Abstract
It has been reported that the two major types of RNA interference triggers, the classical Dicer-generated small RNAs (siRNAs), which function with all members of the Argonaute (Ago) protein family in mammals, and the Ago2-sliced small RNAs (sli-siRNAs), which function solely through Ago2, have similar potency in target cleavage and repression. Here, we show that sli-siRNAs are generally more potent than siRNAs in silencing mismatched targets. This phenomenon is usually more apparent in targets that have mismatched nucleotides in the 3′ supplementary region than in targets with mismatches in the seed region. We demonstrate that Ago2 slicer activity is a major factor contributing to the greater silencing efficiency of sli-siRNA against mismatched targets and that participation of non-slicing Agos in silencing mismatched siRNA targets may dilute the slicing ability of Ago2. The difference in length of the mature guide RNA used in sli-RISCs and si-RISCs may also contribute to the observed difference in knockdown efficiency. Our data suggest that a sli-siRNA guide strand is likely to have substantially stronger off-target effects than a guide strand with the same sequence in a classical siRNA and that Dicer and non-slicing Agos may play pivotal roles in controlling siRNA target specificity.
INTRODUCTION
The biogenesis of most microRNAs (miRNAs), a class of endogenous small RNAs, involves the enzyme Dicer, which processes precursor-miRNA (pre-miRNA) hairpins in the cytoplasm to create 21- to 23-nucleotide (nt) RNA duplexes with 3′ overhangs. Dicer also chops double-stranded RNAs (dsRNAs) into canonical small interfering RNAs (siRNAs) that exist as guide strand/passenger strand duplexes with a 19-base pair dsRNA stem and an overhang of two nts at the 3′ end of each strand (Figure 1A). These siRNAs are loaded onto Argonaute proteins (Ago1, 2, 3 and 4 in mammals) to form RNA-induced silencing complexes (RISCs) that cleave fully complementary RNA targets and repress partially complementary RNA targets (1–4).
Figure 1.
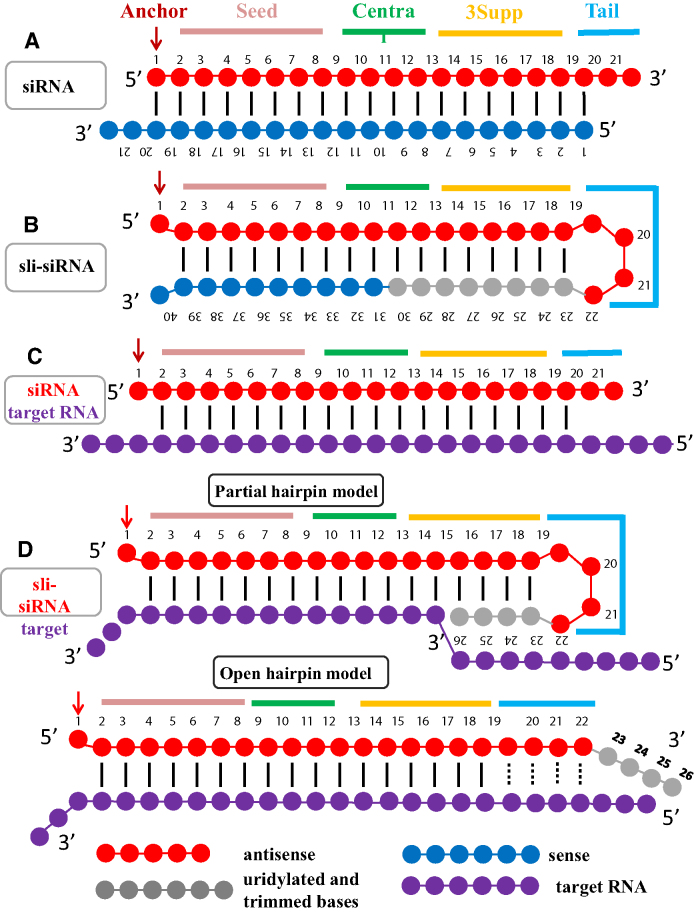
Schematic illustrations of siRNA and sli-siRNA molecules and guide RNAs base-pairing with target RNA. (A) Model molecule of a 21-mer siRNA. (B) Model molecule of a 40-mer sli-siRNA. (C) The siRNA guide strand base-pairing with target RNA. (D) The proposed partial hairpin and open hairpin models of sli-siRNA guide strand base-pairing with target RNA. The partial hairpin model posits that some sli-siRNA isoforms, especially 23- to 26-mers may only use the nts 2 to 14 for target binding. The open hairpin model posits that after passenger strand slicing, the partial hairpin opens during seed-mediated target binding, allowing nts 19 to 22 base pair with targets.
This canonical siRNA RNA interference (RNAi) pathway involves at least three major steps: 1) guide strand/passenger strand duplex generation by Dicer, 2) guide strand selection and loading of the duplex onto an Ago to form the pre-RISC, and 3) conformational changes of the Ago to transform a pre-RISC to an active RISC (5,6). Interestingly, a few miRNAs, such as miR-451, use an elegant one-step RNAi mechanism, in which both guide strand generation and maturation are coupled with RISC formation and activation. All processes are mediated solely by Ago2, without needing a Dicer processing step (7–10). This one-step slicing mechanism has been used to design sliced-siRNAs (sli-siRNAs) and other miR-451 mimics that are mainly processed by Ago2 and have significantly reduced sense strand activity (Figure 1b) (10–16).
Despite the fact that sli-siRNAs function solely through Ago2, whereas siRNAs can function through all Agos, both types of RNAi triggers show similar potency in cleaving fully complementary targets and repressing partially complementary targets (10,12). This raises an intriguing question: why has nature not evolutionarily selected the one-step, single-factor RNAi mechanism that uses Ago2 as the sole RNAi processor and eliminated the multi-step, multi-factor mechanism involving Dicer and all Agos? In fact, most miRNAs use Dicer-generated RNA duplex intermediates that can be loaded onto all Agos, while few miRNAs use the seemingly simpler Ago2 processing pathway (7–9).
In this study, we found that sli-siRNAs and classical siRNAs exhibit differential knockdown efficiency, with sli-siRNAs exhibiting higher potency than siRNAs for mismatched targets. We found that the increased efficiency of sli-siRNAs compared to siRNAs is usually more apparent for targets with mismatched nts located in the 3′ supplementary (3supp) base pair region, compared to targets with mismatches located in the seed region. We also found that Ago2 slicer activity is a major factor that contributes to the higher efficiency of sli-siRNAs for mismatched targets and that the involvement of non-slicing Agos (Ago1, 3 and 4) may reduce this effect in siRNA RISCs (si-RISCs). Additionally, we found that activated sli-siRNA RISCs (sli-RISCs) contain Ago2 loaded with guide RNAs of variable lengths, mostly 23- to 26-mer, due to the trimming/tailing process at the 3′end of the guide RNAs. We propose that sli-RISCs loaded with longer isoforms of guide RNAs may base-pair with their target RNA differently than si-RISCs do (Figure 1C and D).
Our work has revealed an important aspect regarding the specificity of RNAi. Although sli-siRNAs can dramatically reduce off-target activity conferred by the passenger strand, the guide strand of a sli-siRNA could cause substantially stronger off-target effects than the guide strand of an siRNA with the same nt sequence. This differential targeting specificity is regulated by non-slicing Agos, which are selective for siRNA duplexes processed by Dicer. Therefore, the generation of duplex RNAs by Dicer is critical for regulating RNAi targeting specificity.
MATERIALS AND METHODS
Cell lines and cell culture
Human embryonic kidney cell line HEK-293, human colon cancer cell line HCT-116, human cervical carcinoma cell line HeLa, HeLa miR-21 knockout cells, and mouse embryonic fibroblast (MEF) cells were maintained in high glucose (4.5 g/l) DMEM supplemented with 2 mM glutamine, 10% FBS and 2 mM penicillin/streptomycin. Cells were incubated at 37°C, 5% CO2.
Human Ago expression plasmids
For co-transfection of RNAi molecules with human Ago2 (hAgo2), we used a pIRESneo-FLAG/HA-Ago2 plasmid (Addgene #10822) to generate slicing and non-slicing forms of hAgo2. We first cloned FLAG/HA-Ago2 as a Hind III and Spe I fragment into Hind III and Xba I sites in pcDNA3.1-neo (Thermo Fisher Scientific, Grand Island, NY, USA) to generate pcDNA-FLAG/HA-hAgo2; then we used gBlocks (Integrated DNA Technologies [IDT], San Diego, CA, USA) or PCR to generate hAgo2-dLS (truncated before the N domain) and hAgo2-LS-mut (with five amino acids in hAgo2 mutated to match the amino acids of hAgo1 at the same locations: P27R, D30G, R36K, Q41L, F45Y). Catalytically defective hAgo2 (hAgo2-CD, with D597A and D669A mutations) was generated using the QuikChange Lightning Site-Directed Mutagenesis Kit from Agilent (Santa Clara, CA, USA). S5A and S5E were generated by replacing a Kpn I to EcoRI fragment containing the S824–S834 region with Kpn I to EcoRI gBlock sequences (IDT). hAgo1, hAgo3, and hAgo4 expression plasmids were pIRESneo-FLAG/HA-Ago1 (Addgene #10820), pIRESneo-FLAG/HA-Ago3 (Addgene #10823), and pIRESneo-FLAG/HA-Ago4 (Addgene #10824), respectively. pcDNA3.1-neo was used as the control vector in cotransfection with Agos.
Dual-luciferase reporter assay
All reporter assays were performed using psiCheck 2.0-based, dual-luciferase reporters from Promega (Madison, WI) that express both firefly luciferase (Fluc) and Renilla luciferase (Rluc). Reporters that carried target sequences complementary to the RNAi molecules of interest were constructed by inserting annealed oligonucleotides into the Xho I/Spe I sites of the 3′ UTR of the Rluc gene in a psiCheck2.2 vector (10).
RNAi triggers and reporter constructs were co-transfected into cells using Lipofectamine 2000 (Invitrogen), as previously reported (10). For each experiment, at least three independent transfections were performed in duplicate in 24-well plates. Cell were grown to 75–85% confluency in 500 μl medium and were transfected with luciferase reporter (50 ng) and varying amounts of siRNA or sli-siRNA (100 ng of stuffer DNA, plus 1μl of stock siRNA at 1.6–1000 nM concentrations, and 1 μl of Lipofectamine 2000).
Forty-eight hours after transfection, cells were lysed with 100 μl passive lysis buffer (Promega) and luciferase levels from 20 μl of lysate were determined with the Dual-Luciferase Reporter Assay System and GloMax 96 Microplate Luminometer (Promega). Changes in expression of Rluc (target) were normalized to Fluc (internal control) and then calculated as a percentage relative to the Rluc/Fluc ratio of a scrambled siRNA control. The relative ratios of Rluc/Fluc were used to determine the efficiency of silencing. Data were averaged from at least three independent transfections, and each transfection had at least two replicates. Error bars indicate the standard deviation of the mean.
Stable cell lines
A stable HCT-116 cell line that expresses agshRNA-1148, an Ago2-processed small hairpin RNA (shRNA) that targets the M2 subunit of ribonucleotide reductase gene RRM2, was described in a previous report (10). The same agshRNA expression vector and lenti-viral expression vector with a puromycin selection marker were used to construct a sliced-miR-21 (sli-miR-21) expression lenti-viral vector. HeLa miR-21 knockout cells were transduced with the sli-miR-21 expression lentiviruses and puromycin selection was used to generate a stable sli-miR-21 expression HeLa cell line (17).
RNA isolation and northern blot analysis
RNA isolation and Northern blot analysis were carried out as previously described (10,18). Briefly, total RNA was extracted using TRIzol® (Thermo Fisher Scientific). Total RNA (20 μg) was separated on 12% SDS-PAGE/8M urea gels, and gels were blotted onto positively charged nylon membranes. DNA oligonucleotide probes complementary to the target RNA sequences were labeled with γ-32P-ATP. The probes were hybridized to the membranes overnight in PerfectHyb Plus hybridization buffer (Sigma, St. Louis, MO, USA), after which membranes were washed once in 6× SSPE/0.1% SDS for 10–30 min and twice in 6× SSC/0.1% SDS for 10–30 min each. U2 small nucleolar RNAs were used as RNA loading controls.
Small RNA deep sequencing and reads processing
Small RNA deep sequencing was carried out by a customized bias reduction protocol previously reported (19,20). Briefly, 1.0 μg of total RNA was used to construct small RNA libraries for single reads, flow cell cluster generation, and 42 cycle (42-nt) sequencing. To construct small RNA libraries, the 5′ adaptor used in the Illumina (San Diego, CA, USA) Truseq small RNA deep sequencing protocol was replaced with a customized 5′ adaptor by adding three random nts at the 3′end of the original 5′ adaptor to reduce bias in small RNA sequencing results (19,20). The 3′ adaptor provided in the kit was used for 3′ end ligation and sample barcoding.
Small RNA sequencing reads were processed as reported and further analyzed using miRge and Microsoft Excel (19–21). To reconstruct the possible profile of the mature sequences with and without 3′ end trimming/tailing (the 3′ end nts as the potential trimmed/tailed nts), we extracted all reads perfectly aligned to a 10-nt sequence: the 4th to the 13th nts of each siRNA/miRNA. We found that the read count fetched using this 10-nt sequence of miR-21 was comparable to miR-21 read counts using miRge. Most importantly, this method reduced the workload compared to analysis all miR-21 reads generated by small RNA sequencing. To further reduce the workload and to avoid low count reads that are generally considered unreliable, we eliminated reads that had counts below 10. The remaining 10-nt matched reads of each siRNA/miRNA were modified at their 5′ ends if they were not perfectly matched at the first 3 nts: correcting mutations, adding deleted nts, or removing extra nts and residual 5′ adaptors. For example, reads from sli-miR-451 with nts N, NN, or BBB (N: any nt; B: any non-A nt) before the fourth nt were changed to AAA. In this way, we created a read profile that perfectly matched the first 13 nts at the 5′ end of a siRNA/miRNA with all sorts of 3′ end variations. Next, these reads were summarized and classified by read-length distribution as reads with 3′ end trimming/tailing. Then, to create a reads profile of potential mature reads without 3′ end trimming/tailing, the above reads with 3′ end trimming/tailing were modified at their 3′ ends based on their alignment to the full sli-siRNA/miRNA sequences by correcting mutations, adding back deleted nts, and removing tailed nts and residual 3′ end adaptors. Therefore, these reads were perfectly matched to a segment of the full sli-siRNA/miRNA sequences. But, using this method we cannot know if a read was trimmed to the last nt or if the last nt was tailed to the nt in front of it.
qPCR
The BIO-RAD (Hercules, CA) iTaq™ Universal SYBR® Green One-Step Kit was used for qPCR. Briefly, total RNA was isolated by TRIzol® followed by DNase treatment. In each reaction, 500 ng of DNase-treated total RNA was used, and all other reagents were used as specified in the protocol provided in the kit. We also followed the qPCR program suggested by the vendor. Fluc gene expression in the reporter was used as a normalization control to calculate ΔCt (ΔCt = CtRluc– Ct Fluc) for each sample. The normalized relative Rluc/Fluc ratio, calculated as 2−ΔΔCt for each sample, where ΔΔCt = ΔCtsiRNA- ΔCtctrl, was used to measure Rluc RNAi knockdown. Data were averaged from at least three replicates.
Oligonucleotides
All DNA and RNA oligonucleotides were synthesized by IDT; sequences are listed in Supplementary Tables S4-S6.
RESULTS
Reporters to test the silencing efficiency of RNAi triggers against mismatched targets
Previous studies have reported that sli-siRNAs can be as potent as classical siRNAs for both cleavage of fully complementary targets and repression of partially complementary targets (10,15). However, because different Ago-mediated RISCs are involved in sli-siRNA versus siRNA targeting, we hypothesized that these two types of RNAi molecules silence genes differently. Moreover, because maturation of the sli-siRNA could be ongoing when the guide strand binds the target, we hypothesized that sli-RISCs may use different functional mechanisms than si-RISCs, (Fig. 1b) (7–9,22). We further hypothesized that mismatched targets may be tolerated differently by sli- and si-RISCs, leading to differences in the decision to execute target cleavage versus repression and overall silencing efficiency.
In siRNA targeting, the sequence of siRNA and its base pairing region on a target can be divided into three regions: the seed, central, and 3supp regions (Figure 1a) (23). Many lines of evidence have shown that siRNAs/miRNAs use the ‘seed sequence’ to nucleate their binding to targets and use the 3supp region to stabilize the RISC for action (23–29). To test the silencing efficiency of sli-siRNAs and siRNAs against mismatched targets, we introduced mutations into RNAi target sequences to create reporters mismatched to the RNAi molecules at nts 3 to 18. We produced wild type (WT, fully complementary) and mutant reporters for 1) siRNA-887 (si-887) and sli-siRNA-887 (sli-887) that target the RRM2 gene (10); 2) siRNA-ARX (si-ARX) and sli-siRNA-ARX (sli-ARX) that target the ARX gene; and 3) siRNA-451 (si-451) and sli-siRNA-451 (sli-451), a mimic of mouse pre-miR-451 (mmiR-451). We generated reporters for mismatches in all three target regions, as well as reporters with G:U wobble pairs for comparison (Figure 2a and Supplementary Figure S1). In order to avoid a synergistic or combinatorial effect from multiple RISCs, reporters were designed to only carry one copy of the target sequence. Most reporters had a single nt mismatch with the RNAi molecules, and a few reporters carried 2-nt mismatches; one to two mutations were created for each nt position within a target sequence, such that the sequences for mutations at the same position differed by one nt and sequences for mismatched reporters for two different positions differed by two nts.
Figure 2.
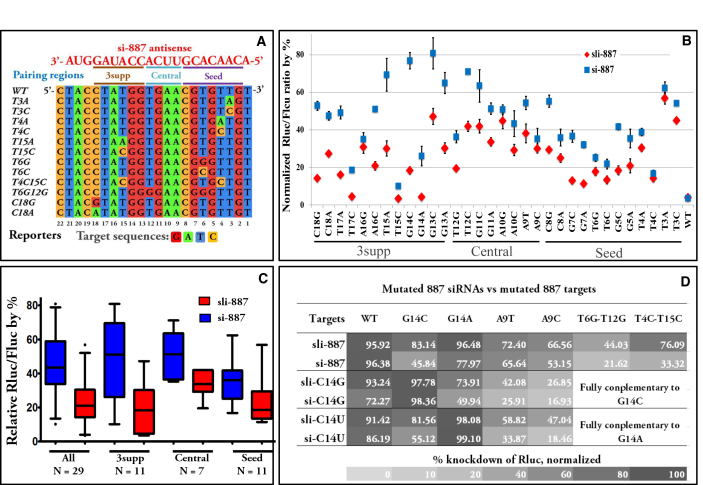
Silencing of mismatched targets by sli-887 versus si-887. A. Examples of mismatched reporter sequences. Wild type (WT) and example mismatched si-887 reporter sequences are aligned and presented 5′ to 3′. Sequences were numbered according to the mismatched nt position in the RNAi sequence. B. Knockdown of mismatched, G:U paired (reporters A10G and A16G), and WT targets by sli -887 and si-887 (both at a final concentration of 0.4 nM). Normalized Rluc/Fluc ratios are plotted according to the position of the mutations in the sli-siRNA or siRNA. Error bars represent standard deviation of the mean. C. Box plots of mismatched reporter data (excluding wobble pairs A10G and A16G) from panel b. Data are presented for all mutations (All), as well as grouped by mutation location in the seed region (Seed), the central region (Central), and the 3supp region (3supp). D. Heat map of normalized relative percent knockdown of Rluc for mismatched targets by sli-887 and si-887 mutants (all at a final concentration of 0.4 nM). Reporter G14C perfectly complements sli-siRNA-887-C14G and siRNA-887-C14G. Reporter G14A perfectly complements sli-siRNA-887-C14U and siRNA-887-C14U. A9T and A9C do not perfectly complement any of the sli-siRNAs or siRNAs.
In order to clearly observe the differences in silencing between each set of WT targets and corresponding mutants and to avoid saturation of the reporter system, we titrated the concentrations of reporters and RNAi reagents to knockdown a WT reporter between 80% and 95%, and compared knockdown efficiency for the corresponding mismatched reporters under the same conditions.
Sli-siRNAs have higher tolerance for mismatched targets than classical siRNAs
After optimizing the concentrations of sli-887 and si-887 needed to knockdown the fully complementary WT reporter by about 95%, we tested all 887 mutant reporters under the same conditions (Figure 2b, 200 pM of sli-887 or si-887 was used).
We observed sequence context-dependent effects on silencing, with almost all mismatches and the two G:U wobble pairs (A10G and A16G in Figure 2b) negatively affecting target silencing by both types of RNAi molecules. Overall, sli-887 tolerated mismatches in the target much better than si-887, as measured by the difference in silencing between sli-887 and si-887 (via relative Rluc/Fluc; P < 0.0001). For some mismatched reporters (G13A, G14C, T15A, T17A, C18G in Figure 2b), sli-887 had a knockdown efficiency more than 2-fold that of si-887. Furthermore, the silencing effect was only greater for sli-887 compared to si-887 for mismatched targets, as sli-887 and si-887 had a similar knockdown effects on both G:U wobble pair reporters (A10G and A16G in Figure 2b).
In order to identify the differential effects of mismatch in various regions of the target, mismatches were grouped by their location: seed region (nts 3 to 8, numbered according to the nt position in the siRNA), central region (nts 9 to 12), or 3supp region (nts 13 to 18). Despite large sequence context-dependent variation in silencing—especially in si-887 knockdown of reporters carrying mismatches in the 3supp region—when reporters were grouped by target regions, the difference in the silencing of mismatched targets by sli-887 compared to si-887 was most obvious (by magnitude and statistical significance) for targets mutated in the 3supp region (Figure 2c; Table 1). In contrast, differences in silencing of mismatches grouped by targeted regions (seed vs. central vs. 3supp) compared within si-887 or sli-887 were much less significant (P > 0.004 for all pair-wise comparisons) than the differences between si-887 and sli-887 over all regions (P < 0.0008) (Figure 2b–c; Table 1 and Supplementary Table S1).
Table 1.
Differences in silencing of mismatched targets by sli-siRNA (sli-) versus siRNA (si-). P-values were determined by two-tailed paired Student's t-tests with 95% confidence intervals
| Difference (sli- efficiency) – (si- efficiency) | P-value | |||||
|---|---|---|---|---|---|---|
| Region of Target Mismatch | 887 | ARX | 451 | 887 | ARX | 451 |
| All regions | 20.40 | 32.60 | 18.50 | <0.0001 | <0.0001 | <0.0001 |
| Seed | 13.36 | 13.62 | 12.84 | 0.0003 | <0.0097 | <0.0001 |
| Central | 17.23 | 38.83 | 21.07 | 0.0008 | <0.0001 | <0.0001 |
| 3supp | 30.10 | 47.42 | 19.95 | <0.0001 | <0.0001 | <0.0001 |
To further confirm that the differences between sli-887 and si-887 in the silencing of mismatched targets are strong for targets mismatched in the 3supp region, we mutated the C at nt 14 (14C) in sli-887 to a G or U, creating the mutants sli-887-C14U and sli-887-C14G (their WT targets are mismatched sli-887 reporters G14A and G14C, respectively; Supplementary Figure S1a). Sli-887-wt, sli-887-C14U, and sli-887-C14G, and the corresponding classical si-887-wt, si-887-C14U, and si-887-C14G, had similar silencing efficiencies for their perfectly matched targets. However, all sli-siRNAs were more potent than siRNAs at silencing mismatched targets (Figure 2d). Sli-887 was also more potent than si-887 in silencing reporters that carried two nt mismatches (T6G-T12G and T4C-T15C; Figure 2a, d). These results indicate that sli-siRNAs have a higher tolerance than siRNAs for mismatched base pairing with targets.
Next, we tested the above observations using a set of reporters for si-ARX and sli-ARX (Supplementary Figure S1b). Based on the differences of sli-887 and si-887 in silencing mismatched targets, to observe a clearer difference between siRNAs and sli-RNAs in silencing mismatched targets, we titrated the RNAi triggers and selected low concentrations of sli-ARX and siRNA-ARX to knockdown the WT reporter by about 80% (Figure 3a).
Figure 3.
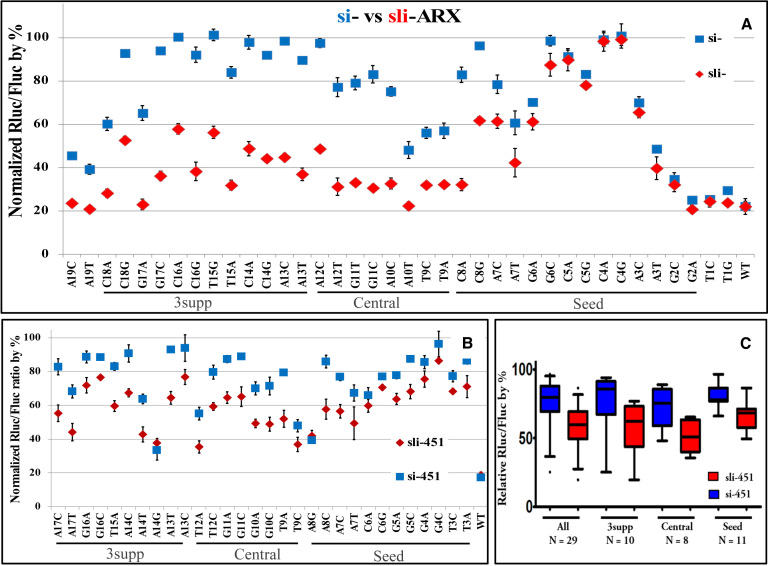
Knockdown of mismatched targets by sli-ARX and sli-451. A. Mismatched target knockdown by sli -ARX and si-ARX. A serial dilution of sli-ARX or si-ARX was used to determine the concentration that knocked down the fully complementary target to about 80% (27.5 pM for si-ARX and 20 pM for sli-ARX), which was used in subsequent transfection for all reporters. Normalized Rluc/Fluc ratios are plotted according to the position of the mutations in the sli-siRNA (sli-) or siRNA (si-). Error bars represent standard deviation of the mean. B. Knockdown of mismatched and G:U paired reporters (A8G and A14G) by sli-siRNA-451 (sli-451) and siRNA-451 (si-451). A serial dilution of sli-451 or si-451 was used to determine the concentration that knocked down the fully complementary target to about 80% (10 pM for si-451 and 13 pM for sli-451), which was used in subsequent transfection for all reporters. The normalized relative Rluc/Fluc ratios are plotted according to the position of the mutations in the sli-siRNA or siRNA. Error bars represent standard deviation of the mean. C. Box plots of mismatched reporter data (excluding wobble pairs A8G and A14G) from panel b. Data are presented for all mutations (All), as well as grouped by mutation location in the seed region (Seed), the central region (Central), and the 3supp region (3supp).
The data clearly show that sli-ARX tolerated mismatches in the target much better than si-ARX across all mismatches (P < 0.0001). The difference in knockdown efficiency between sli-ARX and si-ARX were greatest when the mismatches were located in the 3supp region versus in the seed region or the central region (Table 1). The differences in silencing mismatched targets grouped by base pairing regions (seed vs. central vs. 3supp) compared within si-ARX or sli-ARX were much smaller (P > 0.001 for all pair-wise comparisons) than the difference between si-ARX and sli-ARX over all regions (P < 0.0001) (Figure 3a and Table 1 and Supplementary Table S1). When mismatches were located in the seed region, the differences in knockdown efficiency between sli-ARX and si-ARX were relatively small (13.62% and P < 0.01), whereas the knockdown efficiency of sli-ARX was almost two-fold that of si-ARX for mismatches located in the 3supp region, and the difference was much more significant (47.42% and P < 0.0001) (Figure 3a and Tables 1 and Supplementary Table S1).
The above sli-siRNAs, which target RRM2 and ARX, were artificially designed sequences that both showed higher knockdown potency than classical siRNAs for mismatched targets but also exhibited sequence context-dependent differences in silencing efficiency (Figures 2b and 3a). Whether the miR-451 sequence, which was evolutionarily selected to use the one-step sli-RISC mechanism, demonstrates similar properties was unknown.
We repeated the above experiments using mismatched reporters for miR-451 (Supplementary Figure S1c). Our results showed that the ability of sli-451, designed based on mouse pre-miR-451 (mmiR-451), to tolerate mismatches was significantly higher than that of an siRNA mimic of mature miR-451 (si-451) across all target regions (P < 0.0001). This difference was larger for targets that had mismatches in the central or 3supp region than in the seed region of the target sequence. In contrast, we observed almost identical knockdown efficiency by sli-451 and si-451 of two targets with G:U wobble pair (A8G, A14G), and there were no significant differences in silencing of mismatched targets within si-451 or sli-451 when grouped by target regions (P > 0.3) (Figure 3B and C and Table 1 and Supplementary Table S1).
Taken together, our results indicate that sli-siRNAs are generally more potent than siRNAs for targets mismatched by 1 or 2 nts and that this phenomenon is sequence context-dependent. Furthermore, the extent of the difference in the silencing potency between sli-siRNA and siRNA depends on the general location of the mismatches within the target sequence.
Sli-siRNAs have higher potency against mismatched targets than Dicer-processed siRNAs
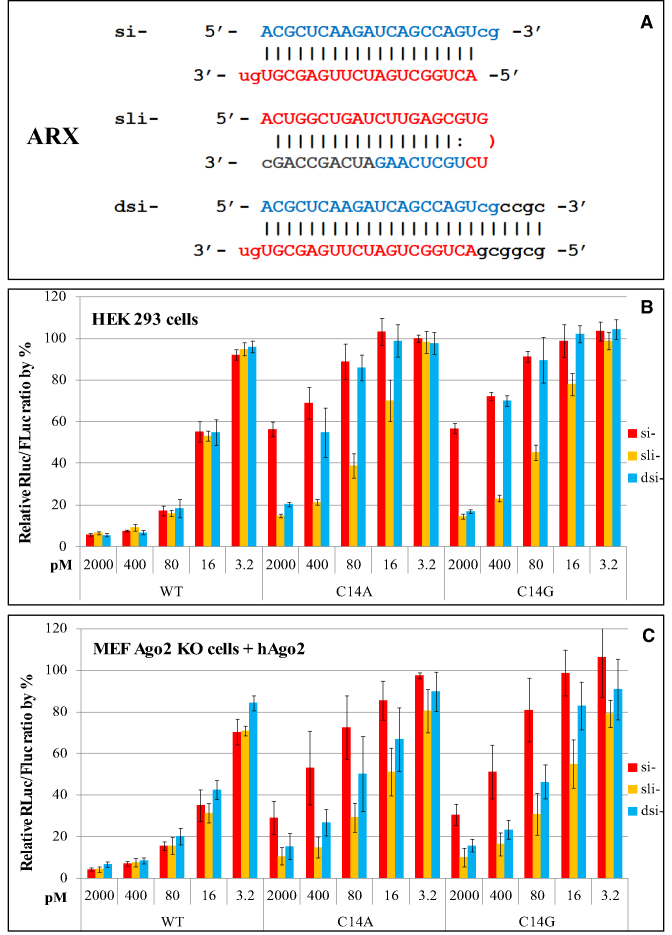
The siRNAs used in the above experiments are mimics of Dicer-generated 21-mer products with 2-nt 3′ end overhangs and do not require Dicer to function (10). To test whether the observed difference in silencing potency was directly related to Dicer processing or if the effect is specific to the synthetic artificial 21-mers, we compared the silencing effects of sli-ARX with that of si-ARX and a 27/25-mer Dicer substrate siRNA (dsi-), a class of 27-mer siRNAs that need Dicer processing to be functional (30,31) (Figure 4A).
Figure 4.
Knockdown of mismatched targets by sli-siRNA-ARX, siRNA-ARX and dsiRNA-ARX at five different concentrations. (A) Nucleotide sequences of siRNA-ARX (si-), sli-siRNA-ARX (sli-) and dsiRNA-ARX (dsi-) molecules. (B) Knockdown efficiency of the fully complementary WT reporter and two position 14C mutated reporters (C14A and C14G) by sli-siRNA-ARX (sli-), siRNA-ARX (si-) and dsiRNA-ARX (dsi-) in HEK-293 cells. (C) Knockdown of WT, C14A and C14G reporters co-transfected with an hAgo2 expression plasmid, and RNAi reagents into MEF Ago2 KO cells. Normalized relative Rluc/Fluc ratios are plotted against the concentrations of each RNAi in pM. Error bars represent standard deviation of the mean.
Using the same reporters from the above ARX reporter assay in HEK-293 cells, our data showed that si-, sli- and dsi-ARX molecules similarly silenced the fully complementary reporter, but almost all concentrations (3.2–2000 pM) of sli-ARX showed higher target knockdown efficiency for targets carrying mutations located in the 3supp region (C14A and C14G) than si-ARX and dsi-ARX (Figure 4B). We also observed similar results in Ago2 knockout (KO) mouse embryonic fibroblast (MEF) cells transfected with an hAgo2 expression vector (32) (Figure 4C). These results demonstrate that our Dicer substrate siRNA and classical 21-mer siRNA generally had comparable silencing potency for both fully complementary targets and mismatched targets and corroborate the finding that Dicer-generated siRNAs have different knockdown effects on mismatched targets than Ago2-processed siRNA.
siRNA overhangs minimally reduce silencing potency for mismatched targets
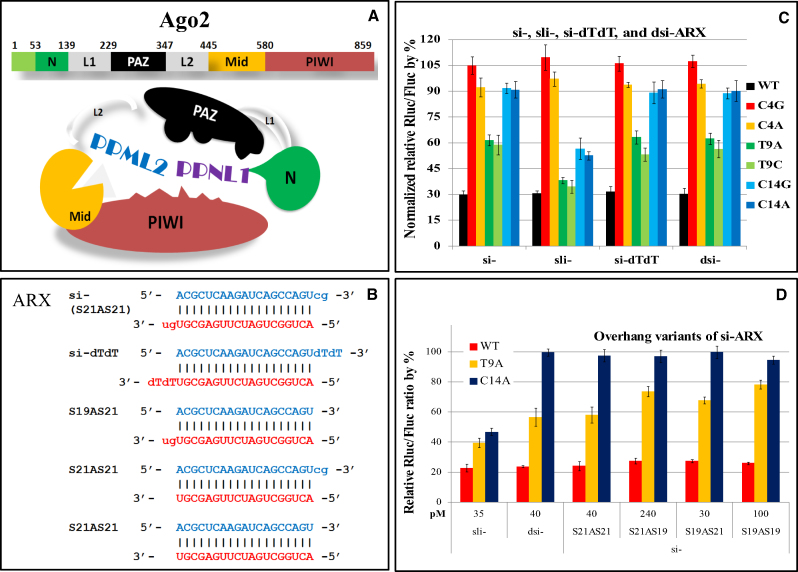
Dicer-processed siRNAs have a 2-nt 3′ overhang and the 3′ overhang of the guide strand is anchored into the pocket in the PAZ domain of Agos (33,34). Unlike the duplex siRNAs, sli-siRNAs form a small 4-nt loop from nts 19 to 22 (Figure 1B). It is unlikely that the 4-nt loop region will fit into the 3′ overhang anchoring pocket in the PAZ domain, instead, it may reside in a groove formed by the PIWI, PAZ and N domains and L1 linker (PPNL1 groove) (Figure 5A).
Figure 5.
Knockdown of mismatched targets by sli- ARX, si-ARX and other siRNA-ARX variants. (A) A schematic representation of Ago2. A full-length Ago2 molecule contains L1 and L2 linkers, as well as N, PAZ, Mid and PIWI domains. These domains form two major grooves: the PPNL1 groove formed by PAZ-L1-N-PAZ and the PPML2 groove formed by PIWI-Mid-L2-PAZ. (B) Nucleotide sequences of si-ARX variants. (C) Knockdown of ARX reporters by 21-mer siRNA (si-), sli-siRNA (sli-) and the 27/25-mer Dicer substrate siRNA (dsi-). (D) Knockdown of mismatched targets by sli-, dsi-, and the four combinations of siRNA 3′ overhang variants from panel C in HEK-293 cells. Normalized relative Rluc/Fluc ratios are plotted against the concentrations of each RNAi in pM. Error bars represent standard deviation of the mean.
To test the potential effect of the presence of the overhangs on the potency of siRNAs against mismatched targets, we compared the knockdown efficiency of sli-ARX, si-ARX, and dsi-ARX to that of si-ARX-dTdT (si-dTdT), containing a deoxythymidine dinucleotide (dTdT) overhang, a popular design standard for classical siRNAs (Figure 5B). First, our results demonstrated that mismatches in the seed region (reporters C4G and C4A) abolished the silencing effect of all four types of RNAi triggers. Secondly, sli-ARX had a better target knockdown effect than any of the three siRNA variants for targets carrying mutations located in the central region (reporters T9A and T9C) or 3supp region (reporters C14G and C14A) (Figure 5C). These results correlated well with q-PCR results (Supplementary Figure S2), although the q-PCR data showed smaller differences in knockdown efficiency, probably because q-PCR only measures knockdown efficiency at the RNA level, whereas the reporter assay measures luciferase activity at the protein level, which reflects the outcomes of both RNA cleavage and translational repression. The data also imply that the siRNA with the 2-nt (dTdT) overhang that cannot base pair with the target and the siRNA with the 2-nt overhang that can base pair with the target have similar potency for both fully complementary targets and mismatched targets.
siRNA potency against fully complementary targets is not a major factor contributing to the lower silencing of mismatched targets by si-RISCs
A simple way to manipulate the potency of an siRNA is to manipulate the overhangs. Overhang manipulations can dramatically affect the potency of classical siRNAs, but they will not change the chemical properties of nts in the stem region of the RNA duplex, which are the nts that base pair with target during silencing (Figure 1A).
To test whether siRNA variants with altered potency against the fully complementary target also have altered potency for mismatched targets, we compared dsi-ARX, sli-ARX and four different overhang combinations of siRNA-ARX with 2-nt 3′ overhangs on one, both, or neither strand of the duplex siRNA (Figure 5B). S21AS21 was made by annealing the 21-mer passenger, or sense (S), strand with the 21-mer guide or antisense (AS), strand—the equivalent of si-ARX, the 21-mer siRNA with two 2-nt 3′ end overhangs; S21AS19 was made by annealing the 21-mer S to the 19-mer AS (without the 2-nt overhang); S19AS21 was made by annealing the 19-mer S (without the 2-nt overhang) to the 21-mer AS; S19AS19,the blunt-ends 19-mer siRNA, was made by annealing the 19-mer S to the 19-mer AS (both without 2-nt overhangs).
Our results showed that overhangs can clearly affect siRNA potency, as different concentrations are needed to achieve a similar knockdown effect for the same fully complementary targets: 40 pM for S21AS21-ARX (the equivalent of si-ARX; 21-nt S and AS strands compete for loading onto RISC), 240 pM for S21AS19-ARX (the 21-nt S strand is favored for RISC loading), 30 pM for S19AS21-ARX (the 21-nt AS strand is favored for RISC loading), and 100 pM for S19AS19-ARX (neither 19-nt strand is favored for RISC loading; S and AS strands compete for loading onto RISC) to knockdown the WT reporter by ∼80%. When the above concentrations were used, overhang variants and the canonical siRNA achieved similar knockdown of the two mismatched targets (T9A and C14A), which were lower than knockdowns by sli-ARX for the same targets (Figure 5D).
Therefore, although the potency of RNAi molecules may contribute to the difference in silencing of mismatched targets between si- and sli-RNAs, it is unlikely that the potency of an siRNA acts as a major factor in lower silencing of mismatch silencing by si-RISCs.
Ago2 slicer activity is important for the greater silencing of mismatched targets by sli-RISCs
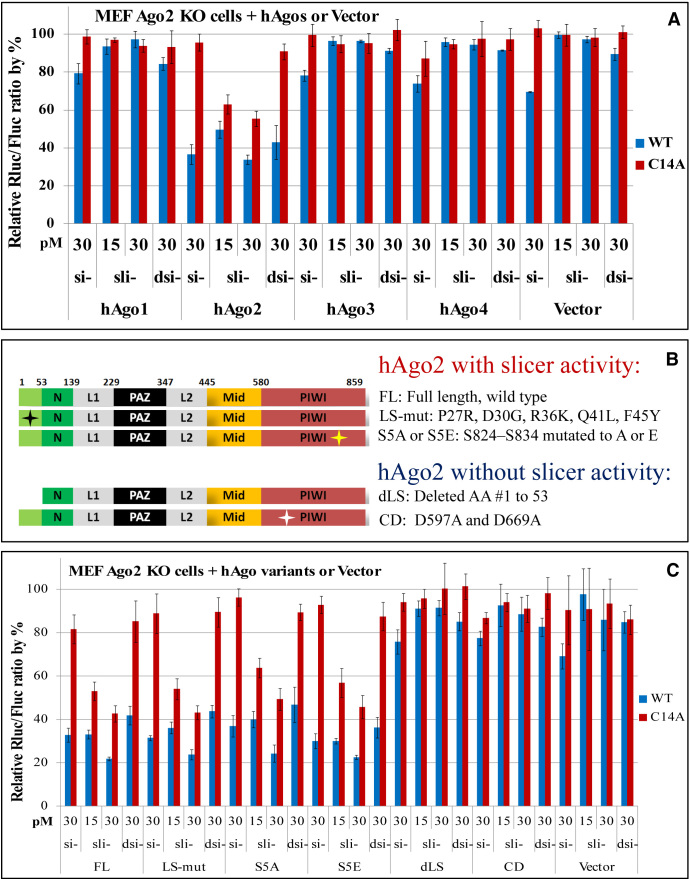
The greater potency of sli-siRNAs compared to siRNAs at knocking down mismatched targets results from the total silencing effect from a combination of target cleavage and target repression. Fully complementary targets of both sli-siRNAs and siRNAs are sliced by Ago2, and the cleaved RNAs are degraded. In contrast, partially complementary targets can be repressed by all Agos. The Ago-formed RISCs bind targets, then transport and store the repressed RNA in P-bodies for degradation or reentry into translation (35,36). In order to tease out the contributions of individual Agos to the higher silencing potency of sli-RISCs for mismatched targets, we performed experiments in MEF Ago2 KO cells transfected with the expression vectors of hAgos 1 through 4.
We first tested new batches of diluted si-, sli-, and dsi-ARX, and they showed similar potency for the fully complementary targets in HEK-293 cells (Supplementary Figure S3A). Then, the same batches of diluted si-, sli- and dsi-ARX were tested in MEF Ago2 KO cells transfected with an hAgo2-expressing vector. When co-transfected with hAgo2, the knockdown of the fully complementary WT reporter using 30 pM si- or dsi-ARX was in between that using 15 pM and 30 pM sli-ARX (Supplementary Figure S3B).
To compare knockdown efficiency for mismatched targets, 15 and 30 pM of sli-siRNAs or 30pM of siRNAs were co-transfected with hAgo2 or the non-slicing hAgos (hAgo1, hAgo3 or hAgo4) into MEF Ago2 KO cells. Co-transfection of si-ARX with hAgo2 in MEF Ago2 KO cells resulted in the greatest knockdown (∼60%) of the WT reporter, compared to co-transfection with hAgo1, hAgo3 or hAgo4. In contrast, co-transfection with an empty vector led to only ∼30% knockdown of the WT reporter by si-ARX. Therefore, in our assay, about 50% of the knockdown efficiency of si-ARX for the WT reporter comes from Ago2-mediated silencing (Figure 6A). When we co-transfected si-ARX with hAgo1, hAgo3 or hAgo4 into the MEF Ago2 KO cells, knockdown of the WT reporter dropped by 7% compared to co-transfection with an empty vector, indicating that the excess of non-slicing Agos may reduce the potency of si- toward fully complementary targets slightly, as measured by our reporter assay. The same set of data also showed that, although si- or dsi-ARX could repress the WT target in the absence of Ago2 (20–30%), they were much less potent in repressing the C14A 3supp mismatch reporter (<10%). When hAgo2—but not hAgo1, hAgo3, hAgo4 or an empty vector—was transfected into MEF Ago2 KO cells, sli-ARX (at both 15 and 30 pM) knocked down C14A more efficiently than 30 pM si- or dsi-ARX (Figure 6A).
Figure 6.
Knockdown of the ARX reporters in Ago2 KO MEF cells co-transfected with non-slicing Agos and Ago2 mutant expression plasmids. (A) Knockdown of WT and C14A ARX reporters in Ago2 KO MEF cells by si-, sli- and dsi-ARX. Reporter and RNAi reagents were co-transfected with hAgo1, hAgo2, hAgo3, and hAgo4 expression plasmids, or the cloning vector plasmid. (B) Schematic representations of Ago2 variants. (C) Knockdown of WT and C14A ARX reporters in MEF Ago2 KO cells by si-, sli- and dsi-ARX. Reporter and RNAi reagents were co-transfected with Ago2 mutant expression plasmids or a vector plasmid. Normalized relative Rluc/Fluc ratios are plotted against the concentrations of each RNAi in pM. Error bars represent standard deviation of the mean.
We were surprised that excessive non-slicing Agos slightly reduced the potency of si- and dsi-RNAs toward fully complementary targets, so we performed the same transfection in HeLa cells. Again we observed that excessive non-slicing Agos reduce the knockdown efficiency of si-, sli- and dsi-ARX toward fully complementary targets from 50% to ∼40%, whereas overexpression of Ago2 slightly enhanced the knockdown of fully complementary targets by si- sli-, and dsi-ARX from 50% to 60% (Supplementary Figures S3C and D). These results support the conclusion that a large part of the observed difference in mismatched target silencing by sli-siRNA and siRNA comes from Ago2-mediated target cleavage or repression, whereas the participation of non-slicing Ago in target silencing may reduce this effect.
In order to further support the above conclusion that Ago2 makes sli-siRNAs more potent against mismatched targets, we performed several reporter assays in MEF Ago2 KO cells that were transfected with full length (FL) or catalytically defective hAgo2. Several hAgo2 variants were constructed for this study: 5 conserved amino acids in the leader sequence of Ago2 were mutated to match the corresponding amino acids in the same region of hAgo1 to generate LS-mut (37); hAgo2 multi-site phosphorylation mutants S5A (five residues S824–S834 mutated to A) and S5E (five residues S824–S834 mutated to E) have enhanced slicer activity (38); truncated dLS lacks the leader sequence (before the N domain, containing a motif required for slicer activity) and does not exhibit slicer activity (39–41); and the catalytically defective hAgo2 mutant (CD) carries double mutations of D597A and D669A (Figure 6B).
When co-transfected with any of the RNAi molecules, the hAgo2 variants retaining slicer activity (LS-mut, S5A, and S5E) exhibited potent knockdown of the ARX WT reporter, similar to that of the wild type full length (FL) hAgo2. In contrast, the slicer activity-deficient hAgo2 mutants (dLS and CD) had reduced knockdown effects for WT reporter, similar to that of an empty vector. sli-ARX exhibited much more potent knockdown of 3supp-mismatched target reporter C14A than did si- and dsi-ARX in cells transfected with the slicer activity-competent hAgo2 variants (Figure 6C). These results further support the idea that the slicer activity of Ago2 mediated target cleavage, not Ago2 mediated target repression, plays an important role in the observed difference between sli-siRNA and siRNA in silencing mismatched targets.
sli-RISCs use 23- to 26-mer guide RNAs for function
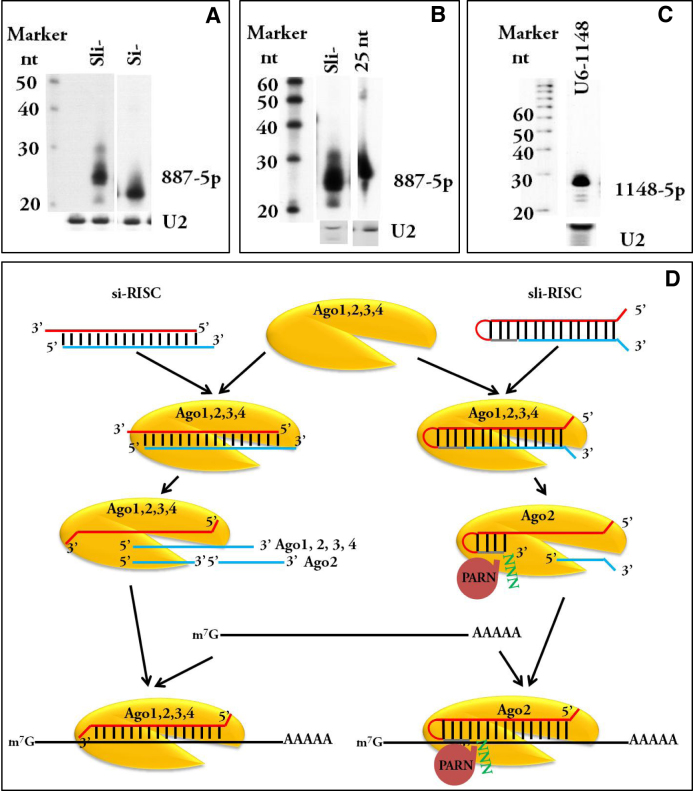
The one-step silencing pathway of Ago2-miR-451 mimics also involves trimming/tailing processes at the 3′ end of the guide strand after Ago2 has sliced the passenger strand (7–9,12). The poly(A)-specific ribonuclease (PARN) has been identified as the enzyme responsible for 3′-5′ trimming of pre-miR-451 mimics that have been sliced by Ago2 (22). Data in the same publication also suggest that the trimming/tailing step per se is dispensable for the silencing function of pre-miR-451 in vivo and that sli-RISCs can use intermediate guide RNAs that are longer than 22-mer mature guide RNA for target silencing. This concept is supported by our previously published data: agshRNA-1148, the stably expressed human U6-driven sli-siRNA-1148 that targets RRM2, can reduce RRM2 protein levels by over 50% in Western blot analysis, even though the mature form mainly exists in a band around 27-nt, not 22-nt, according to Northern blot analysis (10).
We analyzed deep sequencing reads of both human and mouse miR-451 isoforms documented in the microRNA sequence database miRBase and found that the 23- to 26-nt forms comprise almost 70% of mouse miR-451 isoforms (or isomiRs; reflected as reads without tailing/trimming nts) (Supplementary Table S2). Published Northern blot data and in vitro processing data of pre-miR-451 mimics indicate that sli-siRNA also exists mainly as 23- to 26-nt processed products (10,12,22). Therefore, unlike si-RISCs that are mixed assemblies incorporating Agos that are loaded primarily with uniform 21-mer guide RNAs, sli-RISCs could contain Ago2 loaded with different isoforms, mainly 23- to 26-mers, of guide RNAs.
These data prompted us to perform deep sequencing of sli-887, -1148 and -1354, which target RRM2 (10); sli-ARX; and the sli-siRNA versions—sliced miRNAs (sli-miRs)—of human, mouse, and zebra fish pre-miR-451 (hsli-, msli- and dsli-miR-451). In order to perform a side-by-side comparison of an isomiR of a Dicer-processed native miRNA with a miRNA that was artificially expressed as an miR-451 mimic, we generated a miR-21 knockout HeLa cell line that stably expresses sli-miR-21 (17). The first base U in canonical miR-21 was changed to A to facilitate expression of sli-miR-21 by a modified human U6 promoter (10).
The reads matched to each sli-siRNA/miRNA were analyzed either with tailing/trimming nts (Table 2) or without tailing/trimming nts (Supplementary Table S3) (see Materials and Methods for detailed description). The length distribution of reads without tailing/trimming nts agrees well with the length distribution of reads with tailing/trimming nts. The length distribution of reads with tailing/trimming nts showed that hsli-, msli-, and dsli-miR-451 resembled the miR-451 isoforms documented in miRBase: ∼40% reads are 19- to 22-nt forms and ∼50% are 23- to 26-nt forms, with 21-, 23- and 26-nt forms as the three major isoforms. sli-887 mainly exists in its 23- and 24-nt forms; sli-1148 is one of the molecules for which most reads are 25- to 30-nts, suggesting that the trimming for this sli-RNA has very low efficiency; sli-1354 mainly exists in its 23-, 24- and 27-nt forms; and sli-ARX also exhibited low efficiency trimming, mainly existing in its 25- and 30-nt forms with a high percentage of 26- to 29-nt forms. About 75% sli-miR-21 exists in 23- to 25-nt forms. In contrast, about 80% of native iso-miR-21 in HeLa cells exists in 22- and 23-nt forms (Tables 2 and Supplementary Table S3). The read abundances of the processed isoforms of chemically synthesized sli-887 that were transfected into HEK-293 cells and of stably expressed sli-1148 in HCT116 cells correspond well with northern blot analysis (Figure 7A–C and Supplemental Figure S4) (10).
Table 2.
Deep sequence read length distribution—reads without trimming/tailing. Isoforms were classified by read length. Listed are the proportions of read counts of each read length to the total counts of all isoforms.
| Length (nt) | sli-miR-451 | sli-siRNA | miR-21 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Human | Mouse | Fish | sli-887 | sli-1148 | sli-1354 | sli-ARX | sli-miR-21 | miR-21 | |
| 16 | 0.13 | 0.13 | 0.14 | 0.00 | 0.01 | 0.00 | 0.01 | 0.09 | 0.13 |
| 17 | 0.05 | 0.03 | 0.01 | 0.00 | 0.00 | 0.00 | 0.06 | 0.01 | 0.02 |
| 18 | 0.19 | 0.04 | 0.05 | 0.00 | 0.01 | 0.07 | 0.09 | 0.18 | 0.22 |
| 19 | 4.53 | 3.87 | 4.60 | 0.07 | 0.05 | 0.26 | 0.82 | 0.07 | 0.11 |
| 20 | 7.14 | 4.91 | 4.96 | 0.19 | 0.41 | 0.34 | 4.57 | 0.14 | 0.15 |
| 21 | 20.55 | 20.72 | 17.28 | 0.57 | 1.44 | 1.94 | 1.82 | 2.84 | 4.37 |
| 22 | 9.59 | 8.92 | 8.18 | 1.58 | 0.92 | 0.72 | 0.43 | 13.15 | 27.89 |
| 23 | 15.97 | 11.31 | 19.31 | 74.68 | 1.69 | 69.49 | 2.27 | 24.66 | 51.93 |
| 24 | 9.54 | 9.47 | 12.95 | 20.27 | 3.27 | 4.85 | 3.76 | 25.25 | 14.56 |
| 25 | 7.19 | 5.22 | 7.99 | 0.80 | 4.56 | 0.89 | 15.64 | 23.69 | 0.14 |
| 26 | 18.13 | 27.50 | 18.06 | 0.46 | 3.60 | 1.06 | 5.19 | 2.59 | 0.03 |
| 27 | 1.13 | 1.41 | 0.91 | 0.09 | 9.28 | 8.66 | 2.01 | 2.69 | 0.01 |
| 28 | 0.56 | 0.65 | 0.32 | 0.03 | 7.58 | 1.48 | 4.47 | 0.43 | 0.01 |
| 29 | 0.99 | 1.26 | 0.49 | 0.04 | 6.55 | 0.50 | 4.08 | 1.61 | 0.00 |
| 30 | 0.23 | 0.35 | 0.18 | 0.04 | 3.73 | 5.29 | 17.36 | 1.68 | 0.00 |
| 31 | 0.57 | 0.79 | 0.34 | 0.43 | 26.57 | 3.18 | 19.36 | 0.06 | 0.00 |
| 32 | 0.04 | 0.06 | 0.02 | 0.09 | 12.74 | 0.41 | 5.65 | 0.00 | 0.00 |
| 33 | 0.03 | 0.05 | 0.02 | 0.01 | 5.70 | 0.02 | 3.23 | 0.00 | 0.00 |
| 34 | 0.00 | 0.00 | 0.00 | 0.00 | 3.15 | 0.00 | 2.93 | 0.00 | 0.00 |
| 35 | 0.00 | 0.00 | 0.00 | 0.00 | 1.72 | 0.00 | 2.46 | 0.01 | 0.00 |
| 36 | 0.00 | 0.00 | 0.00 | 0.00 | 0.85 | 0.00 | 1.18 | 0.00 | 0.00 |
| 37 | 0.00 | 0.00 | 0.00 | 0.00 | 0.23 | 0.00 | 0.44 | 0.00 | 0.00 |
| 38 | 0.00 | 0.00 | 0.00 | 0.00 | 0.09 | 0.00 | 0.16 | 0.00 | 0.00 |
| 39 | 0.00 | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | 0.06 | 0.00 | 0.00 |
| 40 | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 | 0.01 | 0.02 | 0.00 | 0.00 |
| Others | 3.45 | 3.32 | 4.19 | 0.66 | 5.80 | 0.83 | 1.93 | 0.84 | 0.42 |
Figure 7.
Proposed function of sli-RISCs versus si-RISCs. (A) Northern blotting to detect chemically synthesized sli-887 and si-887. Processed products of sli-887 are longer than the 21-mer si-887. (B) Northern blotting to detect sli-887 and the 25-nt form of sli-887 (nts 1 to 25). Processed products of sli-887 are shorter than the 25-mer form. (C) Northern blotting to detect processed sli-1148 from a stably expressed sli-1148 driven by a U6 promoter, which mainly exists in an approximately 30-mer isoform, in HCT116 cells. (D) Proposed model of sli-RISC versus si-RISC function: si-RISCs can involve three non-slicing Agos (Ago1, Ago3, and Ago4) and slicing Ago2 loaded with uniform 21-mer guide RNAs; sli-RISCs contain the slicing Ago2 loaded with guide RNAs with various 3′ end lengths, and silencing may couple with 3′ end trimming/tailing by PARN. After the passenger strand is sliced, the hairpin might open during target binding, allowing nts 19 to 22 to base pair (open hairpin hypothesis), or the loop from nts 19 to 22 might remain, allowing only nts 2 to 14 to be used for target binding (partial hairpin hypothesis).
These profiling data showed that the trimming and tailing process at the 3′ end during the maturation of the guide strand of miR-451 mimics is sequence-dependent and that RISCs formed by miR-451 mimics may contain a high percentage of 23- to 26-mers guide RNAs.
DISCUSSION
In this study, we designed a reporter assay to measure RNAi activity against mismatched targets. Our assay in Ago2 KO MEFs showed that our reporters were adequate for measuring the target cleavage activity of RISCs, especially at low concentrations of RNAi reagents. The same data also showed that repression activity by non-slicing Ago-mediated RISCs under our experimental conditions contributed only a small portion of the total RNAi activity measured by our assay.
We showed that, in most cases, a one-nt mismatch between an RNAi molecule and its target, anywhere from nts 3 to 18 in the RNAi molecule (5′ to 3′), negatively affects RNAi silencing efficacy. We also found that sli-siRNAs are generally more tolerant than classical siRNAs for mismatches in the target sequence. This tolerance is sequence context-dependent and usually greater for mismatches located in the 3supp region than those in the seed region or central region of the target. We further showed, si-RISCs formed with Ago2 or non-slicing Agos (including slicer activity-defective Ago2) repressed mismatched targets less than they did fully complementary targets. However, Ago2 slicer activity is primarily responsible for the difference in siRNA and sli-siRNA silencing potency toward mismatched targets, and participation of non-slicing Agos in si-RISC-mediated silencing may dilute this effect.
Although it has been reported that non-slicing Agos can be loaded with pre-miR-451 mimics to form pre-sli-RISCs, only the Ago2-loaded form can be further processed to mature sli-RISCs. It has been proposed that non-slicing Ago-loaded sli-siRNAs are released and reloaded with Ago2 for maturation and function (11,16). Therefore, most sli-RISCs are slicing-competent Ago2-RISCs, whereas classical si-RISCs can be activated with both slicing and non-slicing Agos. The competitive loading of slicing and non-slicing-Agos on siRNAs reduces the amount of siRNAs loaded with Ago2. This reduction of Ago2-si-RISCs may dilute the role of Ago2 slicer activity in siRNA-mediated gene silencing when RNAi is limited.
Non-slicing Agos may dilute the slicing-Ago effect in two ways: (i) non-slicing RISC-mediated repression is much weaker than slicing RISC-mediated target cleavage and (ii) non-slicing RISC-repressed target RNA may be protected from degradation, enabling reentry into translation and even target upregulation under some circumstances (42,43). The target overexpression that we observed in HeLa cells (Supplementary Figures S3C and D) may have resulted from both of these effects, whereas the 7% reduction in target repression in Ago2 KO MEF cells (Figure 6A) may have occurred solely due to the second, target-protection effect. However, because the effects of non-slicing Agos on repression contributed to only a small portion of the total RNAi activity measured by our reporter assay, we conclude that Ago2-mediated target cleavage contributes the most to the greater silencing activity of sli-RNA than si-RNA for mismatched targets.
Our data also showed that the difference between sli-siRNAs and siRNAs in knocking down mismatched targets is usually larger when mismatches are located in the 3supp region. Using three sli-siRNA/siRNA pairs, we showed that the knockdown of targets carrying mismatches in 3supp region is significantly greater for sli-siRNAs and siRNAs (P < 0.0001) (Table 1). Although the differences in silencing efficacy across grouped regions of target mismatches (seed vs. central vs. 3supp) compared within an siRNA or sli-siRNA were generally not as significant as comparisons between sli-siRNAs and siRNAs over all target regions, the comparisons within sli-siRNAs were more significant (by P-value) than those within siRNAs. Three comparisons within sli-siRNAs were significant (P < 0.01 for seed vs. 3supp and seed vs. central in sli-ARX and for central vs. 3supp in sli-887), compared to none within siRNAs (Table 1 and Supplementary Table S1). These data suggest that, in addition to the different amounts of Ago2-loaded guide RNAs in sli-RISCs versus si-RISCs, there could be other factors involved in the differential actions of sli-RISCs and si-RISCs on mismatched targets. Indeed, there is another major difference between activated si-RISCs and sli-RISCs: whereas both ends of the guide strand in si-RISCs are pocketed by the Mid and PAZ domains of Agos so that nts 2 to 19 can fully pair with the target (33,34,44–47), sli-RISC generation is coupled with trimming/tailing at the 3′ end of the guide strand, so sli-RISCs may be loaded with guide sli-siRNAs of various lengths (7–9,22). Activated si-RISCs usually contain uniform 21-mer guide RNAs, whereas activated sli-RISCs are loaded mostly with variable guide RNAs of 23 to 26 nts.
An interesting question is how sli-RISCs use longer guide RNAs to silence targets. After Ago2 passenger strand slicing and seed binding, do the remaining nts of the sli-siRNA guide strand maintain a loop structure or is the loop opened during target base pairing (Figure 1D). In a study on short shRNAs, Dallas et al. proposed an open hairpin model. According to their model, after the passenger strand is sliced, as the guide strand binds to the target, the partially paired loop region of the guide strand opens up by branch migration, enabling the nts of the loop to also base pair with the target (16). Here, we propose the partial hairpin model, an alternative in which the guide RNA remains self-paired, maintaining a stem-loop structure at the apical stem region during targeting.
According to the open hairpin model, sli-RISCs function similarly to si-RISCs in that seed binding causes the Ago structure to change, promoting base pairing with the 3supp region and the partial hairpin region that has opened up by branch migration. But unlike nts 20 and 21 of siRNAs that bind to the Ago PAZ domain and do not base pair with targets, nts 19 to 22 of sli-RNAs may not bind to the PAZ domain and are free to base pair with targets. The additional pairing of nts 19 to 22 may enhance 3supp base pairing between sli-RNAs and their targets, conferring greater tolerance of the sli-RISC for mismatches in the 3supp region (Figure 1D). However, overall RNAi efficacy depends on the balance between target binding and RISC release. Additional base pairs will not only increase RNAi/target binding but will increase the dwell time of the guide RNA on a target, reducing the RISC turnover rate. Therefore, additional base pairs may not necessarily lead to overall higher potency against matched or mismatched targets.
The open hairpin model also suggests that mismatches in both types of RISCs will have similar effects on Ago2 structure and slicer activity. However, although there are no differences in the silencing of GU wobble paired targets between si-RISCs and sli-RISCs, the difference in the silencing of mismatched targets by si-RISCs and sli-RISCs is sequence context-dependent. These results suggest that mismatches between the RNAi molecules and their targets affect Ago2 slicer activity differently in si-RISCs and sli-RISCs, causing differential tolerance for mismatched base pairing with targets. Because the components of Ago2 si-RISCs and sli-RISCs are otherwise the same, we believe, in opposition to the open hairpin model, that the difference in RISC activity likely arises due to changes in Ago2 slicer activity upon binding to differing structures of the guide RNAs in sli-RISCs and si-RISCs.
According to our partial hairpin model, the guide RNA maintains its 4-nt loop region between base-paired nts 19 and 22, as well as a 3 to 4 base-paired apical stem region (nts 15–18 paired with nts 26–23, respectively), an unpaired region formed by nts 1 to 14 or 15, and possibly some tailing bases at 3′end, forming a cane (δ)-shaped guide RNA (Figures 1D and 7D, and Supplementary Figure S5). The δ-shaped guide siRNA may only use the first 14 nts for base pairing with target inside the RISC (Figures 1C, D and 7D), resulting in a lower energy barrier for sli-RNA/target base pairing, higher RISC turnover rate, and greater tolerance for mismatches in the 3supp region compared to siRNAs.
However, according to the partial hairpin model, the tailed/trimmed bases at the 3′ end of the guide strand are inside Ago2. Therefore, target silencing could not likely occur simultaneously with tailing/trimming, as the current and previously published data suggest (10,22). Alternatively, the open hairpin model suggests that the 3′ end of the guide RNA is outside of Ago2, facilitating concurrent silencing and 3′ end tailing/trimming (Figures 1D and 7D). Our findings are not inconsistent with the partial hairpin model, however, because some guide RNAs may be silencing targets while other guide RNAs are being tailed/trimmed on separate sli-RISCs. Therefore, both the partial hairpin model and the open hairpin model remain working hypotheses, and future studies are necessary to elucidate the functional mechanisms of sli-siRNAs in vivo.
Our partial hairpin model posits that sli-siRNAs function through a one-step RISC activation model, in which the target-silencing function of sli-RISC is activated during cleavage of the sli-siRNA passenger strand. We hypothesize that, in sli-RISCs loaded with δ-shaped guide siRNAs, the loop region of the guide RNAs is confined inside the niche formed by PAZ, N, and PIWI domains, maintaining the sli-RISC in a slicing-competent conformation (Figures 1D, 5A and 7D, and Supplemental Figure S5A). The miR-451 maturation mechanism implies that the small 4-nt loop structure in sli-siRNAs does not affect Ago2 slicer function. Our previous study on the properties of sli-siRNA also supports the δ-shaped guide RNA sli-RISC hypothesis (10). In contrast to the tolerated mutations in the 3supp region of a target in the current study, we previously observed that mismatches in the 3supp region of the sli-siRNA guide RNA stem (i.e. self-paired with nts in its trimming/tailing region in the apical stem region) had a stronger effect on both sli-siRNA processing and silencing potency than mismatches in the basal stem (i.e. in the seed region, base-paired with the last 10 nts of the sli-siRNA) (Figure 1B). This result was demonstrated in agshRNA-1148 processing. While a GU pair in the 3supp region has a dramatic effect, a mismatch in the seed region has almost no effect on agshRNA-1148 processing (Supplementary Figure S5c). It seems that base pairs in the apical stem region of a sli-siRNA are critical for maturation and maintenance of the sli-RISC in a catalytically competent conformation, whereas base pairs in the 3supp region between sli-siRNA and targets are less important for silencing. This observation suggests that the PPNL1 groove of Ago2, where the 3supp region of sli-siRNAs resides, is important for maintaining the sli-RISC in a catalytically competent conformation (Figure 1D and Supplementary Figure 5A). This role of PPNL1 is consistent with the finding that the domains at the N terminus (N-L1-PAZ) of Ago2 are critical for correctly aligning the target RNA with the Ago2 catalytic center for slicing. The PPML2 groove—formed by the PAZ, PIWI and Mid domains and L2 linker—where the seed region resides, has more flexibility for mismatches or wobble base pairs and may facilitate the release of short fragments and target binding (Figure 5A) (33,34,39–41,47,48).
Unlike sli-RISCs, si-RISCs can be activated without concurrent cleavage of the passenger strand. si-RISCs can be loaded with both intact and segmented passenger strands, and in any case, non-slicing Ago-formed si-RISCs are not capable of cleaving the passenger strand (49,50). Instead, si-RISCs require a multi-step process: the duplex siRNA is loaded, depending on the thermodynamic properties of the ends of the RNA duplex, and the 5′ phosphate group of the siRNA is anchored to the Mid domain of an Ago; the guide strand is selected, which involves opening of the duplex by the N domain of the siRNA, and the passenger strand is ejected (51); and once a mature si-RISC is formed, the seed of the guide strand nucleates its binding to the target, which promotes 3supp region binding to the target and initiates silencing (50,51).
The activated si-RISC carries a guide RNA that resembles a horizontal ς-shaped RNA inside the binding grooves of the Ago: the 5′ end of the strand is anchored to the Mid domain, and the 3′ end of the strand is tethered to the PAZ domain (Figures 1C and 7D, and Supplemental Figure S5C). Therefore, we proposed that after binding to the seed region of a target, both the PAZ and N domains of si-RISCs are rotated away from the Mid-PIWI domains to facilitate 3supp region base-pairing with target for gene silencing (52–56). Our one-step working model of sli-RISCs suggests that the rotation of both the PAZ and N domains away from the Mid-PIWI domains that enables 3supp binding in si-RISCs is not necessary for sli-RISC silencing function (53).
The current study suggests that guide strands from sli-siRNAs could have more off-target effects than guide strands from siRNAs. 3supp pairing was previously reported as a major determinant of Ago target specificity, which has been further supported by deep sequencing of miRNA-target chimeras (23,28,29). Our results revealed a previously unknown pivotal role of Dicer and non-slicing Agos in determining target specificity. The Dicer processing step in siRNA biogenesis seems to play multiple roles: producing siRNA duplexes that can be loaded onto non-slicing Agos, which affect RISC function and alter siRNA target specificity. Because target repression is the dominant gene silencing strategy used in animals, it is conceivable that the dominance of the classical multi-step siRNA pathway was driven by selection pressure for greater target specificity. The short length of the seed region broadens the target spectrum at the cost of reduced silencing potency. However, the reduced potency could be overcome by using multiple seed sites to enhance on-target effects and achieve synergy in both silencing efficacy and specificity.
Our study was carried out in a reporter system to measure target silencing, in which targets are saturated and the amount of siRNAs is limited. However, for both research and clinical applications, it is necessary to use siRNAs at high concentrations to achieve high-level silencing. Therefore, in contrast with our experimental conditions, siRNAs are often saturated while the amount of targets is usually limited by their biological expression levels. A carefully designed siRNA will avoid mismatched targets, which are often uncommon. It is the genes with matched seed sequences, which usually exist in the hundreds or thousands that really need to be considered for off-target effects in RNAi applications (29,57,58). To this end, off-target effects from the guide strand are not avoidable for sli-siRNAs and siRNAs. However, off-target effects from passenger strands can be reduced by siRNA design (e.g. the S19AS21 form in Figure 5d) or using miR-451 mimics. The passenger strand activity from sli-siRNAs is usually 100- to 1000-fold lower than that of siRNAs (7–10,15). Therefore, overall, sli-siRNAs will have fewer off-target effects than siRNAs and may be a preferable option for research and clinical applications.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our City of Hope colleagues Drs Kerin Higa and Keely Walker for expert editing of this manuscript, as well as Kaniel Cassady for his critical reading and correction of this manuscript. We thank Dr Gregory Hannon for generously providing the Ago2 knockout MEFs. G.S., Y.Y. and A.R. conceived of and designed the experiments and drafted the manuscript. All authors contributed to the experiments and the final version of the manuscript. All authors declare no competing financial interests. Authors would like to thank Louise Shively and Natalie Rosas for their help in this project.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
City of Hope (to A.D.R.); Taiwan Ministry of Health and Welfare Surcharge of Tobacco Products [MOHW105-TDU-B-212-134001 CERC to Y.Y.]. Funding for open access charge: City of Hope (to A.D.R.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Ha M., Kim V.N.. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell. Biol. 2014; 15:509–524. [DOI] [PubMed] [Google Scholar]
- 2. Foulkes W.D., Priest J.R., Duchaine T.F.. DICER1: mutations, microRNAs and mechanisms. Nat. Rev. Cancer. 2014; 14:662–672. [DOI] [PubMed] [Google Scholar]
- 3. Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R.. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005; 436:740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J.. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001; 409:363–366. [DOI] [PubMed] [Google Scholar]
- 5. Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D.. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003; 115:199–208. [DOI] [PubMed] [Google Scholar]
- 6. Khvorova A., Reynolds A., Jayasena S.D.. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003; 115:209–216. [DOI] [PubMed] [Google Scholar]
- 7. Cheloufi S., Dos Santos C.O., Chong M.M., Hannon G.J.. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010; 465:584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cifuentes D., Xue H., Taylor D.W., Patnode H., Mishima Y., Cheloufi S., Ma E., Mane S., Hannon G.J., Lawson N.D. et al. . A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010; 328:1694–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang J.S., Maurin T., Robine N., Rasmussen K.D., Jeffrey K.L., Chandwani R., Papapetrou E.P., Sadelain M., O’Carroll D., Lai E.C.. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:15163–15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun G., Yeh S.Y., Yuan C.W., Chiu M.J., Yung B.S., Yen Y.. Molecular properties, functional mechanisms, and applications of sliced siRNA. Mol. Ther. Nucleic Acids. 2015; 4:e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dueck A., Ziegler C., Eichner A., Berezikov E., Meister G.. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 2012; 40:9850–9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang J.S., Maurin T., Lai E.C.. Functional parameters of Dicer-independent microRNA biogenesis. RNA. 2012; 18:945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Y.P., Schopman N.C., Berkhout B.. Dicer-independent processing of short hairpin RNAs. Nucleic Acids Res. 2013; 41:3723–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ge Q., Ilves H., Dallas A., Kumar P., Shorenstein J., Kazakov S.A., Johnston B.H.. Minimal-length short hairpin RNAs: the relationship of structure and RNAi activity. RNA. 2010; 16:106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma H., Zhang J., Wu H.. Designing Ago2-specific siRNA/shRNA to avoid competition with endogenous miRNAs. Mol. Ther. Nucleic Acids. 2014; 3:e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dallas A., Ilves H., Ge Q., Kumar P., Shorenstein J., Kazakov S.A., Cuellar T.L., McManus M.T., Behlke M.A., Johnston B.H.. Right- and left-loop short shRNAs have distinct and unusual mechanisms of gene silencing. Nucleic Acids Res. 2012; 40:9255–9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen B., Chen X., Wu X., Wang X., Wang Y., Lin T.Y., Kurata J., Wu J., Vonderfecht S., Sun G. et al. . Disruption of microRNA-21 by TALEN leads to diminished cell transformation and increased expression of cell-environment interaction genes. Cancer Lett. 2015; 356:506–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun G., Yan J., Noltner K., Feng J., Li H., Sarkis D.A., Sommer S.S., Rossi J.J.. SNPs in human miRNA genes affect biogenesis and function. RNA. 2009; 15:1640–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun G., Wu X., Wang J., Li H., Li X., Gao H., Rossi J., Yen Y.. A bias-reducing strategy in profiling small RNAs using Solexa. RNA. 2011; 17:2256–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun G., Cheng Y.W., Lai L., Huang T.C., Wang J., Wu X., Wang Y., Huang Y., Wang J., Zhang K. et al. . Signature miRNAs in colorectal cancers were revealed using a bias reduction small RNA deep sequencing protocol. Oncotarget. 2016; 7:3857–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baras A.S., Mitchell C.J., Myers J.R., Gupta S., Weng L.C., Ashton J.M., Cornish T.C., Pandey A., Halushka M.K.. miRge - A Multiplexed Method of Processing Small RNA-Seq Data to Determine MicroRNA Entropy. PLoS One. 2015; 10:e0143066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoda M., Cifuentes D., Izumi N., Sakaguchi Y., Suzuki T., Giraldez A.J., Tomari Y.. Poly(A)-specific ribonuclease mediates 3′-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013; 5:715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wee L.M., Flores-Jasso C.F., Salomon W.E., Zamore P.D.. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012; 151:1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lewis B.P., Burge C.B., Bartel D.P.. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005; 120:15–20. [DOI] [PubMed] [Google Scholar]
- 25. Schirle N.T., Sheu-Gruttadauria J., MacRae I.J.. Gene regulation. Structural basis for microRNA targeting. Science. 2014; 346:608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B.. Prediction of mammalian microRNA targets. Cell. 2003; 115:787–798. [DOI] [PubMed] [Google Scholar]
- 27. Brennecke J., Stark A., Russell R.B., Cohen S.M.. Principles of microRNA-target recognition. PLoS Biol. 2005; 3:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moore M.J., Scheel T.K., Luna J.M., Park C.Y., Fak J.J., Nishiuchi E., Rice C.M., Darnell R.B.. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 2015; 6:8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P.. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007; 27:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim D.H., Behlke M.A., Rose S.D., Chang M.S., Choi S., Rossi J.J.. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005; 23:222–226. [DOI] [PubMed] [Google Scholar]
- 31. Rose S.D., Kim D.H., Amarzguioui M., Heidel J.D., Collingwood M.A., Davis M.E., Rossi J.J., Behlke M.A.. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005; 33:4140–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J.. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004; 305:1437–1441. [DOI] [PubMed] [Google Scholar]
- 33. Ma J.B., Ye K., Patel D.J.. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004; 429:318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lingel A., Simon B., Izaurralde E., Sattler M.. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol. 2004; 11:576–577. [DOI] [PubMed] [Google Scholar]
- 35. Rehwinkel J., Behm-Ansmant I., Gatfield D., Izaurralde E.. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005; 11:1640–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R.. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005; 7:719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharma C., Mohanty D.. Sequence- and structure-based analysis of proteins involved in miRNA biogenesis. J. Biomol. Struct. Dyn. 2017; 1–13. [DOI] [PubMed] [Google Scholar]
- 38. Golden R.J., Chen B., Li T., Braun J., Manjunath H., Chen X., Wu J., Schmid V., Chang T.C., Kopp F. et al. . An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature. 2017; 542:197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hauptmann J., Kater L., Loffler P., Merkl R., Meister G.. Generation of catalytic human Ago4 identifies structural elements important for RNA cleavage. RNA. 2014; 20:1532–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hauptmann J., Dueck A., Harlander S., Pfaff J., Merkl R., Meister G.. Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nat. Struct. Mol. Biol. 2013; 20:814–817. [DOI] [PubMed] [Google Scholar]
- 41. Faehnle C.R., Elkayam E., Haase A.D., Hannon G.J., Joshua-Tor L.. The making of a slicer: activation of human Argonaute-1. Cell Rep. 2013; 3:1901–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vasudevan S., Tong Y., Steitz J.A.. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007; 318:1931–1934. [DOI] [PubMed] [Google Scholar]
- 43. Vasudevan S., Steitz J.A.. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007; 128:1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frank F., Fabian M.R., Stepinski J., Jemielity J., Darzynkiewicz E., Sonenberg N., Nagar B.. Structural analysis of 5′-mRNA-cap interactions with the human AGO2 MID domain. EMBO Rep. 2011; 12:415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Frank F., Sonenberg N., Nagar B.. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010; 465:818–822. [DOI] [PubMed] [Google Scholar]
- 46. Boland A., Tritschler F., Heimstadt S., Izaurralde E., Weichenrieder O.. Crystal structure and ligand binding of the MID domain of a eukaryotic Argonaute protein. EMBO Rep. 2010; 11:522–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gu S., Jin L., Huang Y., Zhang F., Kay M.A.. Slicing-independent RISC activation requires the argonaute PAZ domain. Curr. Biol. 2012; 22:1536–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kwak P.B., Tomari Y.. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol. 2012; 19:145–151. [DOI] [PubMed] [Google Scholar]
- 49. Bramsen J.B., Laursen M.B., Damgaard C.K., Lena S.W., Babu B.R., Wengel J., Kjems J.. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res. 2007; 35:5886–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Park J.H., Shin C.. Slicer-independent mechanism drives small-RNA strand separation during human RISC assembly. Nucleic Acids Res. 2015; 43:9418–9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kawamata T., Tomari Y.. Making RISC. Trends Biochem. Sci. 2010; 35:368–376. [DOI] [PubMed] [Google Scholar]
- 52. Tomari Y., Zamore P.D.. Perspective: machines for RNAi. Genes Dev. 2005; 19:517–529. [DOI] [PubMed] [Google Scholar]
- 53. Jinek M., Doudna J.A.. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009; 457:405–412. [DOI] [PubMed] [Google Scholar]
- 54. Rashid U.J., Paterok D., Koglin A., Gohlke H., Piehler J., Chen J.C.. Structure of Aquifex aeolicus argonaute highlights conformational flexibility of the PAZ domain as a potential regulator of RNA-induced silencing complex function. J, Biol. Chem. 2007; 282:13824–13832. [DOI] [PubMed] [Google Scholar]
- 55. Wang Y., Juranek S., Li H., Sheng G., Wardle G.S., Tuschl T., Patel D.J.. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009; 461:754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zander A., Holzmeister P., Klose D., Tinnefeld P., Grohmann D.. Single-molecule FRET supports the two-state model of Argonaute action. RNA Biol. 2014; 11:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Saxena S., Jonsson Z.O., Dutta A.. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J. Biol. Chem. 2003; 278:44312–44319. [DOI] [PubMed] [Google Scholar]
- 58. Jackson A.L., Burchard J., Schelter J., Chau B.N., Cleary M., Lim L., Linsley P.S.. Widespread siRNA ‘off-target’ transcript silencing mediated by seed region sequence complementarity. RNA. 2006; 12:1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.