Abstract
Facioscapulohumeral dystrophy (FSHD) is the third most prevalent muscular dystrophy. A progressive disease, it presents clinically as weakness and wasting of the face, shoulder and upper arm muscles, with later involvement of the trunk and lower extremities. FSHD develops through complex genetic and epigenetic events that converge on a common mechanism of toxicity with mis-expression of the transcription factor double homeobox 4 (DUX4). There is currently no treatment available for FSHD. However, the consensus that ectopic DUX4 expression in skeletal muscle is the root cause of FSHD pathophysiology has allowed research efforts to turn toward cultivating a deeper understanding of DUX4 biology and the pathways that underlie FSHD muscle pathology, and to translational studies aimed at developing targeted therapeutics using ever more sophisticated cell and animal-based models of FSHD. This review summarizes recent advances in our understanding of FSHD, including the regulation and activity of DUX4 in its normal developmental roles as well as its pathological contexts. We highlight how these advances raise new questions and challenges for the field as it moves into the next decade of FSHD research.
Introduction
Facioscapulohumeral dystrophy (FSHD) affects approximately 1 in 10 000–20 000 individuals worldwide (1–3). Symptoms typically appear during the second or third decade of life as asymmetric weakness of the facial (facio), shoulder (scapulo) and upper arm (humeral) muscles, and progress to affect nearly all skeletal muscle groups (4). However, there are also more acute pediatric onset forms of FSHD (5). Non-muscular symptoms are rare but include high-frequency hearing loss and retinal vascular disease (6,7), and are usually associated with the more severe forms of FSHD (8). Disease progression and severity is widely variable, with ∼20% of mutation carriers asymptomatic while another ∼20% require use of a wheelchair (8).
There are two clinically indistinguishable but genetically distinct forms of FSHD. The most common, FSHD type 1 (FSHD1), is autosomal dominantly inherited and caused by deletion of a subset of D4Z4 macrosatellite repeats on chromosome 4q35 that result in an array of fewer than 11 units (9,10). Additionally, approximately 5% of individuals have FSHD type 2 (FSHD2), which results from mutations in regulators of the D4Z4 repeat array, such as SMCHD1 or DNMT3B (11,12). Both forms of FSHD require a specific permissive haplotype of chromosome 4 that contains a polymorphism that creates a polyadenylation signal distal to the D4Z4 array (13). Despite the different genetic etiology, both FSHD1 and FSHD2 are caused by a common pathophysiological mechanism: epigenetic de-repression of the D4Z4 array allowing mis-expression of the double homeobox 4 (DUX4) retrogene encoded within each unit of the D4Z4 repeat (8,14).
DUX4 is a transcription factor that is expressed in the luminal cells of the testis, most likely the spermatogonia and primary spermatocytes, but silenced in most somatic tissues, including skeletal muscle, through repeat-mediated epigenetic repression (15,16). Although expression of DUX4 in skeletal muscle and many other cell types causes cell death (17–20), questions remain concerning the pathophysiologic consequences of DUX4 expression and the regulation of DUX4 expression in development and in FSHD. Further understanding of these mechanisms should guide new potential therapeutic targets for FSHD.
Previous reviews have detailed the history of mapping the FSHD loci, the underlying genetic mutations and their epigenetic consequences, and the determination that mis-expression of DUX4 in skeletal muscle is the cause of FSHD (8,14,16,21–27). This review will focus on the regulation and activity of DUX4 (Fig. 1), particularly work done in the last 2 years that has revealed its normal developmental roles and further illuminated the pathological function of DUX4 in FSHD.
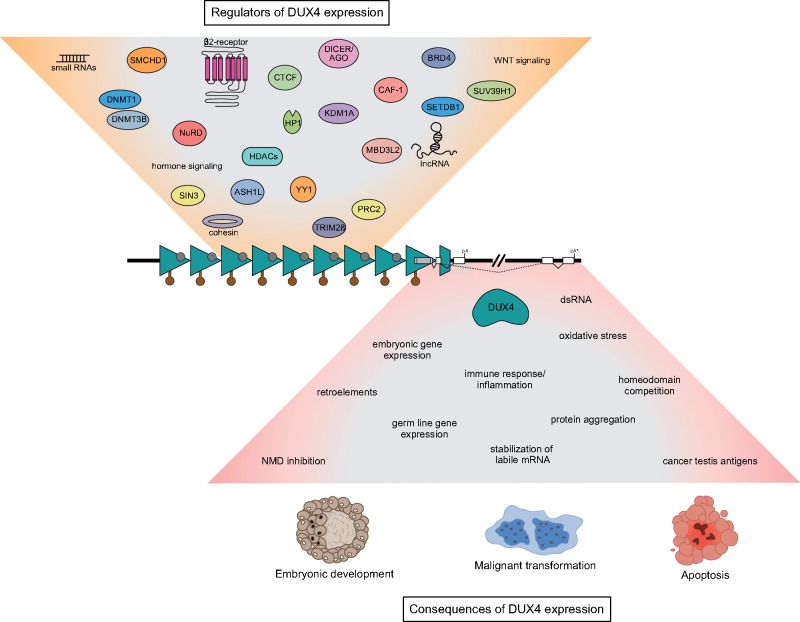
Figure 1.
Causes and effects of DUX4 expression. The DUX4 gene (white boxes with the open reading frame in gray) is encoded within each repeat unit (teal triangles) of the D4Z4 macrosatellite array. In healthy individuals the array is marked by high levels of CpG DNA methylation (brown lollipops) and transcriptionally repressive histone modifications (gray circles), which contribute to DUX4 silencing and are reduced in people affected by FSHD. Other factors and pathways known to regulate DUX4 expression are depicted in the tan funnel above the D4Z4 array. Production of stable DUX4 mRNA depends on the presence of a permissive haplotype located immediately distal to the last D4Z4 repeat that contains a functional polyadenylation site (pA), or on a polyadenylation sequence (pA*) found in downstream exons that is likely utilized in the germline. The expression of DUX4 protein is associated with many cellular phenotypes, which are outlined in the inverted pink funnel below the D4Z4 array. Depending on the context, DUX4 may or may not lead to normal development, inappropriate apoptosis or malignant transformation.
A Conserved Function for DUX4
In individuals unaffected by FSHD, the DUX4 gene is transcriptionally silenced in most tissues of the body (8). However, our group reported in 2010 that DUX4 is normally expressed in the testis (15), and others have shown low levels of DUX4 in the thymus (28). More recent studies revealed that DUX4 is transiently expressed in four-cell human embryos where it functions to activate a cleavage-stage transcriptional program and might be a key regulator of zygotic genome activation (ZGA) (29,30). The mouse ortholog of DUX4 (Dux) was also shown to be expressed in the two-cell embryo co-incident with murine ZGA (31). Dux activates the mouse cleavage-stage gene program (30) and is necessary for proper embryonic development, as Dux-deficient embryonic stem cells show reduced conversion to the totipotent ‘2C-like’ state (29) and Dux-depleted embryos fail to reach the morula stage (31). Overall, these new findings strongly support a functionally important role for human DUX4 and mouse Dux in early development. They also raise questions such as: what are the mechanisms by which DUX4/Dux are briefly activated and then rapidly silenced during cleavage-stage development? Are these same pathways utilized in other contexts where DUX4 is present (such as testis, thymus and FSHD muscle)? Why are DUX4/Dux not toxic in the embryo? And, do ancestral or related DUX genes (e.g. DUXA, DUXB, DUXC) function similarly?
The conserved function of DUX4 and Dux in early development is consistent with their analogous origins as retroposition events of the ancestral DUXC gene (32–34). However, DUX4 and Dux have diverged considerably at the sequence level, including within the homeodomains, resulting in slightly different DNA-binding motifs and the selective transcriptional induction of species-specific retrotransposons (30). This suggests an evolutionary pressure to maintain activation of the core ancestral gene network while allowing for divergence away from the activation of potentially deleterious retroelements. Understanding these complexities will inform the generation of FSHD model systems, which have generally expressed human DUX4 in non-primate species.
As has been suggested before (24), it is possible that improper expression of the DUX4 early developmental transcriptional regulatory network might be poorly tolerated in skeletal muscle, providing a link between the normal function of DUX4 and its role in the pathogenesis of FSHD. Indeed, it had already been well established that DUX4 activates germline genes, retroelements and immune modulators that define the pre-implantation embryo when mis-expressed in muscle (18,35,36). Whether or how this transcriptional activity might lead to cellular toxicity, and determining the precise mechanisms of toxicity, are areas of active research.
DUX4-Mediated Toxicity
The first report of DUX4 toxicity was from Alberto Rosa’s group (17) and subsequent studies have demonstrated that DUX4 expression causes apoptosis in a multitude of human and mouse cells, including skeletal muscle, both in vitro and in vivo (18–20,37–40). However, the precise pathways responsible for DUX4-induced cytotoxicity are as yet unclear and several studies have identified different mechanisms, all of which might contribute to DUX4 toxicity.
p53
Earlier findings showed that DUX4 toxicity depends on p53 in mouse muscle (20). More recently, it was shown that this requirement for p53 is at least partly context dependent, since p53 knockout in an inducible DUX4 mouse model and P53 ablation in human myoblasts failed to mitigate DUX4-mediated toxicity (19,41). Due to differences in systems used and measurements of toxicity, however, the basis of the cellular context for the role of P53 in DUX4 cytotoxicity remains to be determined.
PAX3/PAX7
Another proposed mechanism of DUX4 toxicity is the impairment of myogenesis by competition of DUX4 with the homeodomain transcription factors PAX3 and PAX7, which are important for muscle development (38,42). In accordance with this hypothesis, it was recently shown that Pax3 or Pax7 overexpression, but not the overexpression of a panel of other related homeodomain factors, could inhibit DUX4 toxicity in mouse cells (43); and that suppression of PAX7 target genes is a hallmark of FSHD (44). Interestingly, a chimeric DUX4 protein with a Pax7 homeodomain substitution retained the ability to provoke cytotoxicity (43). As DUX4 toxicity requires both the ability to bind DNA as well as activate transcription (20,45,46), this finding suggests that there may be a core target or set of targets bound by both DUX4 as well as the DUX4-Pax7 chimeric protein that elicit cellular toxicity.
MYC
A recent genetics-based screen (19) identified two pathways that modulate DUX4-mediated toxicity: the MYC-mediated apoptosis pathway and the double-stranded RNA (dsRNA) antiviral pathway that includes EIF2AK2/PKR and RNASEL (discussed below). Intriguingly, MYC, which is known to cause apoptosis in certain contexts (47), was dramatically upregulated in DUX4-expressing cells via mRNA stabilization (19). This mechanism was not specific to MYC, however, and appears to occur with other labile mRNAs such as FOSB and JUN (19), suggesting that DUX4 might regulate a set of genes involved in broadly altering a metabolic immediate-early response. As a consequence of MYC stabilization, DUX4 might also inhibit mitochondrial activity through the activation of MYC-targeted genes EGR1 and the gamma isoform of BCL2L11/BIM (BIMγ) (19). The enhanced half-life of normally labile mRNAs also suggests that DUX4 can provoke a general effect of globally decreased RNA quality control mechanisms, potentially related to the DUX4-mediated inhibition of the nonsense-mediated decay (NMD) pathway (48). This phenomenon of decreased RNA quality control may further play a role in DUX4-associated RNA/protein aggregation, described below. Whether DUX4 has similar effects in early development remains to be determined, but elevation of MYC and expression of BIMγ has been reported in germline cells (49,50) and pre-implantation embryos (51,52), and MYC RNA levels increase dramatically in human cleavage-stage embryos (see Supplementary Table 2 in 29).
Antiviral response pathways and RNA/protein aggregation
The genetic screen also demonstrated that knockdown of the dsRNA responsive, pro-apoptotic EIF2AK2/PKR and RNASEL at least partially rescued DUX4 toxicity (19). Further investigation revealed that DUX4-induced dsRNA forms aggregates that co-localize with the NMD-associated exon junction component EIF4A3, indicating that DUX4-induced dsRNAs might be spliced, and that EIF4A3 sequestration to DUX4-induced dsRNAs may partially explain NMD inhibition by DUX4, perhaps acting in concert with the DUX4-mediated UPF1 degradation described previously (48). Of interest, aggregation of a potential dsRNA-binding protein, TDP-43 (53) and FUS was also observed in DUX4-expressing cells (54), which parallels protein aggregation in other neuromuscular diseases such as amyotrophic lateral sclerosis. Together, these findings suggest that there may not be a specific target or set of targets that elicit DUX4 cytotoxicity, but rather that stabilization of otherwise labile mRNAs and/or the accumulation of dsRNAs by DUX4 results in RNA/protein aggregation and ultimately the activation of apoptotic pathways. An important next step will be to identify the transcripts which make up DUX4-induced dsRNAs and to explore the consequences of the subset of mRNAs preferentially stabilized following DUX4 expression.
Conservation of toxicity
It was recently shown that human DUX4 and mouse Dux expression, but not the expression of other related homeodomain family members DUX1, DUX5, DUXA or Duxbl, is toxic to human and mouse cells (55). Because DUX4 and Dux are both toxic via mechanisms that depend on their transactivation and DNA-binding domains (20,45,46,55), it follows that a shared target gene, or set of target genes, may be necessary for the induction of DUX4/Dux toxicity. One option for uncovering the pathways necessary for DUX4 toxicity would therefore be to intersect downstream genes commonly targeted by each transcription factor. A recent study took this approach, comparing DUX4 and Dux binding genome-wide via chromatin immunoprecipitation coupled to high-throughput sequencing (55). With a multitude of shared targets, it remains difficult to ascribe a single target, or set of targets, as necessary for DUX4 and Dux toxicity (30,55). More perplexing is that DUX4 expression is toxic in distant, non-mammalian species such as zebrafish and Drosophila melanogaster (56–58). Because the double homeodomain family of genes arose in placental mammals (34), these species evolved without DUX factors and therefore likely have no conserved DUX4 target genes. Thus, it is currently unclear whether a specific DUX4 target gene or set of target genes is necessary for the multi-species DUX4 toxicity, or whether the potent transcriptional activity of DUX4 leads to an abundance of aberrant transcripts and proteins that form toxic aggregates.
DUX4 Mis-Expression in Non-FSHD Diseases
The FSHD modifiers SMCHD1 and DNMT3B not only regulate DUX4, but also have pleiotropic impacts when mutated. Missense mutations in SMCHD1 were recently found in bosma arhinia microphthalmia (BAM) syndrome (59,60), and recessive mutations in DNMT3B are found in patients with immunodeficiency, centromeric instability and facial abnormalities (ICF) syndrome (61,62). In cases of ICF, mutant DNMT3B was associated with a hypomethylated D4Z4 array, making ICF patients more susceptible to DUX4 mis-expression and FSHD if they have the permissive 4q haplotype (12).
D4Z4 translocations to the IgH locus that result in the mis-expression of a truncated DUX4 protein in B-cells have recently emerged as the leading cause of acute lymphoblastic leukemia (ALL) in adolescents and young adults and contribute to leukemogenesis in mice (63–68). In a subset of sarcomas, a translocation fuses the DNA-binding portion of capicua (CIC) with the carboxy-terminal activation domain of DUX4 and results in aberrant gene expression (69). It is interesting to note that the DUX4 protein expressed in these two cancers is different than that found in normal development or in FSHD. In the cases of ALL, the truncated DUX4 transcript encodes a protein that lacks the DUX4 carboxy-terminal activation domain, an isoform that is largely transcriptionally inactive when expressed in skeletal muscle cells (35). In contrast, in the sarcomas, it is the activation domain of DUX4 that is fused to the DNA-binding domain of CIC, presumably activating the expression of genes that are normally repressed by CIC.
Regulators of DUX4 Expression
In most somatic tissues, the D4Z4 arrays appear to be silenced via multiple mechanisms, including DNA methylation, histone modifications and repressive chromatin proteins, which are disrupted in FSHD (11,12,56,70–77) (Fig. 1). Specifically, the D4Z4 repeats are typically CpG hypermethylated, wrapped in histones that contain tri-methylated histone H3 lysine 9 (H3K9me3) and histone H3 lysine 27 (H3K27me3) (repressive chromatin marks associated with heterochromatin), and bound by proteins (like YY1, HP1γ, EZH2 and cohesin) known to silence gene expression. However, a holistic understanding of DUX4 gene regulation is lacking. For example, it is as yet unclear how DUX4 repression is established and maintained in somatic tissues, or how DUX4 is activated in the early embryo.
Recent work used CRISPR-based locus-specific proteomics to characterize proteins that bind the D4Z4 array (77). This approach, combined with gene depletion experiments, revealed that the nucleosome remodeling deacetylase (NuRD) and chromatin assembly factor 1 (CAF-1) complexes function to repress DUX4 expression in human skeletal muscle and induced pluripotent stem cells. Furthermore, these studies uncovered a role for the DUX4-induced expression of the MBD3L family of methyl-CpG-binding proteins (MBD3L2–5) in relieving NuRD complex silencing activity and amplifying DUX4 levels in FSHD muscle cells. Together with the earlier studies, it is becoming clear that D4Z4 silencing is multi-factorial and might have similarities to the silencing of other repetitive elements in the genome, such as mechanisms that repress retroelements (78).
Models of FSHD
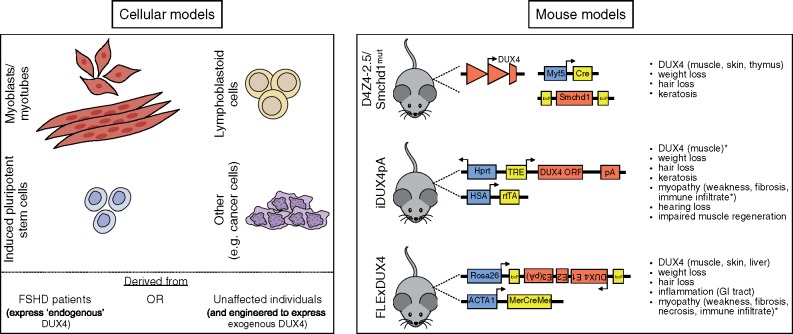
Cell and animal models are indispensable for generating mechanistic understandings of human disease and for testing potential therapeutic approaches in anticipation of clinical trials. Vexingly, the unique complexity of FSHD has slowed the development of model systems that recapitulate the totality of the disease. However, models of specific aspects of FSHD have been developed that can be used to advance pre-clinical studies (8,79) (Fig. 2). The major transcriptional signature associated with FSHD muscle compared to controls is composed of genes regulated by DUX4 (80). Recent studies identifying the transcriptional signature in FSHD myocytes and the DUX4 signature in muscle cell lines engineered to express DUX4 showed a high similarity (18,37), and validates the tissue culture systems for aspects of the study of DUX4 biology and FSHD therapeutic development. Also, 114 lymphoblastoid cell lines from multi-generational FSHD families were recently characterized and present a possible new cellular model for FSHD investigation (81), as does the use of human pluripotent stem cells derived from FSHD patients (15,82) (Fig. 2A).
Figure 2.
Cellular and murine models of FSHD. The normal and pathological functions of DUX4 are currently being investigated using the depicted cell (A) and murine (B) models. Note that DUX4 can be exogenously expressed in cell culture systems via transfection, transduction or stable integration of inducible transgenes. TRE, tetracycline response element; ORF, open reading frame; pA, polyadenylation signal; rtTA, reverse tetracycline-controlled transactivator; E1, exon 1; E2, exon 2; E3, exon 3; MerCreMer, Cre recombinase protein fused to two mutant estrogen-receptor ligand-binding domains; *, phenotype only apparent when DUX4 expression is induced.
There have also been recent advances in the generation of DUX4 mouse models, including modifications which build upon previous systems in an attempt to more closely mimic FSHD pathology (79,83–85). The three most recent murine models, summarized in Figure 2B, all show skin phenotypes, and two show mosaic DUX4 expression and immune infiltrates in muscle as well as muscle wasting, recapitulating several features seen in FSHD. In vivo mouse models provide an opportunity to study cell non-autonomous effects (such as the role of the immune system), making them invaluable for testing therapies. However, one major caveat for the current murine models is the use of human DUX4, which relies on the assumption that the divergence between human DUX4 and mouse Dux has maintained cross-species regulation of the FSHD-relevant mechanisms of pathology despite divergence in binding site motifs and cross-species transcriptional programs (30,55). Because of this divergence, it is possible that the expression of mouse Dux in mouse skeletal muscle might better recapitulate the full scope of the disease transcriptome and mechanisms and provide a model that more broadly recapitulates FSHD.
FSHD Therapeutics
There are currently no effective treatments for FSHD, but therapeutic modalities are being actively explored (Table 1). Consequently, there is an urgent need to establish measurable clinical outcomes that are feasible for a trial timescale. The slowly progressive nature of FSHD has made finding functional outcomes that change predictably and rapidly enough a challenge. Tissue biomarkers may provide one practical approach, and a set of the DUX4 target genes most robustly expressed in FSHD muscle has been identified as candidate biomarkers that require further validation and correlation with clinical severity and progression (80). Serum biomarkers are also being explored (86,87). Additionally, there is much recent effort around using magnetic resonance imaging (MRI) to track FSHD disease progression, and studies have shown a correlation between short TI inversion recovery (STIR) positivity, likely a marker of inflammation, and progression to fibro-fatty infiltration (88–93), suggesting that MRI might be a valuable non-invasive mechanism to identify muscles with active disease.
Table 1.
FSHD therapeutic development
| Therapeutic | Mechanism(s) of action | Tested in patients? | References |
|---|---|---|---|
| Anti-inflammatory | Immunosuppression | Yes | (101,102) |
| Inhibit pathologic processes downstream of DUX4 | |||
| Antioxidant | Prevent oxidative stress | Yes | (103–106) |
| Inhibit pathologic processes downstream of DUX4 | |||
| Antisense RNA | Enhance D4Z4 repression | No | (19,39,76,77,94–96,107,108) |
| Inhibit DUX4 expression | |||
| Inhibit pathologic processes downstream of DUX4 | |||
| BET bromodomain inhibitor | Enhance D4Z4 repression | No | (97) |
| Inhibit DUX4 expression | |||
| Beta-2 adrenergic agonist | Increase muscle strength/mass | Yes | (97,109–111) |
| Enhance D4Z4 repression | |||
| Inhibit DUX4 expression | |||
| Calcium channel blocker | Restore calcium dysregulation | Yes | (112,113) |
| Inhibit pathologic processes downstream of DUX4 | |||
| Exercise | Increase muscle strength/mass | Yes | (114–116) |
| GSK3β inhibitor | Enhance D4Z4 repression | No | (40) |
| Inhibit DUX4 expression | |||
| Myostatin inhibitor | Increase muscle strength/mass | Yes | (117,118) |
| Steroid | Increase muscle strength/mass | Yes | (119) |
| Tissue transplantation | Enhance muscle regeneration | Yes | (120,121) |
| Block DUX4 spreading | |||
| Tyrosine kinase inhibitor | Enhance muscle regeneration | No | (98) |
| Inhibit pathologic processes downstream of DUX4 | |||
| Unknown | Enhance D4Z4 repression | No | (77,99) |
| Inhibit DUX4 expression | |||
| Block DUX4 activity | |||
| Block DUX4 spreading | |||
| Inhibit pathologic processes downstream of DUX4 |
Prior to the consensus that mis-expression of DUX4 is the root cause of FSHD, clinical trials investigated several drug classes hypothesized to improve overall FSHD muscle function but none showed a robust benefit to patients. Current approaches are more targeted, with the ultimate goal of pharmacological control of either DUX4 gene expression or the activity of the DUX4 protein, and include antisense oligonucleotides that target DUX4 mRNA (76,94–96) and high-throughput screens to identify small molecules that block DUX4 expression or activity (97). Endeavors to inhibit downstream effectors of DUX4 (19,98), enhance DUX4-repressive proteins such as SMCHD1 and NuRD (77,99), inhibit activators of DUX4 such as ASH1L and MBD3L2 (70,77), and block the spread of DUX4 through the myofiber (48,77,100) are also avenues of future therapeutic progress. Those targets which are specific to repeat elements or are relatively narrowly expressed present the best options for development. Overall, having multiple FSHD therapeutic candidates in the drug development pipeline makes it imperative to actively prepare for clinical trials, including those that are designed to assess multiple therapies relative to each other.
Conclusions
The last two years have seen remarkable advances in our understanding of DUX4 biology. In addition to illuminating FSHD pathophysiology, the work highlighted here has significant implications for early embryo development, cellular reprogramming and cancer biology. We are hopeful that integrating the insights from scientists in these diverse fields will ultimately lead to a treatment or cure for FSHD.
Acknowledgements
We thank the FSHD families whose participation has been critical for progress. We apologize to those whose work was not cited or discussed due to space limitations.
Conflict of Interest statement. None declared.
Funding
Work on this review was supported by National Institutes of Health R01AR045203 (S.J.T.) and F31NS101773 (R.R.).
References
- 1. Flanigan K.M., Coffeen C.M., Sexton L., Stauffer D., Brunner S., Leppert M.F. (2001) Genetic characterization of a large, historically significant Utah kindred with facioscapulohumeral dystrophy. Neuromuscul. Disord., 11, 525–529. [DOI] [PubMed] [Google Scholar]
- 2. Mostacciuolo M.L., Pastorello E., Vazza G., Miorin M., Angelini C., Tomelleri G., Galluzzi G., Trevisan C.P. (2009) Facioscapulohumeral muscular dystrophy: epidemiological and molecular study in a north-east Italian population sample. Clin. Genet., 75, 550–555. [DOI] [PubMed] [Google Scholar]
- 3. Deenen J.C., Arnts H., van der Maarel S.M., Padberg G.W., Verschuuren J.J., Bakker E., Weinreich S.S., Verbeek A.L., van Engelen B.G. (2014) Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology, 83, 1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Padberg G.W., Lunt P.W., Koch M., Fardeau M. (1991) Diagnostic criteria for facioscapulohumeral muscular dystrophy. Neuromuscul. Disord., 1, 231–234. [DOI] [PubMed] [Google Scholar]
- 5. Goselink R.J.M., Voermans N.C., Okkersen K., Brouwer O.F., Padberg G.W., Nikolic A., Tupler R., Dorobek M., Mah J.K., van Engelen B.G.M. (2017) Early onset facioscapulohumeral dystrophy - a systematic review using individual patient data. Neuromuscul. Disord., 27, 1077–1083. [DOI] [PubMed] [Google Scholar]
- 6. Lutz K.L., Holte L., Kliethermes S.A., Stephan C., Mathews K.D. (2013) Clinical and genetic features of hearing loss in facioscapulohumeral muscular dystrophy. Neurology, 81, 1374–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Statland J.M., Sacconi S., Farmakidis C., Donlin-Smith C.M., Chung M., Tawil R. (2013) Coats syndrome in facioscapulohumeral dystrophy type 1: frequency and D4Z4 contraction size. Neurology, 80, 1247–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tawil R., van der Maarel S.M., Tapscott S.J. (2014) Facioscapulohumeral dystrophy: the path to consensus on pathophysiology. Skelet. Muscle, 4, 12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deutekom J.C.T.V., Wljmenga C., Tlenhoven E.A.E.V., Gruter A.-M., Hewitt J.E., Padberg G.W., Ommen G-JBv., Hofker M.H., Fronts R.R. (1993) FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet., 2, 2037–2042. [DOI] [PubMed] [Google Scholar]
- 10. Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.-M., Hofker M.H., Moerer P., Williamson R.. et al. (1992) Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet., 2, 26–30. [DOI] [PubMed] [Google Scholar]
- 11. Lemmers R.J., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W., Amell A.M., van der Vliet P.J., Almomani R., Straasheijm K.R.. et al. (2012) Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet., 44, 1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van den Boogaard M.L., Lemmers R., Balog J., Wohlgemuth M., Auranen M., Mitsuhashi S., van der Vliet P.J., Straasheijm K.R., van den Akker R.F.P., Kriek M.. et al. (2016) Mutations in DNMT3B Modify Epigenetic Repression of the D4Z4 Repeat and the Penetrance of Facioscapulohumeral Dystrophy. Am. J. Hum. Genet., 98, 1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lemmers R.J., van der Vliet P.J., Klooster R., Sacconi S., Camano P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W.. et al. (2010) A unifying genetic model for facioscapulohumeral muscular dystrophy. Science, 329, 1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van der Maarel S.M., Tawil R., Tapscott S.J. (2011) Facioscapulohumeral muscular dystrophy and DUX4: breaking the silence. Trends Mol. Med., 17, 252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Snider L., Geng L.N., Lemmers R.J., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J.. et al. (2010) Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet., 6, e1001181.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Daxinger L., Tapscott S.J., van der Maarel S.M. (2015) Genetic and epigenetic contributors to FSHD. Curr. Opin. Genet. Dev., 33, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kowaljow V., Marcowycz A., Ansseau E., Conde C.B., Sauvage S., Matteotti C., Arias C., Corona E.D., Nunez N.G., Leo O.. et al. (2007) The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord., 17, 611–623. [DOI] [PubMed] [Google Scholar]
- 18. Rickard A.M., Petek L.M., Miller D.G. (2015) Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum. Mol. Genet., 24, 5901–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shadle S.C., Zhong J.W., Campbell A.E., Conerly M.L., Jagannathan S., Wong C.J., Morello T.D., van der Maarel S.M., Tapscott S.J. (2017) DUX4-induced dsRNA and MYC mRNA stabilization activate apoptotic pathways in human cell models of facioscapulohumeral dystrophy. PLoS Genet., 13, e1006658.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wallace L.M., Garwick S.E., Mei W., Belayew A., Coppee F., Ladner K.J., Guttridge D., Yang J., Harper S.Q. (2011) DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol., 69, 540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tawil R., Van Der Maarel S.M. (2006) Facioscapulohumeral muscular dystrophy. Muscle Nerve, 34, 1–15. [DOI] [PubMed] [Google Scholar]
- 22. de Greef J.C., Frants R.R., van der Maarel S.M. (2008) Epigenetic mechanisms of facioscapulohumeral muscular dystrophy. Mutat. Res., 647, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Statland J.M., Tawil R. (2011) Facioscapulohumeral muscular dystrophy: molecular pathological advances and future directions. Curr. Opin. Neurol., 24, 423–428. [DOI] [PubMed] [Google Scholar]
- 24. van der Maarel S.M., Miller D.G., Tawil R., Filippova G.N., Tapscott S.J. (2012) Facioscapulohumeral muscular dystrophy: consequences of chromatin relaxation. Curr. Opin. Neurol., 25, 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hewitt J.E. (2015) Loss of epigenetic silencing of the DUX4 transcription factor gene in facioscapulohumeral muscular dystrophy. Hum. Mol. Genet., 24, R17–R23. [DOI] [PubMed] [Google Scholar]
- 26. Wang L.H., Tawil R. (2016) Facioscapulohumeral Dystrophy. Curr. Neurol. Neurosci. Rep., 16, 66.. [DOI] [PubMed] [Google Scholar]
- 27. Himeda C.L., Jones T.I., Jones P.L. (2015) Facioscapulohumeral muscular dystrophy as a model for epigenetic regulation and disease. Antioxid. Redox. Signal., 22, 1463–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Das S., Chadwick B.P. (2016) Influence of repressive histone and DNA methylation upon D4Z4 transcription in non-myogenic cells. PLoS One, 11, e0160022.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hendrickson P.G., Dorais J.A., Grow E.J., Whiddon J.L., Lim J.W., Wike C.L., Weaver B.D., Pflueger C., Emery B.R., Wilcox A.L.. et al. (2017) Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet., 49, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Whiddon J.L., Langford A.T., Wong C.J., Zhong J.W., Tapscott S.J. (2017) Conservation and innovation in the DUX4-family gene network. Nat. Genet., 49, 935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. De Iaco A., Planet E., Coluccio A., Verp S., Duc J., Trono D. (2017) DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet., 49, 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leidenroth A., Clapp J., Mitchell L.M., Coneyworth D., Dearden F.L., Iannuzzi L., Hewitt J.E. (2012) Evolution of DUX gene macrosatellites in placental mammals. Chromosoma, 121, 489–497. [DOI] [PubMed] [Google Scholar]
- 33. Leidenroth A., Hewitt J.E. (2010) A family history of DUX4: phylogenetic analysis of DUXA, B, C and Duxbl reveals the ancestral DUX gene. BMC Evol. Biol., 10, 364.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clapp J., Mitchell L.M., Bolland D.J., Fantes J., Corcoran A.E., Scotting P.J., Armour J.A., Hewitt J.E. (2007) Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am J Hum. Genet., 81, 264–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R.. et al. (2012) DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell, 22, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Young J.M., Whiddon J.L., Yao Z., Kasinathan B., Snider L., Geng L.N., Balog J., Tawil R., van der Maarel S.M., Tapscott S.J. (2013) DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet., 9, e1003947.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jagannathan S., Shadle S.C., Resnick R., Snider L., Tawil R.N., van der Maarel S.M., Bradley R.K., Tapscott S.J. (2016) Model systems of DUX4 expression recapitulate the transcriptional profile of FSHD cells. Hum. Mol. Genet., 25, 4419–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bosnakovski D., Xu Z., Gang E.J., Galindo C.L., Liu M., Simsek T., Garner H.R., Agha-Mohammadi S., Tassin A., Coppee F.. et al. (2008) An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J., 27, 2766–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vanderplanck C., Ansseau E., Charron S., Stricwant N., Tassin A., Laoudj-Chenivesse D., Wilton S.D., Coppee F., Belayew A. (2011) The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS One, 6, e26820.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Block G.J., Narayanan D., Amell A.M., Petek L.M., Davidson K.C., Bird T.D., Tawil R., Moon R.T., Miller D.G. (2013) Wnt/beta-catenin signaling suppresses DUX4 expression and prevents apoptosis of FSHD muscle cells. Hum. Mol. Genet., 22, 4661–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bosnakovski D., Gearhart M.D., Toso E.A., Recht O.O., Cucak A., Jain A.K., Barton M.C., Kyba M. (2017) p53-independent DUX4 pathology in cell and animal models of facioscapulohumeral muscular dystrophy. Dis. Model. Mech., 10, 1211–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bosnakovski D., Daughters R.S., Xu Z., Slack J.M., Kyba M. (2009) Biphasic myopathic phenotype of mouse DUX, an ORF within conserved FSHD-related repeats. PLoS One, 4, e7003.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bosnakovski D., Toso E.A., Hartweck L.M., Magli A., Lee H.A., Thompson E.R., Dandapat A., Perlingeiro R.C.R., Kyba M. (2017) The DUX4 homeodomains mediate inhibition of myogenesis and are functionally exchangeable with the Pax7 homeodomain. J. Cell Sci., 130, 3685–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Banerji C.R.S., Panamarova M., Hebaishi H., White R.B., Relaix F., Severini S., Zammit P.S. (2017) PAX7 target genes are globally repressed in facioscapulohumeral muscular dystrophy skeletal muscle. Nat. Commun., 8, 2152.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Geng L.N., Tyler A.E., Tapscott S.J. (2011) Immunodetection of human double homeobox 4. Hybridoma (Larchmt), 30, 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Corona E.D., Jacquelin D., Gatica L., Rosa A.L. (2013) Multiple protein domains contribute to nuclear import and cell toxicity of DUX4, a candidate pathogenic protein for facioscapulohumeral muscular dystrophy. PLoS One, 8, e75614.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McMahon S.B. (2014) MYC and the control of apoptosis. Cold Spring Harb. Perspect. Med., 4, a014407.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Feng Q., Snider L., Jagannathan S., Tawil R., van der Maarel S.M., Tapscott S.J., Bradley R.K. (2015) A feedback loop between nonsense-mediated decay and the retrogene DUX4 in facioscapulohumeral muscular dystrophy. Elife, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu J.W., Chandra D., Tang S.H., Chopra D., Tang D.G. (2002) Identification and characterization of Bimgamma, a novel proapoptotic BH3-only splice variant of Bim. Cancer Res., 62, 2976–2981. [PubMed] [Google Scholar]
- 50. Percharde M., Wong P., Ramalho-Santos M. (2017) Global hypertranscription in the mouse embryonic germline. Cell Rep., 19, 1987–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boumela I., Assou S., Aouacheria A., Haouzi D., Dechaud H., De Vos J., Handyside A., Hamamah S. (2011) Involvement of BCL2 family members in the regulation of human oocyte and early embryo survival and death: gene expression and beyond. Reproduction, 141, 549–561. [DOI] [PubMed] [Google Scholar]
- 52. Scognamiglio R., Cabezas-Wallscheid N., Thier M.C., Altamura S., Reyes A., Prendergast A.M., Baumgartner D., Carnevalli L.S., Atzberger A., Haas S.. et al. (2016) Myc depletion induces a pluripotent dormant state mimicking diapause. Cell, 164, 668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Saldi T.K., Ash P.E., Wilson G., Gonzales P., Garrido-Lecca A., Roberts C.M., Dostal V., Gendron T.F., Stein L.D., Blumenthal T.. et al. (2014) TDP-1, the Caenorhabditis elegans ortholog of TDP-43, limits the accumulation of double-stranded RNA. EMBO J., 33, 2947–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Homma S., Beermann M.L., Boyce F.M., Miller J.B. (2015) Expression of FSHD-related DUX4-FL alters proteostasis and induces TDP-43 aggregation. Ann. Clin. Transl. Neurol., 2, 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Eidahl J.O., Giesige C.R., Domire J.S., Wallace L.M., Fowler A.M., Guckes S.M., Garwick-Coppens S.E., Labhart P., Harper S.Q. (2016) Mouse Dux is myotoxic and shares partial functional homology with its human paralog DUX4. Hum. Mol. Genet., 25, 4577–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Snider L., Asawachaicharn A., Tyler A.E., Geng L.N., Petek L.M., Maves L., Miller D.G., Lemmers R.J., Winokur S.T., Tawil R.. et al. (2009) RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum. Mol. Genet., 18, 2414–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mitsuhashi H., Mitsuhashi S., Lynn-Jones T., Kawahara G., Kunkel L.M. (2013) Expression of DUX4 in zebrafish development recapitulates facioscapulohumeral muscular dystrophy. Hum. Mol. Genet., 22, 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jones T.I., Parilla M., Jones P.L. (2016) Transgenic drosophila for investigating DUX4 and FRG1, two genes associated with facioscapulohumeral muscular dystrophy (FSHD). PLoS One, 11, e0150938.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gordon C.T., Xue S., Yigit G., Filali H., Chen K., Rosin N., Yoshiura K.I., Oufadem M., Beck T.J., McGowan R.. et al. (2017) De novo mutations in SMCHD1 cause bosma arhinia microphthalmia syndrome and abrogate nasal development. Nat. Genet., 49, 249–255. [DOI] [PubMed] [Google Scholar]
- 60. Shaw N.D., Brand H., Kupchinsky Z.A., Bengani H., Plummer L., Jones T.I., Erdin S., Williamson K.A., Rainger J., Stortchevoi A.. et al. (2017) SMCHD1 mutations associated with a rare muscular dystrophy can also cause isolated arhinia and Bosma arhinia microphthalmia syndrome. Nat. Genet., 49, 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hansen R.S., Wijmenga C., Luo P., Stanek A.M., Canfield T.K., Weemaes C.M., Gartler S.M. (1999) The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl. Acad. Sci. U.S.A., 96, 14412–14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jin B., Tao Q., Peng J., Soo H.M., Wu W., Ying J., Fields C.R., Delmas A.L., Liu X., Qiu J.. et al. (2008) DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum. Mol. Genet., 17, 690–709. [DOI] [PubMed] [Google Scholar]
- 63. Dong X., Zhang W., Wu H., Huang J., Zhang M., Wang P., Zhang H., Chen Z., Chen S.J., Meng G. (2018) Structural basis of DUX4/IGH-driven transactivation. Leukemia, doi: 10.1038/s41375-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Marincevic-Zuniga Y., Dahlberg J., Nilsson S., Raine A., Nystedt S., Lindqvist C.M., Berglund E.C., Abrahamsson J., Cavelier L., Forestier E.. et al. (2017) Transcriptome sequencing in pediatric acute lymphoblastic leukemia identifies fusion genes associated with distinct DNA methylation profiles. J. Hematol. Oncol., 10, 148.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yasuda T., Tsuzuki S., Kawazu M., Hayakawa F., Kojima S., Ueno T., Imoto N., Kohsaka S., Kunita A., Doi K.. et al. (2016) Recurrent DUX4 fusions in B cell acute lymphoblastic leukemia of adolescents and young adults. Nat. Genet., 48, 569–574. [DOI] [PubMed] [Google Scholar]
- 66. Zhang J., McCastlain K., Yoshihara H., Xu B., Chang Y., Churchman M.L., Wu G., Li Y., Wei L., Iacobucci I.. et al. (2016) Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat. Genet., 48, 1481–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lilljebjorn H., Henningsson R., Hyrenius-Wittsten A., Olsson L., Orsmark-Pietras C., von Palffy S., Askmyr M., Rissler M., Schrappe M., Cario G.. et al. (2016) Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat. Commun., 7, 11790.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liu Y.F., Wang B.Y., Zhang W.N., Huang J.Y., Li B.S., Zhang M., Jiang L., Li J.F., Wang M.J., Dai Y.J.. et al. (2016) Genomic profiling of adult and pediatric B-cell acute lymphoblastic leukemia. EBioMedicine, 8, 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kawamura-Saito M., Yamazaki Y., Kaneko K., Kawaguchi N., Kanda H., Mukai H., Gotoh T., Motoi T., Fukayama M., Aburatani H.. et al. (2006) Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4; 19)(q35; q13) translocation. Hum. Mol. Genet., 15, 2125–2137. [DOI] [PubMed] [Google Scholar]
- 70. Cabianca D.S., Casa V., Bodega B., Xynos A., Ginelli E., Tanaka Y., Gabellini D. (2012) A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell, 149, 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Huichalaf C., Micheloni S., Ferri G., Caccia R., Gabellini D. (2014) DNA methylation analysis of the macrosatellite repeat associated with FSHD muscular dystrophy at single nucleotide level. PLoS One, 9, e115278.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. van Overveld P.G., Lemmers R.J., Sandkuijl L.A., Enthoven L., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. (2003) Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet., 35, 315–317. [DOI] [PubMed] [Google Scholar]
- 73. Zeng W., de Greef J.C., Chen Y.Y., Chien R., Kong X., Gregson H.C., Winokur S.T., Pyle A., Robertson K.D., Schmiesing J.A.. et al. (2009) Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet., 5, e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gabellini D., Green M.R., Tupler R. (2002) Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell, 110, 339–348. [DOI] [PubMed] [Google Scholar]
- 75. Ottaviani A., Rival-Gervier S., Boussouar A., Foerster A.M., Rondier D., Sacconi S., Desnuelle C., Gilson E., Magdinier F. (2009) The D4Z4 macrosatellite repeat acts as a CTCF and A-type lamins-dependent insulator in facio-scapulo-humeral dystrophy. PLoS Genet., 5, e1000394.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lim J.W., Snider L., Yao Z., Tawil R., Van Der Maarel S.M., Rigo F., Bennett C.F., Filippova G.N., Tapscott S.J. (2015) DICER/AGO-dependent epigenetic silencing of D4Z4 repeats enhanced by exogenous siRNA suggests mechanisms and therapies for FSHD. Hum Mol Genet, 24, 4817–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Campbell A.E., Shadle S.C., Jagannathan S., Lim J.W., Resnick R., Tawil R., van der Maarel S.M., Tapscott S.J. (2018) NuRD and CAF-1-mediated silencing of the D4Z4 array is modulated by DUX4-induced MBD3L proteins. Elife, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yang B.X., El Farran C.A., Guo H.C., Yu T., Fang H.T., Wang H.F., Schlesinger S., Seah Y.F., Goh G.Y., Neo S.P.. et al. (2015) Systematic identification of factors for provirus silencing in embryonic stem cells. Cell, 163, 230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lek A., Rahimov F., Jones P.L., Kunkel L.M. (2015) Emerging preclinical animal models for FSHD. Trends Mol. Med., 21, 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yao Z., Snider L., Balog J., Lemmers R.J., Van Der Maarel S.M., Tawil R., Tapscott S.J. (2014) DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum. Mol. Genet., 23, 5342–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jones T.I., Himeda C.L., Perez D.P., Jones P.L. (2017) Large family cohorts of lymphoblastoid cells provide a new cellular model for investigating facioscapulohumeral muscular dystrophy. Neuromuscul. Disord., 27, 221–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Caron L., Kher D., Lee K.L., McKernan R., Dumevska B., Hidalgo A., Li J., Yang H., Main H., Ferri G.. et al. (2016) A human pluripotent stem cell model of facioscapulohumeral muscular dystrophy-affected skeletal muscles. Stem Cells Transl. Med., 5, 1145–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. de Greef J.C., Krom Y.D., den Hamer B., Snider L., Hiramuki Y., van den Akker R.F.P., Breslin K., Pakusch M., Salvatori D.C.F., Slutter B.. et al. (2018) Smchd1 haploinsufficiency exacerbates the phenotype of a transgenic FSHD1 mouse model. Hum. Mol. Genet., 27, 716–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bosnakovski D., Chan S.S.K., Recht O.O., Hartweck L.M., Gustafson C.J., Athman L.L., Lowe D.A., Kyba M. (2017) Muscle pathology from stochastic low level DUX4 expression in an FSHD mouse model. Nat. Commun., 8, 550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jones T., Jones P.L. (2018) A cre-inducible DUX4 transgenic mouse model for investigating facioscapulohumeral muscular dystrophy. PLoS One, 13, e0192657.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Statland J., Donlin-Smith C.M., Tapscott S.J., van der Maarel S.M., Tawil R. (2014) Multiplex screen of serum biomarkers in facioscapulohumeral muscular dystrophy. J. Neuromuscul. Dis., 1, 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Petek L.M., Rickard A.M., Budech C., Poliachik S.L., Shaw D., Ferguson M.R., Tawil R., Friedman S.D., Miller D.G. (2016) A cross sectional study of two independent cohorts identifies serum biomarkers for facioscapulohumeral muscular dystrophy (FSHD). Neuromuscul. Disord., 26, 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mul K., van den Boogaard M.L., van der Maarel S.M., van Engelen B.G. (2016) Integrating clinical and genetic observations in facioscapulohumeral muscular dystrophy. Curr. Opin. Neurol., 29, 606–613. [DOI] [PubMed] [Google Scholar]
- 89. Mul K., Vincenten S.C.C., Voermans N.C., Lemmers R., van der Vliet P.J., van der Maarel S.M., Padberg G.W., Horlings C.G.C., van Engelen B.G.M. (2017) Adding quantitative muscle MRI to the FSHD clinical trial toolbox. Neurology, 89, 2057–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ferguson M.R., Poliachik S.L., Budech C.B., Gove N.E., Carter G.T., Wang L.H., Miller D.G., Shaw D.W.W., Friedman S.D. (2017) MRI change metrics of facioscapulohumeral muscular dystrophy: stir and T1. Muscle Nerve, doi: 10.1002/mus.26038. [DOI] [PubMed] [Google Scholar]
- 91. Friedman S.D., Poliachik S.L., Otto R.K., Carter G.T., Budech C.B., Bird T.D., Miller D.G., Shaw D.W. (2014) Longitudinal features of STIR bright signal in FSHD. Muscle Nerve, 49, 257–260. [DOI] [PubMed] [Google Scholar]
- 92. Tasca G., Monforte M., Iannaccone E., Laschena F., Ottaviani P., Leoncini E., Boccia S., Galluzzi G., Pelliccioni M., Masciullo M.. et al. (2014) Upper girdle imaging in facioscapulohumeral muscular dystrophy. PLoS One, 9, e100292.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tasca G., Pescatori M., Monforte M., Mirabella M., Iannaccone E., Frusciante R., Cubeddu T., Laschena F., Ottaviani P., Ricci E. (2012) Different molecular signatures in magnetic resonance imaging-staged facioscapulohumeral muscular dystrophy muscles. PLoS One, 7, e38779.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ansseau E., Vanderplanck C., Wauters A., Harper S.Q., Coppee F., Belayew A. (2017) Antisense oligonucleotides used to target the DUX4 mRNA as therapeutic approaches in faciosscapulohumeral muscular dystrophy (FSHD). Genes (Basel), 8, 93.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chen J.C., King O.D., Zhang Y., Clayton N.P., Spencer C., Wentworth B.M., Emerson C.P. Jr., Wagner K.R. (2016) Morpholino-mediated knockdown of DUX4 toward facioscapulohumeral muscular dystrophy therapeutics. Mol. Ther., 24, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Marsollier A.C., Ciszewski L., Mariot V., Popplewell L., Voit T., Dickson G., Dumonceaux J. (2016) Antisense targeting of 3′ end elements involved in DUX4 mRNA processing is an efficient therapeutic strategy for facioscapulohumeral dystrophy: a new gene-silencing approach. Hum. Mol. Genet., 25, 1468–1478. [DOI] [PubMed] [Google Scholar]
- 97. Campbell A.E., Oliva J., Yates M.P., Zhong J.W., Shadle S.C., Snider L., Singh N., Tai S., Hiramuki Y., Tawil R.. et al. (2017) BET bromodomain inhibitors and agonists of the beta-2 adrenergic receptor identified in screens for compounds that inhibit DUX4 expression in FSHD muscle cells. Skelet. Muscle, 7, 16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Moyle L.A., Blanc E., Jaka O., Prueller J., Banerji C.R., Tedesco F.S., Harridge S.D., Knight R.D., Zammit P.S. (2016) Ret function in muscle stem cells points to tyrosine kinase inhibitor therapy for facioscapulohumeral muscular dystrophy. Elife, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Balog J., Thijssen P.E., Shadle S., Straasheijm K.R., van der Vliet P.J., Krom Y.D., van den Boogaard M.L., de Jong A., F Lemmers R.J.L., Tawil R.. et al. (2015) Increased DUX4 expression during muscle differentiation correlates with decreased SMCHD1 protein levels at D4Z4. Epigenetics, 10, 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Tassin A., Laoudj-Chenivesse D., Vanderplanck C., Barro M., Charron S., Ansseau E., Chen Y.W., Mercier J., Coppee F., Belayew A. (2013) DUX4 expression in FSHD muscle cells: how could such a rare protein cause a myopathy? J. Cell Mol. Med., 17, 76–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Walker G., Butterfield R., Mathews K., Servais L., Day J., Gidaro T., Shukla S., Maggi L. (2017) Results of a phase 1b/2 study of ATYR1940 in adolescents and young adults with early onset facioscapulohumeral muscular dystrophy (FSHD) (ATYR1940-C-003). Neuromuscul. Disord., 27, S199. [Google Scholar]
- 102. Gershman A., Chiang K., Do M., Abbink E., Harbers V., Audebert C., Campana-Salort E., Monforte M., Iyadurai S., Carey L.. et al. (2016) A randomized, double-blinded, placebo-controlled, multiple ascending dose study to evaluate the safety, tolerability, pharmacokinetics, immunogenicity, and biological activity of ATYR1940 in adult patients with facioscapulohumeral muscular dystrophy (FSHD). Neuromuscul. Disord., 26, S167. [Google Scholar]
- 103. Passerieux E., Hayot M., Jaussent A., Carnac G., Gouzi F., Pillard F., Picot M.C., Bocker K., Hugon G., Pincemail J.. et al. (2015) Effects of vitamin C, vitamin E, zinc gluconate, and selenomethionine supplementation on muscle function and oxidative stress biomarkers in patients with facioscapulohumeral dystrophy: a double-blind randomized controlled clinical trial. Free Radic. Biol. Med., 81, 158–169. [DOI] [PubMed] [Google Scholar]
- 104. van der Kooi E.L., de Greef J.C., Wohlgemuth M., Frants R.R., van Asseldonk R.J., Blom H.J., van Engelen B.G., van der Maarel S.M., Padberg G.W. (2006) No effect of folic acid and methionine supplementation on D4Z4 methylation in patients with facioscapulohumeral muscular dystrophy. Neuromuscul. Disord., 16, 766–769. [DOI] [PubMed] [Google Scholar]
- 105. Bosnakovski D., Choi S.H., Strasser J.M., Toso E.A., Walters M.A., Kyba M. (2014) High-throughput screening identifies inhibitors of DUX4-induced myoblast toxicity. Skelet. Muscle, 4, 4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Dmitriev P., Bou Saada Y., Dib C., Ansseau E., Barat A., Hamade A., Dessen P., Robert T., Lazar V., Louzada R.A.N.. et al. (2016) DUX4-induced constitutive DNA damage and oxidative stress contribute to aberrant differentiation of myoblasts from FSHD patients. Free Radic. Biol. Med., 99, 244–258. [DOI] [PubMed] [Google Scholar]
- 107. Wallace L.M., Liu J., Domire J.S., Garwick-Coppens S.E., Guckes S.M., Mendell J.R., Flanigan K.M., Harper S.Q. (2012) RNA interference inhibits DUX4-induced muscle toxicity in vivo: implications for a targeted FSHD therapy. Mol. Ther., 20, 1417–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bortolanza S., Nonis A., Sanvito F., Maciotta S., Sitia G., Wei J., Torrente Y., Di Serio C., Chamberlain J.R., Gabellini D. (2011) AAV6-mediated systemic shRNA delivery reverses disease in a mouse model of facioscapulohumeral muscular dystrophy. Mol. Ther., 19, 2055–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kissel J.T., McDermott M.P., Mendell J.R., King W.M., Pandya S., Griggs R.C., Tawil R., Group F.-D. (2001) Randomized, double-blind, placebo-controlled trial of albuterol in facioscapulohumeral dystrophy. Neurology, 57, 1434–1440. [DOI] [PubMed] [Google Scholar]
- 110. Payan C.A., Hogrel J.Y., Hammouda E.H., Lacomblez L., Ollivier G., Doppler V., Eymard B., Attarian S., Pouget J., Desnuelle C.. et al. (2009) Periodic salbutamol in facioscapulohumeral muscular dystrophy: a randomized controlled trial. Arch. Phys. Med. Rehabil., 90, 1094–1101. [DOI] [PubMed] [Google Scholar]
- 111. van der Kooi E.L., Vogels O.J., van Asseldonk R.J., Lindeman E., Hendriks J.C., Wohlgemuth M., van der Maarel S.M., Padberg G.W. (2004) Strength training and albuterol in facioscapulohumeral muscular dystrophy. Neurology, 63, 702–708. [DOI] [PubMed] [Google Scholar]
- 112. Lefkowitz D.L., Lefkowitz S.S. (2005) Fascioscapulohumeral muscular dystrophy: a progressive degenerative disease that responds to diltiazem. Med. Hypotheses, 65, 716–721. [DOI] [PubMed] [Google Scholar]
- 113. Elsheikh B.H., Bollman E., Peruggia M., King W., Galloway G., Kissel J.T. (2007) Pilot trial of diltiazem in facioscapulohumeral muscular dystrophy. Neurology, 68, 1428–1429. [DOI] [PubMed] [Google Scholar]
- 114. Bankole L.C., Millet G.Y., Temesi J., Bachasson D., Ravelojaona M., Wuyam B., Verges S., Ponsot E., Antoine J.C., Kadi F.. et al. (2016) Safety and efficacy of a 6-month home-based exercise program in patients with facioscapulohumeral muscular dystrophy: a randomized controlled trial. Medicine, 95, e4497.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Olsen D.B., Orngreen M.C., Vissing J. (2005) Aerobic training improves exercise performance in facioscapulohumeral muscular dystrophy. Neurology, 64, 1064–1066. [DOI] [PubMed] [Google Scholar]
- 116. Voet N., Bleijenberg G., Hendriks J., de Groot I., Padberg G., van Engelen B., Geurts A. (2014) Both aerobic exercise and cognitive-behavioral therapy reduce chronic fatigue in FSHD: an RCT. Neurology, 83, 1914–1922. [DOI] [PubMed] [Google Scholar]
- 117. Wagner K.R., Fleckenstein J.L., Amato A.A., Barohn R.J., Bushby K., Escolar D.M., Flanigan K.M., Pestronk A., Tawil R., Wolfe G.I.. et al. (2008) A phase I/IItrial of MYO-029 in adult subjects with muscular dystrophy. Ann. Neurol., 63, 561–571. [DOI] [PubMed] [Google Scholar]
- 118. Acceleron Pharma. A phase 2 randomized, double-blind, placebo-controlled study of ACE-083 in patients with facioscapulohumeral muscular dystrophy. Available from: https://clinicaltrials.gov/ct2/show/record/NCT02927080. ClinicalTrials.gov Identifier: NCT02927080. Accessed April 20, 2018.
- 119. Tawil R., McDermott M.P., Pandya S., King W., Kissel J., Mendell J.R., Griggs R.C.. FSH-DY Group (1997) A pilot trial of prednisone in facioscapulohumeral muscular dystrophy. Neurology, 48, 46–49. [DOI] [PubMed] [Google Scholar]
- 120. Dib C., Bou Saada Y., Dmitriev P., Richon C., Dessen P., Laoudj-Chenivesse D., Carnac G., Lipinski M., Vassetzky Y.S. (2016) Correction of the FSHD myoblast differentiation defect by fusion with healthy myoblasts. J. Cell Physiol., 231, 62–71. [DOI] [PubMed] [Google Scholar]
- 121. Royan Institute. Intramuscular transplantation of autologous muscle derived stem cell (MDSC) and adipose derived mesenchymal stem cells (AD-MSC) in patients with facioscapulohumeral dystrophy (FSHD). Available from: https://clinicaltrials.gov/ct2/show/study/NCT02208713. Accessed April 20, 2018.