Abstract
Despite the key role of the human ribosome in protein biosynthesis, little is known about the extent of sequence variation in ribosomal DNA (rDNA) or its pre-rRNA and rRNA products. We recovered ribosomal DNA segments from a single human chromosome 21 using transformation-associated recombination (TAR) cloning in yeast. Accurate long-read sequencing of 13 isolates covering ∼0.82 Mb of the chromosome 21 rDNA complement revealed substantial variation among tandem repeat rDNA copies, several palindromic structures and potential errors in the previous reference sequence. These clones revealed 101 variant positions in the 45S transcription unit and 235 in the intergenic spacer sequence. Approximately 60% of the 45S variants were confirmed in independent whole-genome or RNA-seq data, with 47 of these further observed in mature 18S/28S rRNA sequences. TAR cloning and long-read sequencing enabled the accurate reconstruction of multiple rDNA units and a new, high-quality 44 838 bp rDNA reference sequence, which we have annotated with variants detected from chromosome 21 of a single individual. The large number of variants observed reveal heterogeneity in human rDNA, opening up the possibility of corresponding variations in ribosome dynamics.
INTRODUCTION
According to the messenger RNA (mRNA) hypothesis, as first reviewed by Jacob and Monod, each ribosome has been considered equivalent to all others, a tabula rasa instructed by mRNAs to form corresponding proteins (1). To sustain the growth of any cell, about half of all RNA synthesis is therefore ribosomal RNA and variable rates of transcription are maintained in part by the relative activity of multiple copies of rDNA in the cell nucleus. In humans, ∼400 rDNA repeats are distributed among five nucleolar organizer regions (NORs) on the short arms of the acrocentric chromosomes 13, 14, 15, 21 and 22 (2,3,4). Most of the repeats are organized as tandem arrays. The number of rDNA repeats in individual human NORs is in the range of 1–3 to more than 140 (4,5,6), and only 20–50% of all RNA genes are transcriptionally active in most human cells (7). An rDNA repeat unit encodes a copy of 18S, 5.8S and 28S rRNA sequences separated by internal transcribed spacer sequences and flanked by external transcribed spacers (ETSs) and an ∼30 kb intergenic spacer (IGS). The precursor 45S pre-rRNA transcript is synthesized by polymerase I and processed into the mature rRNA species (8,9).
Intra-genomic variation in the ribosomal RNA genes is well-documented in several species (10,11,12,13). For human, subcloned rDNA fragments sequenced from several individuals found early evidence of variation—primarily single nucleotide variants (SNVs)—in the transcribed regions (14,15,16,17) compared to a reference sequence (18). However, none of the observed variants was validated in uncloned DNA, and in the intervening years, there has been no comprehensive census of rDNA variants or their frequency within and between individuals.
Large structural variants within human NORs, including palindromic rDNA repeats, have also been noted (19). But again, there has been no study at sequence resolution of the structure or frequency of such variants, or the degree to which they are shared in individuals or populations. In fact, the only reported systematic study of sequences proximal (centromeric) and distal (telomeric) to rDNA arrays (20), based on the analysis of bacterial artificial chromosome (BAC) and fosmid clones available in GenBank, concluded that these sequences are nearly identical among all five acrocentric chromosome pairs.
Unfortunately, computational assembly of the NORs from shotgun sequencing of total human DNA is hampered by the size and similarity of the rDNA tandem repeat units (21). Assembling rDNA repeats from whole-genome data results in a consensus representation that is suitable for inter-species comparisons but masks variation within species and individuals (22). The assembly problem is simplified via cloning, which can isolate a few copies of rDNA for sequencing, but each individual copy still contains internal repeats that are difficult assemble. Recent advances in sequencing technology have resulted in read lengths of greater than 100 kb (23), capturing entire rDNA units in individual reads and thereby greatly simplifying sequence assembly. Although the error rate of long-read sequencing is relatively high, technologies such as PacBio single-molecule sequencing produce largely random error that can be statistically corrected with sufficient sequencing depth, resulting in near-perfect (>99.999%) consensus accuracy (24). Thus, targeted, long-read sequencing has enabled highly accurate reconstruction of individual rDNA units for the first time.
Using a combination of transformation-associated recombination (TAR) cloning and multiple sequencing technologies, we report a pilot characterization of variants in rDNA repeats from a single chromosome of one individual. We first isolated and sequenced individual rDNA units using long reads to identify candidate variants. These variants were then validated and their frequencies assessed using long-read and short-read sequencing of whole genomes and transcriptomes sampled from multiple individuals. From this analysis, an improved rDNA reference sequence is presented along with a new catalog of rDNA variants assessed with high confidence.
MATERIALS AND METHODS
Cell line and media
Mouse/human monochromosomal hybrid cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA, USA) and 800 ug/ml G418 (InvivoGen) at 37°C in 5% CO2. The A9 (21–16) cell line (mouse A9 cells containing human chromosome 21) was obtained from Dr Mitsuo Oshimura (25).
Construction of ribosomal DNA transformation-associated recombination (TAR) cloning vectors and rDNA clone recovery
The TAR vectors, TAR- #2, - #6, - #11, -#16 and - #22, were constructed using the shuttle vector pJYB, containing a yeast cassette (CEN6 and HIS3) and a BAC cassette carrying the F-factor origin of replication (Supplementary Figure S1). These vectors contain combinations of four 130–170 bp targeting sequences (hooks) H1, H2, H3 and H4, chosen from rDNA 5′ ETS region (see Figure 1A). These targeting sequences were polymerase chain reaction (PCR)-amplified from the BAC CH507-528H12 (GI: FP236383.15). Sequences of the primers and their location on rDNA sequence are presented in Supplementary Table S1. In three vectors, #6, #11 and #16, both hooks have the same orientation corresponding to that in the reference rDNA (Figure 1B). For these vectors, the expected size of the targeted genomic fragment is ∼43 kb or two to three times bigger if two or three rDNA units will be cloned. In two other vectors, #2 and #22, one of the hooks is inverted (Figure 1C). The hooks were cloned into the pJYB vector as either SalI–AscI or NotI–XbaI fragments. The TAR vectors were linearized with and AscI and NotI (the sites are flanked by pBR322 origin sequence) before transformation to yield a molecule bounded by the desired targeting sequences. Detailed physical maps of the TAR vectors, are shown in Supplementary Figure S1.
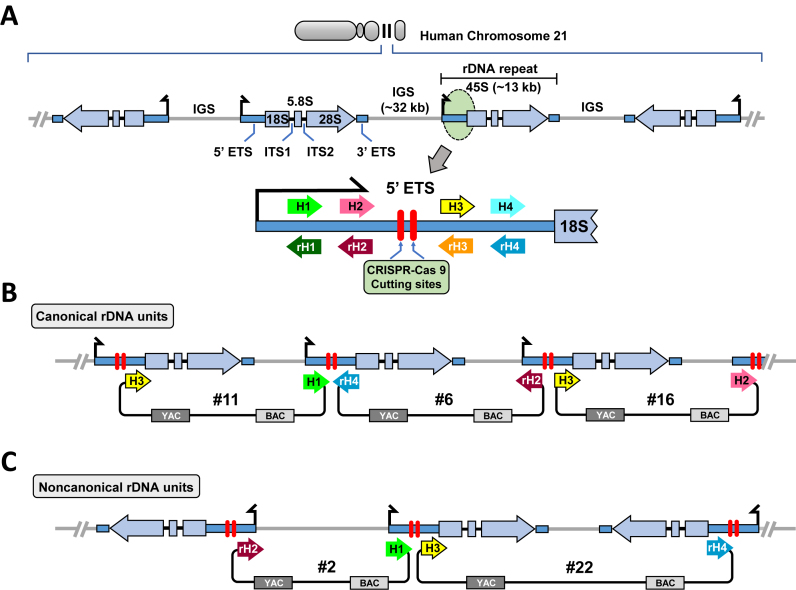
Figure 1.
Scheme for isolation of human ribosomal DNA from mouse/human hybrid cell line as circular YAC/BAC. (A) Schematic representation of rDNA cluster in human acrocentric chromosomes 21. Both canonical rDNA units organized as a tandem repeats and rDNA units forming palindromic structure are shown. Each unit is composed of an ∼13 kb transcribed region encoding 45S rRNA (5′ ETS, 18S, ITS1, 5.8S, ITS2, 28S and 3′ ETS) and an ∼32 kb IGS. All four targeting sequences for TAR vectors, hooks (H1, H2, H3 and H4), were chosen from the 5′ ETS region that is ∼1.4 kb upstream of the 18S rRNA. Human rDNA could be cleaved at the two positions between H2 and H3 by the Cas9–gRNA complexes (red bars). (B) Scheme for TAR cloning of rDNA organized as tandem repeats. Three TAR vectors, #11, #6 and #16, with canonical orientation of the hooks are shown. Homologous recombination between the targeting sequences in the vectors and a targeted human rDNA fragment leads to the establishment of a circular YAC/BAC. (C) Scheme illustrating TAR cloning of rDNA sequences organized as a palindrome. Two TAR vectors, #2 and #22, in which one hook is inverted (rH2 or rH4) were constructed. Homologous recombination between the targeting sequences in the vectors and the targeted human rDNA fragment may lead to the establishment of a circular YAC/BAC only for regions with a palindromic structure. (ETS: External transcribed spacer; ITS: Internal transcribed spacer).
To TAR clone human rDNA, each vector and genomic DNA gently isolated from the A9 (21–16) hybrid cells were presented to yeast spheroplasts and His+ transformants were selected as described (see below). In some experiments, genomic DNA was pre-treated with CRISPR-Cas9 nucleases to generate double-strand breaks near the targeted genomic region (Supplementary Figure S2) to promote an increase in gene-positive colonies (26).
For transformations, the highly transformable Saccharomyces cerevisiae strain VL6-48 (MATα, his3-Δ200, trp1-Δ1, ura3-52, lys2, ade2-101, met14) that has HIS3 deleted was used (27). For TAR cloning of the rDNA regions, 2–3 μg of genomic DNA isolated from the mouse/human monochromosomal hybrid cell line was mixed with a AscI/NotI-linearized TAR vector (∼1 μg) and presented to freshly prepared yeast spheroplasts. Yeast transformants were selected on synthetic complete medium plates lacking histidine. In total, six to eight transformation experiments were carried out for each TAR vector.
To identify rDNA-containing clones, the transformants were combined into pools, each containing 30 transformants and tested with diagnostic primers (Supplementary Table S1) for unique sequences specific for the targeted human rDNA unit. Individual clones obtained by four different TAR cloning vectors were designated as JH2–JH18. Supplementary Table S2 shows which vector has been used for isolation of each clone and the size of each cloned region.
Physical characterization of TAR clones
To prove the presence of the predicted genomic sequences in TAR/YAC isolates, DNA from the clones was examined by PCR (Supplementary Table S1) using primers covering the rDNA reference sequence. Analysis of one isolated clone (JH5) is shown in Supplementary Figure S3C. The TAR-isolated YAC/BACs were moved to Escherichia coli by electroporation. In brief, yeast chromosome-size DNAs were prepared in agarose plugs and, after melting and agarase treatment, the DNAs were electroporated into DH10B competent cells (Gibco/BRL) by using a Bio-Rad Gene Pulser as previously described (28). To determine the size of the BACs, each of the clones was analyzed by contour-clamped homogeneous electric field (CHEF) gel electrophoresis. Supplementary Figure S3D illustrates CHEF gel analysis of six rDNA-containing BAC isolates.
Fluorescence in situ hybridization (FISH) analysis
Two probes were prepared. The first probe targets human chromosome 21 alphoid DNA derived from a pYB 21α-18 BAC containing ∼18 kb of human chromosome 21 alphoid type-I sequence (Supplementary Figure S4A). The second probe is derived from an ∼53 kb rDNA BAC clone that contains only human IGS sequence (JH10, Supplementary Table S2). Both probes were used to confirm the presence of human chromosome 21 in hybrid cells (Supplementary Figure S4A and B). BAC DNAs were Cyanine 3-UTP (Enzo) labeled using a nick-translation kit (Abbot Molecular Ins.). The probes were denatured at 78°C for 10 min and kept to 37°C before use. Slides were incubated at 72°C for 2 min before overnight incubation at 37°C. After washes with 0.4 × Saline-sodium citrate (SSC) + 0.3% Tween 20 at 72°C followed by a wash with 2 × SSC + 0.1% Tween 20 at room temperature, standard procedures were used to visualize probes. Slides were mounted with VectaShield and screened for the presence of the human chromosome 21.
Fiber-FISH analysis
The A9 (21–16) cells were trypsinized and resuspended in phosphate-buffered saline (PBS). 1 × 105 Cells were then embedded in a pulsed-field gel electrophoresis agarose plug to prepare high-molecular-weight genomic DNA. After proteinase K digestion, the agarose plug was washed with TE buffer, melted in 0.1 mM MES (pH 6.5) and digested using 2 μl of β-agarase (Biolabs). The DNA solution was poured into a Teflon reservoir and DNA was combed onto silanized coverslips (Microsurfaces) using a combing machine. The coverslips were baked at 65°C for an hour and then denatured with 0.5 M NaOH for 20 min. PBS-washed coverslips were dehydrated with a 70, 90 and 100% ethanol series and dried 10 min before hybridization. The biotin labeled rDNA IGS spacer DNA probe and DIG labeled rDNA coding region probe were synthesized by Bio-prime labeling kit (Invitrogen) and DIG DNA labeling kit (Roche), respectively. A total of 300 ng of labeled DNA probes/sample were boiled for 5 min in hybridization buffer, immediately chilled on ice for 10 min and then loaded onto the combed DNA coverslip and incubated overnight at 37°C in a moisture chamber. After hybridization, coverslips were washed twice with 50% FA/2 × SSC and twice with 2 × SSC at room temperature. For detection of ssDNA, biotin, and DIG, we used mouse anti-ssDNA antibody (Millipore), Alexa555 conjugated streptavidin and anti-DIG-FITC (Roche) as the primary antibody, respectively. Following the wash, coverslips were incubated with chicken anti-mouse Alexa 647, biotin conjugated streptavidin and rabbit anti-FITC secondary antibodies. To amplify the biotin and DIG signals, additional incubations were performed with Alexa 555 conjugated streptavidin and goat anti-rabbit FITC, respectively. Images were taken using an OLYMPUS IX70 (60 × /1.40 oil Ph3 PlanApo, Retiga-SRV FAST1394) controlled by IPlab pathway 4.0, and 1 μm was estimated to cover 2 kb, as previously reported using image J software (29).
Illumina sequencing
DNA from 13 BACs was purified using a Qiagen protocol, resulting in BAC clones with sizes ranging from 50–170 kb with concentrations of 75–900 ng/μl. These BACs were further sheared using a BioRuptor for 7.5 min at high intensity with 30 s on-off cycles between two and five cycles were used depending on the initial BAC size and DNA concentration, achieving an average sheared fragment size of 308 bp. A total of 50 ng of sheared DNA was used to generate sequencing libraries using the Illumina ChIPSeq library generation kit. In short, the ends of the sheared DNA fragments were first repaired using a mix of DNA polymerase and ligase, then purified using Agencort Ampure XP magnetic beads. The 3′ ends were adenylated and Illumina specific indexed adaptors were ligated. These ligation products were purified and separated on a 9% agarose gel, and fragments between 300–450 bp were purified. The isolated libraries were enriched for fragments containing ligated adaptors on both ends using 18 cycles of PCR followed by a final cleanup with magnetic beads. The final sequencing libraries had a size range of 280–650 bp and an average size of 388 bp. Sequencing was performed using the Illumina HiSeq 2500 in rapid run mode with on-board cluster generation. The total raw read count was 514 million with ∼440 million passing the quality filter and 92% of the reads with a Q score >30. The 18 indexed samples ranged from 1.8 to 11.9% of total reads with an average of ∼5.3%. This gave read counts for each BAC in the range of 8–52 million (Supplementary Table S2). The raw bcl files were demultiplexed and converted to fastq files using bcl2fastq software provided by Illumina.
PacBio sequencing and assembly
DNA from 13 BACs was isolated, mechanically sheared and size selected. Separate libraries were constructed for each BAC and sequenced on a Pacific Bioscience RSII instrument using two SMRTcells each, generating thousands of fold coverage per BAC (Supplementary Table S2). Sequence assemblies were generated with Canu 1.3 using default parameters (30). Valid assemblies were expected to contain the vector sequence, a degree of circular overlap and the hook sequences used for cloning. Vector sequence and circular overlaps were identified using Nucmer (31) and removed, and the assemblies were re-oriented toward the first base after the removed vector. The assemblies were then polished using Quiver (24) with the raw PacBio data to maximize accuracy. Illumina polishing using Pilon (32) was also attempted. However, the Illumina-polished assemblies yielded lower validation rates versus whole-genome data, so the PacBio-polished assemblies were used for all analyses.
Nanopore sequencing
Following the strategy of Jain et al. (33), a standard Oxford Nanopore protocol was modified to enable the sequencing of entire BACs in individual ∼100 kb reads. Specifically, using the Nanopore SQK-RAD001 ‘rapid’ sequencing kit, a series of titration experiments were carried out to determine an optimal concentration of the transposase-based reagent such that a fraction of the circular BACs would be cut at a single location. This simultaneously linearizes the BACs and adds the necessary sequencing adapters. Results of the titration experiments were evaluated by CHEF gel electrophoresis (Supplementary Figure S5). Satisfactory results were obtained by incubating 2 μl of Fragmentation mix ( FRM) with 400 ng of BAC DNA in a total volume 20 μl for 1 min at 30°C. Nanopore sequencing libraries were constructed using this modified protocol and were sequenced on FLO-MIN104 (R9) flow cells according to the manufacturer’s instructions. The BAC-length nanopore reads were aligned against the PacBio assemblies for validation using bwa-mem (https://arxiv.org/abs/1303.3997).
Whole-genome validation samples
Whole-genome validation experiments were based on four samples for which both high-coverage Illumina and PacBio data were available (34,35,36,37,38,39): AK1 (Korean ancestry), HX1 (Chinese ancestry), NA12878 (Caucasian ancestry) and CHM1 (Caucasian ancestry) (see Data Access).
Whole-genome short-read 45S validation
All rDNA-like regions in GRCh38 were masked and replaced by a single reference sequence. To mask rDNA-like regions in the reference, 500 × coverage of 2 × 100 bp reads were simulated from U13369.1 and mapped against GRCh38 using bwa-mem. All 500 bp reference windows (non-overlapping) hit by ≥2 simulated reads were masked (∼313 kb of sequence in total, 144 kb of which are on chromosome 21). In addition, reference contigs GL000220.1 and KI270733.1, which contain near-complete matches to U13369.1, were also removed, (removing an additional 224 kb of sequence).
A multiple sequence alignment containing all 45S sequences extracted from the TAR-cloned BACs was constructed using mafft (parameter-auto) (40) and manually curated. Illumina reads from the four validation samples were mapped to the modified reference genome, separately for each validation sample. Using samtools (41) mpileup (parameters -q13 -Q10), high-confidence read alleles and their coverages were extracted in a column-wise fashion and projected onto the 45S multiple sequence alignment. To count as ‘validated’, an allele required at least 20 supporting read alleles in at least one sample. Variants in the spacer region were assessed in an analogous manner, employing a multiple sequence alignment containing the new reference sequence and sequences homologous to the new reference sequence extracted from the BACs.
Within-genome variant allele frequencies
Within-genome variant allele frequency was approximated by dividing variant allele coverage by total column coverage. These data were collected during whole-genome short-read validation. Variant allele frequencies are presented only for alleles that passed whole-genome validation.
45S expression analysis
Following Zentner et al. (42), nucleolar RNA-seq sequencing data of the cell line K562 was obtained from ENCODE (see Data Acess), including both ‘long RNA’ (2 × 76 bp Illumina reads) and ‘short RNA’ (1 × 36 bp Illumina reads) datasets generated by the Gingeras lab (43,44). RNA-seq data were also available for the AK1 sample (2 × 100 bp Illumina). Using STAR (44) and GENCODE v25 annotations (45), RNA-seq reads were mapped to the same masked reference genome used for validation of 45S variants, separately for each dataset. Using samtools mpileup (parameters -q13 -Q10), high-confidence RNA-seq read alleles were extracted in a column-wise fashion. The total number of reads mapped to the modified reference sequence was 2 179 533 (short K562), 84 693 198 (long K562) and 1 910 753 (AK1). Analogous to the steps described for the validation of 45S variants from DNA, the extracted sample alleles were projected onto the multiple sequence alignment of BAC-derived sequences. Only alleles with source column total coverage ≥100 were evaluated, and variants with >10% relative allele frequency were counted as ‘validated’.
Whole-genome long-read structural validation
FASTA sequences representing all BAC assemblies were added to the GRCh38 human reference genome. PacBio sequencing reads from the four validation samples were mapped to this extended reference genome using bwa-mem (option -x pacbio). We defined breakpoints coordinates (Supplementary Table S3) and extracted primary alignments spanning these breakpoints plus an additional 2500 bp on each side. Of note, there are differences in read length between the four PacBio samples (average read alignment length: 7.7 kb for CHM1; 6.8 kb for AK1; 5.5 kb for HX1; 3.4 kb for NA12878). Consistent with this, the total number of reported spanning reads was lowest for NA12878. All validating long reads and their breakpoint sequence alignments were confirmed by manual inspection using Nucmer (31).
Location of variants within higher order RNA structure
RNA secondary structures were predicted using methods based on global and local free energy estimation or minimum free energy consensus structure for aligned RNA sequences. Allelic variants of mature and pre-rRNAs were computationally folded and the minimum free energy of the secondary structure was calculated for different window sizes, as in Zuker (46).
Energy minimization was performed using the dynamic programming method and Afold algorithm for evaluation of internal loops (47). Local free energy was estimated for pairs of highly similar sequences, extracted from pairwise alignments with length windows of 100, 350 and 500 nucleotides (48). Structural index (Dist) was defined as the Euclidean base pairing distance.
The RNAalifold program was used to predict RNA consensus structures based on multiple sequence alignments (49) (Vienna package— http://rna.tbi.univie.ac.at). z-scores and empirical P-values were also estimated based on a base-pair distance measure. Base-pair distance was computed as the average distance of two single sequences and a consensus sequence derived by RNAalifold. Two hundred random samples were used to estimate empirical P-values. RNAsnp software (50) was applied to SNVs to detect local RNA secondary structure changes.
The free-energy penalty associated with breaking (opening) of local secondary structure (target structure opening, ΔG kcal/mol) was estimated considering local disruption of secondary structure in windows of different length. Free-energy changes were approximated with nearest-neighbor free-energy parameters using the Afold program (47) and the OligoWalk program (51). Local structure was considered for a set of suboptimal structures.
Monte Carlo simulation and analysis of randomized sequences (52,53) was used to estimate significant differences between target structure opening (ΔG) or structural index (Dist) of allelic variants. The free-energy penalty associated with local secondary structure opening (ΔG kcal/mol) or structural index (Dist) for all random sequences was calculated for local disruption of secondary structure in the given window. P-values for randomizations and for difference between variant alleles were determined by the MWW test.
RESULTS
TAR Isolation of rDNA sequences specific for NOR on human chromosome 21
Individual rDNA units were isolated using TAR cloning, which permits selective targeted isolation of genomic regions from complex genomes (26,54). This method has been successfully applied to close several gaps in the human genome sequence (55). To avoid cross-contamination with rDNA derived from other acrocentric chromosomes, we used genomic DNA from the mouse-human hybrid cell line A9 (21–16), which contains a single human chromosome 21 in a mouse cell background (56). The presence of a single human chromosome 21 was confirmed by fluorescence in situ hybridization (FISH) (Supplementary Figure S4). DNA fiber analysis revealed that human rDNA units in this cell line form a single cluster on human chromosome 21 with no detectable insertions of non-rDNA sequences (Supplementary Figure S6). Approximately 50 copies of human rDNA are present based on qPCR analysis of the human-specific IGS (Supplementary Figure S6).
For the selective isolation of rDNA units from the hybrid cells, five TAR cloning vectors were designed (#2, #6, #11, #16 and #22 in Figure 1). Each vector contains a pair of targeting sequences (hooks) derived from the 5′ ETS ∼1.4 kb upstream 18S rRNA gene. The chosen region of ETS has no homology to rodent ETS in order to exclude targeting mouse rDNA units during TAR cloning. In three vectors, #6, #11 and #16, both hooks have the same orientation corresponding to that in the reference rDNA (Figure 1B). Therefore, it is expected that with these vectors in vivo recombination in yeast will rescue rDNA regions organized as tandem repeats. In two other vectors, #2 and #22, one of the hooks is inverted. Vectors with such hook orientation cannot target tandem rDNA repeats, but are suitable for isolation of rDNA sequences in palindromic orientation (Figure 1C), which have previously been inferred to exist in human DNA (19). The inclusion of an F-factor origin in the TAR vector (Supplementary Figure S1) permits transfer of the yeast artificial chromosomes (YACs) into E. coli cells, in which they can propagate as BACs, facilitating purification of the clones for sequencing.
To TAR clone human rDNA, each vector and genomic DNA isolated from the A9 (16–21) hybrid cells were mixed with yeast spheroplasts. In some experiments, genomic DNA was pre-treated with CRISPR-Cas9 nucleases to generate double-strand breaks near the targeted genomic region (Supplementary Figure S2). This provided a 3-fold increase in rDNA-positive colonies (26, Supplementary Figure S3B). To identify YACs containing human rDNA, ∼3400 yeast clones were tested by PCR for the presence of sequences corresponding to the 30 kb IGS. Among the analyzed yeast transformants, 13 had sequences of a human rDNA unit (Supplementary Figure S3). The YACs containing human rDNA were transferred into E. coli cells by electroporation and propagated as BACs. To check the fidelity of YACs after transfection into bacterial cells, BAC DNA was isolated from two to three independent E. coli transformants and analyzed by CHEF gel electrophoresis. For most TAR isolates, circular molecules of uniform identical size were observed, suggesting that there were no obvious rearrangements of circular YAC/BACs during electroporation (data not shown). In total, 12 independent rDNA-containing clones with inserts of 44–89 kb were chosen for further analysis (Supplementary Table S2).
Assembly of rDNA clones isolated from human chromosome 21
Eight of the newly isolated and sequenced BACs contained at least one full-length rDNA unit (Figure 2). Those containing only a single rDNA unit included JH5, JH8 and JH12 (isolated with TAR vector #6); and JH15 and JH18 (vector #16). Longer BACs included JH11 (vector #6), which contained two tandem rDNA units; JH4 (vector #2), which contained one rDNA unit preceded by a truncated IGS; and JH14 (vector #11), which contained a truncated rDNA unit followed by three IGS fragments. The four remaining BACs contained only head-to-head oriented IGS fragments, including JH2 and JH3 (vector #2); and JH6 and JH10 (vector #6).
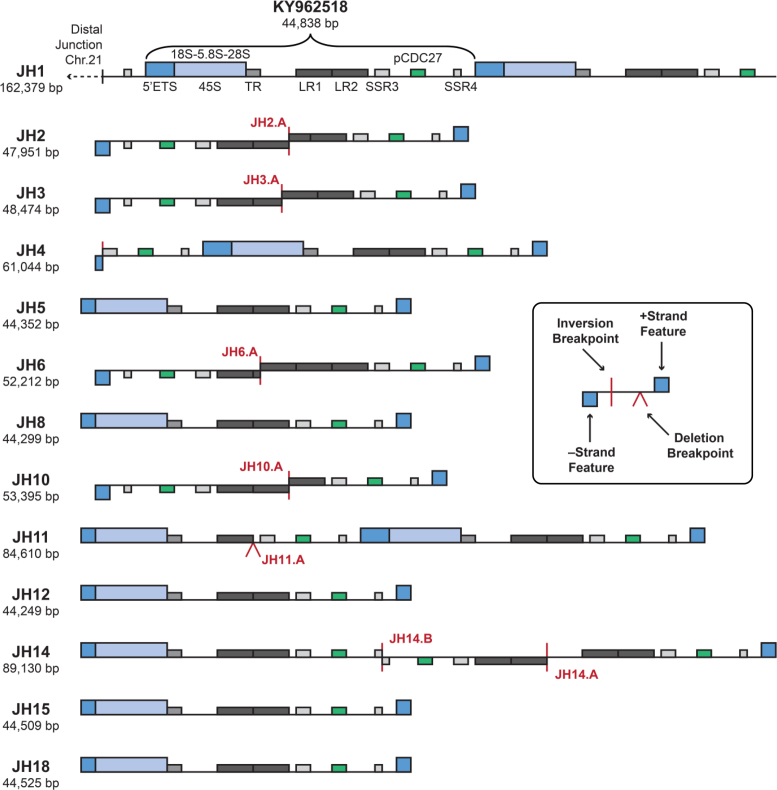
Figure 2.
Schematic structural organization of rDNA-containing BAC clones isolated by TAR from the human chromosome 21. Structures of the individual isolated BACs are illustrated, highlighting rDNA coding and IGS regions. The bracket spans the new reference rDNA sequence derived from JH1, starting from the 5′ETS, deposited as GenBank Accession KY962518. Clones JH2 and JH3, isolated with TAR vector #2 (in which one of the hooks is inverted; see Figure 1C) contain two IGS sequences in a head-to-head configuration consistent with the presence of palindromic structure in rDNA arrays. Another clone with palindromic rDNA sequences, clone JH14, was isolated with TAR vector #11, with the hooks in the same orientation. As illustrated, this clone contains a double palindromic structure that resulted in a final ‘correct’ orientation of the targeted sequences in rDNA array. Notably, this highly rearranged region contains the pseudogene CDC27 (green bar). Vertical red lines indicate inversion break points. Exact coordinates of break points and inversions are listed in Supplementary Table S3. Carats indicate an IGS truncation by deletions. Annotated BAC sequences can be downloaded from GenBank under the Project Accession# PRJNA38015. TR: Tandem repeat; LR1 and LR2: Long repeats; SSR3 and SSR4: Simple sequence repeats; pCDC27: CDC27 pseudogene. SSR1 and SSR2 are nested within LR1 and LR2 and are omitted here for clarity (see Figure 3).
In addition to these new BACs, we also sequenced and assembled a previously annotated BAC, CH507-528H12. The new version sequenced using PacBio long reads is labeled here as JH1. This BAC had been isolated from another mono-chromosomal hybrid cell line carrying chromosome 21, also using a TAR cloning vector (57). It includes two tandem rDNA repeats along with 74 469 kb of sequence upstream of the rDNA repeats with homology to the distal junction of the chromosome 21 rDNA locus (20). Our new long-read assembly is 942 bp longer than the published version (FP236383.15), and a pairwise alignment showed that the two versions differ by 1324 gaps and 144 mismatches, with most of these differences falling within the repeated rDNA regions. Because our new assembly was constructed using very high-coverage of long, single-molecule reads (452-fold coverage in reads >20 kb), the differences are likely caused by incomplete separation of the two rDNA copies in the original Sanger-based assembly. To test this hypothesis, we aligned the newly generated JH1 PacBio reads to both the old and the new assembly, and measured agreement between the aligned reads and the assemblies at the positions of single-nucleotide differences between the assemblies. Consistent with the PacBio sequencing error rate, we found that 86% of aligned read alleles matched our PacBio assembly at the evaluated positions; by contrast, the match rate for the previously published Sanger assembly was only 34%, indicating likely assembly error at the evaluated positions. Based on this evaluation, the PacBio version (JH1) was selected as the representative assembly. These results demonstrate the ability of long reads to correctly resolve complex rDNA array structures.
In total, 13 rDNA clones were assembled and analyzed, including 11 full copies of the 45S transcribed region and 6 non-canonical IGS structures. The schematic structure of these clones is presented in Figure 2 and Supplementary Figure S7A, and the annotated assemblies are available from GenBank (Supplementary Table S2).
Validation of clone assembly by Oxford Nanopore sequencing
To ensure the integrity of the PacBio assembly process in the presence of complex repeat structures, we selected four BACs for validation: three BACs containing palindromic IGS fragments (JH2, JH10, JH14), and one BAC with two distinct rDNA units (JH11). Using a custom Oxford Nanopore protocol capable of producing single-molecule reads spanning an entire BAC sequence, we successfully validated the structure of the three inversion-containing assemblies (NCBI BioProject; PRJNA380105). Low sequencing coverage, combined with the large size of the BAC, prevented nanopore validation of JH11, but the deletion observed in the second rDNA copy of this BAC was successfully validated using long-read population data from whole genomes (see below).
A modified human rDNA reference sequence
For a detailed characterization of rDNA variation with respect to known sequences, we compared our new assemblies to U13369.1, the GenBank sequence that has served as an rDNA reference. All but one of our BACs shared an ∼2 kb insertion in the IGS relative to U13369.1 between positions 22703 and 22714, indicative of population polymorphism or a possible error in the U13369.1 sequence (Supplementary Figure S8). The inserted sequence transforms a degenerate repeat sequence in the spacer region into a near-perfect tandem repeat (Supplementary Figure S7B). The presence of this extra 2 kb sequence, as well as better validation results for the 45S region (see below), identified the first rDNA copy of JH1 as a suitable reference replacement (Figure 2, GenBank accession KY962518).
Validation using short-read whole-genome data confirmed all bases in the 45S region for our new reference sequence (100% of 45S bases in KY962518; Supplementary Table S4), while an analogous analysis for U13369.1 yielded a lower validation rate of 99.77%. Furthermore, long reads from a high-quality whole-genome sample, CHM1 (39), also agreed better with the new reference, especially across the new 2 kb insertion (Supplementary Figure S7C).
The current human reference genome GRCh38 (58) also contains three rDNA units assigned to chromosome 21. The most similar is nearly identical to our revised reference with the exception of 621 and 19 bp deletions toward the end of the IGS; a single C/G SNP in ITS-2; and tens of single base insertion or deletions.
Figure 3 shows all repeats in the new JH1-based 44 838 bp reference sequence along with the locations of structural breakpoints in the clones. We also provide a pairwise sequence alignment between the new reference sequence KY962518 and U13369.1 (Supplementary Data S1). Comparison of the rDNA promoter sequence of U13369.1 and the new reference sequence showed significant differences (16 SNPs and INDELs), and in all cases the whole-genome validation datasets confirmed the new reference. These results are also in agreement with previously published rDNA promoter sequences (59,60).
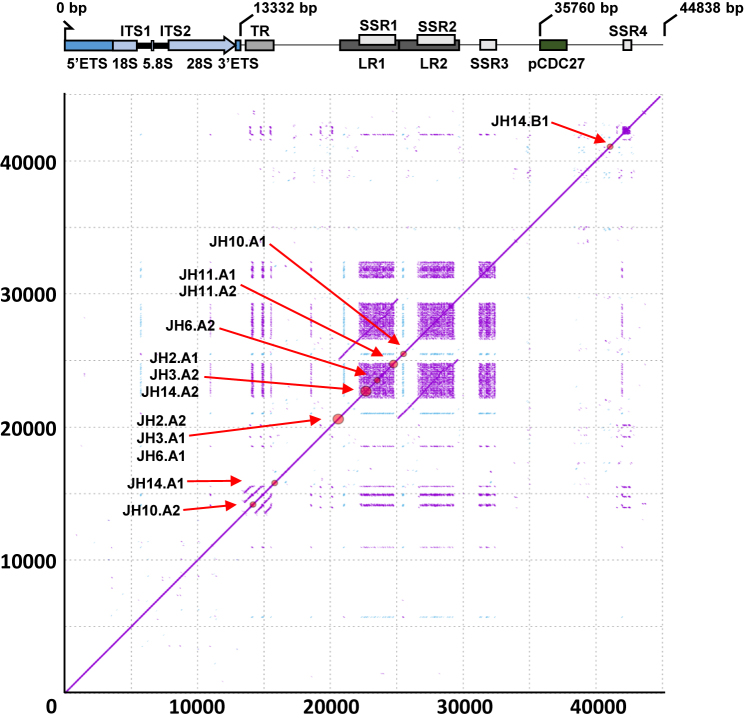
Figure 3.
Schematic diagram illustrating structure and sequence features of the new rDNA reference sequence. High-resolution (k = 15) self-similarity dot plot of the new reference sequence used for structural variant analysis. Purple dots indicate forward-strand matches, blue dots indicate reverse-complement matches. Red circles on the diagonal and labels show the positions of the identified structural variant breakpoints (clustered if distance ≤500 bp; see Supplementary Table S3 for precise coordinates and whole-genome validation status). Gene and sequence feature annotations are displayed below the x-axis. ETS: External transcribed spacer; 18S, 5.8 and 28S: core ribosomal DNA genes; ITS: Internal transcribed spacer; TR: Tandem repeats; LR1-2: Long repeats; SSR1-4: Simple sequence repeats; pCDC27: Pseudogene CDC27. Note that LR1 and LR2 are highly homologous, and that SSR1-4 are composed of near-identical sequence motifs.
Structural analysis and validation of IGS
In addition to the 2 kb insertion relative to U13369.1, we identified seven additional structural variants (six inversion events and one deletion; Supplementary Table S3 and Figure S7A). We anticipated that some inversion/palindromic regions might be recovered, both because they had been inferred to be present in uncloned DNA in a previous report (19), and because we observed apparent head-to-head cloning of rDNA regions in fiber-FISH DNA analyses (Supplementary Figure S6). Indeed, clones JH2 and JH3 were recovered with vector #2, which specifically targeted palindromic segments using inverted hooks for cloning. Two other clones (JH6 and JH10) also contained palindromic segments but were recovered with normally oriented hooks (vector #6 with a truncated rH4 hook). Isolation of such clones may be explained by homologous recombination of one hook and non-homologous DNA end-joining or non-homologous recombination of another hook during TAR cloning (54). A fifth clone, JH14, was recovered with vector #11, and contained both a repeat unit and a divergent palindromic IGS segment.
Most of the inverted segments map to the large tandem repeat central to the IGS. Closer examination of the junction sequences between the IGS repeats in clones JH2, JH3, JH6, JH10 and JH14 reveals that each deletion did indeed occur between homologous repetitive elements, with different pairs of repeats in each clone (Figure 3; Supplementary Figure S9 and Table S3). This suggests that they are independent events, and that the cloned sequences in the different YAC/BACs are from different sites across the chromosome 21 rDNA locus. However, the nature of these non-canonical regions (palindromic sequences with high repeat content) prevented PCR validation in the cell line, and so the possibility that they arose artefactually during or after cloning cannot be fully ruled out (see ‘Discussion’ section).
To assess the extent to which these non-canonical structures are present in population genomes, we used high-quality, long-read sequencing data. Specifically, we mapped PacBio long reads from four whole-genome samples (one Korean; one Chinese; two Caucasian) to a reference genome supplemented with our assembled BAC sequences, and extracted read alignments spanning the breakpoints and an additional 2.5 kb on either side. All identified breakpoints and their mapped PacBio read support are detailed in Supplementary Table S3. The IGS insertion found in the BACs compared to U13369.1, as well as the IGS deletion event in JH11, were robustly covered by high-identity read alignments and inferred to be present in human populations. However, support for palindromic structures was weak in the whole-genome data, with no supporting reads found for the JH6.A and JH14.A inversions, and only a few supporting reads found for inversions JH2.A, JH3.A, JH10.A and JH14.B. This analysis is complicated by the fact that both PacBio and Nanopore sequencing produce a small fraction of palindromic reads at random, and so it is difficult to separate signal from noise with low coverage. Thus, the inference of palindromic structures remains qualified, and again the possibility of cloning artifacts cannot be excluded.
Small sequence variants in 45S and intergenic spacer region
For a combined analysis of 45S variants, we examined the 11 new 45S sequences presented here and identified 101 variant alleles between them (full list: Supplementary Table S5; summary: Supplementary Table S6). Of these 101 variants, 25 are SNVs and 76 are INDELs (short insertions or deletions). Our call set comprises both private variants (called in only 1 or 2 45S sequences; n = 56) and ubiquitous variants (called in 9 or 10 45S sequences; n = 10) (Supplementary Table S7). An analysis of the spatial distribution of variants showed broadly comparable variant frequencies in the ETS and ITS spacer regions; no variants in 5.8S; only a few in 18S; and peak frequencies in the distal half of 28S (Figure 4A). For each subregion, the numbers of variants observed, the numbers of variants/kb, and the types of variants seen are summarized in Table 1.
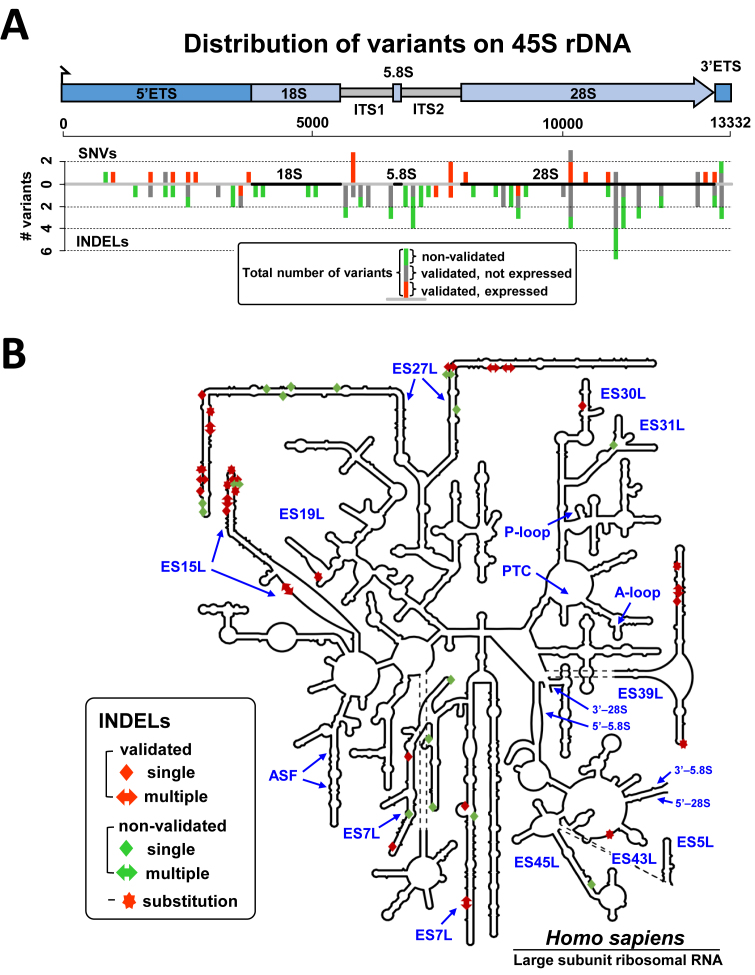
Figure 4.
Variants of 45S rDNA sequence identified in BAC clones isolated from human chromosome 21. (A) Spatial distribution of 45S variant calls indicated separately for SNVs (above the line) and INDELs (below the line). Bars are stacked with validated and expressed variants shown in green; validated and non-expressed variants shown in dark gray; non-validated variants shown in blue. Calls from different BACs are counted separately. A variant is classified as validated if it is supported by at least 20 high-quality Illumina read alleles in at least one of the four WGS validation samples. (B) Schematic secondary structures of the human 28S rRNAs. The 28S rRNA diagrams were taken from the RiboVision Server (65). The diagram was modified to reflect the final rRNA models (66). Functional regions and variable elements of the rRNA, including ES numbering (in blue), are labeled. The consensus secondary structure was not accurately predicted for several ES loci, and the ES27L segment, with a major portion of identified variants, is not shown to scale. INDELs found in isolated human rDNA units are depicted as rectangles or arrows. Single nucleotides are shown as rectangles, multiple nucleotides are shown as arrows. Substitutions are shown as stars. Non-validated variants are shown in green and validated variants are shown in red. Additional information on identified variants in the 28S mature rRNA (primarily SNVs and short INDELs) is present in Supplementary Table S5.
Table 1.
Numbers and types of sequence variants observed in TAR-cloned rDNA loci
| A. Variants in rDNA | |||||
|---|---|---|---|---|---|
| Validated by deep sequencing | |||||
| Features | Total variants (variants/kb) | All | Any | Any RNA-seq | |
| 5′ ETS | 19 (5.2) | 9 | 12 | 7 | |
| 18S | 4 (2.1) | 0 | 0 | 0 | |
| ITS-1 | 14 (13.1) | 5 | 11 | 3 | |
| 5.8S | 0 (0) | 0 | 0 | 0 | |
| ITS-2 | 13 (11.1) | 5 | 5 | 4 | |
| 28S | 43 (8.5) | 18 | 27 | 7 | |
| 3′ ETS | 8 (22.2) | 1 | 5 | 1 | |
| 45S (total) | 101 (7.6) | 38 | 60 | 22 | |
| IGS (total) | 235 (7.5) | 108 | 148 | 3 | |
| B. Variant breakdown into types | |||||
| Deletions | Insertions | ||||
| Features | SNPs | Single nucleotide | Multiple nucleotide | Single nucleotide | Multiple nucleotide |
| 5′ ETS | 8 | 5 | 2 | 3 | 1 |
| 18S | 0 | 4 | 0 | 0 | 0 |
| ITS-1 | 3 | 6 | 0 | 3 | 2 |
| 5.8S | 0 | 0 | 0 | 0 | 0 |
| ITS-2 | 2 | 9 | 0 | 2 | 0 |
| 28S | 9 | 16 | 2 | 6 | 10 |
| 3′ ETS | 3 | 0 | 2 | 2 | 1 |
| 45S (total) | 25 | 40 | 6 | 16 | 14 |
| IGS (total) | 88 | 14 | 59 | 8 | 66 |
(A) A summary of distribution of variants in the 45S pre-rDNA and IGS regions and their frequencies per kilobases are shown. The number of variants validated in all deep sequenced individuals versus any deep-sequenced individuals are also listed. The last column lists variants validated in deep-sequenced RNA samples. (B) Variants categorized as SNPs, insertions and deletions are listed for transcribed rDNA and IGS regions.
In contrast to the non-canonical structural variants, the smaller SNVs could be confidently validated using whole-genome data. Both to validate the existence of identified variants in uncloned human DNA and to assess the extent to which the identified variants may be present in human populations, we examined high-coverage, high-quality Illumina sequencing data from the same four whole-genome sequencing (WGS) samples used for structural validation. In total, we recovered 59% (60/101) of the variants we had called, requiring at least 20 supporting reads; the recovery rate for SNVs and insertions (92 and 87%, respectively) was much higher than the recovery rate for deletions (24%; Supplementary Table S6). Consistent with the Japanese origin of our DNA source for TAR cloning, the highest per-sample combined recovery rate (55%) was observed for the Korean sample AK1 (Supplementary Table S6 and ‘Discussion’ section).
Outside of the 45S transcript unit, 235 variants were detected in the IGS, including a larger number of deletion and insertion structural variants (Supplementary Table S8). For convenience, a multiple alignment of the BAC sequences, including the IGS, is provided and can be displayed in JalView to visualize all variants (61) (Supplementary Data Files S2 and 3).
Expression analysis of 45S variants
To assess the extent to which the identified 45S variants are expressed, we examined RNA-seq data. Specifically, we assessed allele recovery rate in three available RNA-seq datasets, including one from the AK1 cell line and two earlier nucleolar datasets, enriched for pre-rRNA, from the K562 cell line. Consistent with results reported by Zentner et al. (42), we find that millions of RNA reads align to the 45S region of our new reference sequence. With conservative thresholds for allele recovery (>100-fold coverage and >10% supporting reads), we validated 22/79 (22%) of callable alleles from the three RNA-seq datasets, combined (Supplementary Table S6).
We extended the analysis of expression further in AK1, because it offered both high quality sequencing data for both RNA and DNA. Focusing on the 56 variant alleles validated in AK1 WGS genomic DNA, we could assess expression for 37 variants in AK1 RNA-seq data. Of these, 18 (49%) showed evidence for expression. Conversely, of the 45 variant alleles not recovered from AK1 DNA sequencing data, none showed evidence for expression in the AK1 RNA-seq data. In other words, variants are consistent in the DNA and RNA sequences from the same individual (see ‘Discussion’ section).
Location of variants within higher order RNA structure
Of the variants located in the 45S pre-RNA region, including 47 in mature 18S and 28S rDNA sequences, few were predicted to alter RNA structure (‘riboSNitches’). Rather, most variants are structurally synonymous and located in expansion segments (ESs), which are defined as sequence-variable regions across different species (62,63). The relative paucity of variants mapped to 18S rRNA (Table 1 and Figure 4A) is in agreement with the greater evolutionary conservation of 18S compared to 28S rRNA (64,65,66,67). In the 28S, 42 variants map within the ESs (Figure 4B), with the highest densities in ES27L and several predicted to be riboSNitches (Supplementary Figure S10A).
Consensus RNA folding of the ES27L infers highly stable conserved hairpin structures with nucleotides under strong selection for pairing (Supplementary Figure S10B). Two hot spots of variation are located on the opposite strands of a stem-loop structure, and INDELs jointly occur frequently in both spots. The inserted segments are neighbors in RNA folding despite their separation by ∼175 nucleotides in primary sequence (Supplementary Figure S10B). These results are in agreement with recent evidence for concerted conformational dynamics of ES27L and ES31L that enables communication between the mRNA exit site on the 40S subunit and the tunnel exit site on the 60S subunit (62).
ES7L and ES15L also have variants in a region where the evolutionary insertion of helix ES15L-A creates an enlarged internal loop in human ribosomes, leading to new contacts with ribosomal proteins L6e and L30 as well as with ES7L, ES9L and ES10L (62) (Supplementary Figure S11). Whether any of these variants are neutral or modulate ribosome function in human remains to be tested (see ‘Discussion’ section).
Notably, sequence variants in the 5′ ETS lie near processing sites (e.g. sites 01 and 1 in 5′ETS and between sites 3 and E in ITS1; Supplementary Figure S12) and might affect pre-rRNA processing through modification of RNA folding (59). In addition, in the pre-rRNA promoter region, several variants modify CpG sites that show differential methylation status in some tumor samples (60) (Supplementary Figure S12). The rDNA promoter region shows some differences between U13369.1 and new reference sequence, but in the sequenced clones we observe less variation in the core promoter than in ES regions (Supplementary Data S2).
DISCUSSION
Candidate rDNA variants and their validation
Early studies suggested the presence of rDNA sequence variants (14,15,16,17,18), but their frequency within individuals or populations has been unknown. In our pilot effort, we identified and analyzed variants in a number of rDNA units isolated by TAR cloning from a single NOR. Our initial attempts to assemble these units using paired-end Illumina data failed, due to the complex repeat structures. This block was overcome by the use of long, PacBio reads for assembly, combined with ultra-long Nanopore reads for validation of the most complex structures.
Analyses of 13 clones isolated from human chromosome 21 revealed multiple candidate variants among the rDNA units. Most variants were SNVs, short INDELs or variable lengths of short repeat motifs. A majority of the small variants discovered were also recovered from independent, WGS data, indicating that they are true variants, and not the result of sequencing error or cloning artifacts. In particular, 60/101 of the 45S variants identified were confirmed to be present in at least one of the four uncloned whole-genome DNA samples. Some of the remaining variants, a vast majority of which were deletions (35/41), could represent sequencing artifacts (Supplementary Table S6). Many of these appear in the context of homopolymer runs, which are a known weakness of PacBio sequencing. Alternatively, a proportion of these variants may be restricted to certain populations or individuals.
Several isolated rDNA clones also contained palindromic structures, and fiber-FISH analyses of the uncloned chromosome 21 DNA (from which the clones were derived) showed similar non-canonical structures (Supplementary Figure S6). It is plausible that such structures could arise from recombination between IGS repeats (Supplementary Figure S9). However, palindromic sequences were not confidently recovered from long-read WGS data, and so their prevalence in uncloned DNA may not be as high as previously suggested (62).
Localization of variants in promoter or mature rRNA sequences
Secondary structure modeling of the rRNA 5′-leader sequence (59) suggest that some of the observed SNVs and INDELs might modulate local configurations. Thus, variants near the transcription start site of the 45S pre-rRNA are candidates for further study of the effect on expression (68,69). As for transcribed sequences, RNA-seq data indicate that at least one-fifth of the identified 45S variants mapped to mature rRNA are transcribed. In the accepted secondary structure of mature rRNA, most of the observed variants are located in 28S ESs. Those regions vary in sequence across species, and recent high resolution cryo-electron-microscopy density maps of human and Drosophila ribosomes inferred dynamic behavior of ESs and co-evolution of ribosomal RNA with ribosomal proteins (62). The quality of the cryo-EM map allows localization and construction of models for all 30 rRNA ESs in the human 80S ribosome, facilitating prioritization of variants for functional study (62).
A new reference sequence for rDNA repeat units
We have suggested a new canonical rDNA reference sequence, based on the criteria reviewed in ‘Results’ section. Our approach of TAR cloning combined with long-read single-molecule sequencing effectively controls the sequence assembly process and false-positive rate of rDNA variant discovery. Legacy assemblies, by contrast, may not faithfully represent the structure of rDNA repeat units, and appear to have higher per-base error rates. For example, the current U13369.1 reference sequence contains an additional 108 variants compared to our clones, but only 8 of these are validated by the whole-genome data. This 7% validation rate is well below the 67% validation rate observed for the variants identified by our method. Similarly, our approach enabled us to improve the assembly of the existing CH507-528H12 (JH1) clone and accurately resolve both rDNA units present on that BAC.
Importantly, in work in progress, we have seen the same overall structure and largely identical sequence in five rDNA units from chromosome 22; and another reference sequence candidate has been suggested in the thesis of Saumya Agrawal (http://hdl.handle.net/10179/5971), analyzing a deposited but previously unannotated BAC sequence isolated from another chromosome 22 (RP11-164K15, GB Acc# AL353644.34). An analysis of its main 45S unit shows the presence of 12 additional variants, 8 of which validate in WGS; and an overall validation rate very similar to that of the BACs analyzed here (13327 of 13331 45S positions). A pairwise sequence alignment between the complete rDNA unit of AL353644.34 and our new reference sequence furthermore shows the presence of similar, but not identical, structures in the spacer region. It is reassuring that all these sequences contain comparable versions of the 2 kb segment missing in the current standard reference. Furthermore, because sequences from different chromosomes in different individuals are very similar, a new reference will be useful for the full range of rDNA across the acrocentric chromosomes.
There are also indications that the similarity of overall structure seen in rDNA repeat units from different chromosomes extends to sequence variants. Supporting such a possibility, a length variation polymorphism was previously found to be shared among ribosomal genes on non-homologous human chromosomes (70,71). In our analyses, we found many of the same variants in both transcribed and intergenic rDNA regions of individuals from several ethnic groups (Supplementary Figure S13 and Table S8). Nineteen variants were found at frequencies >30% both in our pilot sample and in total DNA from four deep-sequenced individuals (Supplementary Table S7). At such high frequencies, alleles are likely present in rDNA copies on more than one chromosome across human populations. As has been demonstrated in Drosophila (72) and suggested for human (73), crossovers between rDNA repeats on different chromosomes might homogenize rDNA variation across acrocentric chromosomes.
How many variants occur in rDNA remains to be seen, but it is interesting that thus far we find an average rate of roughly 7.5 variants per kb, which is comparable to typical estimates of variation across the genome. In the future, ‘ultra-long’ nanopore sequencing (39,61) may enable the complete assembly of human acrocentric chromosomes and cataloging of rDNA variation across chromosomes and populations.
DATA AVAILABILITY
Supplementary Table S9 lists all sources and accessions of data used in the variant analysis.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Alice Young and the NIH Intramural Sequencing Center (NISC) for assistance with PacBio sequencing. This work utilized the computational resources of the NIH HPC Biowulf cluster (https://hpc.nih.gov).
Authors’ contribution: D.S., A.M.P. and V.L. conceived the study. J.H.K., A.T.D., R.N., S.K., H.S.L., D.D., W.W., Y.P., V.N.K. and A.M.P. sequenced and assembled the BAC sequences. A.T.D., A.Y.O., S.A.S., K.U. and R.N. analyzed the data. J.H.K., A.T.D., R.N., S.A.S., D.S., A.M.P. and V.L. wrote the manuscript draft. All authors edited and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Intramural Research Program of the NIH. Intramural Research Program of the National Human Genome Research Institute (to A.T.D., S.K., A.M.P.); Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, USA (to V.L.); National Institute on Aging; US Department of Health and Human Services, Intramural Funds (to the National Library of Medicine) (to A.Y.O., S.A.S.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Jacob F., Monod J.. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961; 3:318–356. [DOI] [PubMed] [Google Scholar]
- 2. Henderson A.S., Warburton D., Atwood K.C.. Location of ribosomal DNA in the human chromosome complement. Proc. Natl. Acad. Sci. U.S.A. 1972; 69:3394–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schmickel R.D., Knoller M.. Characterization and localization of the human genes for ribosomal ribonucleic acid. Pediatr. Res. 1977; 11:929–935. [DOI] [PubMed] [Google Scholar]
- 4. Stults D.M., Killen M.W., Pierce H.H., Pierce A.J.. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2008; 18:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Héliot L., Mongelard F., Klein C., O’Donohue M.F., Chassery J.M., Robert-Nicoud M., Usson Y.. Nonrandom distribution of metaphase AgNOR staining patterns on human acrocentric chromosomes. J. Histochem. Cytochem. 2000; 48:13–20. [DOI] [PubMed] [Google Scholar]
- 6. McStay B. Nucleolar organizer regions: genomic ‘dark matter’ requiring illumination. Genes Dev. 2016; 30:1598–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gagnon-Kugler T., Langlois F., Stefanovsky V., Lessard F., Moss T.. Loss of human ribosomal gene CpG methylation enhances cryptic RNA polymerase II transcription and disrupts ribosomal RNA processing. Mol. Cell. 2009; 35:414–425. [DOI] [PubMed] [Google Scholar]
- 8. Bowman L.H., Rabin B., Schlessinger D.. Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res. 1981; 9:4951–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sylvester J.E., Sylvester J.E., Whiteman D.A., Podolsky R., Pozsgay J.M., Respess J., Schmickel R.D.. The human ribosomal RNA genes: structure and organization of the complete repeating unit. Hum. Genet. 1986; 73:193–198. [DOI] [PubMed] [Google Scholar]
- 10. Wu Z.W., Wang Q.M., Liu X.Z., Bai F.Y.. Intragenomic polymorphism and intergenomic recombination in the ribosomal RNA genes of strains belonging to a yeast species Pichia membranifaciens. Mycology. 2016; 7:102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bik H.M., Fournier D., Sung W., Bergeron R.D., Thomas W.K.. Intra-genomic variation in the ribosomal repeats of nematodes. PLoS One. 2013; 8:e78230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Simon U.K., Weiss M.. Intragenomic variation of fungal ribosomal genes is higher than previously thought. Mol. Biol. Evol. 2008; 25:2251–2254. [DOI] [PubMed] [Google Scholar]
- 13. Tseng H., Chou W., Wang J., Zhang X., Zhang S., Schultz R.M.. Mouse ribosomal RNA genes contain multiple differentially regulated variants. PLoS One. 2008; 3:e1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gonzalez I.L., Sylvester J.E.. Human rDNA: evolutionary patterns within the genes and tandem arrays derived from multiple chromosomes. Genomics. 2001; 73:255–263. [DOI] [PubMed] [Google Scholar]
- 15. Kuo B.A., Gonzalez I.L., Gillespie D.A., Sylvester J.E.. Human ribosomal RNA variants from a single individual and their expression in different tissues. Nucleic Acids Res. 1996; 24:4817–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leffers H., Andersen A.H.. The sequence of 28S ribosomal RNA varies within and between human cell lines. Nucleic Acids Res. 1993; 21:1449–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uemura M., Zheng Q., Koh C.M., Nelson W.G., De Marzo A.M.. Overexpression of ribosomal RNA in prostate cancer is common but not linked to rDNA promoter hypomethylation. Oncogene. 2012; 31:1254–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gonzalez I.L., Sylvester J.E.. Complete sequence of the 43-kb human ribosomal DNA repeat: analysis of the intergenic spacer. Genomics. 1995; 27:320–328. [DOI] [PubMed] [Google Scholar]
- 19. Caburet S., Conti C., Schurra C., Lebofsky R., Edelstein S.J., Bensimon A.. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 2005; 15:1079–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Floutsakou I., Agrawal S., Nguyen T.T., Seoighe C., Ganley A.R., McStay B.. The shared genomic architecture of human nucleolar organizer regions. Genome Res. 2013; 23:2003–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Niranjan N., Mihai P.. Sequence assembly demystified. Nat. Rev. Genet. 2013; 14:157–167. [DOI] [PubMed] [Google Scholar]
- 22. Agrawal S., Ganley A.R.. Complete sequence construction of the highly repetitive ribosomal RNA gene repeats in eukaryotes using whole genome sequence data. Methods Mol. Biol. 2016; 1455:161–181. [DOI] [PubMed] [Google Scholar]
- 23. Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A., Tyson J.R., Beggs A.D., Dilthey A.T., Fiddes I.T. et al. . Nanopore sequencing and assembly of a human genome with ultra-long reads Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018; 36:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chin C.S., Alexander D.H., Marks P., Klammer A.A., Drake J., Heiner C., Clum A., Copeland A., Huddleston J., Eichler E.E. et al. . Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013; 10:563–569. [DOI] [PubMed] [Google Scholar]
- 25. Kazuki Y., Yakura Y., Abe S., Osaki M., Kajitani N., Kazuki K., Takehara S., Honma K., Suemori H., Yamazaki S. et al. . Down syndrome-associated haematopoiesis abnormalities created by chromosome transfer and genome editing technologies. Sci. Rep. 2014; 4:6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee N.C., Larionov V., Kouprina N.. Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast. Nucleic Acids Res. 2015; 43:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kouprina N., Annab L., Graves J., Afshari C., Barrett J.C., Resnick M.A., Larionov V.. Functional copies of a human gene can be directly isolated by TAR cloning with a small 3′ end target sequence. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:4469–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim J-H., Kononenko A., Erliandri I., Kim T-A., Nakano M., Iida Y., Barrett J.C., Oshimura M., Masumoto H., Earnshaw W.C. et al. . Human artificial chromosome (HAC) vector with a conditional centromere for correction of genetic deficiencies in human cells. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20048–20053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fu H., Martin M.M., Regairaz M., Huang L., You Y., Lin C.M., Ryan M., Kim R., Shimura T., Pommier Y. et al. . The DNA repair endonuclease Mus81 facilitates fast DNA replication in the absence of exogenous damage. Nat. Commun. 2015; 6:6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M.. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017; 27:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Delcher A.L., Phillippy A., Carlton J., Salzberg S.L.. Fast algorithms for Large-scale genome alignment and comparision. Nucleic Acids Res. 2002; 30:2478–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walker B.J., Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J. et al. . Pilon:an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014; 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jain M., Olsen H.E., Turner D.J., Stoddart D., Bulazel K.V., Paten B, Haussler D., Willard H.F., Akeson M., Miga K.H.. Linear assembly of a human centromere on the Y chromosome. Nat. Biotechnol. 2018; 36:321–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seo J.S., Rhie A., Kim J., Lee S., Sohn M-H., Kim C-U., Hastie A., Cao H., Yun J-Y., Kim J. et al. . De novo assembly and phasing of a Korean human genome. Nature. 2016; 538:243–247. [DOI] [PubMed] [Google Scholar]
- 35. Shi L., Guo Y., Dong C., Huddleston J., Yang H., Han X., Fu A., Li Q., Li N., Gong S. et al. . Long-read sequencing and de novo assembly of a Chinese genome. Nat. Commun. 2016; 7:12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Steinberg K.M., Schneider V.A., Graves-Lindsay T.A., Fulton R.S., Agarwala R., Huddleston J., Shiryev S.A., Morgulis A., Surti U., Warren W.C. et al. . Single haplotype assembly of the human genome from a hydatidiform mole. Genome Res. 2014; 24:2066–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eberle M.A, Fritzilas E., Krusche P., Källberg M., Moore B.L., Bekritsky M.A., Iqbal Z., Chuang H-Y., Humphray S.J., Halpern A.L. et al. . A reference data set of 5.4 million phased human variants validated by genetic inheritance from sequencing a three-generation 17-member pedigree. Genome Res. 2017; 27:157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pendleton M., Sebra R., Pang A.W, Ummat A., Franzen O., Rausch T., Stütz A.M., Stedman W., Anantharaman T., Hastie A. et al. . Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods. 2015; 12:780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chaisson M. J., Huddleston J., Dennis M.Y., Sudmant P.H., Malig M., Hormozdiari F., Antonacci F., Surti U., Sandstrom R., Boitano M. et al. . Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015; 517:608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Katoh K., Standley D. M.. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013; 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The Sequence Alignment/Map format and SAMtools. 1000 Genome Project Data Processing Subgroup. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zentner G.E., Saiakhova A., Manaenkov P., Adams M.D., Scacheri P.C.. Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res. 2011; 39:4949–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al. . GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 9:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ogurtsov A.Y., Shabalina S.A., Kondrashov A.S., Roytberg M.A.. Analysis of internal loops within the RNA secondary structure in almost quadratic time. Bioinformatics. 2006; 22:1317–1324. [DOI] [PubMed] [Google Scholar]
- 48. Ogurtsov A.Y., Roytberg M.A., Shabalina S.A., Kondrashov A.S.. OWEN: aligning long collinear regions of genomes. Bioinformatics. 2002; 18:1703–1704. [DOI] [PubMed] [Google Scholar]
- 49. Bernhart S.H., Hofacker I.L., Will S., Gruber A.R., Stadler P.F.. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008; 9:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sabarinathan R., Tafer H., Seemann S.E., Hofacker I.L., Stadler P.F., Gorodkin J.. The RNAsnp web server: predicting SNP effects on local RNA secondary structure. Nucleic Acids Res. 2013; 41:475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mathews D.H., Burkard M.E., Freier S.M., Wyatt J.R., Turner D.H.. Predicting oligonucleotide affinity to nucleic acid targets. RNA. 1999; 5:1458–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kondrashov A.S., Shabalina S.A.. Classification of common conserved sequences in mammalian intergenic regions. Hum. Mol. Genet. 2002; 11:669–674. [DOI] [PubMed] [Google Scholar]
- 53. Shabalina S.A, Spiridonov A.N., Ogurtsov A.Y.. Computational models with thermodynamic and composition features improve siRNA design. BMC Bioinformatics. 2006; 7:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kouprina N., Larionov V.. TAR cloning: insights into gene function, long-range haplotypes, and genome structure and evolution. Nat. Rev. Genet. 2006; 7:805–812. [DOI] [PubMed] [Google Scholar]
- 55. Leem S-H., Kouprina N., Grimwood J., Kim J-H., Mullokandov M., Larionov V.. Closing the gaps on human chromosome 19 revealed genes with a high density of repetitive tandemly arrayed elements. Genome Res. 2004; 14:239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kugoh H., Mitsuya K., Meguro M., Shigenami K., Schulz T.C., Oshimura M.. Mouse A9 cells containing single human chromosomes for analysis of genomic imprinting. DNA Res. 1999; 6:165–172. [DOI] [PubMed] [Google Scholar]
- 57. Zeng C., Kouprina N., Zhu B., Cairo A., Hoek M., Cross G., Osoegawa K., Larionov V., de Jong P.. Large-insert BAC/YAC libraries for selective re-isolation of genomic regions by homologous recombination in yeast. Genomics. 2001; 77:27–34. [DOI] [PubMed] [Google Scholar]
- 58. Schneider V.A., Graves-Lindsay T., Howe K., Bouk N., Chen H.C., Kitts P.A., Murphy T.D., Pruitt K.D., Thibaud-Nissen F., Albracht D. et al. . Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017; 27:849–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shiao Y.H., Lupascu S.T., Gu Y.D., Kasprzak W., Hwang C.J., Fields J.R., Leighty R.M., Quiñones O., Shapiro B.A. et al. . An intergenic non-coding rRNA correlated with expression of the rRNA and frequency of an rRNA single nucleotide polymorphism in lung cancer cells. PLoS One. 2009; 4:e7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Karahan G., Sayar N., Gozum G., Bozkurt B., Konu O., Yulug I.G.. Relative expression of rRNA transcripts and 45S rDNA promoter methylation status are dysregulated in tumors in comparison with matched-normal tissues in breast cancer. Oncol. Rep. 2015; 33:3131–3145. [DOI] [PubMed] [Google Scholar]
- 61. Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J.. Jalview version 2—a multiple sequence alignment and analysis workbench. Bioinformatics. 2009; 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Anger A.M., Armache J.P., Berninghausen O., Habeck M., Subklewe M., Wilson D.N., Beckmann R.. Structures of the human and Drosophila 80S ribosome. Nature. 2013; 497:80–85. [DOI] [PubMed] [Google Scholar]
- 63. Doris S.M., Smith D.R., Beamesderfer J.N., Raphael B.J., Nathanson J.A., Gerbi S.A.. Universal and domain-specific sequences in 23S-28S ribosomal RNA identified by computational phylogenetics. RNA. 2015; 21:1719–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gerbi S.A. The evolution of eukaryotic ribosomal DNA. Biosystems. 1984; 19:247–258. [DOI] [PubMed] [Google Scholar]
- 65. Melnikov S., Ben-Shem A., Garreau de Loubresse N., Jenner L., Yusupova G., Yusupov M.. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 2012; 19:560–567. [DOI] [PubMed] [Google Scholar]
- 66. Petrov A.S., Bernier C.R., Hsiao C., Norris A.M., Kovacs N.A., Waterbury C.C., Stepanov V.G., Harvey S.C., Fox G.E., Wartell R.M. et al. . Evolution of the ribosome at atomic resolution. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:10251–10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Petrov A.S., Gulen B., Norris A.M., Kovacs N.A., Bernier C.R., Lanier K.A., Fox G.E., Harvey S.C., Wartell R.M., Hud N.V. et al. . History of the ribosome and the origin of translation. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:15396–15401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Henras A.K., Plisson-Chastang C., O’Donohue M.F., Chakraborty A., Gleizes P.E.. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA. 2014; 6:225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Montellese C., Montel-Lehry N., Henras A.K., Kutay U., Gleizes P.E., O’Donohue M.F.. Poly(A)-specific ribonuclease is a nuclear ribosome biogenesis factor involved in human 18S rRNA maturation. Nucleic Acids Res. 2017; 45:6822–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Arnheim A., Krystal M., Schmicke l R., Wilson G., Ryder O., Zimmer E.. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proc. Natl. Acad. Sci. U.S.A. 1980; 77:7323–7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Krystal M., E’Eustachio P., Ruddle F.H., Arnheim N.. Human nucleolus organizers on nonhomologous chromosomes can share the same ribosomal gene variants. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:5744–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jacobs P.A., Melville M., Ratcliffe S., Keay A.J., Syme J.. A cytogenetic survey of 11,680 newborn infants. Ann. Hum. Genet. 1974; 37:359–376. [DOI] [PubMed] [Google Scholar]
- 73. Jacobs P.A., Mayer M., Morton N.E.. Acrocentric chromosome associations in man. Am. J. Hum. Genet. 1976; 28:567–576. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary Table S9 lists all sources and accessions of data used in the variant analysis.