Abstract
While bacteria and eukaryotes show distinct mechanisms of DNA damage response (DDR) regulation, investigation of ultraviolet (UV)-responsive expression in a few archaea did not yield any conclusive evidence for an archaeal DDR regulatory network. Nevertheless, expression of Orc1-2, an ortholog of the archaeal origin recognition complex 1/cell division control protein 6 (Orc1/Cdc6) superfamily proteins was strongly activated in Sulfolobus solfataricus and Sulfolobus acidocaldarius upon UV irradiation. Here, a series of experiments were conducted to investigate the possible functions of Orc1-2 in DNA damage repair in Sulfolobus islandicus. Study of DDR in Δorc1-2 revealed that Orc1-2 deficiency abolishes DNA damage-induced differential expression of a large number of genes and the mutant showed hypersensitivity to DNA damage treatment. Reporter gene and DNase I footprinting assays demonstrated that Orc1-2 interacts with a conserved hexanucleotide motif present in several DDR gene promoters and regulates their expression. Manipulation of orc1-2 expression by promoter substitution in this archaeon revealed that a high level of orc1-2 expression is essential but not sufficient to trigger DDR. Together, these results have placed Orc1-2 in the heart of the archaeal DDR regulation, and the resulting Orc1-2-centered regulatory circuit represents the first DDR network identified in Archaea, the third domain of life.
INTRODUCTION
Organisms of all three domains of life have to deal with lesions on their chromosomal DNAs generated by environmental and endogenous factors. If left unrepaired, these DNA lesions will either alter the content of the genetic blueprint, giving rise to mutations, or leading to the loss of genome integrity and cell death. DNA damage repair has been studied in great detail in the organisms belonging to the domains of Bacteria and Eukarya, in which distinctive regulatory networks called DNA damage response (DDR) have been revealed (1,2). The bacterial DDR is best represented by the SOS response, which is triggered by ssDNA-RecA filaments, an intermediate of homologous recombination repair (HRR) of double stranded DNA breaks (DSBs). Investigation of the SOS regulation in Escherichia coli has revealed a LexA-dependent regulatory network that is conserved in numerous bacteria (3). Nevertheless, several LexA-independent mechanisms have also been discovered in different bacteria, revealing the diversification of the bacterial DDR regulatory mechanisms (4). DDR regulation in eukaryotes is far more complex than those found in bacteria, involving multiple genome surveillance mechanisms. Two best-known examples are the ataxia telangiectasia-mutated (ATM) and the ATM and Rad3-related (ATR) signal transduction pathways that control, for example, cell cycle checkpoint regulation (5,6). Nevertheless, all these bacterial and eukaryotic DDR mechanisms share some common features; after recognition of DNA damage signal, a series of cellular events occur in a coordinated fashion, including inhibition of DNA replication, cell cycle arrest and activation of the synthesis of various DNA repair enzymes, and such a regulation ensures efficient DNA damage repair to occur in a timely fashion in the cell.
Investigation of UV-responsive genome expression in a few model archaea including Halobacterium NRC-1 (7,8), Sulfolobus solfataricus P2 (9–11) and Sulfolobus acidocaldarius DSM639 (10) has revealed that a number of genes are differentially expressed. Nevertheless, uncertainty about whether these observations reflect the presence of an archaeal DDR regulation persists primarily because of two reasons: (a) The differentially expressed genes (DEGs) exclude those coding for the enzymes responsible for nucleotide excision repair (NER), which are strongly activated by DNA damage in bacteria and eukaryotes (12,13), and (b) archaea do not code for any homologs of bacterial or eukaryotic DDR regulators. As a result, it remains as an open question whether organisms of the archaeal domain possess any DDR regulation, and if so, how the process is controlled.
In this work, we employed S. islandicus, a genetic model in the Crenarchaeota (14) to investigate the DNA damage-responsive genome expression and its regulation. Previous genetic analysis of three orc1 genes in S. islandicus showed that orc1-1 and orc1-3 code for replication initiators responsible for replication initiation from the oriC1 and oriC2 of the chromosome but deletion of orc1-2 gene does not impair the origin usage in this archaeon (15). The strong up regulation of orc1-2 in S. solfataricus and S. acidocaldarius (upon UV irradiation (9,10)) and in S. islandicus (by treatment of 4-nitroquinoline 1-oxide (NQO) and UV light (16)) prompted us to investigate possible functions of Orc1-2 in DNA damage response in this crenarchaeon. We found that Orc1-2 has gained a novel function during evolution, i.e. the factor functions as a global regulator in the DNA damage response network of S. islandicus.
MATERIALS AND METHODS
Growth conditions, transformation and NQO treatment of Sulfolobus
S. islandicus strains used in this work are listed in Supplementary Table S1. Sulfolobus cells were cultured at 78°C in SCV (basic salts plus 0.2% sucrose, 0.2% casamino acids, 1% vitamin solution) medium (17) or in ACV medium (in which sucrose is replaced with d-arabinose). If required, uracil was added to 20 μg/ml. Plasmids were introduced into S. islandicus strains by electroporation as described originally for S. solfataricus (18).
NQO treatment experiments were conducted as previously described (16). A stock of the drug (130 mM) was prepared with DMSO and kept in –20 °C. By the time of NQO experiment, the stock solution was diluted to 1.3 mM with H2O. Then, the diluted NQO solution was added to Sulfolobus cultures of an early exponential growth phase (i.e. absorbance at 600 nm (A600) = ca. 0.2) to the concentrations specified in each experiment. Cell samples were taken during incubation and used for A600 measurement, cell aggregation assay, cell viability assay, RNA extraction, western blot analysis and flow cytometry individually.
Cell viability of Sulfolobus cultures was estimated by determination of their colony formation units (CFU). Cells were collected from 1 ml of culture by centrifugation for each cell sample and re-suspended in the equal volume of pre-warmed SCV medium. Each cell resuspension was diluted in series of dilutions and plated for colony formation using the two-layer plating method previously described (19). Colonies of Sulfolobus cells appeared on plates after 7 days of incubation.
Transcriptome analysis by RNA sequencing and verification of the data by RT-qPCR
Exponentially growing cultures of S. islandicus E233S1 (the wild-type strain, WT,) and Δorc1-2 were diluted to an A600 of ca. 0.2 and grown in the presence or absence of 2 μM NQO for 6 h. Cell mass was then collected by centrifugation and employed for extraction of total RNAs using the Trizol reagent (Ambion, Austin, TX, USA). The quality and quantity of the total RNA preparations were evaluated using NanoDrop 1000 spectrophotometer (Labtech, Wilmington, MA, USA) and 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA sequencing (RNA-Seq) was conducted in Novogene (Beijing, China). About 3 μg of high quality RNA were used for construction of RNA-Seq libraries, which were then subjected to next generation sequencing using an Illumina HiSeq2000. This gave 4.1–4.8 million high quality sequence reads for each RNA sample, which were then mapped to the genome of S. islandicus Rey15A (20). The resulting data were then analyzed by Fragments Per Kilobase of transcript sequence per Million base pairs sequenced (FPKM) analysis (21) to reveal expression levels of all genes in the S. islandicus genome. Differential genome expression analysis (NQO-treated samples versus the corresponding untreated references) was performed using the DEGSeq R package (22). Corrected P-value of 0.005 and log2 (Fold change) of 1 were set as the threshold for significant difference in differential gene expression.
Quantitative reverse transcription PCR (RT-qPCR) was employed to validate the RNA-Seq data. DDR genes chosen for verification included dpo2, upsX, upsA, tfb3, SiRe_1957 and SiRe_1550. First-strand cDNAs were synthesized with total RNA samples using a reverse transcriptase (Thermo-Scientific, Waltham, MA, USA) and random hexamer primers. The resulting cDNA samples were used for estimation of mRNA levels of the above DDR genes by qPCR, using the Maxima SYBR Green/ ROX qPCR Master Mix (Thermo Scientific, Waltham, MA, USA) and gene-specific primers (Supplementary Table S2). PCR was performed in a CFX96 Touch™ real-time PCR detection system (Bio-Rad, Hercules, CA, USA) with the following steps: denaturing at 95°C for 5 min, 40 cycles of 95°C 15 s, 55°C 15 s and 72°C 20 s. Relative amounts of mRNAs were estimated by using the comparative Ct method with 16S rRNA as the reference (23). A correlation between the two sets of data was found to be 0.9705 with an R-value of 0.96 (Supplementary Figure S1).
Cell aggregation assay and flow cytometry
The extent of cell agggregation in S. islandicus cultures was estimated by direct observation of cell aggregates in fresh cultures under a Nikon Eclipse Ti-E inverted microscope (Nikon, Kobe, Japen). Data were collected from at least 12 fields of view images and 500 single cells for each cell sample, and the same analysis was conducted for three independent growth experiments.
Flow cytometry of S. islandicus cells was conducted as previously described (24). Briefly, the archaeal cells were fixed with ice-cold ethanol, stained with 40 μg/ml ethidium bromide (Sigma-Aldrich, St. Louis, MO, USA) and 100 μg/ml mithramycin A (Apollo Chemical, Tamworth, UK) and analyzed in an Apogee A40 cytometer (Apogeeflow, Hertfordshire, UK) equipped with a 405 nm laser. A dataset of at least 60 000 cells was collected for each cell sample.
Construction of S. islandicus orc1-2araS mutant using a CRISPR-based genome-editing method
The genome-editing plasmid pGE-orc1-2araS was constructed by following the strategy previously described (25). A target site was selected on the orc1-2 gene, consisting of a 5′-CCN-3′ (positioned at –28 referring to the start codon of orc1-2), a type I-A protospacer adjacent motif and the immediately downstream 40-nt DNA sequence (protospacer). Two DNA oligonucleotides were then designed based on the protospacer, giving orc1-2araS Spacer F and orc1-2araS Spacer R (Supplementary Table S2). Annealing of the two oligonucleotides yielded a DNA fragment containing the designed spacer. The resulting DNA fragment also contained the 5′-flanking 4 nt protruding ends that are compatible to the ends of the SapI-digested pSe-Rp vector. Therefore, ligation of the spacer fragment and the digested vector yielded the mini-CRISPR plasmid pAC-orc1-2.
The donor DNA fragment was generated by splicing and overlapping extension (SOE)-PCR with Fast Pfu DNA polymerase (TransGene, Beijing, China), following the published procedure (26). First, orc1-2 gene was amplified by PCR from the genome DNA using the primer pair of orc1-2fwd-NcoI/orc1-2rev-XmaI. Insertion of the PCR fragment into pZC1-S-50-orc1-2 yielded pSe-araS-orc1-2 carrying the araS-orc1-2 fusion gene. Then, a 654-bp genomic DNA fragment was obtained from the archaeal genome by PCR with the primer pair of orc1-2araS SOEfwd-SphI/orc1-2araS SOErev whereas the fusion gene was amplified by PCR from pSe-araS-orc1-2 with the primer pair of orc1-2araS SOEfwd/orc1-2araS rev-XhoI. Finally, the two PCR fragments were joined together by PCR using the primer pair of orc1-2araS fwd-SphI/orc1-2araS rev-XhoI. The resulting DNA fragment was digested with SphI and XhoI and inserted into pAC-orc1-2 at the same sites, giving pGE-orc1-2araS. The identity of the plasmid was confirmed by sequence determination of the DNA inserts by DNA sequencing.
pGE-orc1-2araS was then introduced into S. islandicus E233S1 by electroporation, giving transformants that were subjected to pyrEF counter selection to cure the pGE plasmid as previously described (19). The genotype of single colonies obtained on 5-FOA-containing plates were determined by PCR amplification of the insert with orc1-2araS check-fwd/orc1-2araS check-rev primers and subsequent sequencing of the PCR products. One of the verified mutants was designated as S. islandicus orc1-2araS and used in subsequent experiments.
Construction of reporter gene plasmids and report gene assay of DDR gene promoters
Reporter plasmids were constructed using the Sulfolobus-E. coli shuttle vector pSeSD (27) with the S. solfataricus β-glycosidase gene (lacS) (28) as the reporter gene. Four highly up-regulated genes (SiRe_1881: upsA, SiRe_1879: upsE, SiRe_1717: tfb3 and SiRe_1316: cedA1) were selected for the experiment. Promoter fragments of these genes were amplified by PCR with Fast Pfu DNA polymerase (TransGene, Beijing, China) from the genomic DNA of S. islandicus REY15A (20), using the following primer pairs individually, upsA-fwd-SphI/upsA-rev-NdeI, upsE-fwd-SphI/upsE-rev-NdeI, tfb3-fwd-SphI/tfb3-rev-NdeI or cedA1-fwd-SphI/cedA1-rev-NdeI (Supplementary Table S2). The resulting PCR products (ca. 220 bp) were purified with the GeneJET PCR Purification kit (Thermo Scientific, Waltham, MA, USA), and the purified DNAs were digested with SphI and NdeI and purified again. Each purified promoter fragment was then inserted into pSe-lacS (27), yielding pSe-upsA-LacS, pSe-upsE-LacS, pSe-tfb3-LacS and pSe-cedA1-LacS reporter gene plasmids containing the promoters of the wild-type DNA damage responsive element (DDRE) (Supplementary Table S1). Promoter derivatives containing the mutated DDRE (DDREmut) motifs were constructed using the SOE-PCR procedure (29). First, two overlapping primers were designed containing desired mutations in the center of each pair of oligonucleotides (Supplementary Table S2), including upsA-SOE-top/upsA-SOE-bm, upsE-SOE-top/upsE-SOE-bm, tfb3-SOE-top/tfb3-SOE-bm and cedA1-SOE-top/cedA1-SOE-bm. These SOE primers and their corresponding promoter-cloning primers were then employed for SOE-PCR with the corresponding reporter gene plasmid as template. The resulting DDREmut promoter fragments were then inserted into pSe-lacS (27), giving four reporter gene plasmids with DDREmut promoters, i.e. pSe-upsAmut-LacS, pSe-upsEmut-LacS, pSe-tfb3mut-LacS and pSe-cedA1mut-LacS (Supplementary Table S1). Determination of the sequences of the plasmid-borne wild-type and mutated promoters of the four DDR genes by DNA sequencing confirmed the identity of these reporter gene plasmids.
These reporter gene plasmids were then introduced into S. islandicus E233S1 (WT) and Δorc1-2 individually by electroporation. Three colonies of transformants were chosen from each transformation and grown for 6 h in SCV either in the presence or absence of 2 μM NQO. Cell mass was then collected from which cell extracts were prepared by sonication. β-Glycosidase activity in the cell extracts of different S. islandicus strains was determined using the ONPG (ρ-nitrophenyl-b-d-galactopyranoside) method as described previously (30).
Western blotting and hybridization analysis
Cells in 15 ml culture were collected by centrifugation. Cell pellets were re-suspended in 1 ml 10 mM Tris–HCl buffer (pH 8.0). The resulting cell suspensions were sonicated to disrupt Sulfolobus cells. Cell debris was removed by centrifugation 13000 rpm at 4°C for 30 min, yielding cellular extracts for further analysis. Protein concentrations of the samples were determined using Coomassie Protein Assay Kit (Thermo Scientific, Waltham, MA, USA). Similar amounts of protein (ca. 10 μg) were taken from the prepared cell extracts and loaded on a 12% polyacrylamide gel for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Then, fractionated proteins were transferred from the polyacrylamide gel onto a nitrocellulose blotting membrane (GE Healthcare, Waukesha, WI, USA), using the Semi-Dry Electrophoretic Transfer Cell system (Bio-Rad, Hercules, CA, USA) and used for immunoblotting. Briefly, the membrane was incubated in 5% skim milk blocking agent for 1 h, and then incubated with individual primary rabbit antibodies (against Orc1-2 or PCNA3 proteins) and finally with the horseradish peroxidase-labeled goat anti-rabbit antibody (Beyotime, Beijing, China) as described previously (31). Protein bands were visualized using the ECL western blot substrate (Thermo Scientific, Waltham, MA, USA) and recorded by exposure to an X-ray film.
Sequence analysis of promoters of highly activated DDR genes
Promoter sequences (100 bp preceding the start codon) of 21 Orc1-2-depedent NQO-responsive genes (over 16 folds change after NQO treatment, Supplementary Table S6) were individually retrieved from the genome sequence of S. islandicus REY15A (20). These sequences were then used for de novo motif discovery by using MEME (Multiple EM for Motif Elicitation) with the default setting to identify conserved motifs as previously reported (32).
DNase I footprinting assay
Orc1-2 protein was expressed in E. coli Rosetta cells carrying pET-orc1-2. The E. coli strain was cultured in an LB medium containing 30 μg/ml kanamycin at 37°C until A600 = 0.6. Orc1-2 protein synthesis was induced by adding 0.5 mM IPTG, and the induction was for 12 h at 16°C. Cell mass was harvested by centrifugation and re-suspended in the lysis buffer (50 mM phosphate saline buffer, 500 mM NaCl, 20 mM imidazole). Cells were disrupted using a French press at 4°C. After removing cell debris by centrifugation, the supernatant was loaded onto a 1 ml Ni-NTA column (GE Healthcare, Chicago, Illinois, USA). His-tagged recombinant Orc1-2 protein was purified by following the manufacturer's instruction.
DNase I footprinting assay was performed as described previously (33). The upsE and tfb3 original promoter PupsE-DDRE and Ptfb3-DDRE and the mutated promoter PupsE-DDREmut and Ptfb3-DDREmut were amplified individually from the corresponding reporter gene plasmids (Supplementary Table S1) using primers listed in Supplementary Table S2. The resulting PCR products were then cloned into pCR™4-TOPO® (Thermo Scientific, Waltham, MA, USA), yielding plasmids that were used as template for PCR with an M13fwd primer carrying a 5′-end FAM labeling and an M13rev primer carrying a HEX 5′-end labeling. The resulting fluorescence-labeled DNAs were used as DNA probes for DNase I footprinting analysis. The coding strand of the upsE promoter region was labeled by FAM (peaks above the promoter sequences) and the non-coding strand was labeled by HEX (peaks below the promoter sequence). The two DNA strands of the tfb3 promoter region were labeled in the opposite combination: HEX for the coding strand and FAM for the non-coding strand. To ensure efficient binding, 400 ng of labeled DNA and 13.7 μg of Orc1-2 protein were mixed and incubated at 40°C for 20 min in 50 μl binding buffer [20 mM HEPES (pH 7.6), 10 mM (NH4)2SO4, 1 mM DTT, 0.2% Tween-20, 30 mM KCl]. Then, 0.02 U of RNase-free DNase I (Thermo Scientific, Waltham, MA, USA) was added and incubated at 37°C for 2 min to degrade unprotected DNA. Finally, 1/10 (v/v) of 50 mM EDTA was added to each reaction and incubated at 65°C for 10 min to stop the reaction. DNAs in the samples were extracted using GeneJET PCR Purification kit (Thermo Scientific, Waltham, MA, USA) and sent for fragment length analysis by capillary electrophoresis in Eurofins Genomics company (Ebersberg, Germany). Electropherograms were aligned using GeneMapper v2.6.3 (Applied Biosystems).
RESULTS
S. islandicus Δorc1-2 showed hypersensitivity to NQO treatment
First, S. islandicus Δorc1-2 strain constructed previously (15) was investigated for its sensitivity to NQO, a drug that forms stable bulky quinolone adducts on bases of DNA in bacterial and eukaryotic cells, leading to the formation of DSBs (34,35), and it induces programmed cell death in S. islandicus (16). Both Δorc1-2 and its corresponding WT were grown in SCV media containing different concentrations of NQO. Growth of these cultures was monitored by measuring their A600 values. We found that 2.5 μM NQO completely inhibited the growth of the mutant while it required 4 μM NQO to stop the growth of the WT strain (Supplementary Figure S2), indicating that Δorc1-2 is hypersensitive to this drug, in reference to WT.
The sensitivity of WT and Δorc1-2 to NQO was also evaluated by determination of their survival rate after drug treatment. Both strains were again grown in SCV in the presence of different concentrations of the drug for 6 h (hours post treatment, hpt). The number of viable cells in all cultures was then estimated by determination of their colony formation units (CFU), with the results summarized in Table 1. Two features are evident in these data: (a) the number of viable cells in NQO-treated cultures exhibited a strong reverse correlation to the drug concentration for both Δorc1-2 mutant and the WT strain, and (b) the ratio of cell viability between WT and Δorc1-2 changed from 3.8 to 30 folds as the NQO content increased from 1 to 3 μM in the medium. These results confirmed the hypersensitivity of Δorc1-2 to NQO treatment.
Table 1.
Survival rates of S. islandicus wild-type strain (E233S1) and Δorc1-2 after NQO treatment
| Viable cells in % after NQO treatmenta | |||
|---|---|---|---|
| Doses of NQO (μmol/l) | E233S1 | Δorc1-2 | Ratio |
| 0 | 100 | 100 | 1.0 |
| 1 | 42.66 | 11.33 | 3.8 |
| 2 | 11.47 | 1.92 | 6.0 |
| 2.5 | 3.29 | 0.23 | 14.3 |
| 3 | 1.23 | 0.04 | 30.8 |
| 4 | 0.04 | 0.002 | 20.0 |
aExponentially growing cultures were treated with different doses of NQO for 6 h. Cell samples were taken and plated on drug-free SCVU plates for determination of colony formation units (CFU). Survival rates are expressed as % of viable cells in drug-treated cultures relative to those in the corresponding drug-free reference cultures.
Orc1-2 deficiency by gene deletion eliminated the NQO-responsive expression
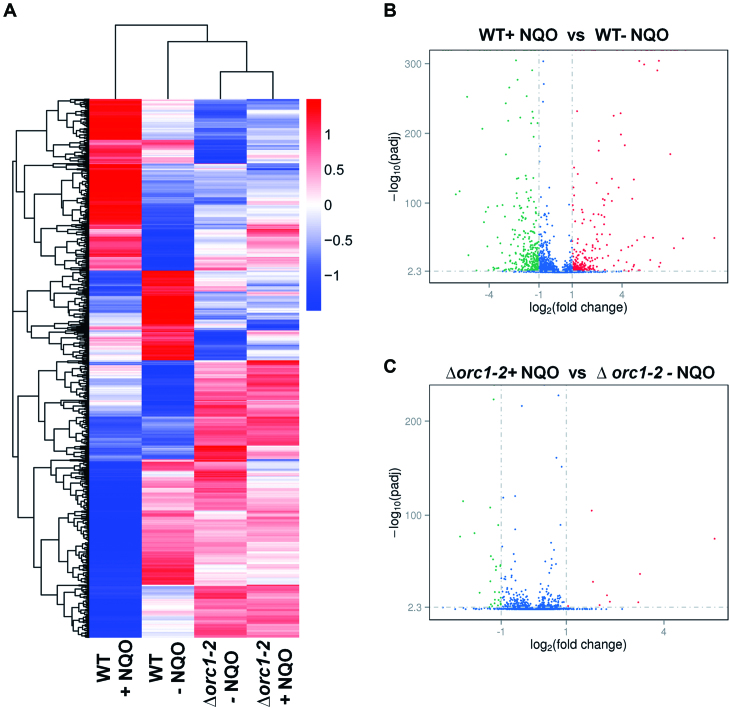
Next, we extracted total RNAs from cell samples of WT and Δorc1-2 cultures (grown in SCV containing 2 μM NQO) at 6 hpt and from the corresponding cell samples of untreated reference cultures. These RNA samples were employed for determination of mRNA abundance of individual genes by RNA-Seq. FPKM plot analysis of the RNA-Seq data revealed a large number of DEGs upon NQO treatment in WT, which did not show NQO-responsive expression in Δorc1-2 (Figure 1A, Supplementary Tables S3, S4 and S5). These include a total of 646 genes among which 290 are up regulated and 356 are down regulated (with a corrected P-value = 0.005, fold change > 2) (Supplementary Table S3, Figure 1B).
Figure 1.
Global transcriptional change mediated by Orc1-2 upon NQO treatment. (A) Heatmap of the genome expression of S. islandicus E233S1 (WT) and Δorc1-2. Both strains were grown in the presence or absence of 2 μM NQO (indicated as + NQO and - NQO, respectively) for 6 h. Cell mass was collected from which total RNAs were prepared and used for RNA-Seq analysis. Genes are clustered with their log10(FPKM+1) values and their expression levels are illustrated with different colors with red colors representing the highest levels of expression whereas blue ones indicating the lowest levels of expression. (B) Volcano plot of differentially expressed genes in the WT strain. (C) Volcano plot of differentially expressed genes in Δorc1-2. X-axis: fold change in gene expression; Y-axis: statistical significance of the fold change. Genes exhibiting >2-fold (i.e. –1 > log2 > +1) up and down regulation are highlighted in red and green, respectively, whereas those that showed a <2-fold change in differential gene expression are shown in blue.
Many up-regulated DEGs could be implicated in DNA damage repair (Supplementary Table S4), including: (a) orc1-2 coding for one of the three Orc1/Cdc6 orthologous proteins in this archaeon (20), (b) genes in the gene operon of UV-inducible pili of Sulfolobus (the Ups system) (36) and genes of the crenarchaeal system for exchange of DNA (the Ced system), the latter of which has recently been shown to be responsible for intercellular DNA transfer in Sulfolobus (37), (c) genes coding for the basal transcriptional factors TFB1 and TFB3 (38,39) and (d) genes coding for proteins involved in homologous recombination repair (40,41).
Several highly repressed DEGs are implicated in different cellular processes including replication initiation (Orc1-1 and Orc1-3) (42–45), genome maintenance and segregation (chromatin protein Sul7d and Cren7 (46) and chromosome segregation proteins SegA and SegB (47)), and cell division (CdvA, ESCRT-III and Vps4 (48,49) as well as several ESCRT-III paralogs (50), Supplementary Table S5). Together, these results suggested that the NQO-induced DNA damage could impose DNA replication inhibition and cell cycle arrest in S. islandicus.
RNA-Seq analysis also revealed that 37 genes showed differential expression in Δorc1-2 (9 up- and 28 down-regulated genes Figure 1C). Among the down regulated genes, 24 showed a <3 folds of repression whereas the remaining four genes had 3- to 5-fold reduction. The relatively low levels of repression suggest that the observed changes may reflect background fluctuation of gene expression in the mutant. The up regulated genes include six genes of 2- to 5-fold activation and three highly activated genes: SiRe_0629 and SiRe_0630 (9–10 folds), SiRe_0655 (47 folds), all of which are of unknown function. To this end, these data indicated that Δorc1-2 is no longer capable of mediating NQO-responsive expression.
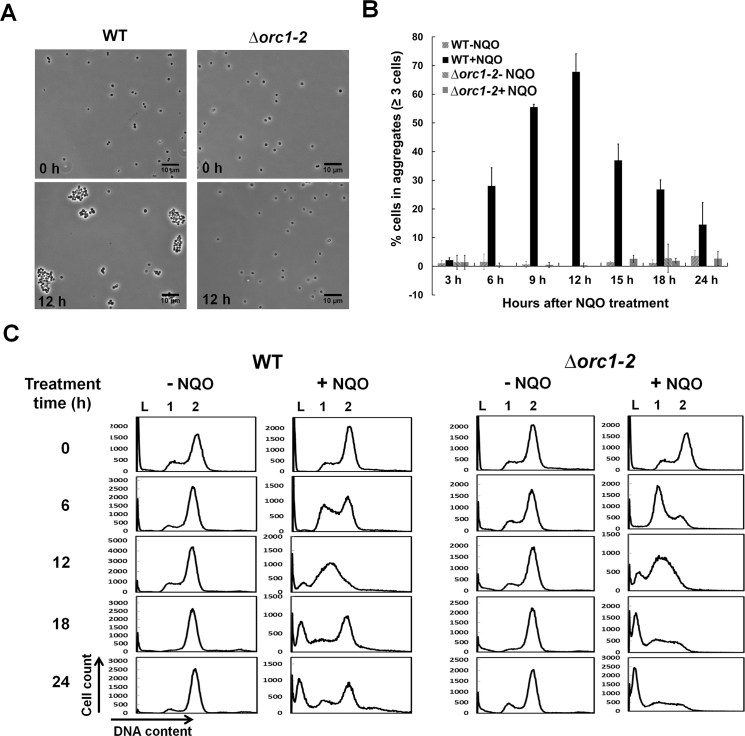
S. islandicus Δorc1-2 lost the capability of cell aggregation and cell cycle regulation
To further study the phenotype of the S. islandicus Δorc1-2, the deletion mutant and WT were grown in the medium containing 2 μM NQO for 24 h. Cell samples were taken during incubation and examined for cell aggregates under microscope. We found that WT formed small cell aggregates of 3–7 cells at 6 hpt, and larger cell aggregates (10–30 cells) appeared at 12 hpt (Figure 2A). In contrast, there was essentially no difference in cell aggregation between the NQO-treated Δorc1-2 cells and their corresponding untreated references since <5% of cells were found in aggregates and that number did not change after drug treatment (Figure 2B). Therefore, these results indicated that Δorc1-2 has lost the capability of cell aggregation.
Figure 2.
Δorc1-2 mutant lost the capability of cell aggregation and cell cycle regulation. (A) Formation of cell aggregates before (0 h) and after treatment with 2 μM NQO (12 h). (B) Quantification of the extent of cell aggregation. (C) Cell cycle profiles of the cultures. DNA contents were divided into 256 arbitrary points on the X-axis, and cell counts (Y-axis) were obtained for each point and used to plot against the DNA content. WT: S. islandicus E233S1; Δorc1-2: orc1-2 deletion mutant derived from the E233S1; DNA-less cells (L); cells containing one chromosome (1), and cells containing two chromosomes (2). Error bars: standard derivations of three independent experiments.
These cell samples were also analyzed by flow cytometry. As shown in Figure 2C, the population of cells with DNA content clustering at 1 chromosome (G1+, comprising of G1 and ‘apparent G1’ cells, the latter of which contain 1 chromosome with fired origins of replication) slightly increased in WT at 6 hpt (<15%), and strikingly, G1+ cells accounted for 75% of the total cell population in the Δorc1-2 cell sample. These results suggested that cell division was inhibited in WT cells, but not in Δorc1-2 cells, consistent with the RNA-Seq data in which the expression of cdvA, cdvB and vps4, which code for the proteins (CdvA, ESCRT, Vsp4) responsible for the ESCRT mode of cell division in Sulfolobus (48,49), was down regulated in NQO-treated WT cells, but their expression was not changed in NQO-treated Δorc1-2 cells (Supplementary Table S5). In addition, the expression of orc1-1, orc1-3, which code for replication initiators responsible for initiation of oriC1 and oriC2 of the S. islandicus chromosome (15), was also inhibited in NQO-treated WT cells but not in NQO-treated Δorc1-2 cells (Supplementary Table S5), and these results suggested that replication initiation could have been inhibited in the WT cells but not in the Δorc1-2 cells. We reasoned that most G1+ cells could be ‘apparent’ G1 cells containing fired origins of replication on their chromosome but the DNA replication could be blocked by NQO-induced DNA lesions on the chromosome in the cells. If so, collapse of stalled replication forks would induce DSBs, leading to cell death. Indeed, continuous incubation of these NQO-treated cultures led to cell death to most Δorc1-2 cells as well as a fraction of NQO-treated WT cells (Figure 2C). To this end, our results indicated that the Orc1-2-depedent DDR regulation is of crucial importance to genome integrity maintenance in this archaeon.
Promoters of DDR genes mediated NQO-responsive expression
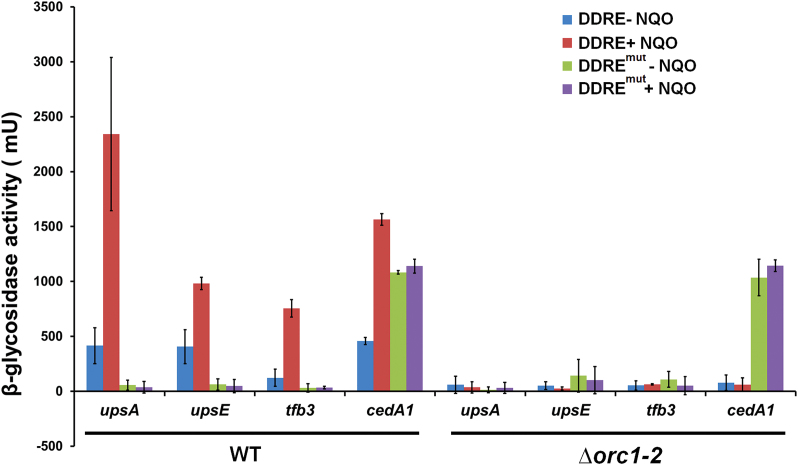
In bacteria and archaea, short DNA segments immediately upstream of genes often contain all promoter elements required for directing the expression of the genes. To test if promoters of DDR genes could also contain DNA motifs required for DDR regulation, promoter fragments of four DDR genes, i.e. upsA, upsE, tfb3 and cedA1, were amplified by PCR using the primers list in Supplementary Table S2, giving a ca. 200-bp DNA fragment (upstream of the start codon) for each promoter. PCR fragments of these promoters were then used to replace the araS-SD promoter in pSeSD (27), yielding reporter gene plasmids for these DDR genes (listed in Supplementary Table S1). All these promoters were found to confer NQO-responsive expression from the reporter gene plasmids in S. islandicus (see below). Then, promoter sequences (100 bp preceding the start codon) of 21 highly up regulated DDR genes (over 18 folds change, Supplementary Table S6) were retrieved from the genome sequence of S. islandicus REY15A (20) and analyzed for conserved sequence motifs using the MEME (Multiple EM for Motif Elicitation) suite (32). A 20 bp consensus (5′-AATAGTTTCRGWDTACTCWS-3′) was identified, containing the DNA motif of 5′-ANTTTC-3′ previously reported for UV-responsive gene promoters of S. acidocaldarius (51). The motif is positioned at –23 to –55 bp upstream of the ATG codon in most identified DDR genes (Supplementary Figure S3).
Next, the hexanucleotide motif (5′-ANTTTC-3′) in four promoters (upsA, upsE, tfb3 and cedA1) was mutated individually by transversion mutation, giving respective mutated promoters. Both the native promoters and their mutated derivatives were analyzed for NQO-responsive expression in WT and Δorc1-2. In WT transformants, mutation of the 5′-ANTTC-3′ motif completely abolished the NQO-responsive expression from each promoter (Figure 3), indicating the motif functions as an NQO-responsive element on these promoters. By contrast, none of the promoters showed the NQO-responsive expression in the Δorc1-2 mutant (Figure 3), suggesting that the 5′-ANTTTC-3′ motif and the Orc1-2 protein could interact with each other to mediate DDR regulation in this archaeon.
Figure 3.
Promoters of NQO-inducible genes dictate the regulation of the NQO-responsive expression. Four highly up regulated genes (upsA, upsE, tfb3 and cedA1) were chosen for the reporter gene assay. S. islandicus strains carrying one of the 8 reporter gene plasmids were grown in SCV in the presence, or absence of 2 μM NQO (denoted as +NQO and –NQO respectively) for 6 h, and cell mass was collected from which cell extracts were prepared and used for determination of β-glycosidase activity. DDRE: reporter gene plasmids of original promoters containing the 5′-ANTTTC-3′ motif; DDREmut: reporter gene plasmids of mutated promoters carrying transversion mutation in the 5′-ANTTTC-3′ motif; WT: the genetic host S. islandicus E233S1; Δorc1-2: S. islandicus orc1-2 deletion mutant. Error bars: standard derivations of three independent experiments.
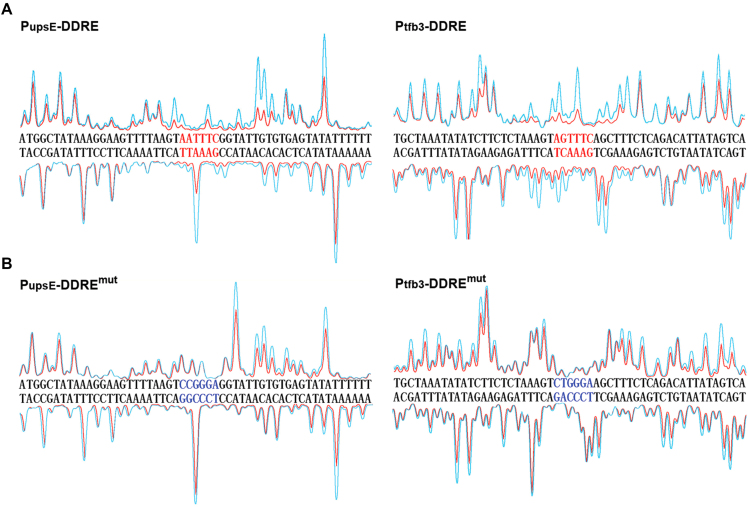
Orc1-2 bound to the conserved motif on DDR gene promoters
To test if Orc1-2 could bind to the 5′-ANTTTC-3′ motif present in the promoter regions of DDR genes, the S. islandicus orc1-2 gene was cloned into pET30a, an E. coli expression vector, giving pET-orc1-2. The expression plasmid was introduced into E. coli for overexpression of Orc1-2 recombinant protein. Highly purified Orc1-2 recombinant protein was obtained (Supplementary Figure S4). DNase I footprinting assay was performed with the original and mutated promoters of upsE and tfb3 genes, in the presence, or absence of Orc1-2 protein, following the procedure described previously (33). We found that Orc1-2 protein protected the sequence of 5′-ANTTTC-3′ motif and its flanking regions on both DNA strands (Figure 4A), and substitution of the 5′-ANTTTC-3′ motif on the mutated promoters completely abolished the protection of the DDRE region by Orc1-2 binding (Figure 4B). These results, together with the report genes assays shown in Figure 3, indicated that Orc1-2 protein binds specifically to 5′-ANTTTC-3′ on these DDR gene promoters and activates their expression.
Figure 4.
Orc1-2 protein bound to a conserved motif on upsE and tfb3 promoters. DNase I footprinting was performed with fluorescence-labeled DNA fragment of PupsE or Ptfb3 promoter (ca. 200 bp) in the presence (red peaks) or absence (cyan peaks) of Orc1-2. DDRE: Promoter fragments containing the 5-ANTTTC-3′ motif (highlighted in red); DDREmut: mutated promoter fragments carrying the transversion mutation of the 5-ANTTTC-3′ motif (highlighted in blue). The promoter sequences shown for PupsE and Ptfb3are positioned from –17 to –69 and from –47 to –99 in reference to their start codons, respectively.
S. islandicus cells stably expressing a low level of Orc1-2 protein exhibited hypersensitivity to NQO treatment
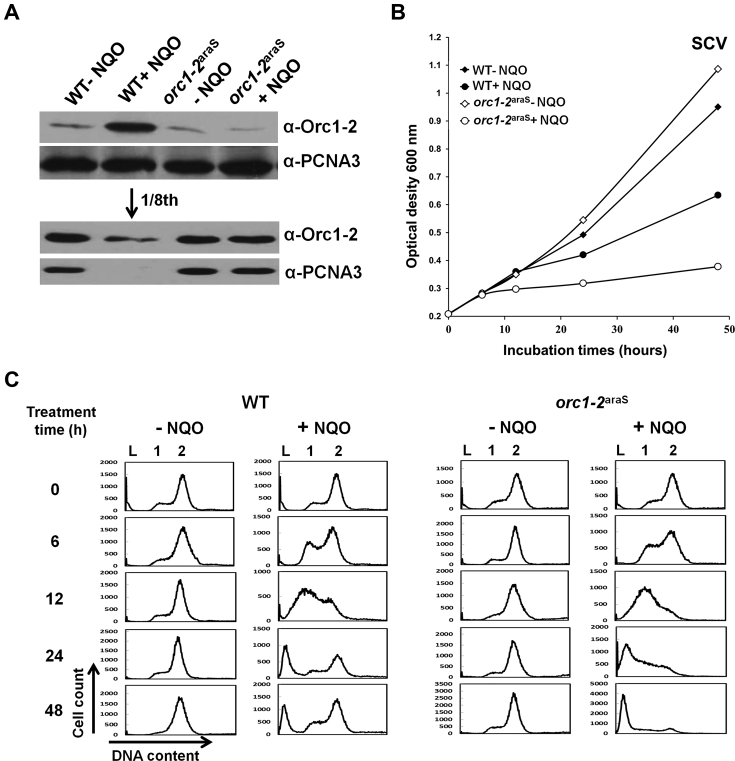
To yield an insight into the DDR regulation by Orc1-2 in this archaeon, we constructed a S. islandicus strain in which the original promoter of the orc1-2 gene was replaced with araS-50, a promoter derivative of the araS gene coding for an arabinose-binding protein. This yielded the promoter-substitution mutant designated orc1-2araS (Supplementary Figure S5). It has been shown that the araS-50 promoter confers arabinose-inducible expression in this archaeon (30). Therefore, when cultured in SCV with sucrose as the carbon source, a non-inducible medium for the orc1-2araS gene, Orc1-2 should be expressed to a constantly low level in the mutant. Then, WT and orc1-2araS were grown in SCV for 48 h either in the presence or absence of 2 μM NQO. Cell samples were taken during incubation and examined for orc1-2 expression, culture growth and cellular DNA content.
Analysis of Orc1-2 protein in the WT and orc1-2araS cells by immunoblotting revealed that the cellular content of Orc1-2 remained constantly low (Figure 5A) in the presence of NQO, consistent with the nature of the orc1-2araS fusion gene. The mutant showed an interesting phenotype: it grew in a similar fashion as for WT in the absence of NQO; however, mutant growth was strongly inhibited in the presence of the drug and the growth inhibition persisted (Figure 5B). These results suggest that orc1-2araS could be deficient in initiating DDR regulation as shown for Δorc1-2 (compare with the data in Figure 2 and Supplementary Figure S2). Indeed, flow cytometry of the cell samples showed that, a large number of orc1-2araS cells became DNA-less cells during incubation in the NQO-SCV medium, and this is in contrast to the WT cells grown in the same medium in which majority of cells were recovered from NQO treatment at 48 hpt (Figure 5C). Together, these results indicated that S. islandicus cells containing a low level of Orc1-2 protein fail to respond to NQO treatment and they are as hypersensitive to NQO treatment as for Δorc1-2 cells.
Figure 5.
S. islandicus Orc1-2araS cells failed to respond to NQO treatment in SCV media. (A) Western analysis of Orc1-2 protein. Only the samples taken at 6 hpt were shown. In the lower panel, only NQO-treated WT sample (lane 2) was diluted for 8 folds. PCNA3 (one of the subunits of the replication clamp) was used as the loading reference. (B) Growth curves based on absorbance at 600 nm. (C) Flow cytometry profile of cell samples. DNA contents were divided into 256 arbitrary points on the X-axis, and cell counts (Y-axis) were obtained for each point and used to plot against the DNA content. WT: S. islandicus E233S1; orc1-2araS: Promoter-substitution mutant containing the araS-50 promoter-orc1-2 fusion gene. DNA-less cells (L); cells containing one chromosome (1), and cells containing two chromosomes (2).
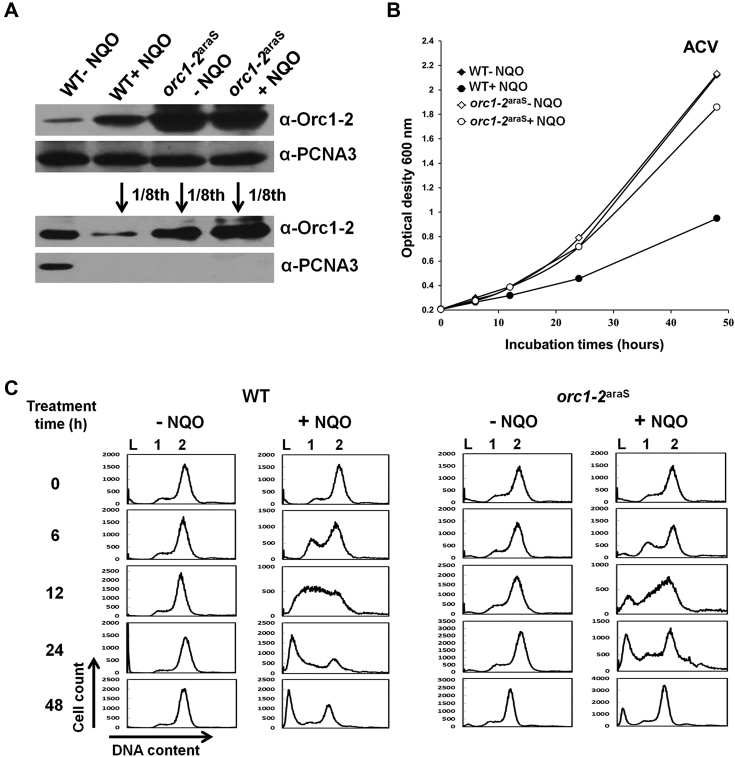
S. islandicus cells stably expressing a high level of Orc1-2 protein responded more promptly to NQO treatment
Next, we investigated how the archaeal cells containing a high level of Orc1-2 could respond to NQO treatment. To do that, both orc1-2araS and WT were grown in ACV with d-arabinose as the carbon source, in the presence or absence of 2 μM NQO. This medium is an inducible medium for the expression of the orc1-2araS gene such that Orc1-2 should be expressed to a constantly high level in orc1-2araS cells. These cultures were grown for 48 h during which cell samples were taken for the analyses as above described. First, immunoblotting confirmed that Orc1-2 was expressed to a high level in ACV-cultured orc1-2araS cells (Figure 6A). Then, growth data revealed that NQO treatment had little influence on the growth of orc1-2araS cells in ACV since very similar growth curves were obtained for NQO-treated versus untreated orc1-2araS cultures (Figure 6B). Nevertheless, flow cytometry detected the increase of the cell population of 1–2 chromosomes in the NQO-treated cultures of both strains, indicating that both WT and orc1-2araS are capable of recovering from NQO-induced DNA damage under this growth condition (Figure 6C). Strikingly, both sets of data (growth curves and flow cytometry profiles) suggest a quick recovery for the promoter substitution mutant, relative to WT (Figure 6). Taken together, these results suggested that a high level of Orc1-2 could probably shorten the time required for execution of DDR regulation in S. islandicus.
Figure 6.
S. islandicus Orc1-2araS cells exhibited a quicker recovery from NQO treatment in ACV media. (A) Western analysis of Orc1-2 protein. Only the samples taken at 6 hpt were shown. In the lower panel NQO-treated WT sample (lane 2), NQO-treated and untreated orc1-2araS samples (lane 3 and 4) were diluted for 8 folds. PCNA3 (one of the subunits of the replication clamp) was used as the loading reference. (B) Growth curves based on absorbance at 600 nm. (C) Flow cytometry profile of cell samples. DNA contents were divided into 256 arbitrary points on the X-axis, and cell counts (Y-axis) were obtained for each point and used to plot against the DNA content. WT: S. islandicus E233S1; orc1-2araS: Promoter-substitution mutant containing the araS-50 promoter-orc1-2 fusion gene. DNA-less cells (L); cells containing one chromosome (1), and cells containing two chromosomes (2).
A constant high level of Orc1-2 protein enabled immediate induction of DDR genes in the archaeon upon NQO treatment
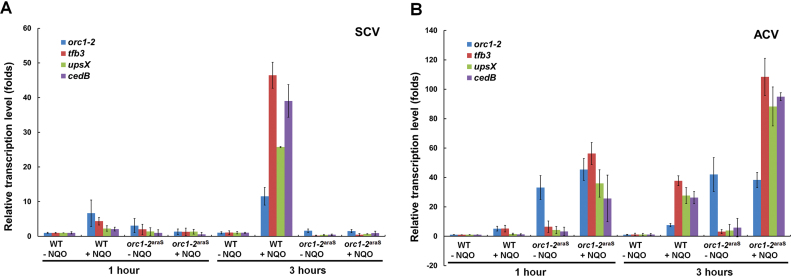
To test that, the activation of gene expression in orc1-2araS and WT cells after NQO treatment was investigated for a few selected DDR genes, including tfb3, upsX and cedB. Cell samples taken at the early stage of NQO treatment (1 and 3 hpt) were used for total RNA extraction, and the extracted RNAs were analyzed for DDR gene expression by RT-qPCR.
As shown in Figure 7A, none of the three DDR genes showed any NQO-responsive expression in the cells expressing a constant low level of Orc1-2 (SCV-cultured orc1-2araS cells), reminiscent of the lack of the DDR regulation in Δorc1-2. By contrast, all three DDR genes were readily activated by NQO treatment in the cells expressing a constantly high level of the regulator (ACV-cultured orc1-2araS cells), and in fact, their expression levels in orc1-2araS cells at 1 hpt are higher than those in WT cells at 3 hpt (Figure 7B). In summary, two sequential events have been identified in the archaeal DDR regulation: (a) activation of the expression of Orc1-2, and (b) the subsequent activation of target genes by Orc1-2. Therefore, Orc1-2 plays a very important role in the DDR regulation in this archaeon.
Figure 7.
High Orc1-2 level in S. islandicus cells prior to NQO treatment enabled earlier responsive expression. (A) Relative expression levels of tfb3, upsX and cedB in WT and orc1-2araS cells at 1 and 3 hpt when grown in SCV in which the expression level of orc1-2 was constantly low. (B) Relative expression levels of tfb3, upsX and cedB in WT and orc1-2araS cells at 1 and 3 hpt when grown in ACV in which the expression level of orc1-2 was constantly high. The expression level of these genes in the cell samples of WT and orc1-2araS was estimated by RT-qPCR with the obtained data normalized to the level of 16S RNA. WT: S. islandicus E233S1; orc1-2araS: Promoter-substitution mutant containing the araS-50 promoter-orc1-2 fusion gene. Error bars: standard derivations of three independent experiments.
DISCUSSION
Here, we show that Orc1-2 functions as a global regulator to mediate DNA damage response in S. islandicus, a hyperthermophilic crenarchaeon. To our knowledge, this represents the first identification of a key regulator in DNA damage response of an archaeal organism.
S. islandicus Orc1-2 is an ortholog of the archaeal/eukaryotic Orc1/Cdc6 superfamily of replication initiators (42–45). The encoding gene is not closely located to any origins of replication on the chromosome of S. islandicus (15), and this genetic organization is conserved in all known species in Sulfolobales (44,45) (Supplementary Figure S6). A previous genetic analysis of orc1-2 function in S. islandicus revealed that the gene does not have a function in replication initiation, in contrast to its orthologs Orc1-1 and Orc1-3 that function as the replication initiator to the adjacent oriC1 and oriC2 individually (15). Here, we show that Δorc1-2 is hypersensitive to NQO, a chemical that yields bulky adducts on bases of chromosomal DNAs, which are to be repaired by NER (35), and the mutant has lost the capability to form NQO-induced cell aggregation and cell cycle regulation (Figure 2). In fact, NQO-responsive expression is completely abrogated in Δorc1-2 (Figure 1, Supplementary Table S3). Together, these findings suggest that, during evolution, Orc1-2 has lost the function as a replication initiator and gained the new function as a primary regulator in DNA damage response. In addition, investigation of bacterial replication initiator DnaA proteins and eukaryotic replication initiators Orc1 and Cdc6 proteins reveals that these factors also function as transcriptional regulator, but these factors have maintained their function in replication initiation (52–54), which is in contrast to the scenario reported for the S. islandicus Orc1-2 here. Therefore, this Orc1-2 protein represents the first example of functional diversification of proteins of the Orc1/Cdc6 superfamily.
Interestingly, it has been reported that archaeal Orc1/Cdc6 proteins form two distinct clades corresponding to Orc1-1 and Orc1-2 clusters in which proteins of the Orc1-2 cluster evolve faster than those of the Orc1-1cluster (44,45). Since S. islandicus Orc1-2 functions as a global DDR regulator, the fast evolution of the Orc1-2 cluster probably reflects the acquisition of a new function for some of Orc1 proteins in this cluster. Furthermore, the conservation of the genetic organization of orc1-2 and its flanking genes in Sulfolobales (Supplementary Figure S6) suggests that these Orc1-2 homologs could also function as a key regulator in archaeal DDR regulation. Moreover, it is also very tempting to investigate whether non-Sulfolobales members of the Orc1-2 cluster could also play a role in DNA damage-induced genome expression as for the S. islandicus Orc1-2.
Our research has gained important insights into the mechanisms of Orc1-2-dependent regulation in S. islandicus. First, a conserved motif (5′-ANTTTC-3′) is present in the promoters of a number of highly up regulated DDR genes, and it is identical to the motif identified in UV-inducible genes of S. acidocaldarius (51). Here, we show that the S. islandicus Orc1-2 protein binds specifically to the motif present in a selected set of DDR gene promoters in vitro (Figure 4), and inactivation of either component (Orc1-2 deficiency or mutagenesis of the DNA binding motif) abolishes the NQO-responsive expression from all tested promoters in reporter gene assay (Figure 3). To this end, the deduced mechanism of transcriptional activation is that Orc1-2 binds to the DDR promoters and facilitates gene expression. Second, a high level of orc1-2 expression is essential but not sufficient to trigger DDR regulation in this archaeon, and this is strongly supported by the following findings: (a) When orc1-2 is constitutively expressed to a constantly low level in orc1-2araS, a promoter-substitution mutant, DDR is not induced by NQO treatment as observed for Δorc1-2. (b) When orc1-2 is expressed to a constantly high level, the mutant does not show any growth delay or retardation in the absence of NQO. Third, orc1-2araS cells grown in ACV media contain a high level of Orc1-2 but they do not exhibit any DDR regulation in the absence of NQO. Nevertheless, these orc1-2araS cells respond more promptly to NQO treatment in DDR initiation than WT cells (Figure 7). In addition, TFB3 is a truncated TFB paralog that also functions as a DDR regulator to a subset of DDR genes (31). However, the TFB3-dependent activation of DDR genes is completely abolished in Δorc1-2, indicating that TFB3 must function downstream of Orc1-2 in the DDR network of this archaeon (Supplementary Table S4). Taken together, these results suggest that Orc1-2 is the primary regulator in the DDR network in this archaeon and that the factor could be activated by posttranslational modifications (PTMs), in analogy to the eukaryotic ATM-dependent activation of DDR regulators, such as p53, an extensively characterized tumor suppressor that regulates cell cycle arrest, DNA repair and apoptosis (55,56).
Genes that are regulated both by Orc1-2 and by TFB3 include those present in the ups operon coding for the proteins involved in the UV-responsive pilus formation (36) and those of the Ced system that mediates intercellular DNA transfer (37). It has been known for a long time that S. acidocaldarius efficiently mediates chromosomal DNA exchange (57) and the process is highly efficient (58). In fact, other Sulfolobus species have also been shown to mediate DNA conjugation although the process has to be induced such as by UV irradiation or DNA damage treatment with bleomycin (36,37,59). Since the two systems have been implicated in repairing UV-induced DNA damage in S. acidocaldarius (36,37,59,60) and NQO-mediated DNA damage in S. islandicus, activation of their gene expression represent one of the DDR network of cellular events in S. islandicus.
The up regulation of known HRR genes including nurA, rad50, mre11 and herA (Supplementary Table S4) by DNA damage agents is consistent with the requirement for DNA repair with high fidelity and the strong activation of DNA transfer activity for importing DNA template for HRR as discussed above. Currently, whether there is any functional connection between the DNA transfer and HRR activity remains to be investigated. Employment of the existing genetic manipulation methods to tackle this problem is challenging since each HRR gene is essential in this organism (61,62). Nevertheless, an efficient CRISPR-based gene knockdown approach has been reported for this crenarchaeon recently (63) and successfully used to dissect the function of the essential topR1 gene coding for a reverse gyrase (24). This approach should be useful to test the hypothesis that the HRR system works in concert with the Ced system in archaeal DNA damage repair.
Among the genes coding for the four DNA polymerases in S. islandicus REY15A (20), only SiRe_0614 and SiRe_0615 that code for DNA polymerase B2 (Dpo2) show NQO-responsive activation, and the regulation is Orc1-2-dependent but TFB3-independent, as demonstrated here and in a previous work. Strikingly, the Sulfolobus Dpo2 has been considered as an inactivated DNA polymerase because there are multiple substitutions in the catalytic residues of its polymerase and exonuclease domains (64). Nevertheless, a recombinant protein of the large subunit of the S. solfataricus Dpo2 is capable of catalyzing DNA synthesis using DNA templates containing a range of DNA lesions, such as 8-oxoguanine, hypoxanthine and uracil (65), suggesting that it could be a translesion DNA polymerase. Since SiRe_0236 coding for the Y-family DNA polymerase (whose homolog in S. solfataricus functions as DNA lesion-bypass enzyme) did not exhibit any up regulation upon NQO treatment, t would be interesting to investigate if Dpo2 could be an active DNA polymerase in DNA damage repair.
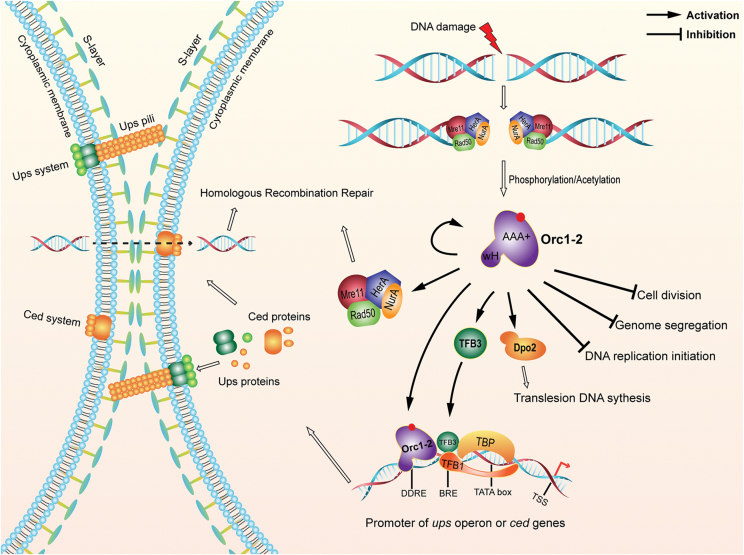
In conclusion, our research has yielded the first picture of the archaeal DDR network for the regulation of cellular processes upon DNA damage, in which the key regulator, Orc1-2 is positioned at the heart of the regulatory network (Figure 8). First, the global regulator has to be activated by DNA damage, and the activation occurs in two aspects: (a) Orc1-2 strongly up regulates its own gene expression, and (b) the factor could be activated by PTMs, such as phosphorylation, acetylation and/or methylation. Then, the activated Orc1-2 exerts either activation or repression to the expression of a large number of DDR genes. The repressed genes include those that mediate cell cycle arrest, including inhibition of cell division, DNA replication initiation and genome segregation. The activated genes are as following: (a) Dpo2 that may function in translesion DNA synthesis, (b) HRR genes that function in DNA repair and (c) TFB3, a secondary DDR regulator responsible for activation of the expression of the ups operon, the ced genes (31), both of which have been implicated in importing DNA fragments for HRR. Finally, (d) the cell aggregation and DNA transfer systems are subjected to dual control by TFB3 and Orc1-2 (Figure 8). These results demonstrate, for the first time, that the strategy of orchestrating a network of cellular events to deal with DNA damage is evolutionarily conserved across the three domains of life.
Figure 8.
An Orc1-2-centered network of DNA damage response in S. islandicus. DNA damage agents yield lesions on DNA that will be converted into double-stranded breaks, which activate the DNA damage signal transduction pathway in this archaeon. Then, the global regulator, Orc1-2 is probably activated by posttranslational modifications, such as phosphorylation and/or acetylation. The activated form of Orc1-2 then recognizes DDRE present in the promoters of DDR genes and activates or represses their expression, including several different cellular processes as well as its own gene. AAA+: ATPases associated with diverse cellular activities; wH: wing-helix DNA binding domain; DDRE: DNA damage-responsive element; BRE: Transcriptional factor B (TFB) recognition element; TATA; TATA box serving as the binding site for TATA-binding protein (TBP); TTS: transcription start site; Ups: UV-responsive pili of Sulfolobus; Ced: Crenarchaeal system for exchange of DNA.
DATA AVAILABILITY
The accession number for the RNA-Seq data in this paper is GEO: GSE101744.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our colleagues in the Archaea Centre, University of Copenhagen and those in the Wuhan laboratory, Huazhong Agricultural University, China for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31771380]; Danish Council for Independent Research [DFF-4181-00274]. Funding for open access charge: National Science Foundation of China. M.S. and X.F. are recipients of PhD studentship from the China Scholarship Council.
Conflict of interest statement. None declared.
REFERENCES
- 1. Baharoglu Z., Mazel D.. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol. Rev. 2014; 38:1126–1145. [DOI] [PubMed] [Google Scholar]
- 2. Giglia-Mari G., Zotter A., Vermeulen W.. DNA damage response. Cold Spring Harb. Perspect. Biol. 2011; 3:a000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Butala M., Zgur-Bertok D., Busby S.J.. The bacterial LexA transcriptional repressor. Cell. Mol. Life Sci. 2009; 66:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kreuzer K.N. DNA damage responses in prokaryotes: regulating gene expression, modulating growth patterns, and manipulating replication forks. Cold Spring Harb. Perspect. Bio.l. 2013; 5:a012674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sirbu B.M., Cortez D.. DNA damage response: three levels of DNA repair regulation. Cold Spring Harb. Perspect. Biol. 2013; 5:a012724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blackford A.N., Jackson S.P.. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell. 2017; 66:801–817. [DOI] [PubMed] [Google Scholar]
- 7. Baliga N.S., Bjork S.J., Bonneau R., Pan M., Iloanusi C., Kottemann M.C., Hood L., DiRuggiero J.. Systems level insights into the stress response to UV radiation in the halophilic archaeon Halobacterium NRC-1. Genome Res. 2004; 14:1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McCready S., Muller J.A., Boubriak I., Berquist B.R., Ng W.L., DasSarma S.. UV irradiation induces homologous recombination genes in the model archaeon, Halobacterium sp. NRC-1. Saline Syst. 2005; 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frols S., Gordon P.M., Panlilio M.A., Duggin I.G., Bell S.D., Sensen C.W., Schleper C.. Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J. Bacteriol. 2007; 189:8708–8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gotz D., Paytubi S., Munro S., Lundgren M., Bernander R., White M.F.. Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol. 2007; 8:R220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Salerno V., Napoli A., White M.F., Rossi M., Ciaramella M.. Transcriptional response to DNA damage in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2003; 31:6127–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Erill I., Campoy S., Barbe J.. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 2007; 31:637–656. [DOI] [PubMed] [Google Scholar]
- 13. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peng N., Han W., Li Y., Liang Y., She Q.. Genetic technologies for extremely thermophilic microorganisms of Sulfolobus, the only genetically tractable genus of crenarchaea. Sci. China Life Sci. 2017; 60:370–385. [DOI] [PubMed] [Google Scholar]
- 15. Samson R.Y., Xu Y., Gadelha C., Stone T.A., Faqiri J.N., Li D., Qin N., Pu F., Liang Y.X., She Q. et al. Specificity and function of archaeal DNA replication initiator proteins. Cell Rep. 2013; 3:485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han W., Xu Y., Feng X., Liang Y.X., Huang L., Shen Y., She Q.. NQO-Induced DNA-Less cell formation is associated with chromatin protein degradation and dependent on A0A1-ATPase in sulfolobus. Front. Microbiol. 2017; 8:1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zillig W., Kletzin A., Schleper C., Holz I., Janekovic D., Hain J., Lanzendorfer M., Kristjansson J.K.. Screening for sulfolobales, their plasmids and their viruses in icelandic solfataras. Syst. Appl. Microbiol. 1994; 16:609–628. [Google Scholar]
- 18. Schleper C., Kubo K., Zillig W.. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:7645–7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deng L., Zhu H., Chen Z., Liang Y.X., She Q.. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles. 2009; 13:735–746. [DOI] [PubMed] [Google Scholar]
- 20. Guo L., Brugger K., Liu C., Shah S.A., Zheng H., Zhu Y., Wang S., Lillestol R.K., Chen L., Frank J. et al. Genome analyses of Icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus-host interaction studies. J. Bacteriol. 2011; 193:1672–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang L., Feng Z., Wang X., Wang X., Zhang X.. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010; 26:136–138. [DOI] [PubMed] [Google Scholar]
- 23. Schmittgen T.D., Livak K.J.. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008; 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 24. Han W., Feng X., She Q.. Reverse gyrase functions in genome integrity maintenance by protecting DNA breaks in vivo. Int. J. Mol. Sci. 2017; 18:E1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Y., Pan S., Zhang Y., Ren M., Feng M., Peng N., Chen L., Liang Y.X., She Q.. Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2016; 44:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Warrens A.N., Jones M.D., Lechler R.I.. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene. 1997; 186:29–35. [DOI] [PubMed] [Google Scholar]
- 27. Peng N., Deng L., Mei Y., Jiang D., Hu Y., Awayez M., Liang Y., She Q.. A synthetic arabinose-inducible promoter confers high levels of recombinant protein expression in hyperthermophilic archaeon Sulfolobus islandicus. Appl. Environ. Microbiol. 2012; 78:5630–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. She Q., Singh R.K., Confalonieri F., Zivanovic Y., Allard G., Awayez M.J., Chan-Weiher C.C., Clausen I.G., Curtis B.A., De Moors A. et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heckman K.L., Pease L.R.. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2007; 2:924–932. [DOI] [PubMed] [Google Scholar]
- 30. Peng N., Xia Q., Chen Z., Liang Y.X., She Q.. An upstream activation element exerting differential transcriptional activation on an archaeal promoter. Mol. Microbiol. 2009; 74:928–939. [DOI] [PubMed] [Google Scholar]
- 31. Feng X., Sun M., Han W., Liang Y.X., She Q.. A transcriptional factor B paralog functions as an activator to DNA damage-responsive expression in archaea. Nucleic Acids Res. 2018; doi:10.1093/nar/gky236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He F., Vestergaard G., Peng W., She Q., Peng X.. CRISPR-Cas type I-A Cascade complex couples viral infection surveillance to host transcriptional regulation in the dependence of Csa3b. Nucleic Acids Res. 2017; 45:1902–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bailleul B., Daubersies P., Galiegue-Zouitina S., Loucheux-Lefebvre M.H.. Molecular basis of 4-nitroquinoline 1-oxide carcinogenesis. Jpn. J. Cancer Res. 1989; 80:691–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Williams A.B., Hetrick K.M., Foster P.L.. Interplay of DNA repair, homologous recombination, and DNA polymerases in resistance to the DNA damaging agent 4-nitroquinoline-1-oxide in Escherichia coli. DNA Repair (Amst.). 2010; 9:1090–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ajon M., Frols S., van Wolferen M., Stoecker K., Teichmann D., Driessen A.J., Grogan D.W., Albers S.V., Schleper C.. UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili. Mol. Microbiol. 2011; 82:807–817. [DOI] [PubMed] [Google Scholar]
- 37. van Wolferen M., Wagner A., van der Does C., Albers S.V.. The archaeal Ced system imports DNA. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:2496–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qureshi S.A., Jackson S.P.. Sequence-specific DNA binding by the S. shibatae TFIIB homolog, TFB, and its effect on promoter strength. Mol. Cell. 1998; 1:389–400. [DOI] [PubMed] [Google Scholar]
- 39. Paytubi S., White M.F.. The crenarchaeal DNA damage-inducible transcription factor B paralogue TFB3 is a general activator of transcription. Mol. Microbiol. 2009; 72:1487–1499. [DOI] [PubMed] [Google Scholar]
- 40. Constantinesco F., Forterre P., Elie C.. NurA, a novel 5′-3′ nuclease gene linked to rad50 and mre11 homologs of thermophilic Archaea. EMBO Rep. 2002; 3:537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Constantinesco F., Forterre P., Koonin E.V., Aravind L., Elie C.. A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea. Nucleic Acids Res. 2004; 32:1439–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bell S.D. Archaeal orc1/cdc6 proteins. Subcell. Biochem. 2012; 62:59–69. [DOI] [PubMed] [Google Scholar]
- 43. Parker M.W., Botchan M.R., Berger J.M.. Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2017; 52:107–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Raymann K., Forterre P., Brochier-Armanet C., Gribaldo S.. Global phylogenomic analysis disentangles the complex evolutionary history of DNA replication in archaea. Genome Biol. Evol. 2014; 6:192–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Makarova K.S., Koonin E.V.. Archaeology of eukaryotic DNA replication. Cold Spring Harb. Perspect. Biol. 2013; 5:a012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Driessen R.P., Meng H., Suresh G., Shahapure R., Lanzani G., Priyakumar U.D., White M.F., Schiessel H., van Noort J., Dame R.T.. Crenarchaeal chromatin proteins Cren7 and Sul7 compact DNA by inducing rigid bends. Nucleic Acids Res. 2013; 41:196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kalliomaa-Sanford A.K., Rodriguez-Castaneda F.A., McLeod B.N., Latorre-Rosello V., Smith J.H., Reimann J., Albers S.V., Barilla D.. Chromosome segregation in Archaea mediated by a hybrid DNA partition machine. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:3754–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Samson R.Y., Obita T., Freund S.M., Williams R.L., Bell S.D.. A role for the ESCRT system in cell division in archaea. Science. 2008; 322:1710–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lindas A.C., Karlsson E.A., Lindgren M.T., Ettema T.J., Bernander R.. A unique cell division machinery in the Archaea. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:18942–18946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu J., Gao R., Li C., Ni J., Yang Z., Zhang Q., Chen H., Shen Y.. Functional assignment of multiple ESCRT-III homologs in cell division and budding in Sulfolobus islandicus. Mol. Microbiol. 2017; 105:540–553. [DOI] [PubMed] [Google Scholar]
- 51. Le T.N., Wagner A., Albers S.V.. A conserved hexanucleotide motif is important in UV-inducible promoters in Sulfolobus acidocaldarius. Microbiology. 2017; 163:778–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scholefield G., Veening J.W., Murray H.. DnaA and ORC: more than DNA replication initiators. Trends Cell Biol. 2011; 21:188–194. [DOI] [PubMed] [Google Scholar]
- 53. Sasaki T., Gilbert D.M.. The many faces of the origin recognition complex. Curr. Opin. Cell Biol. 2007; 19:337–343. [DOI] [PubMed] [Google Scholar]
- 54. Hossain M., Stillman B.. Opposing roles for DNA replication initiator proteins ORC1 and CDC6 in control of Cyclin E gene transcription. Elife. 2016; 5:pii: e12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Beckerman R., Prives C.. Transcriptional regulation by p53. Cold Spring Harb. Perspect. Biol. 2010; 2:a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017; 36:3943–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grogan D.W. Exchange of genetic markers at extremely high temperatures in the archaeon Sulfolobus acidocaldarius. J. Bacteriol. 1996; 178:3207–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jacobs K.L., Grogan D.W.. Spontaneous mutation in a thermoacidophilic archaeon: evaluation of genetic and physiological factors. Arch. Microbiol. 1998; 169:81–83. [DOI] [PubMed] [Google Scholar]
- 59. Frols S., Ajon M., Wagner M., Teichmann D., Zolghadr B., Folea M., Boekema E.J., Driessen A.J., Schleper C., Albers S.V.. UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol. Microbiol. 2008; 70:938–952. [DOI] [PubMed] [Google Scholar]
- 60. van Wolferen M., Ma X., Albers S.V.. DNA processing proteins involved in the UV-induced stress response of Sulfolobales. J. Bacteriol. 2015; 197:2941–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Huang Q., Liu L., Liu J., Ni J., She Q., Shen Y.. Efficient 5′-3′ DNA end resection by HerA and NurA is essential for cell viability in the crenarchaeon Sulfolobus islandicus. BMC Mol. Biol. 2015; 16:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang C., Tian B., Li S., Ao X., Dalgaard K., Gokce S., Liang Y., She Q.. Genetic manipulation in Sulfolobus islandicus and functional analysis of DNA repair genes. Biochem. Soc. Trans. 2013; 41:405–410. [DOI] [PubMed] [Google Scholar]
- 63. Peng W., Feng M., Feng X., Liang Y.X., She Q.. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015; 43:406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rogozin I.B., Makarova K.S., Pavlov Y.I., Koonin E.V.. A highly conserved family of inactivated archaeal B family DNA polymerases. Biol. Direct. 2008; 3:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Choi J.Y., Eoff R.L., Pence M.G., Wang J., Martin M.V., Kim E.J., Folkmann L.M., Guengerich F.P.. Roles of the four DNA polymerases of the crenarchaeon Sulfolobus solfataricus and accessory proteins in DNA replication. J. Biol. Chem. 2011; 286:31180–31193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-Seq data in this paper is GEO: GSE101744.