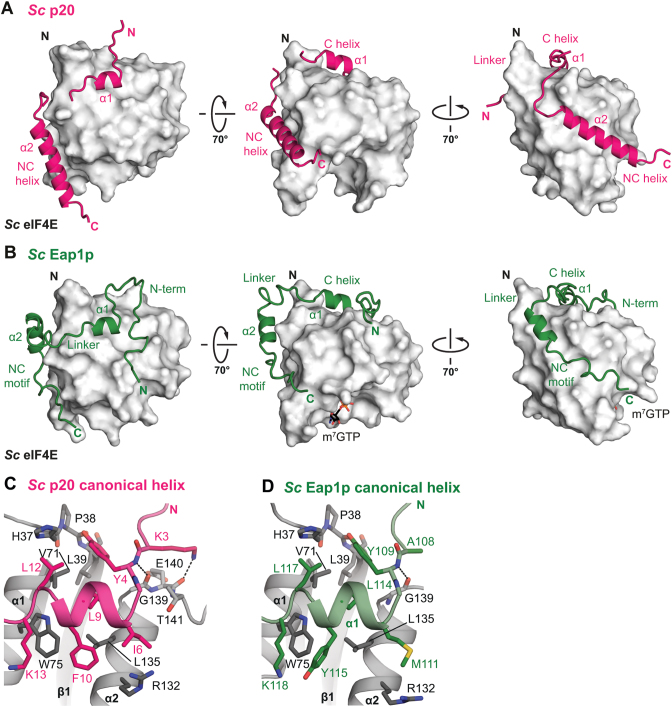
We crystallized the eIF4E-binding regions of Sc p20 (M1–H49) and Sc Eap1p (T91–G150) in complex with Sc eIF4E and refined the structures to 1.75 Å and 1.35 Å resolution, respectively (Figure 4A, B; Table 1). As the wild-type Sc p20– and Eap1p–eIF4E complexes did not crystallize, we employed combinatorial substitutions of surface lysines to alanines in the Sc eIF4E (Supplementary Figure S5H). Analogous to the Ct eIF4E–eIF4G complex, the presence of the cap analog does not influence the conformation of the Sc Eap1p, which was co-crystallized with eIF4E in cap-bound and cap-free states (Supplementary Figure S5F, G; Table 1). The overall architecture of the complexes reveals that the Sc p20 and Eap1p are similar to the Ct eIF4G in that they also contain both canonical and non-canonical motifs binding to the dorsal and lateral surfaces of eIF4E, respectively. Intriguingly, however, neither 4E-BP interacts with the N-terminal extension of eIF4E.
Figure 4.
Structure of Sc 4E-BPs bound to eIF4E. (A) Cartoon representation of the eIF4E-binding region of Sc p20 (fuchsia) in complex with Sc eIF4E (gray) shown as a surface in three orientations. Selected secondary structure elements are labeled in black and fuchsia for eIF4E and p20, respectively. (B) Cartoon representation of the eIF4E-binding region of Sc Eap1p (green) in complex with cap-bound Sc eIF4E (gray) shown as a surface in three orientations. The bound m7GTP cap analog is shown as sticks. (C, D) Close-up view of the canonical helix of Sc p20 (C) and Eap1p (C) bound to the dorsal surface of Sc eIF4E. Selected residues are shown as sticks, and secondary structure elements are labeled and colored as in panels A and B. Dashed lines indicate hydrogen bonds.
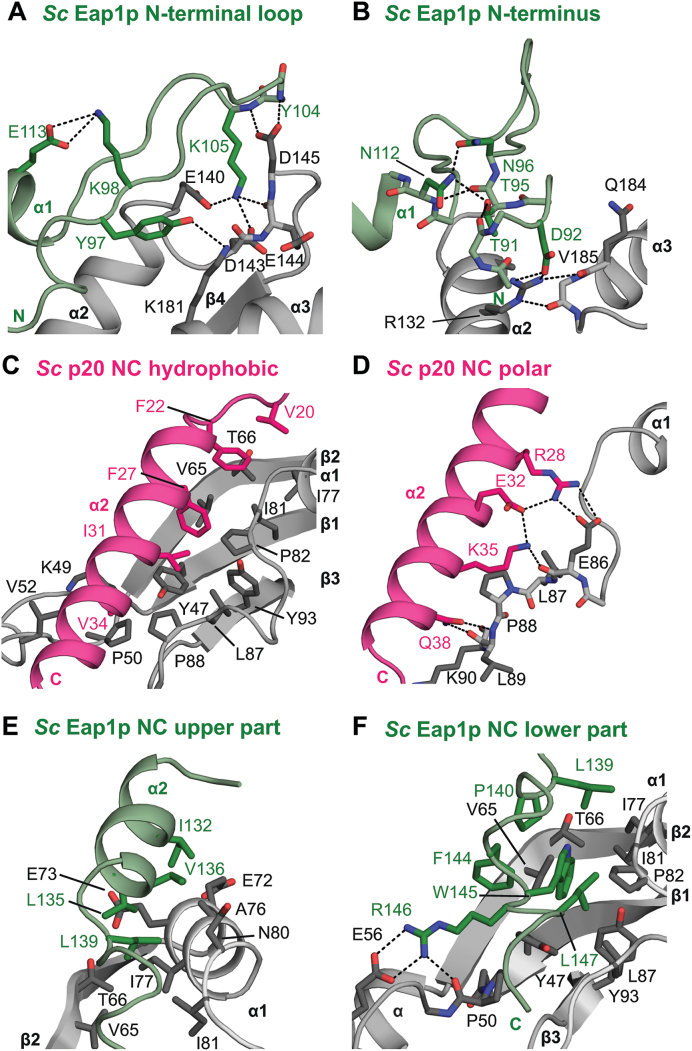
Sc p20 and Eap1p display very similar interaction modes of their canonical motifs with the dorsal surface of the eIF4E (Figure 4C, D) analogous to those previously reported for canonical peptides in complex with eIF4E (5,6,8,12–15,55). A critical difference between the binding modes of the two yeast 4E-BPs, however, is the presence of an N-terminal auxiliary extension in Eap1p, which is absent in p20. This N-terminal extension forms extensive interactions with eIF4E that are unique to this complex and have not been observed in any reported structures of eIF4E complexes with partners (Figure 5A, B). This extension folds into an elongated ‘hairpin’-like conformation on the dorsal surface of eIF4E and significantly increases the buried surface area of the eIF4E–Eap1p interface (2800 Å2 versus 3900 Å2). Key stabilizing contacts are formed by the conserved K105Eap1p at the turn of the ‘hairpin’ that forms salt bridges to E1404E and D1434E as well as a hydrogen bond to the backbone carbonyl of E1444E (Figure 5A; Supplementary Figure S3B). At the very N-terminal end, the Eap1p ‘hairpin’ is anchored to the dorsal surface of eIF4E by a salt bridge formed between D92Eap1p and R1324E (Figure 5B).
Figure 5.
Molecular details of the binding surfaces in the yeast eIF4E–4E-BP complexes. (A, B) Close-up views of the N-terminal extension of Sc Eap1p bound to eIF4E helices α2 and α3 (A) or to an acidic patch in helix α3 (B) of the dorsal surface of eIF4E. Dashed lines indicate hydrogen bonds or salt bridges. Selected residues are shown as sticks and colored in black and green for eIF4E and Eap1p, respectively. (C, D) Close-up views of the non-canonical helix of Sc p20 establishing hydrophobic contacts (C) and polar interactions (D) at the lateral surface of eIF4E. Selected residues are shown as sticks and colored in black and fuchsia for eIF4E and p20, respectively. (E, F) Close-up views of the non-canonical motif of Sc Eap1p contacting the upper (E) or the lower part (panel F; the FWRL motif) of the lateral surface of eIF4E. Residues 145–150 of Sc Eap1p have been omitted for clarity in panel E.
The linker regions of the yeast 4E-BPs are flexible indicating mechanistic differences with human 4E-BP1–3
As observed for the Ct eIF4G, the non-canonical motifs of both Sc 4E-BPs fold into an amphipathic α-helix that binds to the lateral surface of eIF4E (Figure 5C). In p20, the non-canonical α2 helix establishes hydrophobic as well as polar interactions and spans over the complete lateral surface of eIF4E. A triad of invariant hydrophobic residues on this long helix (F27p20, I31p20, V34p20) acts like a ‘zipper’ and mediates the key stabilizing contacts with eIF4E (Figure 5C). In contrast to the exclusively hydrophobic interactions mediated by the non-canonical helix of the Ct eIF4G, p20 participates in a number of polar contacts with eIF4E (Figure 5D). Specifically, R28p20 and K35p20 anchor the non-canonical helix via a salt bridge and a hydrogen bond to E864E. The side chain of the conserved Q38p20 tethers the C-terminal end of the helix to the lateral surface via a hydrogen bond to the backbone of L894E.
The non-canonical α2 helix of Eap1p is notably shorter than in p20, and its principally hydrophobic contacts with the upper region of the lateral surface of eIF4E are less extensive, consistent with the lack of sequence conservation in the non-canonical motifs (Figure 5E; Supplementary Figure S3B). However, the FWRL motif (residues F144–L147), which is C-terminal to the non-canonical helix and highly conserved in Eap1p orthologs (Supplementary Figure S3C), provides critically stabilizing contacts, as evidenced by the Eap1p W145 residue, which is at the center of extensive T-shaped π-π-stacking that also involves F144Eap1p as well as Y474E and Y934E (Figure 5F). The non-canonical motif of Eap1p is anchored at the lateral surface of eIF4E via a salt bridge involving the guanidinium group of a highly conserved R146Eap1p and the carboxylate moiety of E564E.
The canonical and non-canonical helical segments in p20 and Eap1p are connected by flexible linkers, which begin with a Lys residue at position 9 of the extended canonical motif (K13p20, K118Eap1p; Figure 4C, D). Apart from a hydrophobic contact between the aliphatic portion of the Lys and the invariant W754E, there are no contacts between the p20 or Eap1p linkers and the dorsal surface of Sc eIF4E, and the linker conformation is not restricted (Supplementary Figure S5A). In the eIF4E–p20 crystal structure this apparent flexibility is evidenced by the in part very poor electron density of the p20 linker region, whereas in the Eap1p the linker does not engage eIF4E with any specific interactions (Supplementary Figure S5B). The structural superposition of all eIF4E–Eap1p complexes present in the asymmetric unit of the crystal structures further confirms that the linker is conformationally variable (Supplementary Figure S5C). This is in contrast to previous observations in the metazoan 4E-BP1 where the somewhat rigid linker may contribute in conferring a competitive advantage on 4E-BPs over eIF4G for binding to eIF4E via possible restriction of conformational freedom (5,6).
The canonical and non-canonical motifs in 4E-BPs and eIF4G contribute differently to the affinity for eIF4E
Initially, we tested the interaction of the minimal eIF4E-binding regions of p20 containing all binding elements [p20: C+L+NC (M1–H49); Eap1p: N+C+L+NC (K80–G150)], or truncated versions of these proteins with eIF4E in pulldown assays (Figure 6). Both p20 and Eap1p fragments bound to eIF4E. However, the contributions of the different binding elements (N, C and L+NC sequences) to the interaction with eIF4E was strikingly different among the 4E-BPs. In p20, the isolated canonical motif (C) alone efficiently associated with eIF4E, whereas the non-canonical sequences (L+NC) in isolation did not bind eIF4E (Figure 6A). In contrast, the isolated L+NC sequences of Eap1p exhibited a weak interaction with eIF4E and the canonical motif was necessary for strong binding (Figure 6B). The auxiliary segment located N-terminally to the canonical motif in Eap1p was important for the interaction with eIF4E as the N+C fragment was sufficient for binding to eIF4E, which underscores the stabilizing role of the N-terminal sequences observed in the structure of the eIF4E–Eap1p complex. In contrast, the peptide containing only the canonical motif did not bind to eIF4E (Figure 6B, lanes 11 versus 12).
We then examined the thermodynamic contribution of the individual eIF4E-binding elements to the overall affinity of p20 and Eap1p for eIF4E using isothermal calorimetry (ITC; Supplementary Table S2; Supplementary Figure S6). We were unable to determine the affinity of the complete eIF4E-binding regions of both 4E-BPs, as the proteins were unstable in solution when not in complex with eIF4E. Nevertheless, the isolated canonical motif of p20 bound to eIF4E with high nanomolar affinity (KD = 20 ± 1 nM). In contrast, however, the affinity of the canonical motif of Eap1p for eIF4E was more than two orders of magnitude lower (KD = 3400 ± 300 nM) and comparable to that observed for the canonical motif of the Sc eIF4G (KD = 2000 ± 1000 nM; Supplementary Table S2). Interestingly, binding of the p20 and Eap1p canonical motifs to eIF4E was clearly driven by their enthalpic contribution and involved a higher entropic penalty of binding compared to the corresponding motif in eIF4G, which showed both a favorable enthalpic and entropic contribution to binding.
The pronounced difference in affinities of the canonical motifs of p20 and Eap1p despite very similar structures in complex with the eIF4E prompted us to probe the molecular basis of this difference. To this end, we replaced two groups of residues in or next to the canonical helix of Eap1p with their direct p20 equivalents (Supplementary Figure S5D, E). Separately exchanging three residues directly N-terminal to the canonical helix or four residues in the canonical helix increased the affinity of each resulting peptide (Eap1p C YAS* or p20 helix) by 16-fold (Supplementary Table S2). These results are consistent with the notion that p20 achieves high affinity interaction with the eIF4E via several interaction ‘hot spots’ in the canonical helix and the N-terminal flanking sequence.
Interestingly, in the presence of the N-terminal binding element (residues K80–A122; N+C peptide), the affinity of the Eap1p peptide is comparable to that of the canonical motif of p20 (KD = 8 ± 4 nM). The enhanced binding affinity is accompanied by a favorable change in the binding enthalpy (ΔΔH = −5.3 kcal/mol) and by an increase in the entropic penalty of binding [Δ(–TΔS) = 1.7 kcal/mol] compared to the isolated canonical motif of Eap1p. These changes in binding energetics may be indicative of a disorder-to-order transition of the N-terminal sequences of the Eap1p upon binding to eIF4E, as evidenced by the folding of this segment into a ‘hairpin’-like conformation on the surface of eIF4E. The affinity of an Eap1p fragment lacking the eleven N-terminal residues of the extension (T91–A122; Nshort+C), as observed in the crystal structure, is comparable to that of the longer Eap1p (residues K80–A122; KD = 12.9 ± 0.9 nM). This indicates that the structurally characterized fragment of Eap1p is necessary and sufficient for binding to the eIF4E. Consistent with the structure of the complex in which Eap1p does not participate in any interactions with the N-terminal extension of eIF4E, the deletion of the N-terminal residues in the eIF4E (Δ34) did not impact on the affinity of the interaction with Eap1p (T91–A122; KD = 8.8 ± 0.5 nM).
The non-canonical sequences (L+NC) of Eap1p also contribute to the energetics of the interaction with eIF4E. In fact, the affinity of the Eap1p L+NC fragment (residues S123–G150) for eIF4E (KD = 1000 ± 100 nM) was comparable to that of the isolated canonical (C) motif and required a functional lateral surface of eIF4E, as binding to the II-AA mutant was not detected by ITC. Similarly, the affinity of a bipartite fragment of Sc eIF4G (residues V448–N500; C+L+NC) for eIF4E (KD = 70 ± 3 nM) was strongly reduced upon II-AA mutation of the lateral surface (KD = 1020 ± 90 nM), to an affinity comparable to that of the isolated canonical motif (KD = 2000 ± 1000 nM).
Overall, the thermodynamic data support a model in which the individual binding elements of p20 and Eap1p make different contributions to the overall affinity of these proteins toward eIF4E. The canonical motif of p20 makes by far the most critical contribution towards the stability of the complex as evidenced by its high affinity, whereas Eap1p exhibits a more complex binding mode in which several binding elements (N, C and L+NC) contribute to achieve high affinity binding to eIF4E.
DISCUSSION
Binding to the eIF4E lateral surface is conserved in eukaryotic eIF4G proteins
Previous studies raised the question of whether the binding mode of eIF4G to eIF4E diverged during evolution. The eIF4E-binding mode observed in the structures of the fungal complexes in this study contrasts strikingly with an early NMR structure of the Sc eIF4E–eIF4G complex (15) where it was observed that the non-canonical motif does not contact the lateral surface of eIF4E. The differences in the observed conformations of the eIF4G may be explained by the use of CHAPS, a zwitterionic detergent, in the NMR structure determination (15). The use of this detergent may have affected the hydrophobic contacts at the lateral surface of the eIF4E as was previously shown (9). We propose that the binding interface delineated by the canonical and non-canonical motifs of eIF4G, which contact the dorsal and lateral surfaces of eIF4E, as observed in the fungal and the metazoan eIF4E–eIF4G and eIF4E–4E-BPs, and recently also plant eIF4E–eIF4G complexes (5–7,14,55) is the physiologically relevant state required for the initiation of cap-dependent translation and that it is conserved among eukaryotic eIF4G proteins.
Comparative analysis of sequence conservation of the eIF4E-binding region of eIF4G proteins from multiple fungal phyla suggests that the non-canonical motif is present in all eIF4Gs but the precise sequence composition is not conserved (Supplementary Figure S2A, B). In fact, the sequence of the non-canonical motifs of Sc and Ct eIF4G (Ascomycetes) diverges from the analogous sequence of eIF4G proteins present in Basidiomycetes (Ustilago, Sporisorium, Kalmanozyma, Wallemia, Thanatephorus, Pseudozyma) or Glomeromycetes (Rhizophagus), which appear to be more similar to their metazoan counterparts (Supplementary Figure S2B). In addition to the apparent similarity to the metazoan eIF4G non-canonical motifs, the Basidiomycetes and Glomeromycetes non-canonical motifs lack helical propensity, suggesting that in these species the non-canonical motif will not adopt a helical conformation as observed in the Ct eIF4E–eIF4G complex.
A conserved molecular bracelet of eIF4G is a fungal-specific adaptation that enhances binding to eIF4E
A striking feature of the Sc and Ct eIF4E-binding region is their requirement for sequences N-terminal to the canonical motif to stabilize the interaction with the dorsal surface of eIF4E and to achieve high affinity binding (Figure 2 and Supplementary Figure S2A, B). These N-terminal sequences are involved in forming a bracelet-like structure that appears to be a unique feature of fungal eIF4G proteins from Ascomycota (Supplementary Figure S2B). The molecular bracelet binds to eIF4E via an extensive network of inter- and intramolecular interactions (15) (Figure 2B, C). Interestingly, several of these interactions are established with residues that only occur in the N-terminus of certain fungal eIF4Es (Supplementary Figure S1B), thereby rationalizing the conservation of this specific mode of interaction in these fungal eIF4E-eIF4G complexes. Comparing orthologs from metazoa and fungi of distinct phyla, the N-terminus of eIF4E proteins varies in length and sequence and these adaptations may have co-evolved with the concomitant loss of the stabilizing bracelet-like structure in eIF4G.
Yeast 4E-BPs variably utilize their binding elements to modulate affinity toward eIF4E
Our work also reveals the molecular architecture of the yeast 4E-BPs p20 and Eap1p bound to eIF4E (Figure 4). We show that, like eIF4G, these proteins interact with eIF4E using a bipartite binding mode contacting eIF4E dorsal and lateral surfaces to establish an extended interface. However, in contrast to yeast and Ct eIF4G, p20 and Eap1p do not form molecular bracelet-like structures. In p20, a bracelet structure is not possible because the canonical motif is positioned very close to the N-terminus. Thus, p20 displays a minimal eIF4E-binding region, which comprises two helical motifs (canonical and non-canonical) connected by a flexible linker. Interestingly, the p20 canonical motif on its own binds to eIF4E with 100-fold higher affinity than the canonical motifs of either eIF4G or Eap1p (Supplementary Table S2). This high-affinity binding in an isolated binding element may have evolved to compensate for the lack of N-terminal extensions to support the interaction of the canonical motif with the dorsal surface of eIF4E, which are present in both eIF4G and Eap1p, thus providing p20 with the ability to compete effectively for binding to eIF4E. It is probable that the full-length p20 has a substantial competitive advantage over eIF4G for binding to the eIF4E but this could not be directly verified in vitro due to the poor stability of the full-length eIF4E-interacting proteins.
Eap1p employs a different strategy to achieve high affinity for eIF4E: residues located N-terminal to the canonical motif contribute to the interaction with eIF4E, but instead of forming a bracelet, they adopt a defined loop conformation on the dorsal surface of eIF4E. This additional binding element assists the canonical motif of Eap1p in the interaction with the dorsal surface of eIF4E and enhances the affinity by a factor of 400 (Supplementary Table S2).
The molecular details that determine the differences in affinity among conserved canonical motifs are not completely understood. Comparative structural analysis of several eIF4E–4E-BP and eIF4E–eIF4G complexes shows that the same basic interactions are established in all cases. One of the principal contributing factors is the residue immediately preceding the canonical motif (XYX4LΦ) that also forms interactions with the dorsal surface of eIF4E. This is the case in p20, in which a lysine residue (K3) establishes hydrogen bonds with an invariant glutamate located within an acidic patch of the eIF4E dorsal surface (Sc eIF4E E140; Figure 4C). In Eap1p, the directly equivalent interaction is not possible as the lysine is replaced by alanine (A108; Figure 4D), nevertheless another lysine (K105; Figure 5A) on the N-terminal extension projects to this acidic patch on eIF4E and may play a similar role.
The yeast 4E-BPs may stabilize eIF4E on bound transcripts
In both yeast 4E-BPs, the non-canonical motifs are rather large and span over the complete lateral surface of eIF4E, positioning the proteins in close proximity to the eIF4E cap-binding pocket. Interestingly, adjacent to the non-canonical motif, the p20 and Eap1p sequences are rich in positively charged residues (Lys and Arg; Supplementary Figure S3A, B). Given that such sequences are likely to promote non-specific RNA binding (56), it is tempting to speculate that p20 and Eap1p enhance the affinity of eIF4E for the capped mRNA via interactions with the 5′ UTR of the eIF4E-bound mRNA, in a similar way as has been proposed for eIF4G (57,58). Interestingly, both yeast 4E-BPs have also been shown crosslink to or associate with RNA (59–62). This suggests a scenario wherein in addition to competing with eIF4G, the yeast 4E-BPs may stabilize eIF4E on bound transcripts. In support of this notion, whilst this manuscript was in review, it was reported that p20 interacts with synthetic RNA in vitro and substantially enhances the eIF4E affinity for capped RNA when bound to the eIF4E (62).
In summary, our data indicate that a wide diversity of eIF4E complexes with distinct molecular features (N-terminal bracelets, N-terminal loops, flexible or rigid linkers, helical non-canonical motifs or non-canonical loops) can be found in eukaryotes. Although the functional implications of this molecular diversity for translational control are not yet completely understood, the structural and thermodynamic plasticity in the eIF4E binding modes point to species- or protein-specific adaptations to fine-tune translation initiation, which may be exploited in the development of antimycotic therapeutics targeting protein synthesis.
DATA AVAILABILITY
Coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 6FBZ (Ct eIF4E–eIF4G in cap-bound state), 6FC0 (Ct eIF4E–eIF4G), 6FC1 (Sc eIF4E–Eap1p in cap-bound state), 6FC2 (Sc eIF4E–Eap1p) and 6FC3 (Sc eIF4E–p20).
Supplementary Material
ACKNOWLEDGEMENTS
We dedicate this study to the memory of Elisa Izaurralde who passed away much too early whilst this manuscript was in review. She strove always to excel and to meet the highest professional standards in her scientific endeavors and it was her priority to give direction and mentor her trainees and colleagues in the art of scientific communication. We cherish the memory of her kindness, integrity and optimism and the time we had spent with her in the unending pursuit of knowledge.
We thank Tobias Raisch and Gabriele Wagner for setting up crystallization screens, Sigrun Helms and Catrin Weiler for technical assistance, and Oliver Weichenrieder for assistance with X-ray data collection. We are grateful to Ralf-Peter Jansen and Birgit Singer-Krueger for kindly providing yeast genomic DNA. We acknowledge the staff at the PXII beamline of the Swiss Light Source, the P11 beamline of the PETRA III synchrotron and the SWING beamline of SOLEIL synchrotron for excellent support with data collection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Max Planck Society. Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hinnebusch A.G., Lorsch J.R.. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 2012; 4:a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jackson R.J., Hellen C.U., Pestova T.V.. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010; 11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Haghighat A., Mader S., Pause A., Sonenberg N.. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995; 14:5701–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mader S., Lee H., Pause A., Sonenberg N.. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 1995; 15:4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grüner S., Peter D., Weber R., Wohlbold L., Chung M.Y., Weichenrieder O., Valkov E., Igreja C., Izaurralde E.. The structures of eIF4E-eIF4G complexes reveal an extended interface to regulate translation initiation. Mol. Cell. 2016; 64:467–479. [DOI] [PubMed] [Google Scholar]
- 6. Peter D., Igreja C., Weber R., Wohlbold L., Weiler C., Ebertsch L., Weichenrieder O., Izaurralde E.. Molecular architecture of 4E-BP translational inhibitors bound to eIF4E. Mol. Cell. 2015; 57:1074–1087. [DOI] [PubMed] [Google Scholar]
- 7. Sekiyama N., Arthanari H., Papadopoulos E., Rodriguez-Mias R.A., Wagner G., Leger-Abraham M.. Molecular mechanism of the dual activity of 4EGI-1: Dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4036–E4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marcotrigiano J., Gingras A.C., Sonenberg N., Burley S.K.. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell. 1999; 3:707–716. [DOI] [PubMed] [Google Scholar]
- 9. Matsuo H., Li H., McGuire A.M., Fletcher C.M., Gingras A.C., Sonenberg N., Wagner G.. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 1997; 4:717–724. [DOI] [PubMed] [Google Scholar]
- 10. Igreja C., Peter D., Weiler C., Izaurralde E.. 4E-BPs require non-canonical 4E-binding motifs and a lateral surface of eIF4E to repress translation. Nat. Commun. 2014; 5:4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lukhele S., Bah A., Lin H., Sonenberg N., Forman-Kay J.D.. Interaction of the eukaryotic initiation factor 4E with 4E-BP2 at a dynamic bipartite interface. Structure. 2013; 21:2186–2196. [DOI] [PubMed] [Google Scholar]
- 12. Paku K.S., Umenaga Y., Usui T., Fukuyo A., Mizuno A., In Y., Ishida T., Tomoo K.. A conserved motif within the flexible C-terminus of the translational regulator 4E-BP is required for tight binding to the mRNA cap-binding protein eIF4E. Biochem. J. 2012; 441:237–245. [DOI] [PubMed] [Google Scholar]
- 13. Umenaga Y., Paku K.S., In Y., Ishida T., Tomoo K.. Identification and function of the second eIF4E-binding region in N-terminal domain of eIF4G: comparison with eIF4E-binding protein. Biochem. Biophys. Res. Commun. 2011; 414:462–467. [DOI] [PubMed] [Google Scholar]
- 14. Kinkelin K., Veith K., Grunwald M., Bono F.. Crystal structure of a minimal eIF4E-Cup complex reveals a general mechanism of eIF4E regulation in translational repression. RNA. 2012; 18:1624–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gross J.D., Moerke N.J., von der Haar T., Lugovskoy A.A., Sachs A.B., McCarthy J.E., Wagner G.. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003; 115:739–750. [DOI] [PubMed] [Google Scholar]
- 16. Altmann M., Krieger M., Trachsel H.. Nucleotide sequence of the gene encoding a 20 kDa protein associated with the cap binding protein eIF-4E from Saccharomyces cerevisiae. Nucleic Acids Res. 1989; 17:7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Altmann M., Schmitz N., Berset C., Trachsel H.. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 1997; 16:1114–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cosentino G.P., Schmelzle T., Haghighat A., Helliwell S.B., Hall M.N., Sonenberg N.. Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000; 20:4604–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lanker S., Müller P.P., Altmann M., Goyer C., Sonenberg N., Trachsel H.. Interactions of the eIF-4F subunits in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1992; 267:21167–21171. [PubMed] [Google Scholar]
- 20. Blewett N.H., Goldstrohm A.C.. A eukaryotic translation initiation factor 4E-binding protein promotes mRNA decapping and is required for PUF repression. Mol. Cell. Biol. 2012; 32:4181–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chial H.J., Stemm-Wolf A.J., McBratney S., Winey M.. Yeast Eap1p, an eIF4E-associated protein, has a separate function involving genetic stability. Curr. Biol. 2000; 10:1519–1522. [DOI] [PubMed] [Google Scholar]
- 22. Cridge A.G., Castelli L.M., Smirnova J.B., Selley J.N., Rowe W., Hubbard S.J., McCarthy J.E., Ashe M.P., Grant C.M., Pavitt G.D.. Identifying eIF4E-binding protein translationally-controlled transcripts reveals links to mRNAs bound by specific PUF proteins. Nucleic Acids Res. 2010; 38:8039–8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de la Cruz J., Iost I., Kressler D., Linder P.. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:5201–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deloche O., de la Cruz J., Kressler D., Doere M., Linder P.. A membrane transport defect leads to a rapid attenuation of translation initiation in Saccharomyces cerevisiae. Mol. Cell. 2004; 13:357–366. [DOI] [PubMed] [Google Scholar]
- 25. Ibrahimo S., Holmes L.E., Ashe M.P.. Regulation of translation initiation by the yeast eIF4E binding proteins is required for the pseudohyphal response. Yeast. 2006; 23:1075–1088. [DOI] [PubMed] [Google Scholar]
- 26. Mascarenhas C., Edwards-Ingram L.C., Zeef L., Shenton D., Ashe M.P., Grant C.M.. Gcn4 is required for the response to peroxide stress in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2008; 19:2995–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsuo R., Kubota H., Obata T., Kito K., Ota K., Kitazono T., Ibayashi S., Sasaki T., Iida M., Ito T.. The yeast eIF4E-associated protein Eap1p attenuates GCN4 translation upon TOR-inactivation. FEBS Lett. 2005; 579:2433–2438. [DOI] [PubMed] [Google Scholar]
- 28. Meier K.D., Deloche O., Kajiwara K., Funato K., Riezman H.. Sphingoid base is required for translation initiation during heat stress in Saccharomyces cerevisiae. Mol. Biol. Cell. 2006; 17:1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ptushkina M., von der Haar T., Vasilescu S., Frank R., Birkenhager R., McCarthy J.E.. Cooperative modulation by eIF4G of eIF4E-binding to the mRNA 5′ cap in yeast involves a site partially shared by p20. EMBO J. 1998; 17:4798–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rendl L.M., Bieman M.A., Vari H.K., Smibert C.A.. The eIF4E-binding protein Eap1p functions in Vts1p-mediated transcript decay. PLoS One. 2012; 7:e47121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Diebold M.L., Fribourg S., Koch M., Metzger T., Romier C.. Deciphering correct strategies for multiprotein complex assembly by co-expression: application to complexes as large as the histone octamer. J. Struct. Biol. 2011; 175:178–188. [DOI] [PubMed] [Google Scholar]
- 32. Kabsch W. XDS. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Evans P. Scaling and assessment of data quality. Acta Crystallogr. D. Biol. Crystallogr. 2006; 62:72–82. [DOI] [PubMed] [Google Scholar]
- 34. Evans P.R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Papadopoulos E., Jenni S., Kabha E., Takrouri K.J., Yi T., Salvi N., Luna R.E., Gavathiotis E., Mahalingam P., Arthanari H. et al. Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E3187–E3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Langer G., Cohen S.X., Lamzin V.S., Perrakis A.. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008; 3:1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Terwilliger T.C., Grosse-Kunstleve R.W., Afonine P.V., Moriarty N.W., Zwart P.H., Hung L.W., Read R.J., Adams P.D.. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D. Biol. Crystallogr. 2008; 64:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D.. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D. Biol. Crystallogr. 2012; 68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smart O.S., Womack T.O., Flensburg C., Keller P., Paciorek W., Sharff A., Vonrhein C., Bricogne G.. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D. Biol. Crystallogr. 2012; 68:368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Velazquez-Campoy A., Leavitt S.A., Freire E.. Characterization of protein-protein interactions by isothermal titration calorimetry. Methods Mol. Biol. 2004; 261:35–54. [DOI] [PubMed] [Google Scholar]
- 45. Mizoue L.S., Tellinghuisen J.. The role of backlash in the “first injection anomaly” in isothermal titration calorimetry. Anal. Biochem. 2004; 326:125–127. [DOI] [PubMed] [Google Scholar]
- 46. Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J.. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009; 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thomsen M.C., Nielsen M.. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 2012; 40:W281–W287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McWilliam H., Li W., Uludag M., Squizzato S., Park Y.M., Buso N., Cowley A.P., Lopez R.. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013; 41:W597–W600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schneidman-Duhovny D., Hammel M., Sali A.. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic. Acids. Res. 2010; 38:W540–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 51. Sali A., Blundell T.L.. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993; 234:779–815. [DOI] [PubMed] [Google Scholar]
- 52. Slepenkov S.V., Korneeva N.L., Rhoads R.E.. Kinetic mechanism for assembly of the m7GpppG.eIF4E.eIF4G complex. J. Biol. Chem. 2008; 283:25227–25237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marcotrigiano J., Gingras A.C., Sonenberg N., Burley S.K.. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997; 89:951–961. [DOI] [PubMed] [Google Scholar]
- 54. Peter D., Weber R., Kone C., Chung M.Y., Ebertsch L., Truffault V., Weichenrieder O., Igreja C., Izaurralde E.. Mextli proteins use both canonical bipartite and novel tripartite binding modes to form eIF4E complexes that display differential sensitivity to 4E-BP regulation. Genes Dev. 2015; 29:1835–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miras M., Truniger V., Silva C., Verdaguer N., Aranda M.A., Querol-Audi J.. Structure of eIF4E in complex with an eIF4G peptide supports a universal bipartite binding mode for protein translation. Plant Physiol. 2017; 174:1476–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cawley A., Warwicker J.. eIF4E-binding protein regulation of mRNAs with differential 5′-UTR secondary structure: a polyelectrostatic model for a component of protein-mRNA interactions. Nucleic Acids.Res. 2012; 40:7666–7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. von Der Haar T., Ball P.D., McCarthy J.E.. Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′-Cap by domains of eIF4G. J. Biol. Chem. 2000; 275:30551–30555. [DOI] [PubMed] [Google Scholar]
- 58. Yanagiya A., Svitkin Y.V., Shibata S., Mikami S., Imataka H., Sonenberg N.. Requirement of RNA binding of mammalian eukaryotic translation initiation factor 4GI (eIF4GI) for efficient interaction of eIF4E with the mRNA cap. Mol. Cell. Biol. 2009; 29:1661–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Beckmann B.M., Horos R., Fischer B., Castello A., Eichelbaum K., Alleaume A.M., Schwarzl T., Curk T., Foehr S., Huber W. et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 2015; 6:10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Castelli L.M., Talavera D., Kershaw C.J., Mohammad-Qureshi S.S., Costello J.L., Rowe W., Sims P.F., Grant C.M., Hubbard S.J., Ashe M.P. et al. The 4E-BP Caf20p mediates both eIF4E-dependent and independent repression of translation. PLos Genet. 2015; 11:e1005233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Costello J., Castelli L.M., Rowe W., Kershaw C.J., Talavera D., Mohammad-Qureshi S.S., Sims P.F., Grant C.M., Pavitt G.D., Hubbard S.J. et al. Global mRNA selection mechanisms for translation initiation. Genome Biol. 2015; 16:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Arndt N., Ross-Kaschitza D., Kojukhov A., Komar A.A., Altmann M.. Properties of the ternary complex formed by yeast eIF4E, p20 and mRNA. Sci. Rep. 2018; 8:6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 6FBZ (Ct eIF4E–eIF4G in cap-bound state), 6FC0 (Ct eIF4E–eIF4G), 6FC1 (Sc eIF4E–Eap1p in cap-bound state), 6FC2 (Sc eIF4E–Eap1p) and 6FC3 (Sc eIF4E–p20).