Abstract
Alpha satellite is the major repeated DNA element of primate centromeres. Specific evolutionary mechanisms have led to a great diversity of sequence families with peculiar genomic organization and distribution, which have till now been studied mostly in great apes. Using high throughput sequencing of alpha satellite monomers obtained by enzymatic digestion followed by computational and cytogenetic analysis, we compare here the diversity and genomic distribution of alpha satellite DNA in two related Old World monkey species, Cercopithecus pogonias and Cercopithecus solatus, which are known to have diverged about 7 Ma. Two main families of monomers, called C1 and C2, are found in both species. A detailed analysis of our data sets revealed the existence of numerous subfamilies within the centromeric C1 family. Although the most abundant subfamily is conserved between both species, our fluorescence in situ hybridization (FISH) experiments clearly show that some subfamilies are specific for each species and that their distribution is restricted to a subset of chromosomes, thereby pointing to the existence of recurrent amplification/homogenization events. The pericentromeric C2 family is very abundant on the short arm of all acrocentric chromosomes in both species, pointing to specific mechanisms that lead to this distribution. Results obtained using two different restriction enzymes are fully consistent with a predominant monomeric organization of alpha satellite DNA that coexists with higher order organization patterns in the C. pogonias genome. Our study suggests a high dynamics of alpha satellite DNA in Cercopithecini, with recurrent apparition of new sequence variants and interchromosomal sequence transfer.
Keywords: alpha satellite DNA, centromere genomics, chromosomal evolution, higher-order repeats, acrocentric chromosomes, Cercopithecini
Introduction
In eukaryotes, centromeric DNA is made of large tracts of tandemly repeated sequences, also called satellite DNA. Satellite DNAs can differ significantly between closely related species and their evolution is driven by molecular processes that differ from those that affect other parts of genomes. Changes in satellite DNA content and distribution can alter heterochromatin and centromere function and therefore can accompany speciation (Palomeque and Lorite 2008; Plohl et al. 2008). In Primates, the most abundant satellite DNA, called alpha satellite, is made of AT-rich monomers that are about 170 bp in length (Schueler and Sullivan 2006). Alpha satellite monomers represent a very large sequence family that has been classified into distinct subfamilies, which differ in their DNA content but also in their organization and genomic distribution (Waye and Willard 1986; Alexandrov et al. 1988; Vissel and Choo 1991; Shepelev et al. 2009; Hayden 2012; Catacchio et al. 2015). Alpha satellite DNA displays two types of organization in primate genomes: A so-called monomeric organization, where arrays of adjacent monomers belong to the same family, and a higher-order repeats (HORs) organization that involves highly conserved repeated motifs where each motif is made of several monomers, possibly belonging to different families (Schueler and Sullivan 2006).
Several studies have addressed the evolutionary dynamics of alpha satellite DNA, most of them focusing on comparing human DNA sequences with those of great apes (Schueler and Sullivan 2006; Cellamare et al. 2009; Shepelev et al. 2009; Catacchio et al. 2015; Chiatante et al. 2017). The observation of an age gradient when going from centromere toward chromosome arms has led to suggest that, on a single chromosome, alpha satellite families emerge and expand at the centromere core, thereby splitting and displacing older families distally onto each chromosome arm, where they are found in the so-called pericentromeric regions (Schueler et al. 2005). In addition, a certain amount of evidence points to the existence of transfer of alpha satellite DNA between chromosomes. For example, some families are found preferentially on certain subsets of human chromosomes (Alexandrov et al. 1988). Finally, some families and/or HORs are conserved within great apes, but they usually span nonhomologous centromeres (Jorgensen et al. 1987; Archidiacono et al. 1995; Warburton et al. 1996; Rudd et al. 2006; Catacchio et al. 2015). Numerous mechanisms have been called upon to underpin these observations at the molecular level, such as unequal crossing over or sister chromatid exchange, transposition, gene conversion, rolling circle replication and reinsertion, and transposon-mediated exchange (Schindelhauer and Schwarz 2002; Rudd et al. 2006; Palomeque and Lorite 2008; Plohl et al. 2008; Garrido-ramos 2017). Nevertheless, how concerted evolution leads to appearance and accumulation of species-specific sequence variations in short evolutionary periods and drives satellite DNA divergence remains largely unknown (Dover 1982; Pérez-Gutiérrez et al. 2012; Feliciello et al. 2015; Utsunomia et al. 2017).
In contrast to apes, information gathered on alpha satellite families is relatively limited in monkeys (Alkan et al. 2007). Cercopithecini represent a large clade of Old World monkeys containing 35 species that have diverged over the last 10 Myr (Tosi 2008; Guschanski et al. 2013). The numerous chromosomal rearrangements that are observed in this clade can be associated with centromere repositioning or emergence of new centromeres (Dutrillaux et al. 1980; Moulin et al. 2008). This feature makes them interesting models for studying evolution of alpha satellite DNA. In a recent study, we have characterized alpha satellite DNA in Cercopithecus solatus (CSO), using deep sequencing of enzymatically obtained monomers and dimers of alpha satellites, combined with computational and cytogenetic analyzes. Our results provided evidence for the existence of at least four alpha satellite families, termed C1 to C4, that differed from those previously described in the ape lineage (Cacheux et al. 2016).
We present here investigations into the alpha satellite component of another species, Cercopithecus pogonias (CPO), whose genome contains 72 chromosomes, when compared with 60 for CSO (Dutrillaux et al. 1980; Moulin et al. 2008). The experimental strategy was very similar to the one used for CSO. We chose the same sequencing technique, which in principle enables the recovery of full sequence information of monomers up to 400 bp. Although the full sequencing of dimers is theoretically feasible for monomer sizes up to 200 bp, our previous work showed that some technical issues resulted in the recovery of a very low amount of dimer sequences. We therefore decided to use two different restriction enzymes, XmnI and HindIII, that are expected to provide overlapping monomer sequences. A thorough investigation of our data sets allowed us to refine the identification of alpha satellite DNA families and to compare the diversity, structural organization and chromosomal distribution of alpha satellite DNA in both species, thereby providing unprecedented information regarding the dynamics of alpha satellite families during evolution.
Materials and Methods
DNA Collection and Metaphase Preparations
Fibroblast cell samples of CPO (ID: 2001-027, male sample) from a cryo-conserved living cell bank (https://www.mnhn.fr/fr/collections/ensembles-collections/ressources-biologiques-cellules-vivantes-cryoconservees/tissus-cellules-cryoconserves-vertebres) were used for DNA extraction, which was performed using the Omega Biotek Tissue DNA Kit. Fibroblast cell samples of this same specimen were used for metaphase preparations. Cell cultures and metaphase preparations were achieved according to Moulin et al. (2008).
Alpha Satellite DNA Isolation and Sequencing
XmnI or HindIII were used to digest CPO DNA in vitro. 10 µg of CPO genomic DNA were incubated for 6 h at 37 °C with 70 units of XmnI or HindIII (New England Biolabs) in a total volume of 35 µl. The restriction enzymes were then inactivated for 20 min at 65 °C. Both samples were loaded on a 1% agarose gel after addition of 7 µl loading buffer (50% glycerol) and electrophoresis was performed in 0.5× Tris–borate–EDTA buffer, at room temperature for 3 h at 100 V. The gel was briefly stained with ethidium bromide and then imaged by UV transillumination. Bands corresponding to alpha satellite monomers (∼170 bp) were cut and DNA was extracted from the gel with the Omega Biotek Gel extraction kit and resuspended in 100 µl of elution buffer. About 250 ng were obtained for each of the XmnI and HindIII monomers.
Sequencing was performed on a PGM sequencing platform (Ion Torrent technology) using the 400 bp sequencing kit. HindIII DNA sample was blunted according to the Quick Blunting Kit (E1201S, NEB). Two libraries were generated using 50 ng of the two blunt digest pools and the Ion Plus Fragment Library Kit (4471252, Life Technologies) and tagged with Ion Xpress barcode adapters (4471250, Life Technologies). After purification (1.8×) with Ampure XP Beads (A63880, Agencourt Technology), the libraries were quantitated using a Sybr Green qPCR assay (SsoAdvanced supermix, Biorad) based on a custom E. coli reference library. After a dilution of each library down to 26 pM, 0.22 fmol of each library were pooled as templates for the clonal amplification on Ion Sphere particles during the emulsion PCR, performed on a One Touch2 emPCR robot according to the Ion PGM Template OT2 400 Kit user guide (4479878, Life Technologies). The amplification products, tagged and pooled (each sample representing one sixth of the total DNA), were loaded onto an Ion 316v2 chip (4483324, Life Technologies), and subsequently sequenced according to the Ion PGM Sequencing 400 Kit user guide (4482002, Life Technologies). As the chosen chip is expected to provide a maximum of 2 million reads, the maximal number of reads for each sample is expected not to exceed 330,000. After standard filtration of the raw reads (polyclonal and low quality removal), the Ion Torrent sequencing yielded 210,527 sequences for the XmnI sample and 166,099 sequences for the HindIII sample, which represent very good yields.
Alpha Satellite Sequence Filtering
All XmnI sequences with an average Phred score lower than 25, a length outside the range 162–182 bp, and sequences without the XmnI digested sites at the extremities (5′-NNTTC. . .GAANN-3′) were not considered for further analysis. Alpha satellite sequences were identified with a BLAST search against a reference alpha satellite sequence from Chlorocebus aethiops (AM23721) (Altschul et al. 1990). Using default BLAST parameters, all sequences exhibiting a hit longer than 80 bp were considered as alpha satellite sequences and conserved for the following analysis. All sequences were then reoriented if necessary in order to match the orientation of the reference alpha satellite sequence. The orientation information was preserved for investigations regarding reading biases.
All HindIII sequences with an average Phred score lower than 25, a length outside the range 166–186 bp (the blunting step added 4 nucleotides to the classic monomer length), and sequences without the HindIII digested or blunted sites at the extremities (5′-AGCT. . .AGCT-3′) were not considered for further analysis. Alpha satellite sequences were identified with the same BLAST search as above. All sequences were then reoriented if necessary in order to match the orientation of the reference alpha satellite sequence. The four supplementary nucleotides added to the HindIII monomers during the blunting step (5′AGCT3′) were discarded.
Alpha Satellite Sequence Characterization
Monomeric sequences were compared using their 5-mer composition in order to identify putative alpha satellite families without direct alignment. For each set of monomers, the 5-mer frequency table was analyzed using a principal component analysis (PCA) to reduce the space complexity and enable data visualization on the first factorial planes. Sequences were classified into groups by using a hierarchical clustering method (HCA) based on the Ward criterion (Ward 1963) applied to the Euclidean distances calculated from the 100 first principal components of the PCA. Because of the size of the monomer data set, direct classification of the sequences using HCA was not possible. Instead, HCA was applied on 2,500 randomly selected sequences which were used to train a linear discriminant model. This model has been finally used to classify all the other monomers.
Because of the size of the data sets, the consensus sequences and the sequence distance analysis were conducted with different subsets of randomly selected sequences. The selected sequences were aligned using MUSCLE (Edgar 2004) and analyzed with Seaview (Gouy et al. 2010). CENP-B and pJalpha boxes were searched with the patterns TTCGTTGGAARCGGGA and TTCCTTTTYCACCRTAG, respectively (Rosandić et al. 2006) by using the program Fuzznuc (Rice et al. 2000) and allowing two mismatches. All statistical analyses were conducted with R (R Core Team 2014). Our R scripts and other programs are available upon request.
Oligonucleotide Probes
Short oligonucleotide probes (18 or 19 nucleotides) were designed in order to target specifically the different alpha satellite families identified in CPO, by systematic prediction of binding frequencies based on the sequencing results. Sequences and binding frequencies are available in supplementary table S2, Supplementary Material online, which also provides details about the positions of locked nucleic acid (LNA) modifications in the probes. These positions were selected based on previous experience in order to achieve a good binding affinity and specificity (Ollion et al. 2015; Cacheux et al. 2016). When possible, we selected probes that were perfectly complementary to more than 20% of the sequences from the target group and to less than 3% of the sequences from the other groups. Supplementary table S2, Supplementary Material online also provides the expected binding frequencies if hybridization is possible despite the presence of one mismatch between the probe and its targets. Additional probes were used to localize specific sequence variants, such as C2A-G17Del (5′CaTTtTcCcTtCaAgAaTcC3′, 3′Biotin), 158C (5′CaCaAgAaCAgCcTtAgC3′, 3′Digoxygenin) and 158G (5′CaCaAgAaGAgCcTtAgC3′, 3′Biotin). All probes were purchased from Eurogentec (Seraing, Belgium).
Fluorescence In Situ Hybridization Experiments
Fluorescence in situ hybridization (FISH) experiments were performed on metaphase chromosome preparations. Hybridization solutions were prepared by diluting the oligonucleotide probes to a final concentration of 0.1 µM in a hybridization solution consisting of 2× SSC pH 6.3, 50% deionized formamide, 1× Denhardt solution, 10% dextran sulfate, and 0.1% SDS. A 20 µl of the hybridization solution was deposited on each slide and covered with a coverslip. The slides were then heated for 3 min at 70 °C and hybridized for 1 h at 37 °C in a Thermobrite apparatus (Leica Biosystems). Then, each slide was washed twice in 2× SSC at 63 °C. Preparations were then incubated in blocking solution (4% bovine serum albumin [BSA], 1× PBS, 0.05% Tween 20) for 30 min at 37 °C to reduce nonspecific binding. Then, depending on the combination of probes, the following antibodies were used for subsequent revelations: Alexa 488-conjugated streptavidin (1: 200; Life Technologies), Cy5-conjugated streptavidin (1:200; Caltag Laboratories), FITC-conjugated sheep antidigoxigenin (1:200; Roche), and Rhodamine-conjugated sheep antidigoxigenin (1:200; Roche). All antibodies were diluted in blocking solution containing 1× PBS, 0.05% Tween 20, and 4% BSA. Antibody incubation lasted for 30 min at 37 °C. All washings were performed in 2× SSC, 0.05% Tween 20. Chromosomes were counterstained with DAPI (4′,6-diamidino-2-phenylindole) by pipetting 40 µl of a 5 µg/ml solution onto the slides, incubating for 5 min and then briefly washing in 1× PBS. Slides were mounted by adding a drop of Vectashield Antifade Mounting Medium (Vector Laboratories) and covering with a coverslip. Metaphases were imaged using an Axio Observer Z1 epifluorescent inverted microscope (Zeiss) coupled to an ORCA R2 cooled CDD camera (Hamamatsu). The Axio Observer Z1 was equipped with a Plan-Apochromat 63× 1.4 NA oil-immersion objective and the following filters set: 49 shift free for DAPI (G365/FT395/BP445/50), 38 HE shift free for FITC/Alexa488 (BP470/40/FT495/BP525/50), homemade sets for Rhodamine (BP546/10/FF555/BP 583/22) and for Cy5 (BP643/20/FF660/BP684/24). The light source was LED illumination (wavelengths: 365, 470, or 625 nm) except for Rhodamine, for which a metal halide lamp HXP120 was preferred. Immersion oil of refractive index 1.518 at 23 °C was used.
Results
Identification of Alpha Satellite DNA Families from the CPO XmnI Data Set
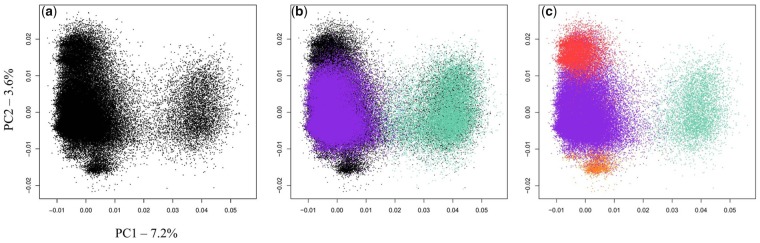
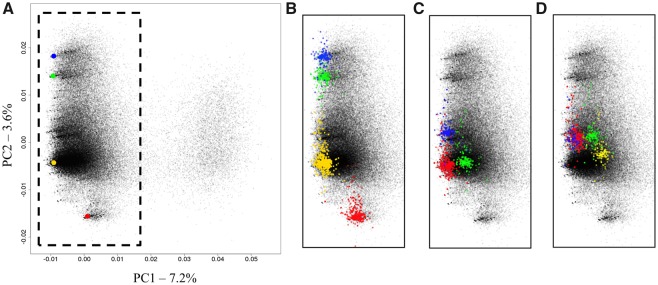
Alpha satellite monomers were isolated from the CPO genome using the XmnI restriction enzyme, then sequenced and parsed as previously described for CSO (Cacheux et al., 2016) (see “Materials and Methods”). The recovered 112,575 sequences were first analyzed with a PCA using the 5-mer nucleotide composition. Visualization of sequences into the plane formed by the two first components of the PCA revealed a pattern that differed slightly from the one obtained for CSO. We distinguished a large group whose structure suggests it could contain several subgroups (left of fig. 1A) and a smaller well separated group (right of fig. 1A). We decided to combine the data from the two species into the same projection space (fig. 1B). The obtained graph shows that the group that appears on the right overlaps quite well with the C2 group of CSO. On the other hand, the group that appears on the left occupies a larger space on the graph, extending both above and below the C1 group of CSO and thereby suggesting that two additional groups of sequences may be present in CPO. We hypothesized that the two previously identified families C1 and C2 coexist in CPO with two new families that we decided to call C5 and C6, respectively (fig. 1C). After having assigned all sequences to each of the four families by using a combination of hierarchical clustering (HCA) and linear discriminant analysis (LDA), as previously described, we confirmed, using phylogenetic trees, the existence of the four families as well as the identities of the C1 and C2 families of CPO and CSO (supplementary fig. S1, Supplementary Material online). The abundance of each family is reported in supplementary table S1, Supplementary Material online together with some of their properties, and their consensus are depicted in figure 2. The consensus sequences of the C1 and C2 families of CPO are identical to those that were established for CSO, at the exception of a single nucleotide at position 167 that was ambiguous in the C2 consensus of CSO. The consensus sequences of both C5 and C6 differed only by three single nucleotide variations from that of C1 (fig. 2), although in the case of C5 the N at position 28 reflects the presence of abundant sequences containing either a G (as in C1) or a T within the data set. The C5 and C6 families exhibit a high sequence homogeneity (95% and 98% mean sequence identity, respectively) which is in the same of order as that of C1 (95%) and much greater than that of C2 (85%). All families contained a pJalpha box and no CENP-B box, as observed for CSO (see supplementary table S1, Supplementary Material online). Some sequences were observed to be present a high number of times in the data set. They will be described in more details further.
Fig. 1.
—Characterization of alpha satellite DNA diversity in the XmnI monomer data set. (A) PCA projection on principal components 1 and 2 of the normalized 5-mer frequency vectors for all sequences from the CPO XmnI monomer data set. Each point represents a monomer sequence. (B) Prediction of the C1 (purple) and C2 (pastel green) sequences from CSO XmnI monomer data set by using the PCA projection of CPO monomers. (C) PCA projection of CPO XmnI monomer data set with sequences colored according to their assignment to the C1 (purple), C2 (pastel green), C5 (red) or C6 (orange) alpha satellite family, based on a hierarchical classification method (see Materials and methods).
Fig. 2.
—Consensus sequences of the alpha satellite families identified in the CPO XmnI data set. The consensus sequences were determined following the alignment of 500 randomly selected sequences for the C1, C2, C5, and C6 families. Each position was considered unambiguous if more than 60% of monomers had the same nucleotide at this position. A point at a position replaces a nucleotide identical to the nucleotide at the homologous position in the C1 consensus.
Chromosomal Distribution of the Alpha Satellite Families Analyzed by FISH
We were next interested in designing oligonucleotide probes for studying the chromosomal distribution of the four families of alpha satellite DNA identified within the CPO monomer data set by FISH. We implemented an in silico probe selection process in order to identify among the most common 18-mer sequences within a group (found in more than 20% of the monomers) those that were specific for this group (found in less than 3% of the monomers of other groups) (Cacheux et al. 2016). This analysis led to the design of probes C5a and C6a that should specifically detect the C5 and C6 families, respectively, thanks to the presence of at least two single nucleotide variations that distinguish their target sites from corresponding sites in the C1 and C2 families (supplementary table S2 and fig. S2, Supplementary Material online). This in silico analysis revealed that the C2a and C2b probes that were previously designed for the specific detection of the C2 family in CSO can also be used for this purpose in CPO (supplementary table S2, Supplementary Material online). On the contrary, there is no oligonucleotide probe design that will allow for the specific detection of the C1 family, that is, that will preclude binding of the probe to sequences from both the C5 and C6 families. Nevertheless, we noticed that the previously designed C1a and C1b probes should detect either both C1 and C5 (for C1a) or C1 and C6 (for C1b) in CPO (supplementary table S2, Supplementary Material online).
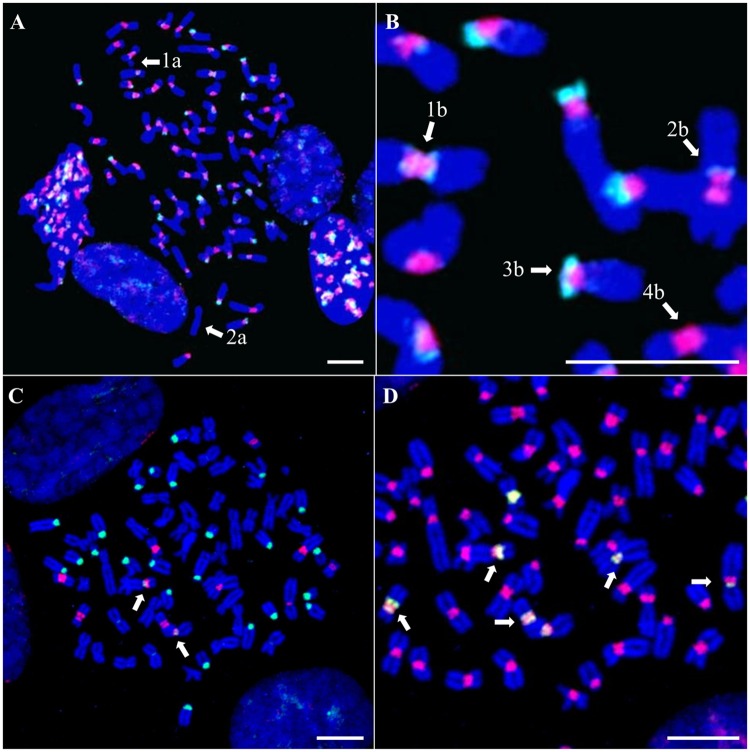
Fluorescence in situ hybridization experiments were performed on metaphases prepared from cells that came from the same male specimen as the one used for the sequencing. The use of the C1a/C2a or C1b/C2b probe sets revealed hybridization patterns on CPO chromosomes that resembled those observed on CSO chromosomes. The probes targeting C1 produced intense signals covering the centromeres of all but two chromosomes (fig. 3A and B). On some chromosomes, the signal appeared very large, extending toward pericentromeric regions. The probes targeting C2 stained intensely the acrocentric chromosomes on their shorter arm and produced weaker signal in the pericentromeric regions of numerous non acrocentric chromosomes (fig. 3A and B;supplementary table S3, Supplementary Material online). The absence of alpha satellite DNA on two chromosomes was confirmed using a probe designed to bind all alpha satellite sequences, and the use of a chromosome banding technique allowed us to identify these chromosomes as the Y and a single chromosome 6 (supplementary fig. S4, Supplementary Material online).
Fig. 3.
—FISH analysis of the C1, C2, C5, and C6 alpha satellite families on CPO chromosomes. CPO metaphase chromosomes are colored in blue. (A, B) Probes C1a, C1b and C2b are hybridized simultaneously. (A) Hybridization of probes C1a and C1b (red) and probe C2b (green). 1a and 2a: Unlabeled chromosomes. (B) Focus on image (A) showing in details the different types of distribution of the C2b signals. 1b: Both pericentromeric regions, 2b: One pericentromeric region toward the long arm, 3b: One pericentromeric region toward the short arm of an acrocentric chromosome, 4b: No signal. (C, D) Probes C1a, C5a, and C6a are hybridized simultaneously. (C) Hybridization of probe C5a (red) and probe C6a (green). Arrows: Two chromosomes where both probes produce signals. (D) Focus on the metaphase shown in (C) but with hybridization of probe C1a (red) and probe C5a (green). Arrows: Pericentromeric hybridization of probe C5a on several chromosomes. Scale bar = 10 µm.
We next investigated the labeling pattern generated using the C5a and C6a probes. Intense signals were observed on 11 chromosomes and 32 chromosomes for the C5a and C6a probes, respectively (fig. 3C and D). Only a single chromosome pair displayed both signals (fig. 3C, see arrows). The identity of these chromosomes was also established using cytogenetic experiments (supplementary figs. S5 and 6, Supplementary Material online): Probe C5a labeled 5 pairs of autosomes and the X chromosome, whereas C6a produced intense signals at the centromere of all 12 pairs of acrocentric chromosomes, slightly lighter signals at the centromere of one pair of submetacentrics, and 6 additional weaker signals which were shown to belong to four chromosome pairs, two of which displaying a heterozygote signal (supplementary fig. S6, Supplementary Material online). Except for one chromosome pair, identified as chromosome 20, the C5a probe provided a signal that was located on both sides of the primary constriction, but absent from the central part, which is still labeled by the C1a probe (see arrows in fig. 3D). This suggests that the C5 family occupies a slightly pericentromeric localization. The C6a probe always provided a signal that was located at the centromere core. The hybridization patterns of C5a and C6a were clearly distinct from each other and from those of the C1 targeting probes, which validates our probe design strategy. As expected, the signals obtained using the C1a and C1b probes were found to overlap with those of C5a and C6a, respectively. Finally, FISH experiments were performed on CSO chromosomes using the C5a and C6a probes. C6a did not provide any signal, as expected from the absence of the C6 family in the CSO genome (not shown). A slight signal was observed using the C5a probe, that was removed by increasing the temperature, suggesting a light nonspecific hybridization (see supplementary fig. S7, Supplementary Material online). Based on all these observations, we conclude that in addition to the C1 and C2 families previously described in CSO, the genome of CPO contains two additional alpha satellite families, named C5 and C6, that display specific chromosomal distribution patterns.
The HindIII Data Set Reveals Additional Families and Organizational Patterns
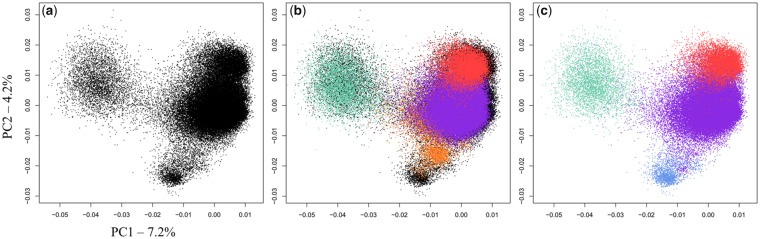
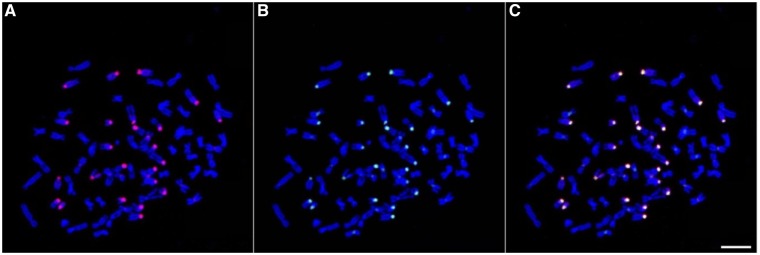
Digestion of CPO DNA using the HindIII restriction enzyme resulted in a ladder pattern similar to the one obtained using the XmnI enzyme (data not shown). The possibility to combine the analysis of monomers obtained using two different restriction enzymes was expected to provide information regarding the organizational pattern of monomers belonging to different families with respect to each other. A monomeric organization should lead to XmnI and HindIII monomers that have a similar nucleotide composition, and therefore a similar 5-mer composition. On the contrary, a higher order organization pattern should lead to monomers with different nucleotide compositions that may be distinguished on the PCA graph because of different 5-mer compositions. We therefore decided to implement on HindIII monomers a similar experimental and analytical approach as the one described for XmnI. A total of 84,485 alpha satellite monomers were recovered. Sequences were visualized into the plane formed by the two first components of the PCA, alone and in combination with the sequences from the XmnI data set (fig. 4A and B). The obtained graph provided evidence for four families and suggested that three of them are identical to those found in the XmnI data set. We decided to name C1′, C2′, and C5′ the three families that overlap with C1, C2, and C5 on the PCA graph (fig. 4C), and C6′ the fourth visible family. Using once again an HCA/LDA approach, all sequences were sorted for their belonging to one of the four families, and consensus sequences were computed. The strict identity of the consensus sequences between C1 and C1′, C2 and C2′, and C5 and C5′, except for a phase shift, suggests a monomeric organization for the C1, C2, and C5 families. On the contrary, the absence, within the HindIII monomer data set, of sequences that have a 5-mer composition identical to C6 demonstrates that C6 cannot have a monomeric organizational pattern. The C6′ family appeared on the PCA graph as a group of points with a size similar to that of C6 and a slightly shifted position (fig. 4B and C). Comparison of the consensus sequences of C6 and C6′ showed that they were identical in the overlapping 106 bp HindIII–XmnI fragment but that they differed by a substitution, C2A, and a deletion, G17Del, within the nonoverlapping but homologous 66 bp XmnI–HindIII fragment (supplementary fig. S8, Supplementary Material online). This feature suggests that monomers from the C6 family may be involved into a higher order organizational pattern where the sequence of the XmnI–HindIII fragment of the monomer adjacent to C6 (on the right side of C6 as shown in supplementary fig. S8, Supplementary Material online) corresponds to the sequence of the XmnI–HindIII fragment of C6′. We designed an oligonucleotide probe targeting the C2A-G17Del variation that distinguishes the C6′ consensus from the C6 consensus. The hybridization pattern of this probe in FISH experiments overlapped quite well with the signals provided by probe C6a (fig. 5). We only noticed additional very weak signals on a few additional chromosomes using the C2A-G17Del probe, which will be discussed further. These results provide strong support for the existence of an HOR structure containing at least two monomers, where a monomer from the C6 family is followed by another monomer whose sequence is only partially known.
Fig. 4.
—Characterization of alpha satellite DNA diversity in the HindIII monomer data set. (A) PCA projection on principal components 1 and 2 of the normalized 5-mer frequency vectors for all sequences from the HindIII monomer data set. Each point represents a monomer sequence. (B) Prediction of the position of the XmnI monomer sequences on the graph shown in (A). Sequences are colored according to their assignment to the C1 (purple), C2 (pastel green), C5 (red) or C6 (orange) alpha satellite families. (C) PCA projection shown in (A) with sequences colored according to their assignment to the C1′ (purple), C2′ (pastel green), C5′ (red), or C6′ (blue) alpha satellite families, using a hierarchical classification method (see Materials and methods).
Fig. 5.
—FISH detection of the C2A-G17Del sequence variant and relative distribution with respect to the C6a probe. Probes C2A-G17Del and C6a are hybridized simultaneously to CPO metaphase chromosomes, which are colored in blue. (A) Hybridization of probe C6a is shown in red. (B) Hybridization of probe C2A-G17Del is shown in green. (C) Combined signals from (A) and (B). Scale bar=10 µm.
Highly Repeated Sequence Variants Provide Insights Into Additional Alpha Satellite Families
In both CSO and CPO, the monomer data sets contained several sequences that were repeated a high number of times. A detailed investigation of the 30 most abundant sequences from CPO was performed for both enzymes. As observed in our previous study (Cacheux et al 2016), several sequences containing a deletion within a homopolymer tract were identified as sequencing errors based on strong biases in the orientation of the sequencing reads (table 1; supplementary fig. S3, Supplementary Material online). These sequences, that are shown with an asterisk in the tables, were not considered in the forthcoming analysis. The most abundant sequence for both enzymes was the exact sequence consensus of the C1 family. Other highly repeated sequences corresponded to sequences that differed from the latter one by a single nucleotide variation (such as in 2, 3, 13, 14, etc.), two single nucleotide variations (such as in 12, 15, 16, etc.), three single nucleotide variations (such as in 5, 10, 11, etc.), and also to sequences combining one or two single nucleotide variations with one single nucleotide deletion (see sequences 7 and 8; all examples are taken from the XmnI data set). Similar variation patterns were observed with both enzymes and in general, identical variations were found with similar frequency within both data sets. Interestingly, the absence in the XmnI data set of a highly repeated sequence from the HindIII data set (number 13) could be explained by nucleotide variations that abolish the cleavage site for XmnI. Although a slight bias for read orientation (see e.g., sequences 10, 13, 16, and 19 in table 1) was sometimes observed, probably due to sequence-dependent differential efficiency of the Ion torrent technology, we reasoned that all these sequences represented homogenous sets of identical sequences directly recovered from the CPO genome.
Table 1.
Analysis of Alpha Satellite Sequences Found in High Copy Number in the CPO XmnI Monomer Data Set
| Id | Sequence | Number | Forward (%) |
|---|---|---|---|
| 1 | Consensus C1 | 2983 | 46 |
| 2 | C158G | 848 | 48 |
| 3 | C116T | 568 | 41 |
| 4 | C114Del | 508 | 1* |
| 5 | C137A-CC149AA | 455 | 34 |
| 6 | T101Del | 323 | 98* |
| 7 | C2A-G17Del | 250 | 66 |
| 8 | C2A-G17Del-C158G | 208 | 70 |
| 9 | C114Del-C158G | 145 | 0* |
| 10 | A3741T-G64A-C158G | 136 | 15 |
| 11 | A40C-C42G | 116 | 44 |
| 12 | C116T-C158G | 112 | 46 |
| 13 | C2A | 103 | 73 |
| 14 | T121A | 100 | 43 |
| 15 | C137A-C158G | 100 | 51 |
| 16 | A3741T-G64A | 100 | 24 |
| 17 | C137A-CC149AA-C114Del | 89 | 1* |
| 18 | C2A-G17Del-C114Del | 81 | 1* |
| 19 | T38G | 77 | 29 |
| 20 | A110G | 76 | 56 |
| 21 | A86T | 74 | 39 |
| 22 | T80Del-T101Del | 67 | 100* |
| 23 | A41G | 65 | 38 |
| 24 | T101Del-C158G | 62 | 98* |
| 25 | G17Del | 59 | 47 |
| 26 | G17C | 58 | 54 |
| 27 | C144A | 57 | 46 |
| 28 | C2A-C158G | 54 | 54 |
| 29 | C137A | 54 | 65 |
| 30 | A40C-C42G-G28T | 53 | 49 |
The sequences are named according to the “Id” column. The “Sequence” column indicates how each sequence variant differs from the consensus sequence of the C1 family, using standard notations. The “Number” column displays the number of identical copies of the sequence in the monomer data set. The “Forward” column displays the percentage of reads obtained in the forward orientation (i.e., the orientation of our reference sequence). Strong biases for read orientation reveal artifactual sequences which are indicated by an asterisk.
We noticed that sequence 5 from the XmnI data set (455 repeats) matched to the consensus of the C6 family, and that sequences 11 (116 repeats) and 30 (53 repeats) matched to the consensus of C5 with a G or a T at position 28, respectively. On the graph showing the two first principal components of the PCA (fig. 6A), the points corresponding to these sequences were located at the left end of elongated groups of points, displaying a “comet-like” structure. For these three highly repeated sequences, we decided to plot sequences from the XmnI data set that were identical to these sequences except for one single nucleotide difference (fig. 6B). Interestingly, the distributions of these sequences overlaps quite well with the beginning of the tails of the comets, suggesting that comets are traces of mutation events that have affected sequences that were initially present in a high number of identical copies. This observation therefore establishes a link between highly repeated sequences, comet-like structures and potential families or subfamilies of alpha satellite DNA. The highly repeated sequence variants can be viewed as the signature of faithful homogenization/amplification events affecting a single alpha satellite monomer, whereas the tails of the comets represent the divergence of sequences following mutation events. In this view, each different amplified sequence variant and the closely related sequences define an independent alpha satellite DNA family. On the graph shown in figure 6, whose transparency was chosen greater than the one shown in figure 1, additional comet-like structures can be distinguished. Repeating the process shown in figure 6B for additional highly repeated sequences, such as 3, 7, and 19 (fig. 6C), or 2, 8, 12, and 15 (fig. 6D), we showed that some of the observed comets seem to derive from identified highly repeated sequences (see e.g., the sequences shown in blue in fig. 6C and D, which correspond to sequences 19 and 12, respectively). As dispersed points corresponding to mutated sequences are observed next to all highly repeated sequence, it is likely that the large cloud of points that was attributed till now to a single C1 family may in fact contain numerous subfamilies, each one deriving from a previously amplified sequence. These subfamilies, which derive from sequences that differ from each other by only few nucleotides, generally overlap on the PCA graph. Interestingly, we were able to detect comet-like structures when we plotted the results of the PCA for the CSO XmnI data set in the CPO axis system (supplementary fig. S9, Supplementary Material online), which shows that the C1 family of CSO has an internal distribution which may be more complex than previously anticipated (Cacheux et al 2016).
Fig. 6.
—Distribution of sequences into comet-like clusters near abundant sequence variants. (A) PCA projection on principal components 1 and 2 of the normalized 5-mer frequency vectors for all sequences from the XmnI monomer data set is shown here with a lower point density than the one shown in figure 1. Sequences corresponding to highly repeated sequences 1 (yellow), 5 (red), 11 (green), and 30 (blue) are highlighted. (B), (C), and (D) Only the region of the PCA projection corresponding to the dotted rectangle (i.e., to the C1 family) is shown. (B) Sequences from the data set that correspond to single nucleotide variations from sequences 1, 5, 11, and 30 are shown using the same color code as in (A). (C) Sequences from the data set that correspond to single nucleotide difference from sequences 3, 7, and 19 are shown in red, green, and blue, respectively. (D) Sequences from the data set that correspond to single nucleotide difference from sequences 2, 8, 12, and 15 are shown in red, green, blue and yellow, respectively.
The comparison of highly repeated sequence variants found in both species reveals that, besides the most abundant sequence variant, that is, sequence 1, only very few sequence variants are found in both species (e.g., sequences 14 and 20). Moreover, numerous relatively abundant sequence variants exist that are found only in one species. One striking observation is that many of the abundant sequence variants of CPO contain the C158G single nucleotide variation (sequences 2, 8, 10, 12, 15, and 28), whereas this variation was barely detected within the CSO sequences (a sequence strictly identical to sequence 2 of CPO was found repeated only 28 times). Using a probe set that was designed in order to distinguish sequences containing a C or a G at position 158 in FISH experiments, we showed that strong signals were observed on all CPO chromosomes using the 158C-detecting probe, whereas the 158G-detecting probe stained only a subset of CPO chromosomes, with strong signals observed at the centromere of all 12 pairs of acrocentric chromosomes while weaker signals were located at the centromere core of a few other chromosomes (supplementary fig. S10, Supplementary Material online). In CSO, FISH experiments had also shown that distribution of one of the highly abundant sequence was limited to four chromosome pairs (Cacheux et al. 2016). These observations support the hypothesis by which amplification events lead to the local accumulation of new sequence variants, whose detailed analysis provides a new approach for the comparative genomics of alpha satellite DNA between species.
Alpha Satellite DNA and Karyotypic Structure in Cercopithecini
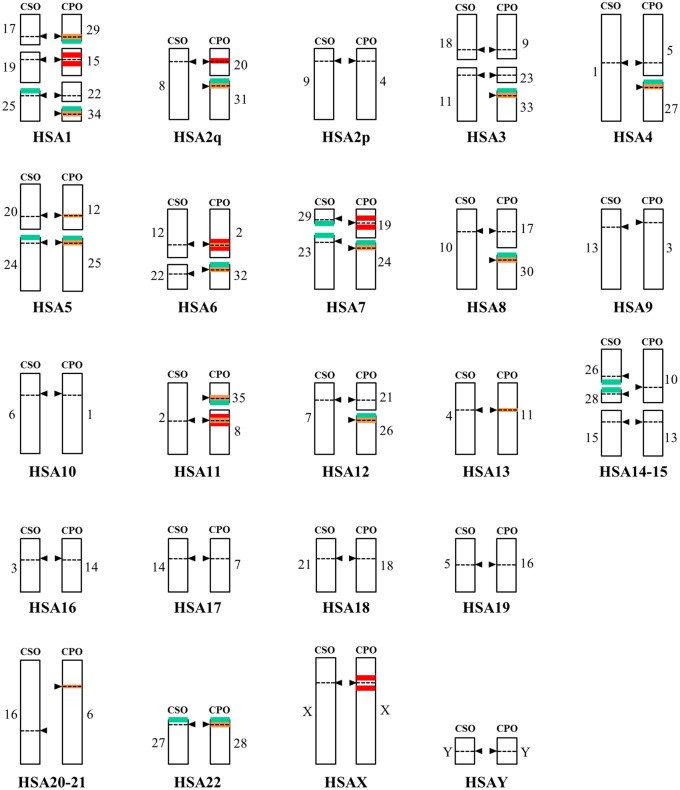
Chromosomal organization of Cercopithecini genomes is often rearranged during evolution, mostly by chromosomal fission/fusion events. Such rearrangements have been previously studied by comparing chromosomes from different species, using cytogenetics techniques (Moulin et al. 2008). The scheme shown in figure 7 represents the alignment of homologous chromosomes from CPO and CSO, which are organized according to their respective homologies to human chromosomes. In order to investigate the conservation of the C2 family on homologous chromosomes, we reported the position of FISH signals obtained with the C2b probe on the scheme from figure 7 (see supplementary fig. S3, Supplementary Material online). The C2 family was found to be very abundant on the short arm of acrocentric chromosomes in both species, and also on some non acrocentric chromosomes, albeit with a lower intensity. From the 12 pairs of acrocentrics in CPO (named 24–35) and the 7 pairs in CSO (named 23–29), only 3 are homologs (CSO23/CPO24, CSO24/CPO25, CSO27/CPO28, see HSA5, HSA7, and HSA22), revealing that C2 sequences carried by nine acrocentrics from CPO and by four acrocentrics from CSO are not found on homologs. Although it is difficult to establish quantitative comparison of signal intensities between homologous metacentric chromosomes, observation of the karyotypes suggests that the distribution of the pericentromeric C2 signals also differs between CPO and CSO on these chromosomes. In particular, strong signals observed on CSO6, CSO9, and CSO10 (see Cacheux et al. 2016) are not found on the homologous CPO1, CPO4, and CPO17 (supplementary fig. S3, Supplementary Material online). All these observations reveal that, during evolution, chromosomes may acquire or lose arrays of C2 sequences in pericentromeric regions, and that the short arms of acrocentrics represent preferential sites for establishing large arrays.
Fig. 7.
—Scheme representing the distribution of alpha satellite families C2, C5, and C6 on CSO and CPO chromosomes. Homologous chromosomes have been aligned using human chromosomes as references. CSO and CPO chromosomes are shown in the left and right hand side, respectively, as indicated. Numbering below each set of chromosomes refers to human chromosome numbers. Homologies are taken from Moulin et al. (2008). Arrows and dotted lines point to centromere positions. Distribution is shown in pastel green for C2, red for C5, and orange for C6. For C2, only the strong signals located on acrocentric chromosomes are shown.
A similar search was not possible for the C5 and C6 families that are found only in CPO. Nevertheless, we also reported the position of FISH signals corresponding to these families on the scheme from figure 7 (see supplementary figs. S5 and 6, Supplementary Material online). The C5 family was found in the centromeric or pericentromeric regions of several metacentric CPO chromosomes, sometimes associated with heterozygosity, and not on acrocentrics, while the C6 family was found in the centromeric regions of all acrocentric chromosomes and of several non acrocentric chromosomes. We noticed that some CPO chromosomes whose global structure is preserved in comparison to CSO (i.e., for which no obvious fusion or fission event has occurred since the last common ancestor of both species) have sometimes integrated sequences from the C5 family (CSO20/CPO12/HSA5 and CSO4/CPO11/HSA13), the C6 family (chromosome X), or both (CSO12/CPO2/HSA6). These observations point toward a high dynamics of centromeric sequences, with possible interchromosomal sequence transfer in the absence of large chromosome reorganization. Interchromosomal exchange of centromeric sequences may be favored between acrocentric chromosomes.
Discussion
Identification of Alpha Satellite DNA Families
In the present study, we have analyzed the content and genomic distribution of alpha satellite DNA in the CPO genome, implementing an experimental strategy that was similar to the one we previously applied to another cercopithecini species, CSO (Cacheux et al. 2016). Our final aim was to compare the diversity and distribution of this important genome component in these two related species. In both species, analysis of XmnI monomers revealed a very abundant family, called C1, and a more diverged and less abundant family, called C2, which have a centromeric and pericentromeric localization, respectively, as shown by FISH experiments using oligonucleotide probes.
Comparing the sequence distribution of both species on PCA graphs led us to distinguish two additional alpha satellite families, which were shown by FISH experiments to be localized only on specific chromosomes from the CPO genome. We also noticed that the axis system that emerged from the PCA analysis of the monomers from CPO revealed comet-like structures in the graphical representation, and that highly repeated sequences were found at the “head” of each comet, whereas the “tail” of the comets contained mutated versions of the highly repeated sequences. We propose here that this pattern reveals the evolutionary processes that underlie the evolution of alpha satellite DNA. At a certain time, a sequence variant is amplified through a recombination-based or rolling-circle mechanism, giving rise to multiple identical copies, which are later modified by mutations, thereby forming a new family. Most of these families differ from each other by only a few nucleotides in their consensus sequence, making their identification through a PCA analysis a highly difficult task. The identification of highly repeated sequence variants provides an alternative approach for their identification. In this view, the so-called C5 and C6 families may represent at least three subfamilies of C1 instead of the initially proposed two independent families.
Structural Organization of Alpha Satellite DNA in Cercopithecini
In our previous study (Cacheux et al. 2016), sequencing of CSO XmnI monomers and dimers had led to the identification of two families of sequences with a monomeric organization (C1 and C2) and of two additional families, called C3 and C4, that were part of a higher order organization. The distribution of these two last families was restricted to the Y chromosome and to the pericentromeres of a few other chromosomes. The low number of dimers that could be analyzed led us to abandon the sequencing of dimers in the present study and to choose instead to perform the global analysis of monomers using two restriction enzymes, with the aim of investigating the potential existence of additional families and of studying the relative organization of monomers from each family relative to each other. The analysis of HindIII monomers did not reveal any important alpha satellite family that would not be cleaved by XmnI. Moreover, the combined analysis of both data sets supports a tandem organization of monomers from the C1, C2, and C5 families. Searching the HindIII data set did not reveal any family of sequences with a nucleotide composition identical to that of C6. On the other hand, it was possible to distinguish within the HindIII data set a family, called C6′, with a nucleotide composition that is not detected among the main families identified in the XmnI data set. These observations demonstrate that C6, as well as C6′, cannot have a monomeric organization.
Alignment of C6 and C6′ showed that they had an identical nucleotide composition over the HindIII–XmnI fragment (using the orientation shown in supplementary fig. S8, Supplementary Material online) but diverged over the XmnI–HindIII fragment. This observation was interpreted as a hint toward the existence of an HOR structure in which monomers from the C6 family are associated on their right side (using the conventional orientation depicted in supplementary fig. S8, Supplementary Material online) to monomers that are known to carry the C2A-G17Del variation but whose complete sequence is not available. The PCA analysis of the XmnI monomer data set did not reveal any obvious alpha satellite family that would carry this variation. Although this observation may simply reflect the absence of an XmnI cleavage site on one side of the associated monomers, observation of table 1 led us to consider an alternative hypothesis: Two highly repeated sequences, namely sequences 7 and 8, do carry this variation. Moreover, the sum of repeat numbers observed for sequences 7 and 8 (458) is very close to the repeat number of sequence 5 (455), which is at the origin of the C6 family. Therefore, our data are compatible with the existence in the CPO genome of two types of HORs, where the C6 family is associated on its right side to either the C7 or C8 family, defined by the fact that they derive from sequences 7 and 8, respectively. Because the high similarity of all the uncovered centromeric families (C1, C5, C6, C7, C8, etc.), the accurate demonstration of these associations as well as the determination of the complete sequence of the C6-containing HOR(s) remains a technical challenge that will only been solved upon successful high throughput sequencing of longer molecules.
The sequences of the proposed HORs, which are located at or very close to centromeres, are very homogenous, as shown by the high sequence identity observed for the C6 family. These features are very different from those of the C3–C4 dimers previously described in CSO, which had a much lower sequence identity and were located in pericentromeric regions. Homogenous alpha satellite HORs have long been considered to be specific to hominoid centromeres before recent studies proved the existence of such organizations in New World monkeys (Terada et al. 2013; Sujiwattanarat et al. 2015). The present observation supports the idea that HORs may be more common and more diverse than initially thought. Whether the newly discovered HORs from the CPO genome are involved in centromere function or not remains to be investigated.
Emergence of New Alpha Satellite DNA Families During Cercopithecini Evolution
CPO and CSO share two main families of alpha satellite DNA, the centromeric C1 family and the pericentromeric C2 family. The sequence identity level is higher in C1 than in C2 for both species, which supports the hypothesis by which C1 has appeared more recently than C2. Interestingly, the structure of the PCA graph led us to postulate the existence in both species of many subfamilies of C1, each one deriving from a highly repeated sequence. The most abundant subfamily, which is derived from a sequence that exactly reflects the consensus of the C1 family, is conserved between both species, whereas most of the others are not conserved. These nonconserved subfamilies have probably emerged after the divergence of the CPO and CSO lineages, that is, in a few million years of evolution.
Our FISH strategy, which makes use of oligonucleotide probes to distinguish localized sequence variations, cannot be used for labeling all these subfamilies specifically, because many of them share one or several nucleotide variations. Moreover, oligonucleotide probes may hybridize to targets despite the presence of a single mismatch in the absence of carefully designed competitors (Cacheux et al. 2016). For example, the nonspecific hybridization of the C2A-G17Del probe on families derived from sequences 13, 25, or 28 may provide an explanation for the observed nonperfect overlap between signals obtained with this probe and those obtained with the C6a probe. Nevertheless, in some cases, FISH experiments could be used for confirming the species-specific distribution of the families and for investigating how this emergence proceeds. In particular, they clearly showed that the distribution of some subfamilies is restricted to a subset of chromosomes. This suggests the existence of local amplification mechanisms which may be eventually followed by interchromosomal transfer. The observation of monomers from the so-called C5 family on both sides of centromeres supports the existence of successive amplification events involving different subfamilies. In this specific case, amplification of C5 monomers would have been followed by amplification or integration, in the middle of the series of C5 monomers, of another sequence variant, which is still detected by the C1a probe but no more by the C5a probe.
Our data therefore point to the existence of recurrent amplification events affecting alpha satellite DNA. The amplification mechanism may lead to a monomeric organization, that is, succession of monomers belonging to the same family, as demonstrated for C5, or to a higher order organization, as shown in the case of C6 and its associated monomers. The sequences that have been amplified never differ from the consensus sequence of the C1 family by more than three or four nucleotides. This property may be caused by the amplification mechanism itself, which would not act on divergent sequences, or from the elimination of amplified sequences that have excessively diverged. The consensus sequence of C1 has been itself the substrate of a major amplification event, probably before the divergence of CSO and CPO, but one cannot exclude that this sequence has been a substrate for amplification mechanisms after this divergence, that is, concomitantly with the amplification of mutated sequence variants. The abundance of a specific variation, C158G, which was found in many of the subfamilies uncovered from the genome of CPO, raises the question of a potential selective pressure that would favor the amplification or maintenance of this variation in the CPO lineage.
Dynamics of Alpha Satellite DNA in Relation to Chromosome Evolution in Cercopithecini
The data we present here provide for the first time the opportunity to compare the chromosomal distribution of various alpha satellite DNA families in two Old World monkey species and to investigate the link between the presence of specific alpha satellite DNA families and specific chromosomal features. The C2 family was found to be highly abundant on the short arm of acrocentric chromosomes in both species, although most of these chromosomes are not homologs. The low level of sequence identity within the C2 family, in comparison to C1, argues against a faithful amplification mechanism that would occur on each chromosome. Therefore, this specific distribution is more likely the result of interchromosomal transfers of alpha satellite arrays. The presence of similar sequences on all acrocentric chromosomes has already been documented in another primate species, the New World monkey Aotus azarae (Prakhongcheep et al. 2013). It has been suggested that the bouquet chromosome configuration, occurring in prophase I, could favor exchange of genetic material between chromosomes that share structural characteristics (Paço et al. 2014). The C2 family was also found in the pericentromeric regions of several non acrocentric chromosomes in both species. Differences in the distribution of these sequences may result from interchromosomal transfer, but another hypothesis would involve the differential elimination of these sequences from pericentromeric regions in different lineages.
The present study shows that new alpha satellite sequences may appear at the centromere of chromosomes whose structure has otherwise not been obviously modified since the divergence between CSO and CPO. We observed some cases of heterozygosity, suggesting some ongoing evolutionary processes where amplified sequences have not been stabilized within the species. Although local amplification mechanisms could explain this observation (see above), the fact that all acrocentric chromosomes of CPO share similar centromeric sequences, that is, those containing the C6 family, also points toward potential interchromosomal transfer. The identification of several C1 subfamilies that were specific for each species underlines the requirement to implement our experimental framework to other species if one wants to investigate the interplay between evolution of alpha satellite DNA and evolution of chromosomal structure in Cercopithecini. Comparing more species should in theory allow one to study the successive events that have affected a specific centromere during evolution. Specific probes for different families may be used for the detection of small rearrangements within the centromeric regions, thereby providing an increased resolution in comparison to classical cytogenetic techniques. In the two species that were investigated, FISH experiments did not reveal the presence of alpha satellite DNA outside of centromeric regions. Nevertheless, such features should be looked for in other species, as chromosomal fissions and fusions are expected to lead to emergence or inactivation of centromeres.
Finally, one unexpected feature was the inability to detect any alpha-satellite repeat on a single chromosome 6. This feature was not observed on another metaphase preparation obtained from a female specimen, where both chromosomes 6 were equally labeled (not shown). This peculiar observation may be the result of a chromosomal rearrangement during cell culture, but may also reflect an heterozygotic individual carrying a chromosome 6 without any satellite DNA at its centromere, as it has been observed for example in orangutan or equids (Piras et al. 2010; Locke et al. 2011). Further sampling will be required to answer this question.
Conclusion
The characterization of the alpha satellite component of the CPO genome provides for the first time the opportunity to compare the diversity and distribution of alpha satellite DNA in two related Old World monkey species, CPO and CSO. The major families of alpha satellite DNA, called C1 and C2, are conserved between both species as well as their gross distribution, but a detailed investigation led us to envision the presence of highly repeated sequences in our data sets as revealing numerous subfamilies of C1 that differ between both species. Each family is the result of evolutionary mechanisms that involve local amplification of a specific sequence variant followed by mutations of the amplified sequences. Although most alpha satellite DNA is characterized by a monomeric organization in both species, we provide evidence for the existence of higher organization patterns that seem to be specific for CPO. Our cytogenomic approach suggests different types of transfer or loss of genetic material that may explain the peculiar distribution of centromeric and pericentromeric sequences. Future work addressing other species within the Cercopithecini clade will help elucidating the evolutionary mechanisms as well as the functional significance of alpha satellite DNA variation.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We are indebted to Florence Anne Richard, to whom we dedicate this manuscript. We thank Anne-Marie and Bernard Dutrillaux for their advice in metaphase preparation and analysis. This research was supported by the Actions Thématiques du Muséum “Génomique et Collections” and “Emergence”.
Authors’ Contributions
C.E. and F.A.R. conceived the study. L.C. and D.G. prepared the samples for sequencing. L.C. and L.P. performed the computational analysis. L.C, F.L., and M.G.-S. performed the FISH experiments and image acquisitions. L.C. and M.G.-S. reconstructed the karyotypes. L.C., L.P., and C.E. drafted the manuscript. All authors except F.A.R., who passed away during the study, read and approved the final manuscript.
Literature Cited
- Alexandrov IA, Mitkevich SP, Yurov YB.. 1988. The phylogeny of human chromosome specific alpha satellites. Chromosoma 96(6):443–453. [DOI] [PubMed] [Google Scholar]
- Alkan C, et al. 2007. Organization and evolution of primate centromeric DNA from whole-genome shotgun sequence data. PLoS Comput Biol. 3(9):1807–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Archidiacono N, et al. 1995. Comparative mapping of human alphoid sequences in great apes using fluorescence. Genomics 25(2):477–484. [DOI] [PubMed] [Google Scholar]
- Cacheux L, Ponger L, Gerbault-Seureau M, Richard FA, Escudé C.. 2016. Diversity and distribution of alpha satellite DNA in the genome of an Old World monkey: Cercopithecus solatus. BMC Genomics 17(1):916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catacchio CR, Ragone R, Chiatante G, Ventura M.. 2015. Organization and evolution of Gorilla centromeric DNA from old strategies to new approaches. Sci Rep. 5(1):14189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellamare A, et al. 2009. New insights into centromere organization and evolution from the white-cheeked Gibbon and marmoset. Mol Biol Evol. 26(8):1889–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiatante G, Giannuzzi G, Calabrese FM, Eichler EE, Ventura M.. 2017. Centromere destiny in dicentric chromosomes: new insights from the evolution of human chromosome 2 ancestral centromeric region. Mol Biol Evol. 34(7):1669–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. 1982. Molecular drive: a cohesive mode of species evolution. Nature 299(5879):111–117. [DOI] [PubMed] [Google Scholar]
- Dutrillaux B, Couturier J, Chauvier G.. 1980. Chromosomal evolution of 19 species of sub-species of Cercopithecinae. Ann Genet. 23(3):133–143. [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 19:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliciello I, Akrap I, Brajkovi J, Zlatar I, Ugarkovi UI.. 2015. Satellite DNA as a driver of population divergence in the red flour beetle Tribolium castaneum. Genome Biol Evol. 7(1):228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-ramos MA. 2017. Satellite DNA: An Evolving Topic. Genes 8, 230. [DOI] [PMC free article] [PubMed]
- Gouy M, Guindon S, Gascuel O.. 2010. SeaView Version 4: a Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol Biol Evol. 27:221–224. [DOI] [PubMed] [Google Scholar]
- Guschanski K, et al. 2013. Next-generation museomics disentangles one of the largest primate radiations. Syst Biol. 62(4):539–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden KE. 2012. Human centromere genomics: now it’s personal. Chromosom Res. 20(5):621–633. [DOI] [PubMed] [Google Scholar]
- Jorgensen AL, Jones C, Bostock CJ, Bak AL.. 1987. Different subfamilies of alphoid repetitive DNA are present on the human and chimpanzee homologous chromosomes 21 and 22. EMBO J. 6:1691–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, et al. 2011. Comparative and demographic analysis of orang-utan genomes. Nature 469(7331):529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin S, Gerbault-Seureau M, Dutrillaux B, Richard FA.. 2008. Phylogenomics of African guenons. Chromosom Res. 16(5):783–799. [DOI] [PubMed] [Google Scholar]
- Ollion J, Loll F, Cochennec J, Boudier T, Escudé C.. 2015. Proliferation-dependent positioning of individual centromeres in the interphase nucleus of human lymphoblastoid cell lines. Mol. Biol. Cell 26(13):2550–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paço A, Adega F, Meštrović N, Plohl M, Chaves R.. 2014. Evolutionary story of a satellite DNA from Phodopus. Genome Biol Evol. 6:2944–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomeque T, Lorite P.. 2008. Satellite DNA in insects: a review. Heredity (Edinb) 100(6):564–573. [DOI] [PubMed] [Google Scholar]
- Pérez-Gutiérrez MA, Suárez-Santiago VN, López-Flores I, Romero AT, Garrido-Ramos MA.. 2012. Concerted evolution of satellite DNA in Sarcocapnos: a matter of time. Plant Mol Biol. 78(1–2):19–29. [DOI] [PubMed] [Google Scholar]
- Piras FM, et al. 2010. Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet. 6(2):e1000845.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plohl M, Luchetti A, Meštrović N, Mantovani B.. 2008. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 409(1–2):72–82. [DOI] [PubMed] [Google Scholar]
- Prakhongcheep O, et al. 2013. Two types of alpha satellite DNA in distinct chromosomal locations in Azara’s owl monkey. DNA Res. 20(3):235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A.. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16(6):276–277. [DOI] [PubMed] [Google Scholar]
- Rosandić M, et al. 2006. CENP-B box and pJα sequence distribution in human alpha satellite higher-order repeats (HOR). Chromosom Res. 14(7):735–753. [DOI] [PubMed] [Google Scholar]
- Rudd MK, Wray GA, Willard HF.. 2006. The evolutionary dynamics of alpha -satellite. Genome Res. 16(1):88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelhauer D, Schwarz T.. 2002. Evidence for a fast, intrachromosomal conversion mechanism from mapping of nucleotide variants within a homogeneous α-satellite DNA array. Genome Res. 12(12):1815–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, et al. 2005. Progressive proximal expansion of the primate X chromosome centromere. Proc Natl Acad Sci U S A. 102(30):10563–10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, Sullivan BA.. 2006. Structural and functional dynamics of human centromeric chromatin. Annu Rev Genomics Hum Genet. 7:301–313. [DOI] [PubMed] [Google Scholar]
- Shepelev VA, Alexandrov AA, Yurov YB, Alexandrov IA.. 2009. The evolutionary origin of man can be traced in the layers of defunct ancestral alpha satellites flanking the active centromeres of human chromosomes. PLoS Genet. 5(9):e1000641.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujiwattanarat P, et al. 2015. Higher-order repeat structure in alpha satellite DNA occurs in New World monkeys and is not confined to hominoids. Sci Rep. 5(1):10315.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2014). R: A Language and Environment for Statistical Computing. Vienna (Austria: ): R Foundation for Statistical Computing; Available from: http://www.R-project.org/. [Google Scholar]
- Terada S, Hirai Y, Hirai H, Koga A.. 2013. Higher-order repeat structure in alpha satellite DNA is an attribute of hominoids rather than hominids. J Hum Genet. 58(11):752–754. [DOI] [PubMed] [Google Scholar]
- Tosi AJ. 2008. Forest monkeys and Pleistocene refugia: a phylogeographic window onto the disjunct distribution of the Chlorocebus lhoesti species group. Zool J Linn Soc. 154(2):408–418. [Google Scholar]
- Utsunomia R, et al. 2017. A Glimpse into the Satellite DNA Library in Characidae Fish (Teleostei, Characiformes). Front Genet. 8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissel B, Choo KH.. 1991. Four distinct alpha satellite subfamilies shared human. Nucleic Acids Res. 19:271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Haaf T, Gosden J, Lawson D, Willard HF.. 1996. Characterization of a chromosome-specific chimpanzee alpha satellite subset: evolutionary relationship to subsets on human chromosomes. Genomics 33(2):220–228. [DOI] [PubMed] [Google Scholar]
- Ward JH. 1963. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 58(301):236–244. [Google Scholar]
- Waye J, Willard H.. 1986. Structure, organization, and sequence of alpha satellite DNA from human chromosome 17: evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol Cell Biol. 6(9):3156–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.