Abstract
mRNA decay is an important strategy by which bacteria can rapidly adapt to their ever-changing surroundings. The 5′-terminus state of mRNA determines the velocity of decay of many types of RNA. In Escherichia coli, RNA pyrophosphohydrolase (RppH) is responsible for the removal of the 5′-terminal triphosphate from hundreds of mRNAs and triggers its rapid degradation by ribonucleases. A diaminopimelate epimerase, DapF, can directly interact with RppH and stimulate its hydrolysis activity in vivo and in vitro. However, the molecular mechanism remains to be elucidated. Here, we determined the complex structure of DapF–RppH as a heterotetramer in a 2:2 molar ratio. DapF-bound RppH exhibits an RNA-favorable conformation similar to the RNA-bound state, suggesting that association with DapF promotes and stabilizes RppH in a conformation that facilitates substrate RNA binding and thus stimulates the activity of RppH. To our knowledge, this is the first published structure of an RNA–pyrophosphohydrolysis complex in bacteria. Our study provides a framework for further investigation of the potential regulators involved in the RNA–pyrophosphohydrolysis process in prokaryotes.
INTRODUCTION
RNA decay plays crucial roles in controlling gene expression and cell responses to changing growth conditions and in the elimination of aberrant mRNAs to recycle nucleotides (1). A pivotal factor governing the RNA lifetime is its 5′-terminus cap or cap-like structure, including variations with N7-methylguanosine (m7G) (2), triphosphate (3), diphosphate (4), nicotinamide adenine dinucleotide (NAD) (5–7) or other nucleotide derivatives (8,9). Generally, the cap/cap-like structure protects mRNAs from cleavage by nucleases, enhancing their stability and extending their lifetime, which results in much higher translation levels (10,11).
Several types of hydrolases remove the 5′ protective structures of RNA. For example, decapping protein 2 (Dcp2) is responsible for m7G cap removal from RNA in eukaryotes (12–14), while in prokaryotes, RNA pyrophosphohydrolase (RppH) and NudC hydrolyze triphosphate and NAD from RNA, respectively (3,15). Dcp2, RppH and NudC all belong to the Nudix hydrolase family, whose members act on substrates of nucleoside diphosphates linked to other moieties X. All members of the enzyme harbor a conserved 23 amino acid sequence (GX5EX7REUXEEXGU) named Nudix motif, which is essential for substrate catalysis (16,17). Moreover, one member of the DXO family, DXO/Rai1, has recently been reported to remove NAD-caps from RNA in humans, releasing NAD and 5′-monophosphate RNAs (7).
In bacteria, most nascent mRNAs are assumed to bear a 5′-triphosphate group, which can subsequently be converted into a 5′-diphosphate group. Both RNA forms are resistant to degradation and can be hydrolyzed by RppH with distinct catalytic efficiency (4). These hydrolysis reactions generate monophosphorylated RNA, which stimulates subsequent ribonuclease cleavage (10). For example, in Escherichia coli, very few rpsT P1 mRNA transcripts are monophosphorylated in cells lacking RppH, and the half-life of these mRNAs is extended 5-fold. Rapid turnover was restored in theΔrppH strain by complementation with the wild-type rppH gene but not by a mutant allele (rppH-E53A). In addition, EcRppH accelerates the degradation of hundreds of transcripts (3). The rppH gene from Bdellovibrio bacteriovorus was able to complement RppH deficiency in E. coli and restore normal RNA degradation, validating its function as an RNA pyrophosphohydrolase in vivo (18). Similar effects were found for Bacillus subtilis and Helicobacter pylori. In the absence of BsRppH, the average measured half-life of the yhxA-glpP transcript increased from 3.7 ± 0.6 min to 8.3 ± 0.6 min, accompanied by an elevation in monophosphorylated RNA (19). In the gastric pathogen Helicobacter pylori, HpRppH has a similar function to EcRppH and triggers at least 63 potential targets, including mRNA and sRNA, for degradation (20). Given the determinant role of RppH in mRNA decay, leveraging activity of this enzyme could be an economic strategy by which microbes can rapidly adjust their cellular mRNA in response to changed nutrient availability or external stimuli. However, very few regulators have been well characterized.
In contrast, regulation of the decapping process in eukaryotes has been extensively explored. Multiple components, including Dcp1, Edc1, Edc3 and PNRC, are reported to participate in the Dcp2-regulatory network (21–24). Dcp1 forms a catalytic core complex with Dcp2 and stimulates Dcp2 activity. The complex structure of the Dcp1–Dcp2 heterodimer suggests that Dcp1 promotes and stabilizes the closed state of Dcp2, which closely resembles the more active/catalytic form (21). In addition, other cofactors employ various strategies to associate with the Dcp1–Dcp2 complex. Edc1 can simultaneously interact with Dcp1 and Dcp2, resulting in a rotation of the Dcp2 catalytic domain. This conformational change promotes cooperative binding of RNA by Dcp2 and Dcp1 and consequently increases the decapping activity of Dcp2 (22). Edc3 extends its Box B helix in Nudix motif of Dcp2 (23). In contrast, PRNC binds solely to Dcp1 (24). The interactions of these factors with the decapping core complex lead to strong substrate binding affinity and accelerate the catalytic step.
Recently, a diaminopimelate epimerase, DapF, which catalyzes the stereoinversion of L,L-DAP to meso-DAP, was shown to strongly associate with RppH and stimulate its activity both in vivo and in vitro (25). The cellular level of DapF affected RppH-catalyzed pyrophosphate removal and the subsequent degradation of target mRNAs. To gain insight into the molecular mechanism by which DapF activates RppH, we determined the crystal structure of RppH in complex with DapF. A structural comparison of DapF-bound RppH with apo RppH and RNA-bound RppH (26) reveals that the conformation of DapF-bound RppH most closely resembles that of the RNA-bound form but differs significantly from that of the apo form, suggesting that DapF stimulates RppH by promoting and/or stabilizing a substrate-favoring conformation, similar to the stimulation of Dcp2 by Dcp1 in eukaryotes (21). Taken together, our results not only reveal a conserved activation mechanism in the processes of removing the 5′-terminus protective structures of RNA, but also provide a framework for the further characterization of new potential regulators involved in RNA pyrophosphohydrolysis in bacteria.
MATERIALS AND METHODS
Molecular cloning, protein expression and purification
Full-length rppH and dapF genes were amplified from the Escherichia coli DH5α strain. rppH was subcloned into pET21b (Novagen) and fused with an 8 × His tag at the C-terminus, while dapF was subcloned into pBB75 (Novagen) without any tag. To obtain well-diffracting crystals, we introduced a C16S mutation into RppH using overlap PCR. The two plasmids were then co-transformed into BL21 (DE3) to construct the DapF–RppH complex. One liter lysogeny broth medium supplemented with 100 μg ml–1 ampicillin and 50 μg ml–1 kanamycin was inoculated with a transformed bacterial pre-culture and shaken at 37°C until the optical density at 600 nm reached 1.0. After being induced with 0.2 mM isopropyl-β-d-thiogalactoside and growing at 16°C for 16 h, the bacterial pellet was collected and homogenized (JNBIO, China) in a buffer containing 25 mM Tris–HCl, pH 8.0 and 500 mM NaCl. After centrifugation at 23 000 g at 4°C, the supernatant was loaded onto a column equipped with Ni2+ affinity resin (Ni-NTA, Qiagen); washed with a buffer containing 25 mM Tris–HCl, pH 8.0, 500 mM NaCl and 15 mM imidazole; and eluted with a buffer containing 25 mM Tris–HCl, pH 8.0, 500 mM NaCl, and 250 mM imidazole. The eluted protein was diluted 10-fold and then applied to a 5 ml heparin column (GE Healthcare), followed by a gradient NaCl elution (up to 1 M) in 25 mM Tris–HCl, pH 8.0. The elution peak was concentrated to 1 ml (∼10 mg ml–1) and digested by elastase (0.05 mg ml–1) at 4°C for 1 h without stopping digestion before being subjected to gel filtration chromatography (Superdex-200 Increase 10/300, GE Healthcare) which was equilibrated with a buffer containing 25 mM Tris–HCl, pH 8.0, 500 mM NaCl and 5 mM 1,4-dithiothreitol (DTT). The peak fractions were collected for crystallization trials.
Variants of DapF and RppH were purified alone for activity assays. dapF was cloned into pET15D (Novagen) and expressed with a 6× His tag fused at the N-terminus. The purification of DapF is the same as that of the DapF–RppH complex, with the minor change that the heparin column was substituted with a Source15Q column. rppH was cloned into pPGH with a GST tag fused at the N-terminus followed by a 3C protease cleavage site. The bacterial pellet was collected and homogenized in a buffer of 25 mM Tris–HCl, pH 8.0, and 500 mM NaCl, followed by centrifugation at 23 000 g at 4°C. The supernatant was loaded onto a GST-4B column (GE Healthcare), washed with the same buffer, eluted with elution buffer (25 mM Tris–HCl, pH 9.0, 500 mM NaCl and 10 mM GSSH), diluted two-fold, and applied to a heparin column for further purification. The GST tag was removed by incubation with 3C protease at 4°C for 2 h, and the product was subjected to gel filtration chromatography (Superdex-200 10/300, GE Healthcare) in a buffer containing 25 mM Tris–HCl, pH 8.0, 500 mM NaCl and 5 mM DTT. The purified proteins were flash frozen in liquid nitrogen and stored at −80°C without further processing.
Full-length meso-diaminopimelate dehydrogenase gene (dapdh) of Corynebacterium glutamicum was synthesized (GENEWIZ, Inc., China) and cloned into pET15D (Novagen). The meso-DAP dehydrogenase (DAPDH) protein with a 6× His tag fused at the N-terminus was expressed and purified in a similar procedure as that of DapF with the only change in buffers containing 150 mM other than 500 mM NaCl. The purified proteins were flash frozen in liquid nitrogen and stored at −80°C.
Crystallization
After digestion by elastase (Sigma-Aldrich), the DapF–RppH (C16S) complex was crystallized by using the hanging-drop vapor-diffusion method at 18°C, and 1 μl of sample was mixed with an equal volume of reservoir solution. Diamond-shaped crystals appeared overnight from a reservoir solution containing 1.0 M potassium sodium tartrate, 0.1 M MES (2- (N-morpholino) ethanesulfonic acid), pH 5.5, 0.2 M sodium iodine and 2% xylitol (Hampton). After 48 h of growth, the crystal ceased growing and was flash frozen in liquid nitrogen. The cryoprotection buffer included 20% ethylene glycol added to reservoir solution. The DapF–RppH complex crystal diffracted beyond 2.3 Å at the Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U1.
Data collection and structure determination
The dataset was collected at the SSRF beamline BL17U1 (27) and processed with the HKL3000 (28). Further processing was performed with the CCP4 suite (29). Data collection and structural refinement statistics are summarized in Table 1. The structure of the DapF–RppH complex was solved by molecular replacement (MR) using the PDB structures 4S2V (RppH) and 4IJZ (DapF) as the search models using the program PHASER (30). The structure was manually and iteratively refined with PHENIX and COOT (31,32). All figures representing structures were prepared with PyMOL (http://www.pymol.org).
Table 1.
Statistics of data collection and refinement
| DapF-bound RppH | |
|---|---|
| Number of crystals | 1 |
| Space group | P41212 |
| Cell dimensions | |
| a, b, c (Å) | 86.42, 86.42, 173.09 |
| α, β, γ (o) | 90, 90, 90 |
| Number of molecules in ASU | 1 |
| Wavelength (Å) | 0.9792 |
| Resolution (Å) | 45–2.30 (2.38–2.30) |
| R merge (%) | 9.8 (121.7) |
| R pim (%) | 3.9 (47.6) |
| I/σI σ | 18.8 (2.4) |
| Completeness (%) | 99.8 (99.8) |
| Number of measured reflections | 418 998 |
| Number of unique reflections | 30 026 |
| Redundancy | 14 (13.9) |
| Wilson B factor (Å2) | 46.4 |
| R work/Rfree (%) | 20.6/23.9 |
| Number of atoms | 3454 |
| Protein | 3355 |
| Main chain | 1696 |
| Side chain | 1659 |
| Water | 79 |
| Other entities | 22 |
| Average B value (Å2) | 65.2 |
| Protein | 65.3 |
| main chain | 63.6 |
| side chain | 67.1 |
| Water | 56.3 |
| Other entities | 72.2 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angle (°) | 1.030 |
| Ramachandran plot statistics (%) | |
| Most favorable | 89.4 |
| Allowed | 10.6 |
| Disallowed | 0 |
The highest-resolution shell is shown in parentheses. Values in parentheses indicate the highest resolution shell. Rmerge=ΣhΣi|Ih,i-Ih|/ΣhΣiIh,i, where Ih is the mean intensity of the i observations of symmetry-related reflections of h. R=Σ|Fobs-Fcalc|/ΣFobs, where Fcalc is the calculated protein structure factor from the atomic model (Rfree is calculated with 5% of the reflections selected).
Preparation of RNA oligonucleotides
The short mRNA was prepared by in vitro transcription with T7 RNA polymerase as previously described (33,34). The double-strand DNA templates were prepared by gradually annealing two single-strand oligonucleotides A8-F (5′-GATCACTAATACGACTCACTATTAGAACAAC-3′; the promoter is shaded gray, and the transcription sequence is underlined) and A8-R (5′-GTTGTTCTAATAGTGAGTCGTATTAGTGATC-3′) in 200 μl at a concentration of 10 μM for each oligonucleotide. The transcription reactions (1 ml) were performed in a buffer containing 100 mM Tris–HCl, pH 8.0, 20 mM MgCl2, 0.01% Triton X-100, 2 mM spermidine, 40 mM DTT, 1 μM DNA template, 5 mM rATP, 2 mM rCTP, 1 mM rGTP (the molar ratio of rNTPs is based on the template sequence), and 0.1 mg ml–1 T7 RNA polymerase at 37°C for 3 h. The reaction sample was centrifuged at 20 000 g for 5 min, and the supernatant was loaded onto a 1 ml monoQ 5/50 GL column (GE Healthcare), followed by gradient elution with 0.5 M NaCl in 25 mM Tris–HCl, pH 8.0. The product peaks were determined by comparing the elution peaks of the sample with that of the reference (lacking T7 RNA polymerase) and further purified by ethanol precipitation, followed by resolving in 50 mM Tris–HCl, pH 8.0. The RNA transcript pppAGAACAAC was verified by mass spectrometry (Ultraflex II MALDI-TOF, Bruker) and applied to the RppH cleavage experiment as a substrate in vitro, otherwise it was flash frozen in liquid nitrogen and stored at −80°C.
The 3 nt triphosphorylated nucleotides were prepared similarly as that of 8 nt. Briefly, after being transcribed by T7 RNA polymerase and purified by monoQ 5/50 GL column, 3 nt triphosphorylated RNA was applied to a 5 ml HiTrap Desalting column (GE healthcare). Pooled fractions were vacuum dried and stored at −80°C.
Determination of RppH activity in vitro
The hydrolysis of RNA was performed in a 100 μl reaction containing 50 mM Tris–HCl, pH 8.0, 20 mM MgCl2, 100 mM NaCl, 0.1 μM RppH and 100 μM substrates, either with or without 0.5 μM DapF. The reactions were incubated at 37°C for 15 min and quenched by the addition of 950 μl of 50 mM sodium acetate trihydrate, pH 5.0, followed by centrifugation at 20 000 g for 5 min and application to ion exchange chromatography (monoQ 5/50 GL column, GE Healthcare). The product (5′-pAGAACAAC-3′) and the substrate (5′-pppAGAACAAC-3′) were eluted at the conductivity 31.1 mS cm-1 and 34.1 mS cm–1 in gradient NaCl (25 mM Tris–HCl, pH 8.0) up to 0.5 M. Elution profiles were recorded at 260 nm, and peaks corresponding to monophosphorylated products or triphosphorylated substrates were integrated using UNICORN software (GE Healthcare). The activity of the protein was defined as the proportion of products generated during the reaction time. The relative activity was calculated by normalizing the measured activity to that of wild-type RppH alone.
Analytical ultracentrifugation (AUC)
The sedimentation velocities (SV) of RppH, DapF, DapF (Y268A), DapF–RppH and DapF (Y268A)-RppH were measured by AUC. The SV-AUC experiments were performed in a Beckman Coulter XL-A/I analytical ultracentrifuge using a one-channel centerpiece. The proteins were prepared in solutions containing 25 mM Tris–HCl, pH 8.0 and 500 mM NaCl. The data were collected by UV detection at a rotor speed of 50 000 rpm at 18°C for 8 h. The SV-AUC data were globally analyzed using Sedfit software to determine the sedimentation coefficients and molecular weights of each fraction.
Isothermal titration calorimetry (ITC) assays
ITC experiments for the interaction of RppH and DapF were performed at 25°C using an Auto-iTC200 titration calorimeter (Malvern). Samples of 50 μM wild-type or mutant RppH (in a volume of 40 μl) were dissolved in reaction buffer containing 25 mM Tris–HCl, pH 8.0, and 500 mM NaCl and titrated against 5 μM wild-type or mutant DapF (in 200 μl volume) in the same buffer. The first injection (0.4 μl) was followed by 19 injections of 2 μl. The heat of dilution values for RppH or DapF were measured by injecting the protein into buffer alone and subtracted from the experimental curves before data analysis. The stirring rate was 750 rpm. MicroCal ORIGIN software supplied with the instrument was used to determine the site-binding model that produced a good fit (low x 2 value) for the resulting data.
ITC experiments for the interaction of 8 nt RNA and proteins were performed at 37°C in 50 mM HEPES, pH 7.5 and 150 mM NaCl. 40 μl of 2 mM RNA was titrated against 200 μl of 50 μM DapF–RppH complex, RppH or DapF, respectively. Method used to perform the titration and data processing was same as previously described (35).
Measurement of enzymatic properties of RppH for RNA hydrolysis in vitro
The hydrolysis of RNA was performed in a 100 μl reaction containing 50 mM Tris–HCl, pH 8.0, 20 mM MgCl2, 100 mM NaCl, 0.1 μM RppH and gradient concentration of substrates (20–240 μM), either with or without 0.5 μM DapF. The reaction was incubated at 37°C for the indicated time, and was terminated by the addition of 950 μl of 50 mM sodium acetate trihydrate, pH 5.0. Velocity of RppH at various concentration of substrate was measured by calculating the product generation in the indicated time. The values of velocity were fitted to the Michaelis–Menten equation equipped in Origin 8.0 to get the Km values of RppH for RNA hydrolysis, in the absence or presence of DapF.
Measurement of enzymatic activity of DapF
The enzymatic activity of DapF was determined by using the modified DapF–DAP dehydrogenase coupled spectrophotometric assay (36,37). Briefly, a premixed reaction contained 100 mM Tris, pH 7.8, 0.2 mM L,L-DAP, 0.88 mM NADP+, 1 mM DTT and 5 μM DAP dehydrogenase, with or without 2.5 μM RppH. DapF was added into the reaction and quickly mixed to homogeneity. Two microliter of the sample was immediately loaded onto a UV spectrophotometer (NanoPhotometer N60, IMPLEN) and the absorbance at 340 nm (A340) was consecutively recorded for 2 min, with an interval of 5 s. The increase in A340 was corresponded to the conversion of meso-DAP derived from L,L-DAP catalyzed by DapF into tetrahydrodipicolinate and the transformation of NADP+ into NADPH. The assay was performed at 25°C in duplicate. The initial velocity (V0) of DapF was represented by the change in A340 at the first 10 s of the reaction.
RESULTS
Crystal structure of the DapF–RppH complex
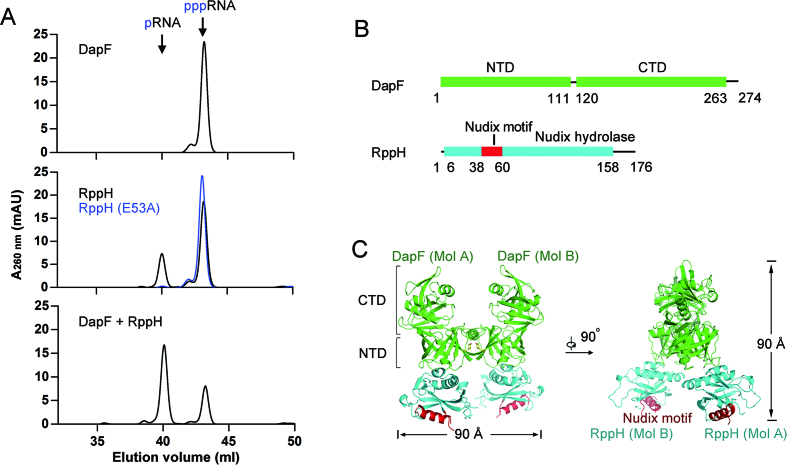
We purified recombinant DapF and RppH to homogeneity and tested their pyrophosphohydrolase activity with a triphosphorylated oligonucleotide (5′-pppAGAACAAC-3′) as a substrate by ion exchange chromatography. Consistent with a previous study (25), DapF has no detectable activity and RppH is capable of hydrolyzing the substrate to monophosphorylatedRNA (5′-pAGAACAAC-3′) (Figure 1A, upper and middle panels). As expected, a catalytically inactive mutant of RppH (E53A) lost its activity (Figure 1A, middle panel). Addition of DapF to the reaction apparently increases the amount of product, indicating that DapF stimulates the hydrolase-activity of RppH (Figure 1A, lower panel). Next, we co-expressed full-length DapF and RppH (Figure 1B). The two proteins eluted as a tightly bound complex in size exclusion chromatography experiments (Supplementary Figure S1). To identify the stable structural core complex, the full-length complex was subjected to digestion with increasing amounts of elastase. After partial digestion, the DapF–RppH binary complex remained as an elastase-resistant core component. The DapF was nearly undigested, but an approximately 4 kDa peptide was removed from RppH (Supplementary Figure S1). Mass spectrometry analysis indicated nearly 15 amino acids on the C-terminus of RppH and the histidine-tag were cleaved by elastase. Finally, we crystallized this protease-resistant complex in the space group P41212. The complex structure was determined by molecular replacement using two available coordinates of RppH (PDB ID: 4S2V) and DapF (PDB ID: 4IJZ) and refined to 2.3 Å resolution with an R-factor of 20.6% and an R-free of 23.9% (Table 1).
Figure 1.
Structure of the DapF–RppH complex. (A) DapF stimulation of RppH activity is detected by ion exchange chromatography. DapF alone has no detectable activity upon triphosphorylated RNA hydrolysis (upper panel). RppH is able to hydrolyze triphosphorylated RNA into monophosphorylated RNA and its catalytic mutant RppH (E53A) is not (middle panel). DapF stimulates the activity of RppH (lower panel). (B) Ribbon diagram representation of DapF and RppH. DapF is colored green and consists of an N-terminal domain (NTD) and a C-terminal domain (CTD). RppH is colored cyan, and the Nudix motif is colored red. The numbers under the ribbons depict the boundaries of motifs or domains. (C) Overall structure of the DapF–RppH complex in two perpendicular views. The H-shaped heterotetramer consists of two DapF and two RppH molecules, approximately 90 Å in width and height. The NTD and CTD of DapF are indicated by square brackets. The tyrosine that sustains the interface of two DapF molecules (MolA and MolB) is represented as yellow sticks. The Nudix motif in the two RppH molecules (MolA and MolB) is indicated in red. All color representations are the same as in diagram (B).
In each biological unit, two DapF molecules bound two RppH molecules and formed a heterotetramer, which resembled the letter ‘H’ in appearance and was approximately 90 Å in width and in height (Figure 1C). The tetramer is composed of two bilateral symmetrical heterodimers, each containing one DapF molecule and one RppH molecule. The DapF protomer consists of an N-terminal domain (NTD, residues 1–111 and 265–271) and a C-terminal domain (CTD, residues 120–263). The RppH protomer consists of a single Nudix domain (residues 6–158), in which resides a Nudix motif (residues 38–60) (Figure 1C). In each heterodimer, the NTD of DapF interacts with RppH, whereas the CTD is oriented far from the interface. One heterodimer interacts with the other via a separate region of the NTD of DapF (residues 265–271) (Figure 2A), leading to a considerable distance between each Nudix motif of RppH (Figure 1C).
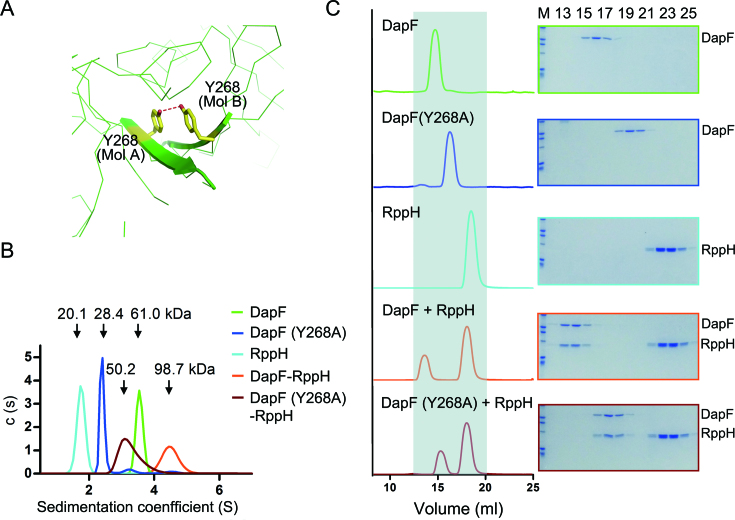
Figure 2.
The 2:2 molar ratio of DapF–RppH complex in solution. (A) Close-up view of the interface of two DapF molecules. The benzene rings of Y268 on each DapF molecule are compactly stacked, with a hydrogen bond formed by two hydroxyl groups buttressing the π-π interaction. The side chain of Y268 is shown as a yellow stick, and the hydrogen bond is indicated by a red dashed line. (B) Molecular mass of DapF, RppH and the DapF–RppH complex measured by analytical ultracentrifugation. A series of colored curves represent the sedimentation coefficient distribution of different samples, with calculated molecular masses indicated above. DapF is colored in green, the Y268 mutant of DapF in blue, RppH in cyan, DapF–RppH complex in orange and DapF (Y268A)-RppH complex in brown. (C) Interaction of DapF and RppH detected by gel filtration. The left panel lists the profiles of samples applied to gel filtration, and the right panel is the corresponding eluted proteins detected by SDS-PAGE and Coomassie brilliant blue staining. Gray shading in the left panel is employed to clarify the shift of the elution peak. All color representations are the same as in diagram (B).
The state of DapF–RppH complex in solution
A previous study reported that residue Y268 on the β-sheet of DapF is essential for its dimerization (36). In our structure of the DapF–RppH complex, we also observed two Y268 residues from distinct DapF protomers stacked together and buttressed by a hydrogen bond (2.8 Å between two hydroxyl oxygen atoms of Y268, Figure 2A). To investigate the oligomerization state of DapF–RppH in solution, we employed analytical ultracentrifugation (AUC) and size exclusion chromatography (SEC) methods to measure their molecular mass. As expected, DapF exists as a dimer in solution and the Y268A mutation disrupts its dimeric state (green and blue lines in Figure 2B). RppH alone exists as a monomer in solution (cyan line in Figure 2B). The DapF–RppH complex is a heterotetramer, as the measured molecular mass (98.7 kDa, orange line in Figure 2B) is consistent with the total molecular mass of two DapF and two RppH molecules (102.2 kDa). Notably, the Y268A mutation of DapF disrupted the DapF–RppH complex from a heterotetrameric to a heterodimeric state, as the measured molecular mass of DapF (Y268A)-RppH (50.2 kDa, brown line in Figure 2B) is nearly identical to the calculated molecular mass of a heterodimer (51.1 kDa).
Our SEC analysis is in line with our AUC observation. DapF eluted at approximately 14.8 ml (green line in Figure 2C), and the DapF mutant (Y268A) eluted at approximately 16.3 ml (blue line in Figure 2C). RppH alone eluted at nearly 18.1 ml (cyan line in Figure 2C). The addition of DapF to excess RppH resulted in co-elution of the two proteins and shifted their elution volume to 13.6 ml, indicating that the DapF–RppH complex formed as a hetrotetramer (orange line and frame in Figure 2C). Interestingly, the monomeric mutant DapF (Y268A) still formed a complex with RppH and shifted both components to 15.3 ml (brown line and frame in Figure 2C), indicating that DapF (Y268A)-RppH exists as a heterodimer. Taken together, these results corroborate the structural oligomerization findings.
Effect of dimeric state of DapF on the activity of RppH
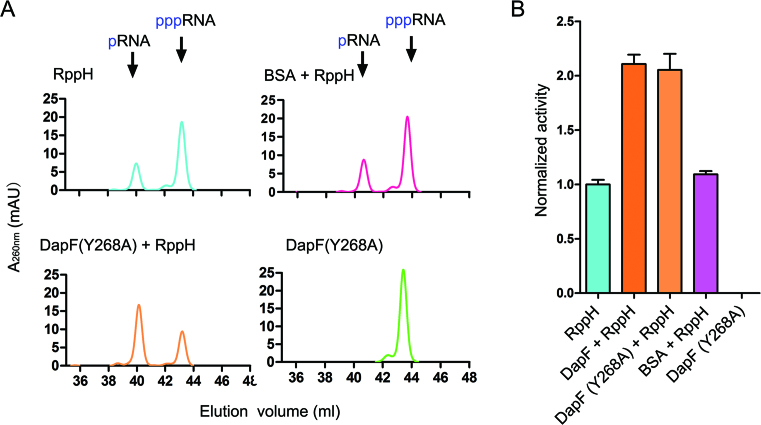
We have shown evidence that DapF stimulates the pyrophosphatase activity of RppH in solution as a dimer (Figures 1A, 2B and 2C), which raises the intriguing question of whether the effect of DapF on RppH depends on its dimerization. To answer this question, we tested the stimulatory effect of DapF (Y268A) on the activity of RppH, since this mutant exists as a monomer by itself but can still interact with RppH to form a heterodimer. The addition of DapF (Y268A) to RppH resulted in a substantial increase in product compared with RppH alone (Figure 3A), which was nearly identical in scale to that of wild-type DapF (Figure 3B). As a control, adding bovine serum albumin (BSA), which is generally considered to be a biological environment crowding molecule, had a negligible effect on the activity of RppH. These results suggest that DapF stimulation of RppH activity has no relationship with its dimeric state.
Figure 3.
Both monomeric and dimeric DapF can stimulate RppH activity. (A) Activity of RppH alone or in the presence of wild DapF, DapF (Y268A) mutant and BSA was detected by ion exchange chromatography. Activities of wild DapF and the DapF (Y268A) mutant are measured as control. (B) Activities of RppH or DapF variants were calculated and normalized to the activity of RppH alone and shown as columns. Each column represents two independent measurements, error bars represent standard deviations.
The interface between DapF and RppH
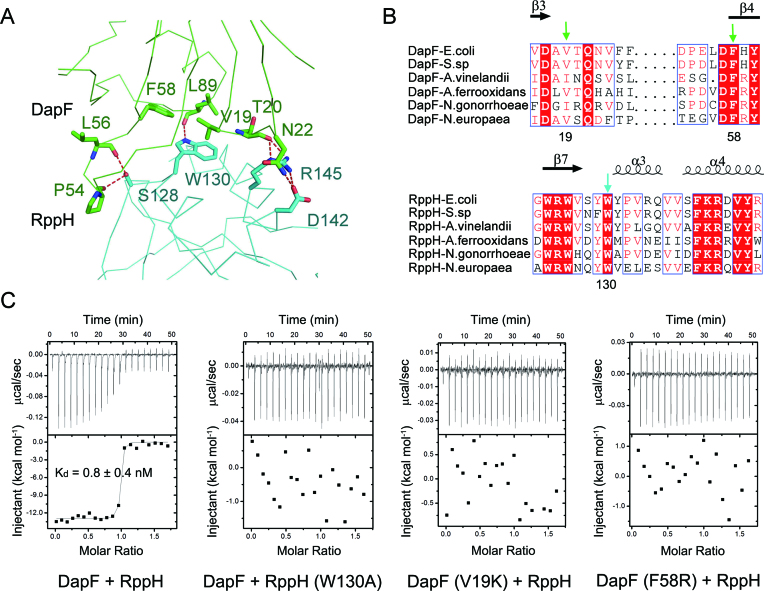
The NTD of one DapF molecule associates with one RppH molecule through extensive hydrophobic and hydrogen bonding interactions, which can be divided into three main parts: first, two carbonyl oxygens on the backbone of DapF (P54 and L56) form two hydrogen bonds with the hydroxyl group of S128 on RppH; second, a hydrophobic core, which consists of three bulky residues (V19, F58 and L89) on DapF and W130 on RppH, is buttressed by a hydrogen bond; third, several polar or charged residues on DapF (T20 and N22) and RppH (D142 and R145) establish a hydrogen bond network (Figure 4A). Among the residues supporting this interface, those comprising the hydrophobic core (V19/F58/L89 of DapF, W130 of RppH) are highly conserved in several Gram-negative bacteria (Figure 4B, Supplementary Figures S2 and S3).
Figure 4.
The interface between DapF and RppH. (A) Close-up view of the interface of DapF and RppH. Residues forming the interface of DapF and RppH are shown as green and cyan sticks, respectively. Red dashed lines represent hydrogen bonds. (B) Partial sequence alignment results of DapF homologues and RppH homologues. Secondary structural elements are indicated above. Completely conserved amino acids are colored in red, and partially conserved amino acids are enclosed in blue boxes. V19 and F58 on DapF, and W130 on RppH are key residues supporting the interface of DapF–RppH compl ex and are indicated by green and cyan arrows, respectively. (C) Interaction of DapF and RppH detected by isothermal titration calorimetry. Wild-type DapF binds RppH with a dissociation constant of ∼0.8 nM. Mutation of either RppH (W130A) or DapF (V19K or F58R) abolishes their interaction.
To explore the contributions of these residues to the interface of DapF and RppH in complex formation, we measured the binding affinity of the DapF–RppH interaction by isothermal titration calorimetry (ITC). The apparent dissociation constant (Kd) value of DapF binding to RppH was approximately 0.8 nM (Figure 4C), roughly in accordance with previous measurements by surface plasmon resonance (∼5.2 nM) (25). Strikingly, incorporating the W130A mutation into RppH completely abolished its interaction with DapF. Single-residue mutations of V19K or F58R on DapF also eliminated its interaction with RppH (Figure 4C). Other alanine-substitution mutations of DapF (T20A/N22A/L89A) or RppH (S128A/D142A/R145A) exhibited no detectable or minor influences on the interaction of DapF and RppH (Table 2).
Table 2.
Interaction of DapF and RppH measured by isothermal titration calorimetry
| RppH (titrant) | DapF (titrate) | Stoichiometry (N) | K d (nM) | ΔH (kcal⋅mol-1) | ΔS (cal⋅mol-1⋅deg-1) |
|---|---|---|---|---|---|
| WT | WT | 0.95 | 0.8 ± 0.4 | −13.0 ± 0.1 | −1.8 |
| S128A | WT | 0.85 | 12.8± 6.0 | −12.5 ± 0.3 | −6.0 |
| R145A | WT | 1.01 | 2.3 ± 1.0 | −14.6 ± 0.1 | −9.3 |
| D142A | WT | 1.05 | 4.8 ± 1.8 | −11.0 ± 0.2 | 1.2 |
| W130A | WT | — | — | — | — |
| WT | V19K | — | — | — | — |
| WT | T20A | 0.97 | 4.2 ± 1.8 | −14.4 ± 0.2 | −9.8 |
| WT | N22A | 0.96 | 0.3 ± 0.1 | −16.4 ± 0.1 | −11.5 |
| WT | F58R | — | — | — | — |
| WT | L89A | 1.06 | 5.2 ± 0.9 | −10.4 ± 0.1 | 3.0 |
— indicates that the interaction was not detectable.
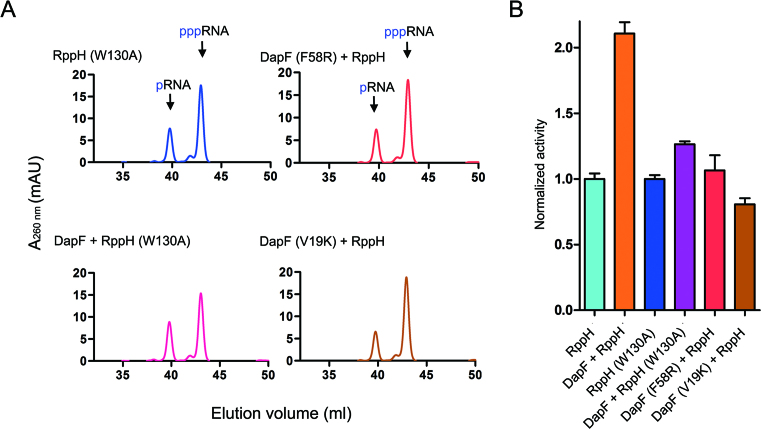
We further tested the effect of DapF on the activity of RppH (W130A), as well as the DapF (V19K) or DapF (F58R) mutants on the activity of RppH, in order to verify the importance of these key interface residues. RppH (W130A) has almost identical activity to wild-type RppH (Figure 5A and B). Wild-type DapF induced only a subtle increase in the activity of RppH (W130A) (<20%) compared with its effect on wild-type RppH (>100%) (Figure 5A and B), which is consistent with the observation that the interaction of DapF and RppH is broken by introducing the W130A mutation into RppH. Meanwhile, neither the DapF (F58R) nor the DapF (V19K) mutant had apparent influences on the activity of RppH (Figure 5A and B), supporting the essential roles of these residues in protein interactions.
Figure 5.
DapF stimulation of RppH hydrolase activity. (A) W130A mutant of RppH is capable of hydrolyzing RNA with 5′ triphosphate termini, generating RNA with 5′ monophosphate termini (upper-left panel). Addition of DapF has only a small influence on the activity of the W130A mutant of RppH (lower-left panel). No increased hydrolase activity of RppH is detected in the presence of DapF mutants (F58R or V19K, right panel). (B) Measured activities are calculated by the increase in products and normalized to that of wild-type RppH. Columns represent the mean value of normalized activity from three independent measurements, and the error bars represent standard deviation.
DapF-bound RppH exhibits RNA-bound conformation
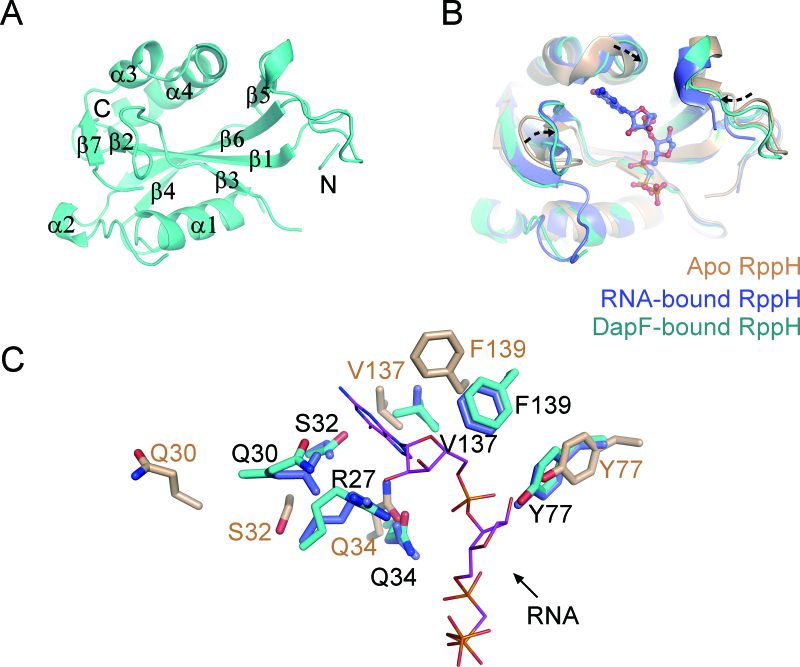
The structure of DapF-bound RppH (MolA, residues 1–158) adopts a canonical Nudix fold, in which seven-stranded mixed-sheets (β1–β7) are sandwiched by three remote perpendicular α-helices (α1, α3 and α4) (Figure 6A and Supplementary Figure S3). We compared the structure of RppH in the DapF-bound state with its structure in the apo state (PDB: 4S2V), which was used as a model for replacement, as well as in the RNA-bound state (PDB: 4S2Y). Overall, the structure of RppH, including the Nudix motif (Supplementary Figure S4), in the three different states exhibits no substantial divergence. The root-mean-square deviation (RMSD) value of DapF-bound and apo RppH is 1.227 Å over 123 Cα atoms, while that of DapF-bound and RNA-bound RppH is 0.455 Å over 119 Cα atoms. However, some regions of RppH in the different states exhibit wobbling (Figure 6B), where DapF-bound RppH is nearly superimposed upon RNA-bound RppH but differs from apo RppH. These regions comprise a long loop between β2 and β3 (residues 26–36), β5 and α2 (residues 75–84), and a portion located near the C-terminal helix (α4) of RppH (residues 135–145, Figure 6A, B and Supplementary Figure S3). In the areas where differences are concentrated, the RMSD value of DapF-bound and apo RppH (1.711 Å over 29 Cα atoms) is larger than that of DapF-bound and RNA-bound RppH (0.288 Å over 28 Cα atoms). Notably, they are all located in the vicinity of its RNA-binding pocket (26), raising the possibility that RppH undergoes substrate-favorable conformational changes upon DapF binding.
Figure 6.
The molecular basis of RppH activation by DapF. (A) A ribbon representation of the DapF-bound RppH (Mol A). The labels on the helices or strands are used to trace the RppH sequence. N-terminal and C-terminal ends are indicated for clarity. (B) Structural alignment of RppH in the apo state (PDB: 4S2V), RNA-bound state (PDB: 4S2Y) and DapF-bound state. Apo RppH is colored in brown, RNA-bound RppH in blue, and DapF-bound RppH in cyan. The diverse regions comprising a wobbling loop and two bent helices are indicated by dashed arrows, which are located in the vicinity of the pocket accommodating substrate RNA. (C) Side chain orientation of residues on the diverse region of RppH. Color representation of residues is the same as in diagram (B). RNA is shown as purple sticks. The residues that may be involved in RNA recognition in DapF-bound RppH nearly superimpose upon those of RNA-bound RppH and differ from those of apo RppH.
We traced residues on the flexible region and found that many of them, including R27/Q30/S32/Q34/Y77/V137/ F139, are distinctly oriented in these three states (Figure 6C), where the position of the residues in DapF-bound RppH is almost identical to that of the RNA-bound state but distinct from that of the apo state. In the structure of the RNA–RppH complex, the R27 side chain in RNA-bound RppH forms a cation–π interaction with the base plane of an RNA guanine, suggesting an important role in substrate recognition (26). Notably, R27 is highly flexible prior to RNA binding, as the electron density of the residue is obscure in the diffracted structure of apo RppH but becomes clear upon substrate binding. The S32 and Q34 side chains on RppH form two water-aided hydrogen bonds with an RNA guanine base. Three bulky residues, Y77, V137 and F139 of RppH, constitute a hydrophobic pocket to accommodate the second RNA base, buttressed by a hydrogen bond formed between the hydroxyl group of Y77 and the phosphate group of the RNA. Upon binding of DapF, these residues all dramatically change toward an orientation that resembles the RNA-bound state (Figure 6C). In contrast, the side chain of catalytic residue (like E53) in the Nudix motif exhibits a similar orientation in various apo/bound forms (Supplementary Figure S4).
To experimentally assess the effect of DapF on the binding of substrate and RppH, we performed ITC to measure the Kd value of RNA and RppH, in the presence or absence of DapF. Without Mg2+, RNA was not hydrolyzed by RppH but might well remain its binding with the enzyme. We successfully measured the Kd value of RNA and DapF–RppH complex binding to be approximately 120 μM (Supplementary Figure S5A). RppH alone appeared to bind with RNA but the enthalpy changes (ΔH) of the reaction is too promiscuous to be fitted so as to obtain a Kd, which prevented us from directly assessing the effect of DapF. In contrast, DapF seemed to have no binding with the substrate (Supplementary Figure S5A). In sample preparation, we observed that high concentration of RNA was prone to trigger the aggregation of RppH and the phenomenon was alleviated by the addition of DapF, suggesting that RppH is less stable than DapF–RppH complex. Then, we measured the Michaelis constant (Km) of RppH for RNA hydrolysis to assess their substrate binding ability in the absence or presence of DapF. The Km value of RppH is ∼163 μM and decreased to 114 μM upon DapF binding (Supplementary Figure S5B). These results suggest that DapF association moderately increases the binding affinity of RppH with the substrate RNA. Taken together, these structural and biochemical analysis suggest that DapF stimulates RppH’s activity by stabilizing it in a conformation that favors RNA binding.
Little effect of RppH on activity of DapF
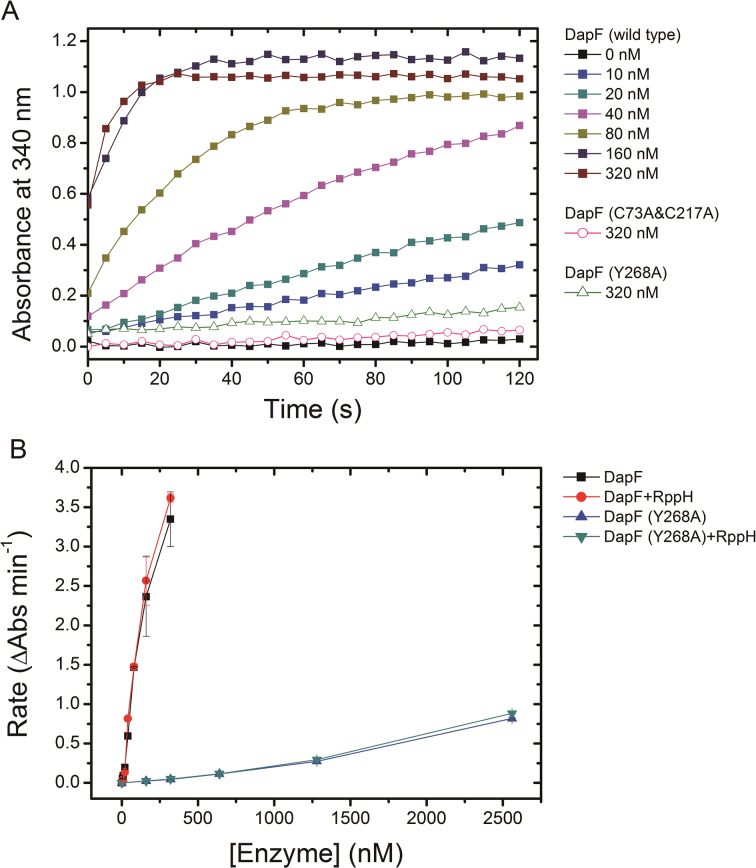
To assess the effect of RppH on epimerase activity of DapF, we tested the enzymatic property of DapF in the presence or absence of RppH. DapF alone has already exhibited distinct activity when its concentration reached 20 nM (Figure 7A). Raising the final concentration of DapF in the reaction (ranging from 0 nM to 320 nM) resulted in elevated velocity converting from L,L-DAP to meso-DAP. In contrast, neither the active site mutant DapF (C73A&C217A) nor the monomeric mutant DapF (Y268A) showed detectable activity even in a high concentration (320 nM) (Figure 7A). These results are consistent with previous report and highlight the importance of DapF dimerization for its activity (36). When RppH was incubated with the wild type DapF (final concentration ranging from 10 to 320 nM), nearly identical velocity of meso-DAP generation was observed (Figure 7B). This is also the case for the monomeric mutant DapF (Y268A). The addition of RppH to DapF (Y268A) (final concentration ranging from 160 nM to 2.5 μM) is unable to change the velocity of meso-DAP production either (Figure 7B). These results suggest that RppH has little effect on the activity of DapF in vitro, in despite of DapF dimerize or not. The activity of DapF was assumed to be related with the distance of two catalytic cysteine residues (Cys73 and Cys217) which was affected by conformational flexibility of the interdomain junction (36). The structural comparison of DapF in apo state and in RppH-bound state revealed a moderate conformational change of DapF upon RppH binding (Supplementary Figure S6). Nevertheless, this conformational change of DapF appears to be not dramatic enough to alter its isomerase activity.
Figure 7.
The effect of RppH on epimerase activity of DapF. (A) The enzymatic activity of DapF was determined using the modified DapF-DAP dehydrogenase coupled spectrophotometric assay. The absorbance at 340 nm (A340) was consecutively recorded for 2 min, with a 5s interval. The filled squares represent A340 of sample including a gradient concentration of wild DapF (ranging from 0 to 320 nM). The pink empty circles represent the active site mutant DapF (C73A&C217A) (320 nM). The green empty triangles are the monomeric mutant DapF (Y268A) (320 nM). (B) The initial change rate of A340 of samples is plotted as a function of DapF concentration, in the presence or absence of RppH. Kinetics were performed at the same conditions for wild-type DapF and the mutant DapF (Y268A). Error bars represent the S.D. of duplicate measurements.
DISCUSSION
Recent research has extended the categories of the 5′-terminal structure of RNA from the classical examples of triphosphate and m7G to include diphosphate and modified nucleotides such as NAD, CoA and m7Gm6A(m). In addition, newly emerging enzymes and cofactors involved in regulation of the deprotection suggest that the complexity of this process may be underappreciated (38). The eukaryotic decapping process executed by Dcp2 and its cofactors has been intensively explored, but research related to the analogous process in prokaryotes is rather limited. DapF is the only identified cofactor that stimulates the activity of RppH in E coli. The molecular mechanism remains to be elucidated. Here, we solved the structure of the DapF–RppH complex at 2.3 Å and demonstrated that DapF stabilizes RppH in a conformation that favors RNA binding and thus stimulates its pyrophosphohydrolase activity.
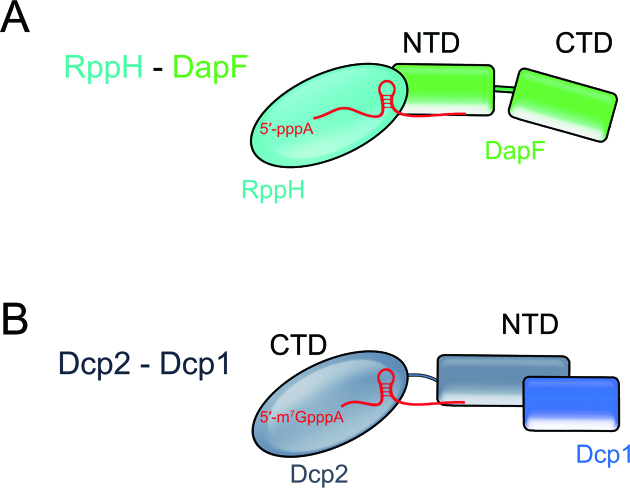
The manner in which RppH is regulated by DapF is reminiscent of that of the eukaryotic decapping enzyme Dcp2. Dcp2 exists in the open or closed state, with the latter closely resembling the active form. Upon Dcp1 binding, Dcp2 can be stabilized in the closed state, resulting in stimulation of the activity of Dcp2 (21). Interestingly, the regulation of both RppH and Dcp2 is consistent with a model in which multiple domains of the enzyme or the cofactor are involved. For example, RppH contains a single Nudix domain. Meanwhile, its cofactor DapF contains two domains, an N-terminal domain (NTD) and a C-terminal domain (CTD). DapF interacts with RppH exclusively via its NTD (Figure 8A). Dcp2 has an NTD for interactions with regulators such as Dcp1 and a CTD, which is responsible for substrate catalysis or recognition (Figure 8B). Importance of other regulators involved in removing the 5′-terminal structure of RNA should also be emphasized. In fact, Dcp1 is far from being sufficient for optimal stimulation of RNA decapping by Dcp2. Other stimulating cofactors (Edc1, Edc3) bind the Dcp1–Dcp2 holoenzyme and significantly change its conformation in closed form (22,23). It may also be the case in the DapF–RppH system, considering that the stimulating effect of DapF on the activity of RppH appears to be less significant (Supplementary Figure S5B). Moreover, RppH has no ‘closed’ conformation or remarkable structural rearrangement upon DapF binding. Thus, whether other regulators involve in the process and trigger notable conformational changes of RppH remains to be further investigated.
Figure 8.
The proposed regulatory model of the RppH-DapF and Dcp2-Dcp1 system. (A) RppH is a solitary domain protein. The cofactor DapF interacts with RppH via its N-terminal domain, leaving the C-terminal domain disengaged. (B) Dcp2 uses its NTD to bind to the regulator Dcp1. The CTD of Dcp2 is responsible for substrate binding and catalysis.
Although RppH is insensitive to the first nucleotide of its substrate RNA, it does have a strong sequence preference for its substrates. In Bacillus subtilis, RppH exclusively catalyzes triphosphorylated RNA bearing guanine in the second nucleotide position, and other positions provide minor contributions to substrate specificity (19). EcRppH has a modest specificity for hydrolysis of triphosphorylated and diphosphorylated RNA, and its efficiency for both of these substrates also depends on the second nucleotide, with a reactivity order of G ≥ A > U ≥ C (4,26,39). An intriguing question is whether DapF binding can alter or affect the substrate specificity of RppH. In the structure of RNA-bound RppH, some key residues directly interact with the second guanine. The side chain of R27 interacts with this guanine via a cation-π interaction, buttressed by a hydrogen bond; the long carbon side chain of K140 is stabilized by the hydrophobic surroundings composed of Y77, V137 and F139, allowing its amino group to form two hydrogen bonds with the guanine; meanwhile, the hydroxyl group of S32 and a water molecule nearby contribute another two hydrogen bonds. In the DapF-bound RppH structure, we also observed that these residues are mostly oriented identical to those in the RNA-bound RppH structure (Figure 6C), implying that the substrate specificity of RppH may not be altered upon binding of DapF. By using a 3 nt triphosphorylated RNA as substrate, we revealed that RppH does not change its preference at the second nucleotide of substrate in the presence of DapF, with similar reactivity order G > A > U ≥ C as that of RppH alone (Supplementary Figure S7). Nevertheless, these results are achieved by using a rather short RNA as substrate which is much different from the authentic RNA existing in cells. The possibility cannot be excluded that DapF may have distinct effects on substrates of different length or secondary structure. The multiple kinds of substrates of RppH in bacteria make the issue complicated, which remains to be systematically explored.
Substrate preference of RppH is also determined by the 5′ phosphate state of RNA, and hydrolysis of diphosphorylated RNA by RppH is ten times faster than that of triphosphorylated RNA (4). Another intriguing issue is whether DapF can selectively stimulate the activity of RppH towards the hydrolysis of triphosphorylated RNA but not diphosphorylated RNA, providing a distinct layer of regulation of RNA decay. Further detailed biochemical assays are required to clarify this possibility.
In this study, we observed that DapF–RppH complex existed as a heterotetramer both in crystal and in solution. Both of dimeric DapF and the monomeric mutant show the similar stimulatory effects on RppH activity. These findings raise a question which is the functional unit of DapF–RppH (ie. a heterodimer or heterotetramer) in cells. In fact, the state of DapF–RppH may depend on the expression level of DapF in vivo. DapF was reported to form a dimer with a dimer-monomer Kd of 22 nM in vitro (36), suggesting DapF is prone to be a dimer. When DapF is highly expressed, dimeric DapF is assumed to be associated with two individual RppH molecules to form a heterotetramer. If the expression of DapF is adjusted to an extremely low level, or in some cases, DapF is mutated to favor monomeric existence, heterodimer of DapF–RppH might be the functional unit. As an enzyme, DapF participates in biosynthesis of lysine and meso-DAP which are important precursors for the synthesis of peptidoglycan and housekeeping proteins. The concentration of DapF in bacteria may be altered in different growth conditions (40). Accordingly, DapF–RppH complex could exist in a dynamic equilibrium of heterotetramer and heterodimer. Considering the determinant role of RppH in RNA degradation, its regulation by DapF should be vital for bacteria growth. Heterotetramer of DapF–RppH complex is equally active as the heterodimer state could serve as a protective mechanism acquired in long term evolution to ensure the continuous regulation of RppH by DapF.
DATA AVAILABILITY
Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 5YGU.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff of the BL17U1/BL19U1 beamline of the National Center for Protein Sciences Shanghai (NCPSS) at the Shanghai Synchrotron Radiation Facility for assistance during data collection and research associates at the Center for Protein Research, Huazhong Agricultural University, for technical support.
Author Contributions: Q.W., D.Z., Z.G., T.Z. and P.Y. designed all experiments. Q.W., D.Z., J.L. and K.P. performed protein expression, purification and crystallization. Z.G. determined all of the structures. Q.W., D.Z., D.L. and J.L. carried out biochemical assays. Q.W., D.Z. and P.Y. wrote the manuscript. All authors discussed the results and commented on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology [2015CB910900]; National Natural Science Foundation of China [31722017 to P.Y., 31770878 for D.Z.]; Fok Ying-Tong Education Foundation [151021]; Fundamental Research Funds for the Central Universities [2662014JQ001, 2662016BC018, 2662017PY031]; Huazhong Agricultural University Scientific & Technological Self-innovation Foundation [2662013RC013]. Funding for open access charge: Huazhong Agricultural University Scientific & Technological Self-innovation Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Condon C. Maturation and degradation of RNA in bacteria. Curr. Opin. Microbiol. 2007; 10:271–278. [DOI] [PubMed] [Google Scholar]
- 2. Furuichi Y., Morgan M., Muthukrishnan S., Shatkin A.J.. Reovirus messenger RNA contains a methylated, blocked 5′-terminal structure: m-7G(5′)ppp(5′)G-MpCp. PNAS. 1975; 72:362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Deana A., Celesnik H., Belasco J.G.. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008; 451:355–358. [DOI] [PubMed] [Google Scholar]
- 4. Luciano D.J., Vasilyev N., Richards J., Serganov A., Belasco J.G.. A novel RNA phosphorylation state enables 5′ End-Dependent degradation in escherichia coli. Mol. Cell. 2017; 67:44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen Y.G., Kowtoniuk W.E., Agarwal I., Shen Y., Liu D.R.. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol. 2009; 5:879–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Walters R.W., Matheny T., Mizoue L.S., Rao B.S., Muhlrad D., Parker R.. Identification of NAD+ capped mRNAs in Saccharomyces cerevisiae. PNAS. 2017; 114:480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiao X., Doamekpor S.K., Bird J.G., Nickels B.E., Tong L., Hart R.P., Kiledjian M.. 5′ end nicotinamide adenine dinucleotide cap in human cells promotes RNA decay through DXO-Mediated deNADding. Cell. 2017; 168:1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kowtoniuk W.E., Shen Y., Heemstra J.M., Agarwal I., Liu D.R.. A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA. PNAS. 2009; 106:7768–7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jaschke A., Hofer K., Nubel G., Frindert J.. Cap-like structures in bacterial RNA and epitranscriptomic modification. Curr. Opin. Microbiol. 2016; 30:44–49. [DOI] [PubMed] [Google Scholar]
- 10. Mackie G.A. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998; 395:720–723. [DOI] [PubMed] [Google Scholar]
- 11. Luciano D.J., Belasco J.G.. NAD in RNA: unconventional headgear. Trends Biochem. Sci. 2015; 40:245–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 2002; 22:8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Z., Jiao X., Carr-Schmid A., Kiledjian M.. The hDcp2 protein is a mammalian mRNA decapping enzyme. PNAS. 2002; 99:12663–12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Dijk E., Cougot N., Meyer S., Babajko S., Wahle E., Seraphin B.. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002; 21:6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cahova H., Winz M.L., Hofer K., Nubel G., Jaschke A.. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2015; 519:374–377. [DOI] [PubMed] [Google Scholar]
- 16. Bessman M.J., Frick D.N., O’Handley S.F.. The MutT proteins or ‘Nudix’ hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 1996; 271:25059–25062. [DOI] [PubMed] [Google Scholar]
- 17. McLennan A.G. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 2006; 63:123–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Messing S.A., Gabelli S.B., Liu Q., Celesnik H., Belasco J.G., Pineiro S.A., Amzel L.M.. Structure and biological function of the RNA pyrophosphohydrolase BdRppH from Bdellovibrio bacteriovorus. Structure. 2009; 17:472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsieh P.K., Richards J., Liu Q., Belasco J.G.. Specificity of RppH-dependent RNA degradation in Bacillus subtilis. PNAS. 2013; 110:8864–8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bischler T., Hsieh P.K., Resch M., Liu Q., Tan H.S., Foley P.L., Hartleib A., Sharma C.M., Belasco J.G.. Identification of the RNA Pyrophosphohydrolase RppH of Helicobacter pylori and Global Analysis of Its RNA Targets. J. Biol. Chem. 2017; 292:1934–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. She M., Decker C.J., Svergun D.I., Round A., Chen N., Muhlrad D., Parker R., Song H.. Structural basis of dcp2 recognition and activation by dcp1. Mol. Cell. 2008; 29:337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Valkov E., Muthukumar S., Chang C.T., Jonas S., Weichenrieder O., Izaurralde E.. Structure of the Dcp2-Dcp1 mRNA-decapping complex in the activated conformation. Nat. Struct. Mol. Biol. 2016; 23:574–579. [DOI] [PubMed] [Google Scholar]
- 23. Charenton C., Taverniti V., Gaudon-Plesse C., Back R., Seraphin B., Graille M.. Structure of the active form of Dcp1–Dcp2 decapping enzyme bound to m7GDP and its Edc3 activator. Nat. Struct. Mol. Biol. 2016; 23:982–986. [DOI] [PubMed] [Google Scholar]
- 24. Mugridge J.S., Ziemniak M., Jemielity J., Gross J.D.. Structural basis of mRNA-cap recognition by Dcp1–Dcp2. Nat. Struct. Mol. Biol. 2016; 23:987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee C.R., Kim M., Park Y.H., Kim Y.R., Seok Y.J.. RppH-dependent pyrophosphohydrolysis of mRNAs is regulated by direct interaction with DapF in Escherichia coli. Nucleic Acids Res. 2014; 42:12746–12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vasilyev N., Serganov A.. Structures of RNA complexes with the Escherichia coli RNA pyrophosphohydrolase RppH unveil the basis for specific 5′-end-dependent mRNA decay. J. Biol. Chem. 2015; 290:9487–9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Q.S., Yu F., Huang S., Sun B., Zhang K.H., Liu K., Wang Z.J., Xu C.Y., Wang S.S., Yang L.F. et al. . The macromolecular crystallography beamline of SSRF. Nucl. Sci. Tech. 2015; 26:12–17. [Google Scholar]
- 28. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 29. Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 1994; 50:760–763. [DOI] [PubMed] [Google Scholar]
- 30. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C.. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 32. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 33. Zhang D., Liu Y., Wang Q., Guan Z., Wang J., Liu J., Zou T., Yin P.. Structural basis of prokaryotic NAD-RNA decapping by NudC. Cell Res. 2016; 26:1062–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C.. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987; 15:8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Turnbull W.B., Daranas A.H.. On the value of c: can low affinity systems be studied by isothermal titration calorimetry. J. Am. Chem. Soc. 2003; 125:14859–14866. [DOI] [PubMed] [Google Scholar]
- 36. Hor L., Dobson R.C., Downton M.T., Wagner J., Hutton C.A., Perugini M.A.. Dimerization of bacterial diaminopimelate epimerase is essential for catalysis. J. Biol. Chem. 2013; 288:9238–9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wiseman J.S., Nichols J.S.. Purification and properties of diaminopimelic acid epimerase from Escherichia coli. J. Biol. Chem. 1984; 259:8907–8914. [PubMed] [Google Scholar]
- 38. Song M.G., Bail S., Kiledjian M.. Multiple Nudix family proteins possess mRNA decapping activity. RNA. 2013; 19:390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Foley P.L., Hsieh P.K., Luciano D.J., Belasco J.G.. Specificity and evolutionary conservation of the Escherichia coli RNA pyrophosphohydrolase RppH. J. Biol. Chem. 2015; 290:9478–9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mengin-Lecreulx D., Michaud C., Richaud C., Blanot D., van Heijenoort J.. Incorporation of LL-diaminopimelic acid into peptidoglycan of Escherichia coli mutants lacking diaminopimelate epimerase encoded by dapF. J. Bacteriol. 1988; 170:2031–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 5YGU.