Abstract
The X chromosome is unique in the genome. In this review we discuss recent advances in our understanding of the genetics and epigenetics of the X chromosome. The X chromosome shares limited conservation with its ancestral homologue the Y chromosome and the resulting difference in X-chromosome dosage between males and females is largely compensated for by X-chromosome inactivation. The process of inactivation is initiated by the long non-coding RNA X-inactive specific transcript (XIST) and achieved through interaction with multiple synergistic silencing pathways. Identification of Xist-interacting proteins has given insight into these processes yet the cascade of events from initiation to maintenance have still to be resolved. In particular, the initiation of inactivation in humans has been challenging to study as: it occurs very early in development; most human embryonic stem cell lines already have an inactive X; and the process seems to differ from mouse. Another difference between human and mouse X inactivation is the larger number of human genes that escape silencing. In humans over 20% of X-linked genes continue to be expressed from the otherwise inactive X chromosome. We are only beginning to understand how such escape occurs but there is growing recognition that escapees contribute to sexually dimorphic traits. The unique biology and epigenetics of the X chromosome have often led to its exclusion from disease studies, yet the X constitutes 5% of the genome and is an important contributor to disease, often in a sex-specific manner.
Introduction to the X Chromosome
The genomes of males and females differ dramatically, with males having a single X chromosome and the sex-determining Y chromosome; whereas females have two X chromosomes, and thus 100 Mb more DNA than males (X: ∼155 Mb, Y: ∼55 Mb). The X and Y are derived from an ancestral pair of autosomes, but the human Y retains functionality for only 17 of the over 600 genes once shared with the X (reviewed in 1). Whether there is upregulation of X-linked genes to maintain equivalence to ancestral autosomal gene dosage remains controversial (reviewed in 2) (3), in part due to the unique gene composition of the X. Both the X and Y have acquired ampliconic gene families that are expressed only in testes, are not well-conserved between humans and mice, are often palindromic and are argued to underlie dramatic selective sweeps on the X (4). The X has also been both recipient and donor for retrogenes, with autosomal copies of X-linked genes compensating for the X silencing that occurs during meiotic sex chromosome inactivation (MSCI) in spermatogenesis (5). miRNA genes are also over-represented on the X, and respond differentially to MSCI during spermatogenesis (6,7). Regardless of whether the X to autosome dosage has equilibrated, there is a dosage imbalance between males and females for the ∼1000 X-linked genes. As hypothesized by Lyon in 1961, compensation for this difference occurs by inactivation of one of the two Xs in early female development.
X-Chromosome Inactivation: Making Two Equal One
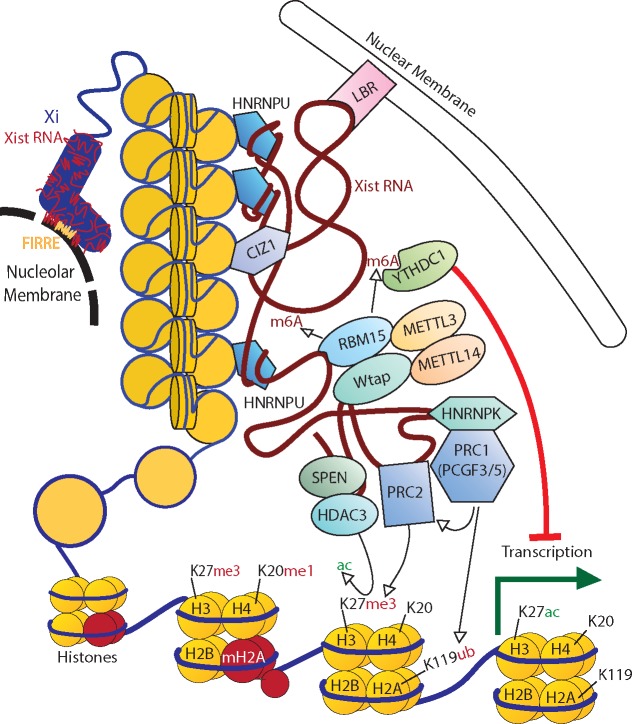
In the over 55 years since the Lyon hypothesis of X-chromosome inactivation (XCI), many features have been shown to distinguish the active X (Xa) from an inactive X (Xi). These features, such as silent chromatin marks, DNA hypermethylation at CpG island-containing promoters and late replication timing, are often shared with other silent, heterochromatic regions of the genome (reviewed in 8). The Xi is comprised of two large superdomains encompassing superloops rather than the compartments (9) and topologically associating domains (TADs) that are seen on other chromosomes (10). In both humans and mice the hinge between these superdomains is the macrosatellite DXZ4/Dxz4, whose presence is critical for this ultrastructure (10–12). The FIRRE/Firre locus serves as one of the superloop bases, and is required for superloop formation (13), nucleolar localization and H3K27me3 retention (see Fig. 1) (14).
Figure 1.
Major factors involved in the inactivation of an X. Xist RNA coats the X and is critical for its localization to either (or both) the nucleolus and nuclear membrane. Localization to the nucleolar membrane requires the lncRNA Firre. Xist acts as a molecular scaffold recruiting various repressive complexes such as PRC1/2 while also acting as a tether between various chromatin and matrix binding factors. Some of these factors, such as PRC2 and HNRNPU are known to be recruited to the Xi in both humans and mice, however most evidence of the interactions and pathways regulating these factors comes from work done in mouse models. H3/H4/H2A/H2B refer to histone subunits, me and ac refer to the methylation and acetylation of histone tails at specified lysines (K).
The initiating event in the heterochromatic and structural conversion from Xa to Xi is the expression of the long non-coding (lnc) RNA, X-inactive specific transcript (XIST), whose locus also serves as a base for the superloops. It is named for its Xi-specific transcription pattern, and, impressively, this long (15–19 kb processed) RNA remains localized to the chromosome from which it is transcribed. The human and mouse XIST/Xist genes are fairly conserved in their exon/intron structure, and both have tandem repetitive regions (generally labelled A–F) with varying extents of conservation; repeat A being well conserved while repeats B–F differ between species. The short isoform of Xist, lacking 5.6 kb of exon 7 downstream of the E repeat, can establish XCI, suggesting that the internal region of Xist exon 7 is functionally dispensable (15). XIST expression is female-specific in somatic tissues of karyotypically normal individuals but is also expressed during spermatogenesis.
Early Events Establishing X Inactivation
During early development, XIST expression from both Xs has been observed in humans but not in mice, suggesting the early events may be species-specific (16–18). Single-cell sequencing suggested dampening of X-linked gene expression when XIST is expressed (17), although alternative analysis suggested that early XCI was the source of decreased expression (19). In human embryonic stem cells (hESCs) the majority of cell lines are found to already have an Xi. Rare populations of XaXa cells are observed, but, problematically for use in regenerative medicine, there are also substantial populations of cells with an eroded Xi (Xe) that has lost XIST expression and XCI (reviewed in 20). Unique to early human XCI is expression of a non-coding RNA expressed from the Xa (named XACT) that has been proposed to compete with XIST (18), and thus may be responsible for the erosion of inactivation in hESCs. Recent work has shown that naïve hESCs have two Xas that express XIST and can undergo XCI upon differentiation; however, most hESCs and human induced pluripotent cell lines retain substantial populations of cells with mono-allelic XIST expression (21,22).
All Xs in excess of one in diploid cells are inactivated, while in triploids there can be more than one X active and in tetraploids there are two Xis and two Xas. Thus, many models of XCI have invoked a dosage-sensitive factor derived from the autosomes that blocks inactivation of one X. Such a blocking factor could not be present on chromosomes (or regions of chromosomes) that are found in females with a trisomy (or segmental duplication), as they would then prevent both Xs from undergoing XCI. Intriguingly, by searching the DECIPHER database for sex-biased duplications, regions of chromosome 19 were found duplicated in males but not females, and include important chromatin regulators and zinc-finger repressors (23). Alternatively, recent mathematical modelling demonstrates that coupled X-chromosomal feedback loops could explain the shift from bi-allelic to mono-allelic XIST as well as the Xi patterns of aneuploids (24).
Epigenetic Silencing of the X: Moving Towards Mechanism
How does an RNA localize to the chromosome and induce silencing? As with other long non-coding RNAs (lncRNAs), it is likely that XIST acts as a scaffold to recruit critical proteins (Fig. 1). Three proteomic studies in mouse utilized antisense probes to capture Xist and its interacting proteins, yielding well over 100 candidate partners, of which 31 proteins were identified in one or more study (25–27) (reviewed in 28). Forward genetic screens to identify proteins involved in XCI have supported and expanded this list of candidates (reviewed in 29), yet their assembly into pathways of silencing is still far from complete. Among the most debated pathways are the polycomb repressive complexes (PRC1/2). Growing evidence suggests that hnRNPK binds the mouse repeat B region to enable PCGF3/5 PRC1 recruitment, which in turn recruits PRC2 (30,31). Binding of the transcriptional repressor SPEN to the A repeat recruits HDAC3 leading to deacetylation of histones, which is thought to be important for PRC2 recruitment (27). The XIST RNA is embedded in the nuclear matrix, and many of the identified interactors are matrix proteins including heterogeneous nuclear ribonucleoprotein U (HNRNPU) (Saf-A) (32) and CIZ1 (33). HNRNPU has been shown to bind the Xist transcript near the C, D repeats as well as exon 7, with a similar distribution across the human transcript (15). The loss of HNRNPU disrupts Xist-induced silencing and localization in cis to the Xi (27). CIZ1 binds to the E repeat of Xist and is enriched on the Xi of humans and mice. While Ciz1-null mice are viable, females develop lymphoproliferative disease and cells show failure to localize Xist (33,34). Interestingly, some naïve T and B cells have been seen to have dispersion of XIST/Xist (35). In these cells, relocalization of Xist requires YY1 (36), a transcription factor with both RNA and DNA binding domains previously reported to be critical for Xist transcription and tethering (37). However, none of the RNA-protein screens identified YY1 binding to the Xist transcript (25–27).
Association of the Xi with the nuclear periphery has long been recognized, and was recently attributed to XIST interaction with Lamin B Receptor (LBR). Repression of LBR prevents gene silencing, perhaps because of failure of polymerase exclusion (27). XIST has more N6-methyladenosine (m6A) modifications than other RNAs, and the proteins involved in establishing this mark (RBM15, WTAP and MTTL3) are thought to be recruited by the A repeat of Xist (38). Knockdown of these factors or the m6A reader YTHDC1 in mouse ESCs prevented gene silencing during XCI (38). Other heterochromatin marks enriched on the Xi, such as a variant of Histone H2A (MACROH2A) and SMCHD1 are thought to be recruited later during XCI and to maintain the stability of silencing on the Xi. Overall, it appears that many of the silencing pathways initiated by XIST, while inter-related also are independent (reviewed in 29) (39,40).
Escapees: Exceptions to Silencing
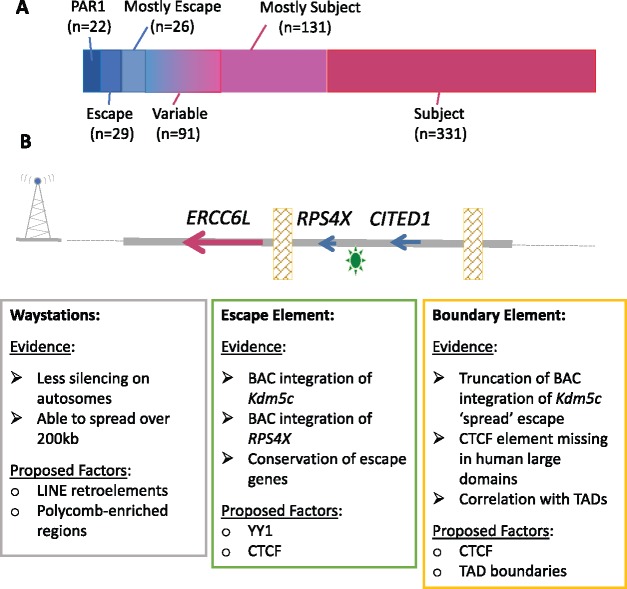
Despite the many independent yet synergistic pathways to establish and maintain inactivation, the Xi is not completely silent. XIST is expressed solely from the Xi, but many other genes continue to be expressed from the Xi in addition to the Xa (Fig. 2A). The pseudoautosomal region (PAR) at the short arm of the X and Y (PAR1) undergoes recombination during male meiosis, thus earning its title by behaving like an autosome. The majority of PAR1 genes escape from XCI but show detectably lower expression in females than males (41), suggesting that expression from the Xi is lower than the Xa, which is true for all escapees—the threshold to call escape generally being at 10% the level of the Xa. Calling escape from XCI has been based on: expression differences between males and females; biallelic expression in single cells or females with non-random XCI; isolation of the Xi in somatic cell hybrids; and assessment of chromatin marks or DNA methylation in males and females [see (42) and the extensive assessment of GTEX data (41)]. Our aggregation of these data assigned a consensus inactivation status to 639 genes (81% of X-linked protein-coding genes expressed in somatic cells), of which 80 genes escape and 93 genes variably escape from XCI (42). In mice, considerably fewer genes escape inactivation, with 17 constitutive escape genes, and at least 20 variable escape genes (reviewed in 8). The difference in number of constitutive escape genes between species is largely due to human escape regions (particularly on the X short arm) containing blocks of up to 15 genes, instead of the 1–2 genes observed for mouse. The blocks of escape genes in humans align (imperfectly) with TADs (8,43). These escape genes are enriched in regions with shorter divergence times from the Y, but also are enriched for regulatory proteins or components of large protein complexes that may be more sensitive to dosage. Indeed, there are 17 human genes that retain ancestral X/Y homology outside the PAR, and the extreme dosage-sensitivity of these genes has been demonstrated by conservation of miRNA target sites (44).
Figure 2.
Escape genes and putative elements involved. (A) Proportion of genes subject to, or escaping from, XCI. Both the escape and the subject category have a subcategory of ‘mostly’ subject/escape which include those genes for which multiple studies tend to agree, but there is an exception. The variable category includes both genes described within studies as variable (n=47), as well as those genes which consistently differ between studies (n=44). (B) Proposed elements involved in a gene’s ability to escape from XCI as applied to the RPS4X escape region in humans. RPS4X and neighbouring gene CITED1 (a variable gene whose expression status had been described as subject or escape) retain the ability to escape from XCI (CITED1 is brain-specific) when present on a BAC integrated into the mouse Hprt locus on the Xi. Evidence and candidates for waystations, boundary and escape elements are described in boxes.
Understanding How Genes Escape X Inactivation
We envision categorization of DNA elements contributing to whether a gene escapes from XCI as waystations, escape elements and boundaries (see Fig. 2B). Waystations are hypothetical elements that boost the silencing signal along the X, and Lyon proposed long interspersed nuclear elements (LINE) repetitive elements could be the waystations (45). Our studies of autosomal bacterial artificial chromosome (BAC) integrations into Hprt had found most to be subject to XCI, suggesting silencing can spread across 200 kb of integrated DNA (46), hence we suggest way stations are able to act across larger distances. Multiple molecular assessments of unbalanced X; autosome translocations (47) as well as mouse Xist transgenes (40) have confirmed that spread of silencing is reduced relative to the extent of silencing seen on the X. In addition to a positive correlation between silencing and presence of LINE elements, the strongest correlation we observed with subject genes was with pre-existing features of heterochromatin, most significantly for EZH2 (47), suggesting waystations may be sequences associated with recruitment of PRC2 (of which EZH2 is a component).
Despite rearrangement of the mouse X relative to the human, 7 of the 17 constitutive escape genes in mice also escape in humans, suggesting evolutionarily conserved cis-acting sequences permitting Xi-expression (8). Evidence for an intrinsic escape element was shown by expression from the Xi of the Kdm5c gene in four random X-linked BAC integrations (48) (reviewed in 49). We have now demonstrated escape from inactivation for the human RPS4X gene in BACs integrated into the X-linked Hprt gene (50). Thus, human elements can be recognized in mouse even though the endogenous mouse Rps4x gene is subject to XCI. Recent studies in mouse have suggested a role for promoter-proximal elements in regulating escapees, particularly CTCF (10). CTCF is well-established as a boundary element; however, additional features must designate which of the close to 2000 CTCF sites on the X chromosome are serving as euchromatic and heterochromatic boundaries (51). Expression from the Xi is likely more complex than three elements, and a full model will need to integrate local and chromosomal ultrastructure. For example, deletion of the Dxz4 hinge region in mouse ESCs impacted expression from the Xi (10).
Mendelian Sexually Dimorphic Traits
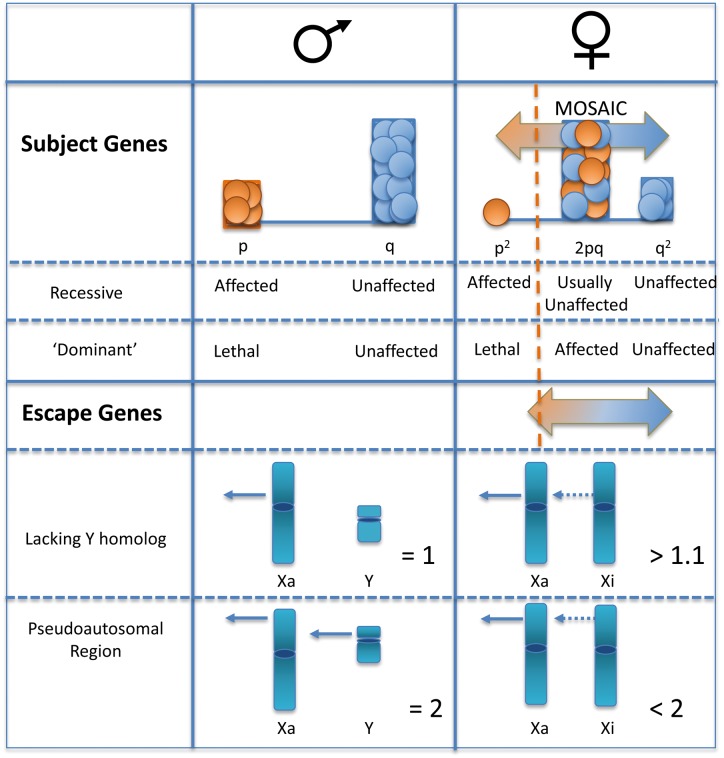
A defining characteristic of X-linked recessive disease inheritance is the overwhelming male bias, as males are hemizygous for the X. For genes that are subject to XCI, heterozygous females will be mosaics of two populations: one with the mutation on the Xa; one with the wild type allele on the Xa (see Fig. 3). Thus, expression in a female will be dependent on the proportion of cells carrying the mutation on the Xa, as well as the cell autonomous nature of the product. Skewed XCI can occur in carriers due to selection against the cells expressing the trait, due to stochastic events, or from selection against other alleles on the X. As with autosomal genes, homozygous X-linked recessive females will also be affected; however, in addition to a carrier mother, a homozygous female would also need to have an affected (hemizygous) father. Similar to most dominant disorders in humans, X-linked dominant disorders are usually co-dominant with male lethality (see Fig. 3). De novo mutations on the paternal X will always be found in daughters not sons (who receive their father’s Y), which can also contribute to a female excess for dominant disorders. With differing epigenetic marks and lack of pairing for the X for much of its length in male meiosis, sex-biased mutation rates are not uncommon. Intellectual disability (ID) is 30% more common in males, and there are over 100 X-linked ID genes identified (52). The number of X-linked genes with mutations continues to increase with sequencing of rare-disease families.
Figure 3.
Impact of an inactive X on disease. For recessive alleles of genes subject to XCI, males will be affected at the population frequency ‘p’. Females will be homozygous at a much lower frequency (p2, so if a trait is seen in 1 in a 1000 males, then it will be seen in 1 in a million females). However, the large heterozygous female population will be mosaic for the trait, with skewing influencing the extent of which each allele is expressed (shaded horizontal arrow). Carriers will only express the trait if a threshold of required product is not met, and an example of a disease threshold is shown as a dotted orange line. For dominant disorders the same frequencies apply; however, generally hemizygous or homozygous individuals do not survive and the disease manifests in carrier females. As escape genes are not fully expressed from the Xi, they will show similar impact of skewing on mosaicism, with a shift towards higher overall expression levels. Again, disease presentation will depend on the threshold of gene product required. Shown below is a comparison of expression levels between males and females for escape genes that retain or lack a Y homologue.
Complex Traits: X Contributions to Sex Differences
Many complex disorders (including cancer) show sexually dimorphic disease rates that are often attributed to differences in sex hormones; however, understanding of the role of the sex chromosomes themselves is growing. A role for the Xi in traits including adiposity, metabolic disease, cardiovascular injury and behaviour has been revealed by mouse breeding schemes that generate XX males and XY females (53). The phenotypic impact of an Xi is likely to be even more pronounced in humans due to the larger number of escapees. Separation of chromosomal effects from hormonal influences is challenging in humans; however, X aneuploidies can offer some insights. A critical role for the second sex chromosome is apparent with over 99% of 45, X conceptuses lost in utero; whereas a supernumerary chromosome increases risk of intellectual impairment and psychological disorders. Through comparisons of risk for individuals with sex chromosome aneuploidies it has been shown that for autism the X may be protective (reviewed in 54); while for lupus gain of an X is a risk factor (55).
The contribution of genes that escape XCI to sexually dimorphic disease is varied. In some cancers, escapees may protect against the impact of de novo mutations, resulting in a male bias (56). In contrast, the heterochromatic nature of the Xi may pre-dispose to mutations, which could have an impact for genes that escape XCI (e.g. somatic DDX3X mutations in female cancer) (57). Furthermore, loss of XCI can be observed in cancers in females (such as breast cancer) resulting in reactivation of X-linked genes (58). The X is often excluded from genome-wide association studies but improved bioinformatic tools that can account for the possibility of escape from inactivation, or skewing of XCI, will hopefully remedy this trend (59) and allow for better identification of X-linked disease associations.
X Inactivation: Opportunities for Therapeutics and Diagnostics
The Xi is classically described as facultative heterochromatin, and the study of XCI offers an outstanding paradigm to understand epigenetic gene regulation. The studies described herein are beginning to address how silencing pathways are integrated via the lncRNA XIST. Recent studies have targeted some of these silencing pathways to explore the potential to reactivate silenced X-linked genes in females with dominant X-linked disorders, particularly Rett syndrome (60). Additionally, XIST itself has been used as a silencing molecule for proof-of-principle for chromosome therapy (61). Reactivation of the Xi (associated with silencing of XIST) can be a marker for re-establishment of pluripotency for human stem cells (22); but care needs to be taken as erosion can appear similar to reactivation (62). The impact of an Xi in females versus a Y in males is far-reaching. There are hormonal differences, ongoing expression of the escapees, and even less explored are the impacts of substantially different amounts of DNA, expression of XIST and differential utilization of epigenetic modifications. Since Lyon first hypothesized XCI we have learned much about the biology and now the impact of the not so inactive Xi.
Acknowledgements
Work in the authors’ laboratory is funded by Canadian Institutes of Health Research (CIHR). T.D-M., B.B. and S.P. have been funded by Natural Sciences and Engineering Research Council of Canada (NSERC) and University of British Columbia.
Conflict of Interest statement. None declared.
References
- 1. Hughes J.F., Page D.C. (2016) The history of the Y chromosome in man. Nat. Genet., 48, 588–589. [DOI] [PubMed] [Google Scholar]
- 2. Veitia R.A., Veyrunes F., Bottani S., Birchler J.A. (2015) X chromosome inactivation and active X upregulation in therian mammals: facts, questions, and hypotheses. J. Mol. Cell. Biol., 7, 2–11. [DOI] [PubMed] [Google Scholar]
- 3. Marin R., Cortez D., Lamanna F., Pradeepa M.M., Leushkin E., Julien P., Liechti A., Halbert J., Brüning T., Mössinger K. (2017) Convergent origination of a Drosophila-like dosage compensation mechanism in a reptile lineage. Genome Res., 27, 1974–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nam K., Munch K., Hobolth A., Dutheil J.Y., Veeramah K.R., Woerner A.E., Hammer M.F., Mailund T., Schierup M.H. (2015) Extreme selective sweeps independently targeted the X chromosomes of the great apes. Proc. Natl. Acad. Sci. USA, 112, 6413–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carelli F.N., Hayakawa T., Go Y., Imai H., Warnefors M., Kaessmann H. (2016) The life history of retrocopies illuminates the evolution of new mammalian genes. Genome Res., 26, 301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sosa E., Flores L., Yan W., McCarrey J.R. (2015) Escape of X-linked miRNA genes from meiotic sex chromosome inactivation. Development, 142, 3791–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Royo H., Seitz H., ElInati E., Peters A.H.F.M., Stadler M.B., Turner J.M.A. (2015) Silencing of X-linked microRNAs by meiotic sex chromosome inactivation. PLoS Genet., 11, e1005461.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Balaton B.P., Brown C.J. (2016) Escape artists of the X chromosome. Trends Genet., 32, 348–359. [DOI] [PubMed] [Google Scholar]
- 9. Wang S., Su J.-H., Beliveau B.J., Bintu B., Moffitt J.R., Wu C.-T., Zhuang X. (2016) Spatial organization of chromatin domains and compartments in single chromosomes. Science, 353, 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Giorgetti L., Lajoie B.R., Carter A.C., Attia M., Zhan Y., Xu J., Chen C.J., Kaplan N., Chang H.Y., Heard E., Dekker J. (2016) Structural organization of the inactive X chromosome in the mouse. Nature, 535, 575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deng X., Ma W., Ramani V., Hill A., Yang F., Ay F., Berletch J.B., Blau C.A., Shendure J., Duan Z., Noble W.S., Disteche C.M. (2015) Bipartite structure of the inactive mouse X chromosome. Genome Biol., 16, 152.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Darrow E.M., Huntley M.H., Dudchenko O., Stamenova E.K., Durand N.C., Sun Z., Huang S.-C., Sanborn A.L., Machol I., Shamim M.. et al. (2016) Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl. Acad. Sci. USA, 113, E4504–E4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barutcu A.R., Maass P.G., Lewandowski J.P., Weiner C.L., Rinn J.L. (2018) A TAD boundary is preserved upon deletion of the CTCF-rich Firre locus. Nat. Commun., 9, 1444.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang F., Deng X., Ma W., Berletch J.B., Rabaia N., Wei G., Moore J.M., Filippova G.N., Xu J., Liu Y.. et al. (2015) The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol., 16, 52.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamada N., Hasegawa Y., Yue M., Hamada T., Nakagawa S., Ogawa Y. (2015) Xist exon 7 contributes to the stable localization of Xist RNA on the inactive X-chromosome. PLoS Genet., 11, e1005430.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okamoto I., Patrat C., Thépot D., Peynot N., Fauque P., Daniel N., Diabangouaya P., Wolf J.-P., Renard J.-P., Duranthon V., Heard E. (2011) Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature, 472, 370–374. [DOI] [PubMed] [Google Scholar]
- 17. Petropoulos S., Edsgärd D., Reinius B., Deng Q., Panula S.P., Codeluppi S., Plaza Reyes A., Linnarsson S., Sandberg R., Lanner F. (2016) Single-cell RNA-Seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell, 165, 1012–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vallot C., Patrat C., Collier A.J., Huret C., Casanova M., Liyakat Ali T.M., Tosolini M., Frydman N., Heard E., Rugg-Gunn P.J., Rougeulle C. (2017) XACT noncoding RNA competes with XIST in the control of X chromosome activity during human early development. Cell Stem Cell, 20, 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moreira de Mello J.C., Fernandes G.R., Vibranovski M.D., Pereira L.V. (2017) Early X chromosome inactivation during human preimplantation development revealed by single-cell RNA-sequencing. Sci. Rep., 7, 10794.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geens M., Chuva De Sousa Lopes S.M. (2017) X chromosome inactivation in human pluripotent stem cells as a model for human development: back to the drawing board? Hum. Reprod. Update, 23, 520–532. [DOI] [PubMed] [Google Scholar]
- 21. Sahakyan A., Kim R., Chronis C., Sabri S., Bonora G., Theunissen T.W., Kuoy E., Langerman J., Clark A.T., Jaenisch R., Plath K. (2017) Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell, 20, 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guo G., von Meyenn F., Rostovskaya M., Clarke J., Dietmann S., Baker D., Sahakyan A., Myers S., Bertone P., Reik W., Plath K., Smith A. (2017) Epigenetic resetting of human pluripotency. Development, 144, 2748–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Migeon B.R., Beer M.A., Bjornsson H.T. (2017) Embryonic loss of human females with partial trisomy 19 identifies region critical for the single active X. PLoS One, 12, e0170403.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mutzel V., Okamoto I., Dunkel I., Saitou M., Giorgetti L., Heard E., Schulz E.G. (2017) Two coupled feedback loops explain random mono-allelic Xist upregulation at the onset of X-chromosome inactivation. BioXRIV. 10.1101/204909.
- 25. Chu C., Zhang Q.C., da Rocha S.T., Flynn R.A., Bharadwaj M., Calabrese J.M., Magnuson T., Heard E., Chang H.Y. (2015) Systematic discovery of Xist RNA binding proteins. Cell, 161, 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Minajigi A., Froberg J.E., Wei C., Sunwoo H., Kesner B., Colognori D., Lessing D., Payer B., Boukhali M., Haas W., Lee J.T. (2015) Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science, 349, aab2276–aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McHugh C.A., Chen C.-K., Chow A., Surka C.F., Tran C., McDonel P., Pandya-Jones A., Blanco M., Burghard C., Moradian A.. et al. (2015) The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature, 521, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moindrot B., Brockdorff N. (2016) RNA binding proteins implicated in Xist-mediated chromosome silencing. Semin. Cell Dev. Biol., 56, 58–70. [DOI] [PubMed] [Google Scholar]
- 29. da Rocha S.T., Heard E. (2017) Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat. Struct. Mol. Biol., 24, 197–204. [DOI] [PubMed] [Google Scholar]
- 30. Pintacuda G., Wei G., Roustan C., Kirmizitas B.A., Solcan N., Cerase A., Castello A., Mohammed S., Moindrot B., Nesterova T.B.. et al. (2017) hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol. Cell, 68, 955–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Almeida M., Pintacuda G., Masui O., Koseki Y., Gdula M., Cerase A., Brown D., Mould A., Innocent C., Nakayama M.. et al. (2017) PCGF3/5–PRC1 initiates polycomb recruitment in X chromosome inactivation. Science, 356, 1081–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kolpa H.J., Fackelmayer F.O., Lawrence J.B. (2016) SAF-A requirement in anchoring XIST RNA to chromatin varies in transformed and primary cells. Dev. Cell, 39, 9–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ridings-Figueroa R., Stewart E.R., Nesterova T.B., Coker H., Pintacuda G., Godwin J., Wilson R., Haslam A., Lilley F., Ruigrok R.. et al. (2017) The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes. Dev., 31, 876–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sunwoo H., Colognori D., Froberg J.E., Jeon Y., Lee J.T. (2017) Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proc. Natl. Acad. Sci. USA, 114, 10654–10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang J., Syrett C.M., Kramer M.C., Basu A., Atchison M.L., Anguera M.C. (2016) Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc. Natl. Acad. Sci. USA, 113, E2029–E2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Syrett C.M., Sindhava V., Hodawadekar S., Myles A., Liang G., Zhang Y., Nandi S., Cancro M., Atchison M., Anguera M.C. (2017) Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic YY1-dependent two-step process in activated B cells. PLoS Genet., 13, e1007050.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jeon Y., Lee J.T. (2011) YY1 tethers Xist RNA to the inactive X nucleation center. Cell, 146, 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Patil D.P., Chen C.-K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R. (2016) m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature, 537, 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kelsey A.D., Yang C., Leung D., Minks J., Dixon-McDougall T., Baldry S.E.L., Bogutz A.B., Lefebvre L., Brown C.J. (2015) Impact of flanking chromosomal sequences on localization and silencing by the human non-coding RNA XIST. Genome Biol., 16, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Loda A., Brandsma J.H., Vassilev I., Servant N., Loos F., Amirnasr A., Splinter E., Barillot E., Poot R.A., Heard E.. et al. (2017) Genetic and epigenetic features direct differential efficiency of Xist-mediated silencing at X-chromosomal and autosomal locations. Nat. Commun., 8, 690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tukiainen T., Villani A.-C., Yen A., Rivas M.A., Marshall J.L., Satija R., Aguirre M., Gauthier L., Fleharty M., Kirby A.. et al. (2017) Landscape of X chromosome inactivation across human tissues. Nature, 550, 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Balaton B.P., Cotton A.M., Brown C.J. (2015) Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol. Sex Dif., 6, 35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marks H., Kerstens H.H.D., Barakat T.S., Splinter E., Dirks R.A.M., van Mierlo G., Joshi O., Wang S.-Y., Babak T., Albers C.A.. et al. (2015) Dynamics of gene silencing during X inactivation using allele-specific RNA-seq. Genome Biol., 16, 149.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Naqvi S., Bellott D.W., Lin K.S., Page D.C. (2018) Conserved microRNA targeting reveals preexisting gene dosage sensitivities that shaped amniote sex chromosome evolution. Genome Res., 10.1101/gr.230433.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lyon M.F. (1998) X-Chromosome inactivation: a repeat hypothesis. Cytogenet. Genome Res., 80, 133–137. [DOI] [PubMed] [Google Scholar]
- 46. Yang C., McLeod A.J., Cotton A.M., de Leeuw C.N., Laprise S., Banks K.G., Simpson E.M., Brown C.J. (2012) Targeting of >1.5 Mb of human DNA into the mouse X chromosome reveals presence of cis-acting regulators of epigenetic silencing. Genetics, 192, 1281–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cotton A.M., Chen C.-Y., Lam L.L., Wasserman W.W., Kobor M.S., Brown C.J. (2014) Spread of X-chromosome inactivation into autosomal sequences: role for DNA elements, chromatin features and chromosomal domains. Hum. Mol. Genet., 23, 1211–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Horvath L.M., Li N., Carrel L. (2013) Deletion of an X-inactivation boundary disrupts adjacent gene silencing. PLoS Genet., 9, e1003952.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carrel L., Brown C.J. (2017) When the Lyon(ized chromosome) roars: ongoing expression from an inactive X chromosome. Philos. Trans. R. Soc. Lond. B Biol. Sci., 372, 20160355.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peeters S.B., Korecki A.J., Simpson E.M., Brown C.J. (2018) Human cis-acting elements regulating escape from X-chromosome inactivation function in mouse. Hum. Mol. Genet., 27, 1252–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ding Z., Ni Y., Timmer S.W., Lee B.-K., Battenhouse A., Louzada S., Yang F., Dunham I., Crawford G.E., Lieb J.D.. et al. (2014) Quantitative genetics of CTCF binding reveal local sequence effects and different modes of X-chromosome association. PLoS Genet., 10, e1004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fieremans N., Van Esch H., Holvoet M., Van Goethem G., Devriendt K., Rosello M., Mayo S., Martinez F., Jhangiani S., Muzny D.M.. et al. (2016) Identification of intellectual disability genes in female patients with a skewed X-inactivation pattern. Hum. Mutat., 37, 804–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Arnold A.P., Reue K., Eghbali M., Vilain E., Chen X., Ghahramani N., Itoh Y., Li J., Link J.C., Ngun T.. et al. (2016) The importance of having two X chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci., 371, 20150113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Printzlau F., Wolstencroft J., Skuse D.H. (2017) Cognitive, behavioral, and neural consequences of sex chromosome aneuploidy. J. Neurosci. Res., 95, 311–319. [DOI] [PubMed] [Google Scholar]
- 55. Sharma R., Harris V.M., Cavett J., Kurien B.T., Liu K., Koelsch K.A., Fayaaz A., Chaudhari K.S., Radfar L., Lewis D.. et al. (2017) Rare X chromosome abnormalities in systemic lupus erythematosus and Sjögren's syndrome. Arthritis Rheumatol., 69, 2187–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dunford A., Weinstock D.M., Savova V., Schumacher S.E., Cleary J.P., Yoda A., Sullivan T.J., Hess J.M., Gimelbrant A.A., Beroukhim R.. et al. (2017) Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat. Genet., 49, 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cheng F., Liu C., Lin C.-C., Zhao J., Jia P., Li W.-H., Zhao Z. (2015) A gene gravity model for the evolution of cancer genomes: a study of 3, 000 cancer genomes across 9 cancer types. PLoS Comp. Biol., 11, e1004497.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chaligné R., Popova T., Mendoza-Parra M.-A., Saleem M.-A.M., Gentien D., Ban K., Piolot T., Leroy O., Mariani O., Gronemeyer H.. et al. (2015) The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer. Genome Res., 25, 488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Skare Ø., Gjessing H.K., Gjerdevik M., Haaland Ø.A., Romanowska J., Lie R.T., Jugessur A. (2017) A new approach to chromosome-wide analysis of X-linked markers identifies new associations in Asian and European case-parent triads of orofacial clefts. PLoS One, 12, e0183772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Carrette L.L.G., Wang C.-Y., Wei C., Press W., Ma W., Kelleher R.J., Lee J.T. (2017) A mixed modality approach towards Xi reactivation for Rett syndrome and other X-linked disorders. Proc. Natl. Acad. Sci. USA, 115, E668–E675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jiang J., Jing Y., Cost G.J., Chiang J.-C., Kolpa H.J., Cotton A.M., Carone D.M., Carone B.R., Shivak D.A., Guschin D.Y.. et al. (2013) Translating dosage compensation to trisomy 21. Nature, 500, 296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kilens S., Meistermann D., Moreno D., Chariau C., Gaignerie A., Reignier A., Lelièvre Y., Casanova M., Vallot C., Nedellec S.. et al. (2018) Parallel derivation of isogenic human primed and naive induced pluripotent stem cells. Nat. Commun., 9, 360.. [DOI] [PMC free article] [PubMed] [Google Scholar]