Abstract
The spatial organization of the genome is essential for the precise control of gene expression. Recent advances in sequencing and imaging technologies allow us to explore the 3D genome and its relationship to gene regulation at an unprecedented scale. In this review, we provide an overview of lessons learned from studying the chromatin structure and their implications in communications between gene promoters and distal cis-regulatory elements, such as enhancers. We first review the current knowledge of general genome organization, followed by the importance of chromatin folding in gene regulation. Next, we proceed to a brief survey of the recently developed chromosome conformation capture technologies, as well as most widely adopted read-outs from such data. We then introduce two emerging models that offer explanations regarding how distal enhancers achieve transcriptional control of target genes in the 3D genome. Last, we discuss the promising prospects of leveraging knowledge in chromatin spatial organization for studying complex diseases and traits.
General Features of Genome Structure
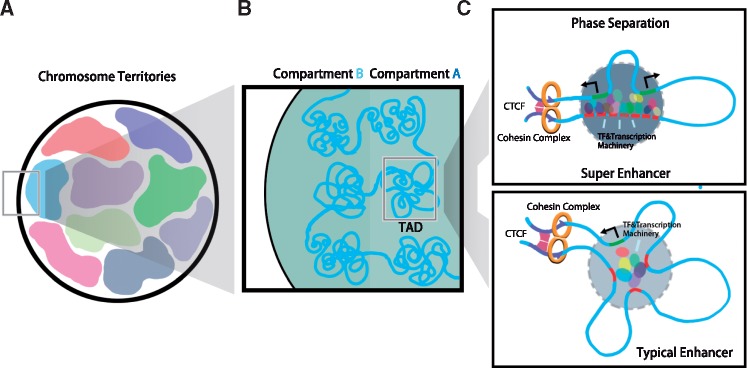
To fit the entire genome of 2-m-long human DNA sequences, containing ∼3 billion nucleotides, in the nucleus of a diameter of ∼10 µm, DNA has to be packed and organized achieving 105× compaction. In vitro structure study of the chromatin showed that every 146 bp of DNA is wrapped around nucleosome (11 nm diameter), providing 5–6× compaction. At the next level, nucleosomes are compacted into the 30 nm chromatin fiber, achieving an additional ∼50× compaction (1). The chromatin is thought to be further compacted into fibers sized several nanometers. However, this hierarchical model of chromosome compaction was challenged by new findings of chromatin structure in nucleus using ChromEMT (2). Unlike in vitro, chromatin is a disorganized chain with diameter 5–24 nm in situ. Chromatin chains are flexible and compacted in different densities with various particle arrangements and conformations (2). Such observation suggests that chromatin fibers are organized at different local chromatin concentrations instead of being folded in a hierarchical order. It remains elusive how precisely such observed chromatin polymer structure determines DNA accessibilities and gene regulation. We learned chromosomes are organized in chromosome territories as the basic structure of genome organization by cell biology studies (3) (Fig. 1A). Chromosome territories are arranged in a somewhat non-random fashion so that small, gene-rich chromosomes tend to locate at the center of the nucleus (4). More recently, molecular approaches with chromatin conformation assays have revealed many insights regarding chromatin structure at different resolutions. For example, the first survey of genome-wide chromatin interaction, at 1 Mb resolution, showed that the genome is partitioned into compartments ‘A’ and ‘B’. ‘A’ is the open and actively transcribed regions and ‘B’ the compacted and repressed regions of the genome (5) (Fig. 1B). Later, several studies were conducted in an attempt to gain a better understanding of spatial organization of genomes by increasing data resolution with in-depth sequencing (6,7). Collective results from these studies demonstrated that the genome is partitioned into Mb-sized topologically associated domains (TADs) containing coregulated genes and cis-regulatory elements (6,8). Disruption of CTCF occupied TAD boundary leads to rearrangement of promoter–enhancer looping and abnormal expression of limb developmental genes (9), for example. Within TADs, chromatin forms loops between enhancer and promoter sequences (Fig. 1C). Such regulatory chromatin loops are cell type-specific, serving as the driving force to determine the cellular identity and linking to many disease-associated variants (10–14). These studies provide a general understanding of the higher-order structure of the genome for coordinated gene regulation and highlight the importance of studying 3D genome structure in cell type-specific transcriptional regulation.
Figure 1.
Schematic diagram of genome organization and gene regulation. Chromosomes are organized into territories in the mammalian genome (A), A/B compartments (transcriptional active versus repressive domains), and TADs (physical constrain of enhancer-promoter interactions) (B). TADs boundaries are insulated by CTCF/cohesin complex (C). Within TADs, enhancer and promoter form chromatin loops with super-enhancers and their target genes tend to have better phase separation (depicted in dark grey) due to the higher concentration of local TFs and transcriptional machinery than typical enhancers (depicted in light grey) (C).
One Primary Motivation: the Mechanism by which Enhancers Achieve Gene Regulation
Enhancers regulate transcription by recruiting transcriptional factors and coactivators that facilitate gene transcription (15,16). For instance, pioneer factors such as FOXA1 are known to initiate chromatin remodeling, leading to histone modifications (e.g. H3K4 methylation) at enhancer sequences in a tissue-specific manner (17). Tissue-specific transcription factors then bind to enhancers in response to signaling, and further recruiting transcriptional machinery to target genes via chromatin looping. Enhancer–promoter (E–P) loops are mediated by CTCF, cohesion and mediator (18). A number of excellent reviews discuss recent research on transcriptional network and chromatin remodeling related to enhancers (19–21).
Large international efforts including ENCODE (22) and Roadmap Epigenomics (23) projects have mainly focused on mapping tissue-specific enhancers via chromatin states and accessibilities. These efforts provide one-dimensional (1D) annotation and have identified hundreds of thousands of putative cis-regulatory elements in the mammalian genome. More recently, the 4D Nucleome Consortium (24) has been assembled with the goal to understand the general principals of spatial and temporal chromatin organization and the roles of chromatin loops in gene regulation and cell function. Enhancers can regulate target genes from distal (25,26), and in some cases, even Mb away (27). Such long-range regulation, combined with high abundance of enhancer elements compared to the promoter sequences in the mammalian genome, renders a dauntingly large number of potential physical interactions, making systematically mapping E–P communications challenging. Because of that, single nulceotide polymorphisms (SNPs) associated with diseases or traits from the genome-wide association studies (GWAS) were conveniently assigned to the nearest gene within certain linear DNA distance (nearest gene model). However, this model has been challenged by examples of SNPs regulating genes further away instead of the closet one(s). For example, FTO intronic variants regulating IRX3 in the brain (28), and IRX3 and IRX5 in adipocytes (29), respectively; an ARL15 intronic variant regulating FST (26); and several others (13,30–32), indicating the complexity and cell type specificity for the functions of regulatory variants.
C-technologies to Study Genome-Wide Chromatin Interactions
It is generally accepted that enhancer and the promoter of its target gene should be brought to close physical proximity mediated by transcription factors and cofactors. Many techniques have been developed to investigate chromatin organization in the nucleus. Chromosome conformation capture (3C) (33), the original C-technology to study one-to-one interactions (i.e. between two pre-defined DNA segments), has successfully identified many E–P interactions including the human beta and alpha globin loci (34,35). Later, 3C derivative methods including 4C (36), 5C (37), Hi-C (5), Chromatin Interaction Analysis by Paired-End Tag Sequencing (38), promoter capture Hi-C (39), HiChIP (40) and Proximity Ligation-Assisted ChIP-seq (41), have been subsequently developed for interrogation of chromatin interactions at different scales and resolutions. The comparison of these different techniques is nicely described in many excellent recent reviews (11,19,20,42).
Common Read-Outs of Genome-Wide Chromatin Structure Data
With Hi-C approach, an ever-increasing deluge of 3D chromatin data has been generated. Efficiently analyzing these data is imperative but, as an understatement, non-trivial (19,20). Hi-C data typically contain 0.1–5 billion raw reads per sample, with various biases and noises. Quality control is therefore challenging, usually involving multi-layer pre-processing (6,7,9,10,12,43–47) and normalization (7,12,48–50) before analysis, as well as reproducibility checks during and after analysis (51,52). Excellent reviews exist (19,20,53) for readers interested in statistical and bioinformatics analysis details. Major read-outs from such genome-wide chromatin structure data include multi-Mb resolution ‘A’ (open and actively transcribed) and ‘B’ (compacted and repressed) compartments, Mb resolution topologically associating domains (TADs), tens-kb resolution Frequently Interacting REgions (FIREs) (12) and high (e.g. kb) resolution chromatin loops and interactions (Fig. 1).
Models for Regulatory Chromatin Interactions Between Promoters and Enhancers
Model 1: enhancer–promoter (E–P) interactions are restricted by the spatial partition of the genome
One possible model for E–P interactions suggested by recent studies of chromatin spatial organization is that TADs, discrete Mb-sized chromatin territories, serve to restrict long-range interactions between enhancers and promoters. TADs are found to be relatively stable over many cell divisions, between different cell types and evolutionarily conserved (6,54). Increasing evidences support the functional roles of TADs, including gene regulation within TAD, DNA replication timing control and propagation of chromatin state along the DNA. For example, genes within the same TAD tend to have coordinated expression dynamics during differentiation, suggesting TAD’s role in concerting the activities of neighboring genes (6,8,54,55). Studies on expression and histone quantitative trait loci (eQTL and hQTL) indicate that co-regulation of regulatory elements tend to occur within the same TAD (56,57). Dynamic and cell type-specific chromatin loops between enhancers and promoter usually occur within TAD (7,58) and mediated by CTCF, cohesion complex and mediator complex (18,59). Disruption of TAD boundaries can lead to ectopic activation of enhancers across the boundaries, subsequently to abnormal gene expression and eventually to noticeable phenotypes or diseases (9,20,44,60). For example, developmental disorders, such as syndactyly and congenital limb malformations, and some pediatric brain disorders, have recently been found to be caused by the disruption of TAD boundaries and consequent dysregulation of disease-causing genes (9,44,60). At domain boundaries, chromatin loops are formed by loop extrusion (61–63). Specifically, a pair of cohesion complexes binds to form a loop extrusion factor, slides along the genome and stops upon contact of domain boundary factors, such as a pair of convergent oriented CTCF motifs, forming an extruded chromatin loop. Such CTCF and cohesion complexes are hallmarks of TAD boundaries (6,59) (Fig. 1B and C). Disrupting CTCF binding or inverting the direction of CTCF motif can lead to aberrant loop formation, and subsequently transcriptional dysregulation. For example, copy number variations overlapping CTCF motif are found to result in enhancer hijacking, which plays a significant role in aberrant gene activation for many types of cancer (60,64,65).
Model 2: phase separation model for enhancer–promoter communication
The second model, phase separation, suggests that cooperative binding (of transcription factors, cofactors and RNA polymerase to enhancer and promoter sequences) leads to a high local density of protein and nucleotide concentration. Such high concentration of multi-molecular assemblies can form a gel-like phase-separated compartment for transcription (66–68). Phase separation is considered to play essential roles in the formation of heterochromatin and super-enhancers, both of high molecular density despite the dramatic difference in functional activities. Such high-density multi-molecule complex fosters separation and creation of relatively self-contained entities, facilitating compartmentalization of biochemical reactions such as transcription (Fig. 1C). Therefore, it is proposed that phase separation occurs more frequently at super-enhancers than at typical enhancers. The phase separation model provides an explanation for the formation of super-enhancers and their sensitivity to various perturbations; the functional mechanisms for enhancers simultaneously regulating multiple genes; how multiple typical enhancers orchestrate gene activation and transcription bursting (67).
The models reviewed above offer insights into enhancers and promoter communication to achieve cell type-specific gene regulation. These two models are not mutually exclusive: enhancers and promoters can be organized and connected by cell type-specific transcription factors which lead to condensation and phase separation within TADs with boundaries stabilized by loop extrusion.
Future perspectives for genetic dissection of human diseases and traits
To date, many GWAS studies have been conducted and identified over >50 000 SNPs significantly associated with various human diseases and traits (69). However, the vast majority of these GWAS SNPs reside in regulatory regions with mostly elusive underlying mechanisms, preventing further leveraging the results for clinical applications. Recently mapped cis-regulatory elements have already started to deliver on the promise of aiding interpretation of GWAS results and generating mechanistic hypotheses at GWAS loci. For example, several recent multi-tissue/cell type studies of chromatin structure (10,12–14) have demonstrated that tissue-specific E–P contacts are essential for elucidating the roles of non-coding variants in disease development.
Cell type-specific cataloging of functional elements is critical for the identification of causal regulatory SNPs and for revealing the underlying biological mechanisms. Compared to the large number of GWAS SNPs identified, functional characterization of GWAS SNPs remains challenging for a number of reasons: first, many regulatory SNPs function in a cell type-specific manner, yet there is still a significant lack of comprehensive annotation of regulatory sequences in physiologically relevant cell types. Second, GWAS variants mostly exert moderate or small effects on phenotypic traits such that their effects are susceptible to confounding or dilution by genetic background and noisy read-outs in validation experiments. Third, functional validations of each genetic variant have been mainly tested individually while the functional regulatory region can easily encompass multiple genetic variants working synergistically. For example, in a recent study of Hirschsprung disease (70), three SNPs were shown to be associated with three independent enhancers, all regulating the same targeting gene RET. Further investigation revealed that these three enhancers were synergistic such that carrying all three risk alleles results in a much higher risk of developing the disease. Lastly, cis-regulatory elements often interact with their cognate genes over long genomic distances, precluding a straightforward, systematic mapping of promoter and enhancer connectivity. Consequently, our ability to interpret and derive mechanistic insights from GWAS results remains limited.
Uncovering chromatin spatial organizations at various resolutions across many tissues and conditions will continue to aid prioritization of critical regulatory regions, in cell type-, disease- and condition-specific manners. Examples of the target genes of GWAS SNPs not being the nearest gene include the well-known obesity-associated FTO intronic variants actually regulating IXR3 gene ∼490 kb away in brain (28), schizophrenia-associated variant regulating FOXG1 gene ∼764 kb away in radial glia cells (13) and an ARL15 intronic variant most likely controlling FST gene ∼522 kb away in liver (26,30). The prioritized variants based on 1D regulatory element annotation can be further evaluated with a combination of functional characterization assays including CRISPR-Cas9 mediated genome editing (71,72), massively parallel reporter assay (MPRA) (73) and imaging technologies such as Fluorescent in situ hybridization. For example, when evaluating a GWAS locus (typically with multiple SNPs identified to be associated with phenotypic trait of interest), besides leveraging tissue-specific 1D epigenomic annotations to identify potential regulatory elements, additional evaluation of SNP(s) based on 3D interactions can help prioritize which one(s) for functional follow-up. Integrating GWAS results with 1D and 3D information, we can then examine the most promising SNP(s) to assess whether the alternative alleles result in disrupted E–P interactions. In the future, integrative approaches combining chromatin spatial organization studies with GWAS studies, along with other multi-omic data, will facilitate revealing genetic mechanisms underlying complex human diseases and traits, ultimately leading to the development of effective preventive method, diagnosis and drug treatment of these diseases.
Conflict of Interest statement. None declared.
Funding
US National Institute of Health (NIH) grants (R01 HL129132 to Y.L., R01 AG057497, R01 EY027789, and UM1 HG009402 to Y.S); UCSF Weill Institute for Neuroscience (to Y.S.); The Hillblom Foundation (to Y.S.); The American Federation for Aging Research (New Investigator Award in Alzheimer’s disease to Y.S.); NIH grant (U54DK107977 to M.H.), in part.
References
- 1. Felsenfeld G., Groudine M. (2003) Controlling the double helix. Nature, 421, 448–453. [DOI] [PubMed] [Google Scholar]
- 2. Ou H.D., Phan S., Deerinck T.J., Thor A., Ellisman M.H., O’Shea C.C. (2017) ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science (New York, N.Y.), 357, eaag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cremer T., Cremer M. (2010) Chromosome territories. Cold Spring Harbor Perspect. Biol., 2, a003889.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Croft J.A., Bridger J.M., Boyle S., Perry P., Teague P., Bickmore W.A. (1999) Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol., 145, 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O.. et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science, 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature, 485, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S.. et al. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell, 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shen Y., Yue F., McCleary D.F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V.V.. et al. (2012) A map of the cis-regulatory sequences in the mouse genome. Nature, 488, 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lupiáñez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R.. et al. (2015) Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell, 161, 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Javierre B.M., Burren O.S., Wilder S.P., Kreuzhuber R., Hill S.M., Sewitz S., Cairns J., Wingett S.W., Várnai C., Thiecke M.J.. et al. (2016) Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell, 167, 1369–1384. e1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krijger P.H., de Laat W. (2016) Regulation of disease-associated gene expression in the 3D genome. Nat. Rev. Mol. Cell Biol., 17, 771–782. [DOI] [PubMed] [Google Scholar]
- 12. Schmitt A.D., Hu M., Jung I., Xu Z., Qiu Y., Tan C.L., Li Y., Lin S., Lin Y., Barr C.L.. et al. (2016) A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep., 17, 2042–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Won H., de la Torre-Ubieta L., Stein J.L., Parikshak N.N., Huang J., Opland C.K., Gandal M.J., Sutton G.J., Hormozdiari F., Lu D.. et al. (2016) Chromosome conformation elucidates regulatory relationships in developing human brain. Nature, 538, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mumbach M.R., Satpathy A.T., Boyle E.A., Dai C., Gowen B.G., Cho S.W., Nguyen M.L., Rubin A.J., Granja J.M., Kazane K.R.. et al. (2017) Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet., 49, 1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Glass C.K., Rosenfeld M.G. (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev., 14, 121–141. [PubMed] [Google Scholar]
- 16. Tjian R., Maniatis T. (1994) Transcriptional activation: a complex puzzle with few easy pieces. Cell, 77, 5–8. [DOI] [PubMed] [Google Scholar]
- 17. Zaret K.S., Carroll J.S. (2011) Pioneer transcription factors: establishing competence for gene expression. Genes Dev., 25, 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S.. et al. (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature, 467, 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schmitt A.D., Hu M., Ren B. (2016) Genome-wide mapping and analysis of chromosome architecture. Nat. Rev. Mol. Cell Biol., 17, 743–755. (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu M., Ren B. (2017) The three-dimensional organization of mammalian genomes. Ann. Rev. Cell Dev. Biol., 33, 265–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gorkin D.U., Leung D., Ren B. (2014) The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell, 14, 762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. ENCODE Project Consortium Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T.. et al. (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature, 447, 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J.. et al. (2015) Integrative analysis of 111 reference human epigenomes. Nature, 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dekker J., Belmont A.S., Guttman M., Leshyk V.O., Lis J.T., Lomvardas S., Mirny L.A., O’Shea C.C., Park P.J., Ren B.. et al. (2017) The 4D nucleome project. Nature, 549, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smallwood A., Ren B. (2013) Genome organization and long-range regulation of gene expression by enhancers. Curr. Opin. Cell Biol., 25, 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin J.S., Xu Z., Reiner A.P., Mohlke K.L., Sullivan P., Ren B., Hu M., Li Y. (2017) HUGIn: Hi-C unifying genomic interrogator. Bioinformatics, 33, 3793–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Laat W., Duboule D. (2013) Topology of mammalian developmental enhancers and their regulatory landscapes. Nature, 502, 499–506. [DOI] [PubMed] [Google Scholar]
- 28. Smemo S., Tena J.J., Kim K.-H., Gamazon E.R., Sakabe N.J., Gómez-Marín C., Aneas I., Credidio F.L., Sobreira D.R., Wasserman N.F.. et al. (2014) Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature, 507, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Claussnitzer M., Dankel S.N., Kim K.-H., Quon G., Meuleman W., Haugen C., Glunk V., Sousa I.S., Beaudry J.L., Puviindran V.. et al. (2015) FTO obesity variant circuitry and adipocyte browning in humans. Q1, 373, 895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Civelek M., Wu Y., Pan C., Raulerson C.K., Ko A., He A., Tilford C., Saleem N.K., Stančáková A., Scott L.J.. et al. (2017) Genetic regulation of adipose gene expression and cardio-metabolic traits. Am. J. Hum. Genet., 100, 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mumbach M.R., Satpathy A.T., Boyle E.A., Dai C., Gowen B.G., Cho S.W., Nguyen M.L., Rubin A.J., Granja J.M., Kazane K.R.. et al. (2017) Enhancer connectome in primary human cells reveals target genes of disease-associated DNA elements. Nat. Genet., 49, 1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iotchkova V., Huang J., Morris J.A., Jain D., Barbieri C., Walter K., Min J.L., Chen L., Astle W., Cocca M.. et al. (2016) Discovery and refinement of genetic loci associated with cardiometabolic risk using dense imputation maps. Nat. Genet., 48, 1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dekker J., Rippe K., Dekker M., Kleckner N. (2002) Capturing chromosome conformation. Science, 295, 1306–1311. [DOI] [PubMed] [Google Scholar]
- 34. Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W. (2002) Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell, 10, 1453–1465. [DOI] [PubMed] [Google Scholar]
- 35. Zhou G.-L., Xin L., Song W., Di L.-J., Liu G., Wu X.-S., Liu D.-P., Liang C.-C. (2006) Active chromatin hub of the mouse alpha-globin locus forms in a transcription factory of clustered housekeeping genes. Mol. Cell. Biol., 26, 5096–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., de Laat W. (2006) Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet., 38, 1348–1354. [DOI] [PubMed] [Google Scholar]
- 37. Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C.. et al. (2006) Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res., 16, 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fullwood M.J., Liu M.H., Pan Y.F., Liu J., Xu H., Mohamed Y.B., Orlov Y.L., Velkov S., Ho A., Mei P.H.. et al. (2009) An oestrogen-receptor-alpha-bound human chromatin interactome. Nature, 462, 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mifsud B., Tavares-Cadete F., Young A.N., Sugar R., Schoenfelder S., Ferreira L., Wingett S.W., Andrews S., Grey W., Ewels P.A.. et al. (2015) Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet., 47, 598–606. [DOI] [PubMed] [Google Scholar]
- 40. Mumbach M.R., Rubin A.J., Flynn R.A., Dai C., Khavari P.A., Greenleaf W.J., Chang H.Y. (2016) HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods, 13, 919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fang R., Yu M., Li G., Chee S., Liu T., Schmitt A.D., Ren B. (2016) Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res., 26, 1345–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bonev B., Cavalli G. (2016) Organization and function of the 3D genome. Nat. Rev. Genet., 17, 661-678. [DOI] [PubMed] [Google Scholar]
- 43. Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W.. et al. (2015) Chromatin architecture reorganization during stem cell differentiation. Nature, 518, 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Franke M., Ibrahim D.M., Andrey G., Schwarzer W., Heinrich V., Schöpflin R., Kraft K., Kempfer R., Jerković I., Chan W.-L.. et al. (2016) Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature, 538, 265–269. [DOI] [PubMed] [Google Scholar]
- 45. Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.-A., Schmitt A.D., Espinoza C.A., Ren B.. et al. (2013) A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature, 503, 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Foracto M., Nicoletti C., Pal K., Livi C.M., Ferrari F., Bicciato S. (2017) Comparison of computational methods for Hi-C data analysis. Nature Methods, 14(7), 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schwarzer W., Abdennur N., Goloborodko A., Pekowska A., Fudenberg G., Loe-Mie Y., Fonseca N.A., Huber W., Haering C., Mirny L.. et al. (2017) Two independent modes of chromatin organization revealed by cohesin removal. Nature, 551, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hu M., Deng K., Selvaraj S., Qin Z., Ren B., Liu J.S. (2012) HiCNorm: removing biases in Hi-C data via Poisson regression. Bioinformatics, 28, 3131–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yaffe E., Tanay A. (2011) Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet., 43, 1059–1065. [DOI] [PubMed] [Google Scholar]
- 50. Imakaev M., Fudenberg G., McCord R.P., Naumova N., Goloborodko A., Lajoie B.R., Dekker J., Mirny L.A. (2012) Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods, 9, 999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang T., Zhang F., Yardımcı G.G., Song F., Hardison R.C., Noble W.S., Yue F., Li Q. (2017) HiCRep: assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res., 27, 1939–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ursu O., Boley N., Taranova M., Wang Y.X.R., Yardimci G.G., Noble W.S., Kundaje A. (2018) GenomeDISCO: A concordance score for chromosome conformation capture experiments using random walks on contact map graphs. Bioinformatics, doi: 10.1093/bioinformatics/bty164. [DOI] [PMC free article] [PubMed]
- 53. Ay F., Noble W.S. (2015) Analysis methods for studying the 3D architecture of the genome. Genome Biol., 16, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J.. et al. (2012) Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature, 485, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Flavahan W.A., Drier Y., Liau B.B., Gillespie S.M., Venteicher A.S., Stemmer-Rachamimov A.O., Suvà M.L., Bernstein B.E. (2016) Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature, 529, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Grubert F., Zaugg J.B., Kasowski M., Ursu O., Spacek D.V., Martin A.R., Greenside P., Srivas R., Phanstiel D.H., Pekowska A.. et al. (2015) Genetic control of chromatin states in humans involves local and distal chromosomal interactions. Cell, 162, 1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Waszak S.M., Delaneau O., Gschwind A.R., Kilpinen H., Raghav S.K., Witwicki R.M., Orioli A., Wiederkehr M., Panousis N.I., Yurovsky A.. et al. (2015) Population variation and genetic control of modular chromatin architecture in humans. Cell, 162, 1039–1050. [DOI] [PubMed] [Google Scholar]
- 58. Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G.L., Lubling Y., Xu X., Lv X., Hugnot J.-P., Tanay A.. et al. (2017) Multiscale 3D genome rewiring during mouse neural development. Cell, 171, 557–572. e524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Phillips-Cremins J.E., Sauria M.E.G., Sanyal A., Gerasimova T.I., Lajoie B.R., Bell J.S.K., Ong C.-T., Hookway T.A., Guo C., Sun Y.. et al. (2013) Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell, 153, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lupianez D.G., Spielmann M., Mundlos S. (2016) Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet., 32, 225–237. [DOI] [PubMed] [Google Scholar]
- 61. Hnisz D., Day D.S., Young R.A. (2016) Insulated neighborhoods: structural and functional units of mammalian gene control. Cell, 167, 1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sanborn A.L., Rao S.S.P., Huang S.-C., Durand N.C., Huntley M.H., Jewett A.I., Bochkov I.D., Chinnappan D., Cutkosky A., Li J.. et al. (2015) Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. USA, 112, E6456–E6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fudenberg G., Imakaev M., Lu C., Goloborodko A., Abdennur N., Mirny L.A. (2016) Formation of chromosomal domains by loop extrusion. Cell Rep., 15, 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Herz H.M. (2016) Enhancer deregulation in cancer and other diseases. BioEssays, 38, 1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kaiser V.B., Semple C.A. (2017) When TADs go bad: chromatin structure and nuclear organisation in human disease. F1000Research, 6, 314.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X., Karpen G.H. (2017) Phase separation drives heterochromatin domain formation. Nature, 547, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., Sharp P.A. (2017) A phase separation model for transcriptional control. Cell, 169, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hult C., Adalsteinsson D., Vasquez P.A., Lawrimore J., Bennett M., York A., Cook D., Yeh E., Forest M.G., Bloom K.. et al. (2017) Enrichment of dynamic chromosomal crosslinks drive phase separation of the nucleolus. Nucleic Acids Res., 45, 11159–11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J.. et al. (2017) The new NHGRI-EBI catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res., 45, D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chatterjee S., Kapoor A., Akiyama J.A., Auer D.R., Lee D., Gabriel S., Berrios C., Pennacchio L.A., Chakravarti A. (2016) Enhancer variants synergistically drive dysfunction of a gene regulatory network in hirschsprung disease. Cell, 167, 355–368. e310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. (2013) RNA-guided human genome engineering via Cas9. Science, 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A.. et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ulirsch J.C., Nandakumar S.K., Wang L., Giani F.C., Zhang X., Rogov P., Melnikov A., McDonel P., Do R., Mikkelsen T.S., Sankaran V.G. (2016) Systematic functional dissection of common genetic variation affecting red blood cell traits. Cell, 165, 1530–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]