Abstract
We report a broadly applicable enzyme digestion strategy for introducing structure-switching functionality into small-molecule-binding aptamers. This procedure is based on our discovery that exonuclease III (Exo III) digestion of aptamers is greatly inhibited by target binding. As a demonstration, we perform Exo III digestion of a pre-folded three-way-junction (TWJ)-structured cocaine-binding aptamer and a stem–loop-structured ATP-binding aptamer. In the absence of target, Exo III catalyzes 3′-to-5′ digestion of both aptamers to form short, single-stranded products. Upon addition of target, Exo III digestion is halted four bases prior to the target-binding domain, forming a major target-bound aptamer digestion product. We demonstrated that target-binding is crucial for Exo III inhibition. We then determine that the resulting digestion products of both aptamers exhibit a target-induced structure-switching functionality that is absent in the parent aptamer, while still retaining high target-binding affinity. We confirm that these truncated aptamers have this functionality by using an exonuclease I-based digestion assay and further evaluate this characteristic in an electrochemical aptamer-based cocaine sensor and a fluorophore-quencher ATP assay. We believe our Exo III-digestion method should be applicable for the generation of structure-switching aptamers from other TWJ- or stem–loop-containing small-molecule-binding aptamers, greatly simplifying the generation of functionalized sensor elements for folding-based aptasensors.
INTRODUCTION
Aptamers are nucleic acid-based affinity reagents that can be isolated via in vitro systematic evolution of ligands by exponential enrichment (SELEX) (1,2) to bind to various targets. They have gained considerable attention as bio-recognition elements with diverse applications in areas such as drug screening (3), medical diagnostics (4,5) and environmental monitoring (6). This is in part because aptamers are chemically stable and can be synthesized at a low cost with high reproducibility (7,8). Additionally, aptamers can be engineered to have tunable target-binding affinities (9,10) or various functionalities (3,11–13). Most aptamer-based assays for small molecule detection utilize structure-switching aptamers, which undergo a conformational change upon target binding (3). However, a majority of aptamers do not have inherent structure-switching functionality, and a multistep process is required to introduce this functionality for sensor development. First, the target-binding domain must be identified, usually through a laborious truncation process (14–16). Afterwards, structure-switching functionality is introduced via sequence engineering (17), splitting (11) or utilization of a complementary strand that partially blocks the aptamer binding site (18). These methods are labor-intensive and require considerable trial and error. Moreover, engineered structure-switching aptamers often have low target-binding affinities, which greatly limits their utility as sensing elements.
We have discovered that exonuclease III (Exo III)-mediated digestion of small-molecule-binding aptamers can be inhibited by target binding, and that the resulting truncation products have structure-switching functionality. Exo III exhibits 3′-to-5′ exonuclease activity on double-stranded DNA, with low activity against single-stranded DNA (19). It has been recently determined that the binding of small molecules, such as minor-groove binders, to double-stranded DNA can stop Exo III digestion a few bases prior to the binding site (20,21). We have confirmed that this also holds true for small-molecule-aptamer complexes, wherein the enzyme preferentially digests non-target-bound aptamers, while digestion of target-bound aptamers is hindered. As a demonstration, we performed Exo III digestion of a pre-folded three-way-junction (TWJ)-structured cocaine-binding aptamer (38-GT) (10) and a stem–loop-structured ATP-binding aptamer (ATP-33) (22). In the absence of target, Exo III catalyzed 3′-to-5′ digestion of both aptamers by the stepwise removal of mononucleotides, forming short, single-stranded products. Upon addition of target, Exo III digestion is halted four bases prior to the target-binding domain. Our results showed that target-binding is crucial for Exo III inhibition, and the concentration of the truncated digestion product increased with increasing concentrations of target. To utilize this concept for practical use, we developed a label-free fluorescence assay to quantify cocaine concentrations based on the extent of aptamer digestion, using SYBR Green I as a signal reporter. This assay achieved a limit of detection of 1 μM cocaine in 10% saliva within 20 min.
Importantly, we observed that the resulting digestion products of both aptamers exhibited a target-induced structure-switching functionality that was absent in the parent aptamer, while still retaining high target-binding affinity. 38-GT and ATP-33 are fully folded even in the absence of target, and both aptamers contain a seven-base-pair stem. Exo III digestion destabilizes both aptamers by truncating their stems to four base-pairs, enabling them to undergo large target-induced conformational changes. This structure-switching functionality was confirmed by digesting the truncated aptamers with exonuclease I (Exo I), a single-strand DNA-specific exonuclease, in the presence and absence of target. We evaluated the functionality of these resulting aptamers by incorporating them into an electrochemical aptamer-based (E-AB) sensor and a fluorophore-quencher assay, both of which demonstrated target-induced conformational changes within the aptamers and achieved excellent sensor performance. We believe that in general, Exo III digestion of existing or future isolated target-bound TWJ- or stem–loop-structured aptamers will consistently be inhibited a few nucleotides prior to their target-binding domain. The resulting truncated aptamers should likewise exhibit structure-switching functionality, enabling serving as ideal biorecognition elements for rapid development of folding-based assays for the detection of various small-molecule targets.
MATERIALS AND METHODS
Reagents
Exonuclease III and exonuclease I were purchased from New England Biolabs. Cocaine hydrochloride, cocaethylene, methylecgonidine, diphenhydramine, nicotine, scopolamine, and adensosine triphosphate (ATP) were purchased from Sigma-Aldrich. Benzoylecgonine tetrahydrate was purchased from Cerilliant Corporation. SYBR Gold and SYBR Green I were purchased from Invitrogen. 2-Amino-5,6,7-trimethyl-1,8-naphthyridine (ATMND) was purchased from Ryan Scientific. All oligonucleotides were ordered from Integrated DNA Technologies, purified with HPLC, and dissolved in PCR water. DNA concentrations were measured using a NanoDrop 2000 (Thermo Scientific). DNA sequences employed in this work are listed below. Mutated nucleotides of 38-GT and ATP-33 are underlined.
38-GC: 5′-GGG AGA CAA GGA AAA TCC TTC AAC GAA GTG GGT CTC CC-3′
38-GT: 5′-GGG AGA CAA GGA AAA TCC TTC AAT GAA GTG GGT CTC CC-3′
35-GT: 5′-GGG AGA CAA GGA AAA TCC TTC AAT GAA GTG GGT CT -3′
31-GT: 5′-GGG AGA CAA GGA AAA TCC TTC AAT GAA GTG G -3′
38-GT-20C: 5′-GGG AGA CAA GGA AAA TCC TCC AAT GAA GTG GGT CTC CC-3′
38-GT-20A: 5′-GGG AGA CAA GGA AAA TCC TAC AAT GAA GTG GGT CTC CC-3′
38-GT-22G: 5′-GGG AGA CAA GGA AAA TCC TTC GAT GAA GTG GGT CTC CC-3′
38-GT-22T: 5′-GGG AGA CAA GGA AAA TCC TTC TAT GAA GTG GGT CTC CC-3′
32-GT-MB: 5′-SH-C6-AGA CAA GGA AAA TCC TTC AAT GAA GTG GGT CT-MB-3′
ATP-33: 5′-CGC ACC TGG GGG AGT ATT GCG GAG GAA GGT GCG-3′
ATP-30: 5′-CGC ACC TGG GGG AGT ATT GCG GAG GAA GGT-3′
ATP-25: 5′-CGC ACC TGG GGG AGT ATT GCG GAG G-3′
ATP-30-FQ: 5′-/IAbRQ/CGC ACC TGG GGG AGT ATT GCG GAG GAA GGT /3Cy5Sp/-3′
ATP-33-M: 5′-CGC ACC TGG GGA AGT ATT GCG GTG GAA GGT GCG-3′
ATP-SW-34: 5′-CCT CCT ACC TGG GGG AGT ATT GCG GAG GAA GGT A-3′
(MB: Methylene blue, SH: thiol, /IAbRQ/: Iowa Black Quencher RQ, /3Cy5Sp/: Cy5)
Aptamer digestion and characterization of digestion products using polyacrylamide gel electrophoresis (PAGE)
Digestion experiments were performed with the following protocol unless specified otherwise. The digestion of the cocaine-binding aptamer and its derivatives were performed using 1 μl of aptamer (final concentration 1 μM) in 44 μl of reaction buffer (10 mM Tris, 0.1 mM MgCl2, 0.1 mg/ml BSA, pH 7.4) containing cocaine (final concentration 250 μM) in a 200 μl PCR tube. The tube was placed in a thermal cycler (Bio-Rad) and incubated at 23°C for 15 min. 5 μl of Exo III (final concentration 0.26 U/μl) or Exo I (final concentration 0.09 U/μl) was then added to initiate digestion. The digestion of ATP-binding aptamers was performed using 1 μl of aptamer (final concentration 1 μM) in 44 μl of reaction buffer (10 mM Tris, 10 mM MgCl2, 0.1 mg/ml BSA, pH 7.4) containing ATP (final concentration 250 μM) in a 200 μl PCR tube. After a 15-min incubation at 23°C, 5 μl of Exo III (final concentration 0.05 U/μl) or Exo I (final concentration 0.14 U/μl) was then added to perform the digestion. For both aptamers, 5 μl digestion products was collected at various time points and stopped with 10 μl formamide loading buffer (71.25% formamide, 10% glycerol, 0.125% SDS, 25 mM EDTA and 0.15% (w/v) bromophenol blue and xylene cyanol). 3 μl of each sample was loaded onto 15% denaturing PAGE gel and separation was carried out at 20 V/cm for 3 hours in 0.5× TBE running buffer. The gel was stained with 1× SYBR Gold for 25 min and imaged using a ChemiDoc MP imaging system (Bio-Rad).
Preparation of a customized DNA ladder
A customized DNA ladder was used to identify the length and concentration of digestion products in gel. We designed ladder components by removing nucleotides from the 3′ end of 38-GT. The final ladder mixture comprised 38-, 35-, 32-, 29-, 26-, 23-, 20-, 17-, 14- and 11-nt fragments in loading buffer at concentrations of 20, 25, 32, 34, 46, 60, 80, 200, 300 and 400 nM, respectively. We generated a plot of each band's intensity divided by oligonucleotide concentration versus its length (Supplementary Figure S1), and used the resulting curve to calculate the concentrations of digestion products. The following oligonucleotides are contained in the ladder:
38-GT: 5′-GGG AGA CAA GGA AAA TCC TTC AAT GAA GTG GGT CTC CC-3′
35-nt: 5′-GGG AGA CAA GGA AAA TCC TTC AAT GAA GTG GGT CT-3′
32-nt: 5′-GGG AGA CAA GGA AAA TCC TTC AAT GAA GTG GG -3′
29-nt: 5′-GGG AGA CAA GGA AAA TCC TTC AAT GAA GT -3′
26-nt: 5′-GGG AGA CAA GGA AAA TCC TTC AAT GA-3′
23-nt: 5′-GGG AGA CAA GGA AAA TCC TTC AA-3′
20-nt: 5′-GGG AGA CAA GGA AAA TCC TT-3′
17-nt: 5′-GGG AGA CAA GGA AAA TC-3′
14-nt: 5′-GGG AGA CAA GGA AA-3′
11-nt: 5′-GGG AGA CAA GG-3′
SYBR Green I fluorescence experiments
After Exo III digestion, 50 μl of digestion products were mixed with 50 μl of 1× SYBR Green I diluted with 10 mM Tris buffer (pH 7.4) including 2 mM EDTA. 80 μl of this mixture was loaded into one well of a 96-well plate. Fluorescence intensities were recorded using a Tecan Infinite M1000 PRO with excitation at 497 nm and emission at 520 nm.
ATMND-displacement experiments
96 μl of binding buffer (10 mM Tris, 0.01 mM MgCl2, pH 7.4) containing 0.25 μM ATMND and 2 μM 38-GT was loaded into wells of a 96-well plate. Then 2 μl of cocaine, cocaine metabolite, or interferents (final concentration: 250 μM) was added into the well and fluorescence was measured using a Tecan Infinite M1000 PRO with excitation at 358 nm and emission at 405 nm.
Isothermal titration calorimetry (ITC) experiments
All ITC experiments were performed at 23°C in each aptamer's respective binding buffer using a MicroCal iTC200 instrument (GE Healthcare). The sample cell contained 20 μM cocaine-binding aptamers, ATP-binding aptamers, or mutant aptamers, while 500 μM cocaine, cocaine metabolite, or cocaine interferents or 800 μM ATP was loaded in the syringe. We introduced 19 total injections of 2 μl, with the first injection being a purge injection of 0.4 μl. Raw data were fitted to single-site or two-site sequential binding module to calculate the dissociation constant (KD) or apparent dissociation constant (K1/2).
Fabrication of an electrochemical aptamer-based (E-AB) sensor for cocaine detection
The E-AB sensor was fabricated by using polycrystalline gold disk electrodes (1.6 mm diameter; BAS). The electrodes were polished and cleaned by following a previously published protocol (23). The clean gold electrode was incubated with a 25 nM solution of 5′ thiolated, 3′ methylene blue-modified 35-GT containing 2 mM tris-(2-carboxyethyl) phosphine hydrochloride in phosphate-buffered saline (PBS) (10 mM phosphate, 1 M NaCl, and 1 mM MgCl2, pH 7.2) for 13 h. The surface was then rinsed with deionized water and passivated with 3 mM 6-mercaptohexanol in the same PBS buffer for 2 h. The electrode was incubated in a Tris buffer (10 mM Tris, 1 mM NaCl, and 0.03 mM MgCl2, pH 7.4) for 1 h prior to the measurements. Sensor performance was evaluated by monitoring the electrode in Tris buffer containing cocaine, benzoylecgonine, cocaethylene, methylecgonidine, diphenhydramine, nicotine or scopolamine using square wave voltammetry (CH Instruments).
Fluorescence assay for ATP detection
1 μl ATP-30-FQ (final concentration 1 μM) and 89 μl of ATP binding buffer (10 mM Tris, 0.1 mM MgCl2, pH 7.4) were mixed with 10 μl of solution containing various concentrations of ATP and added into wells of a 96-well plate. After a 1-min incubation at room temperature, fluorescence emission spectra were recorded from 660 to 850 nm using a Tecan Infinite M1000 PRO at a 648-nm excitation wavelength. Signal gain was calculated by (F0 – F)/F0, where F0 and F is the fluorescence intensity of ATP-free and ATP-containing sample at 668 nm, respectively. Each sample was analyzed in triplicate and means and standard deviations were plotted.
RESULTS AND DISCUSSION
Inhibition of Exo III digestion of a cocaine-binding aptamer upon target binding
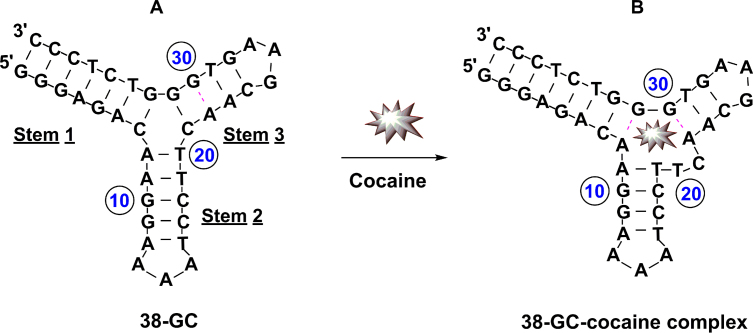
The binding of small molecules to the grooves of double-stranded DNA can modulate the activity of nucleases (20,21). We have now determined that the Exo III-mediated digestion of fully folded aptamers is likewise inhibited by the binding of their small-molecule targets. We demonstrated this using a recently-derived aptamer (38-GC) that binds cocaine with micromolar affinity (10). 38-GC contains a TWJ-structured binding domain and remains predominantly folded even in the absence of target due to the seven Watson-Crick base-pairs in stem 1 (Figure 1A). Upon binding cocaine, 38-GC undergoes a minor conformational change, which relocates a few nucleotides within its TWJ binding domain (9). Specifically, the A8–T20 base-pair in stem 2 and the C21–G31 base-pair in stem 3 are disrupted to form a T20–C21 dinucleotide bulge and an A8–G31 mismatch in the junction (Figure 1B).
Figure 1.
Structure of unbound and cocaine-bound 38-GC. (A) 38-GC undergoes a minor rearrangement of a few nucleotides within its three-way junction upon binding cocaine to form (B) the 38-GC-cocaine complex.
It is believed that the substrate-recognition site of Exo III is located 1 nm away from its catalytic center (24,25). As a result, the activity of Exo III is strongly affected by the structure of DNA three to four bases ahead of its cleavage site. We performed a time-course digestion of 38-GC in the absence and presence of cocaine to investigate whether target binding affects Exo III digestion. The digestion products were characterized by denaturing polyacrylamide gel electrophoresis (PAGE), and their concentrations were calculated relative to a customized DNA ladder loaded in the gel. We observed that Exo III digestion rapidly initiated at stem 1 but stalled 4 bp prior to the binding domain, forming a 35-nt major product regardless of the absence or presence of cocaine. In the absence of cocaine, Exo III continued to non-specifically digest this 35-nt product over time, generating several shorter fragments (Supplementary Figure S2A). However, further digestion of the 35-nt product was strongly inhibited in the presence of cocaine. We believe the digestion of 38-GC is initially hindered because the substrate-recognition site of the enzyme is unable to interact with the TWJ structure. This inhibition intensifies when cocaine is bound to the aptamer, which could physically hinder further DNA digestion by the enzyme. This result is consistent with previous reports demonstrating inhibition of Exo III digestion of double-stranded DNA at positions three to four nucleotides upstream of small-molecule nucleotide adducts (20,21). After a 35-min digestion, we observed an 820 nM concentration of 35-nt product in the presence of cocaine, compared with 326 nM without cocaine (Supplementary Figure S2B).
Effect of TWJ structure on Exo III activity
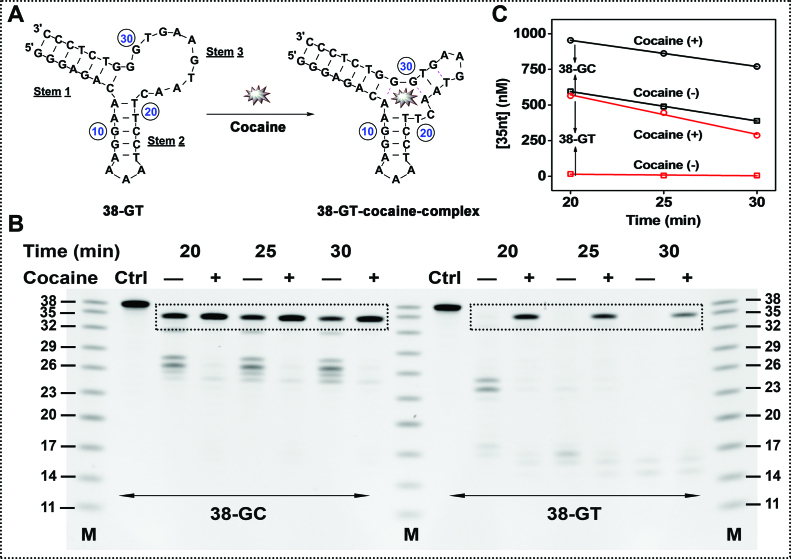
To better elucidate the mechanism of Exo III inhibition, we engineered 38-GT, a variant of 38-GC in which we replaced the matched C24–G28 base-pair with a T24–G28 wobble pair (Figure 2A). Using isothermal titration calorimetry (ITC), we determined that 38-GT binds to cocaine with a similar affinity (KD = 1.6 ± 0.2 μM) to 38-GC (KD = 2.0 ± 0.1 μM) under the same experimental conditions (Supplementary Figure S3). However, stem 3 of 38-GT is less thermally stable compared to 38-GC, and Mfold predicts that this stem predominantly exists in a single-stranded form under our experimental condition (Supplementary Figure S4).
Figure 2.
Time-course of Exo III digestion for 38-GC and 38-GT in the absence (–) or presence (+) of cocaine. (A) Structure of unbound and cocaine-bound 38-GT. (B) PAGE analysis of Exo III-digestion products over time, with (C) a plot of the estimated concentration of the 35-nt product over the course of digestion.
To determine how the TWJ structure affects Exo III digestion, we digested 38-GC and 38-GT under the same experimental conditions with and without cocaine. In the absence of cocaine, 38-GT was completely digested into several short products around 16 nt in length, indicating that the single-stranded stem 3 was digested by Exo III. This was expected, as previous studies have shown that high concentrations of Exo III can non-specifically digest single-stranded DNA (26). In the presence of cocaine, Exo III digestion of 38-GT was effectively inhibited, resulting in the accumulation of a large amount of the 35-nt product (Figure 2B and C). In addition to directly impeding Exo III’s digestion of 38-GT, we believe that cocaine binding to the aptamer also shifts equilibrium from a stem-bulge structure to a TWJ structure that is resistant to Exo III digestion (Figure 2A). After a 25-min Exo III digestion of 38-GC and 38-GT in the presence of cocaine, we respectively observed 800 nM and 460 nM of the 35-nt product (Figure 2B and C). We believe this is due to the greater stability of the TWJ in 38-GC compared to 38-GT, which inhibits Exo III digestion even without binding to cocaine.
Effect of mutations within the binding domain of 38-GT on Exo III activity
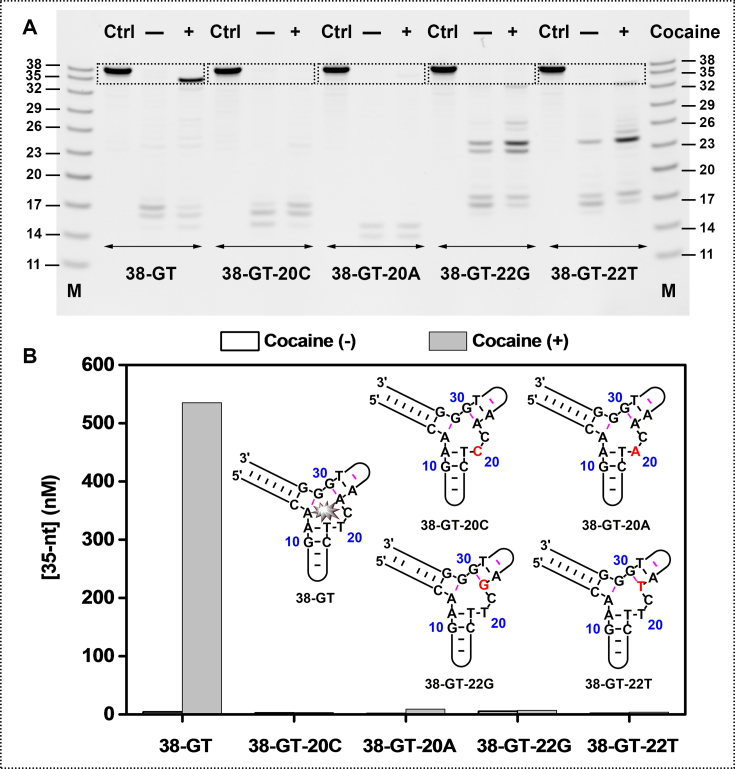
To further confirm that target binding to the aptamer is essential to the inhibition of Exo III digestion, we designed a set of point mutants derived from 38-GT. Two adjacent base-pairs (A8–G31 and A22–G30) and a dinucleotide bulge (T20 and C21) within the binding domain are crucial contributors to the aptamer's binding affinity for cocaine (9,10). Mutations at these positions should profoundly alter binding affinity. We therefore generated four 38-GT mutants in which we replaced the thymine at position 20 with a cytosine (38-GT-20C) or adenine (38-GT-20A), or replaced the adenine at position 22 with a guanine (38-GT-22G) or thymine (38-GT-22T). We expected that these four point mutants would predominately fold in the absence of cocaine, but with impaired target-binding affinity.
Our ITC results confirmed that these mutations prevent cocaine binding (Supplementary Figure S5). We subsequently performed Exo III digestion of 38-GT and these four mutants. Compared to the strong cocaine-induced Exo III inhibition observed with 38-GT, we observed only minimal inhibition with 38-GT-22G and 38-GT-22T, and no inhibition at all with 38-GT-20C and 38-GT-20A (Figure 3). These results confirm that target binding to its aptamer is essential for Exo III inhibition.
Figure 3.
Exo III digestion of 38-GT is inhibited only upon binding to cocaine. (A) PAGE analysis of Exo III digestion for 38-GT and four point mutants after a 25-min reaction. (B) Concentrations of the 35-nt truncation product and structures for 38-GT and four mutants, with mutations highlighted in red.
To investigate the correlation between inhibition of enzyme digestion and target concentration, we performed Exo III digestion of 38-GT and the mutant 38-GT-20C in the presence of different concentrations of cocaine, ranging from 0 to 2000 μM (Supplementary Figure S6). We found that the extent of Exo III digestion of 38-GT decreased with increasing cocaine concentrations, with 600 nM 35-nt product generated in the presence of 2000 μM cocaine. In contrast, only a very small amount of 35-nt product (<20 nM) was produced from 38-GT-20C even in the presence of 2000 μM cocaine, confirming that cocaine itself does not inhibit Exo III digestion. These results demonstrate that the level of Exo III inhibition is target concentration-dependent and clearly correlates with the amount of cocaine-aptamer complex formed.
An Exo III-based, label-free fluorescence assay for sensitive detection of cocaine
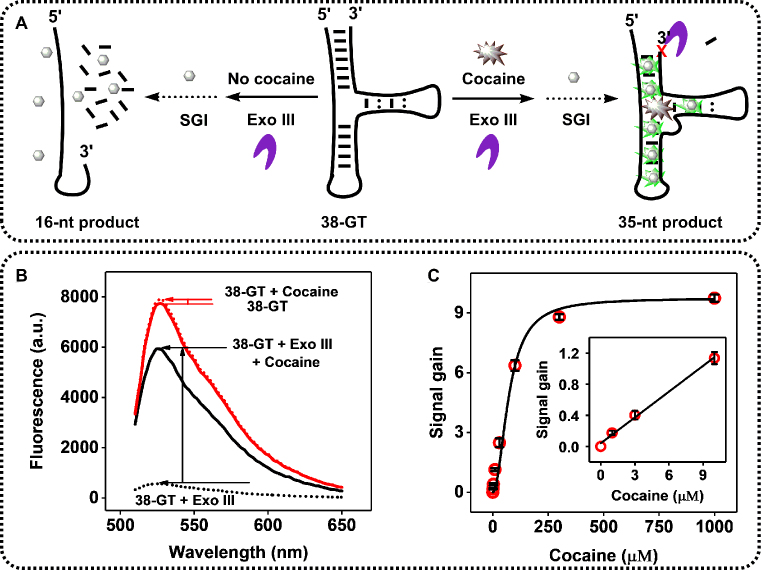
We exploited the fact that Exo III primarily digests unbound rather than target-bound 38-GT to develop a label-free fluorescence assay with SYBR Green I, a commonly used DNA cyanine dye (27) (Figure 4A). In the presence of target, Exo III digestion of 38-GT is halted, generating a 35-nt double-stranded product (Figure 4A, right). We used SYBR Green I to quantify this digestion product, since it preferentially binds double-stranded DNA with 11-fold greater affinity than single-stranded DNA (27), producing a high-intensity fluorescence signal. In the absence of cocaine, Exo III continues to digest the 35-nt product to yield single-stranded 15-nt and 16-nt products (Figure 4A, left), which are unable to bind SYBR Green I, resulting in a low background signal. Accordingly, we observed a large, 9-fold signal gain in the presence of 250 μM cocaine using our Exo III-based assay (Figure 4B). As a control, we monitored the cocaine-induced fluorescence change for 38-GT without Exo III digestion and observed only minimal signal gain, most likely due to minor target-induced conformational changes in the aptamer. We further tested the sensitivity of our assay by generating a calibration curve using different concentrations of cocaine. The fluorescence signal increased proportionally to cocaine concentration, with a linear range from 0 to 100 μM (R2 = 0.9963), and plateaued at 500 μM (Figure 4C). Our Exo III-based assay produced a 5-fold lower measurable limit of detection (1 μM) than a previously reported Exo I-based fluorescence assay (28), which can be attributed to the minimal background in our assay. In our Exo III-based assay, the main source of the background signal is eliminated, as non-target-bound aptamers are completely digested into single-stranded products that cannot be labeled by SYBR Green I. In Lou's Exo I-based assay, some aptamers are folded even in the absence of target, and thus produce high background signal. We subsequently performed our Exo III-based assay for cocaine detection, achieving a measurable limit of detection of 1 μM in 10% saliva (Supplementary Figure S7). Thus, our Exo III-based assay is capable of equally sensitive cocaine detection in body fluids.
Figure 4.
Exo III-based fluorescence assay for cocaine detection. (A) Schematic of cocaine detection based on Exo III inhibition of cocaine-bound aptamers (right) relative to the readily-digested unbound aptamer (left). (B) Fluorescence spectra for Exo III-treated (black line) and untreated (red line) samples with or without 250 μM cocaine. (C) Calibration curve with SYBR Green I (SGI). Inset depicts the signal gain from 0 to 10 μM target.
We subsequently examined whether our Exo III-based assay would allow us to differentiate between the target and other interferents that may bind to the aptamer. To confirm this, we challenged our assay with three cocaine metabolites—benzoylecgonine (BZE), cocaethylene (EC) and methylecgonidine (MEG)—as well as three structurally dissimilar interferent drugs, diphenhydramine (DPH), nicotine (NIC), and scopolamine (SCP). Our assay was not cross-reactive to BZE, MEG, NIC or SCP (Supplementary Figure S8A), which is consistent with ITC data (Supplementary Figure S9) showing that 38-GT does not bind to these interferents. Both EC and DPH were confirmed by ITC to bind to 38-GT (Supplementary Figure S9), but our assay was not cross-reactive to DPH and exhibited only 45% cross-reactivity to EC relative to cocaine (Supplementary Figure S8A). This was unlike the results from an ATMND-displacement assay (10), which generated a false positive in presence of DPH (Supplementary Figure S8B). These results highlight the fact that even though 38-GT can bind to several non-target interferents, our Exo III-based assay introduces an additional measure of specificity through the Exo III digestion process.
Structure-switching functionality adopted into the resulting digestion product
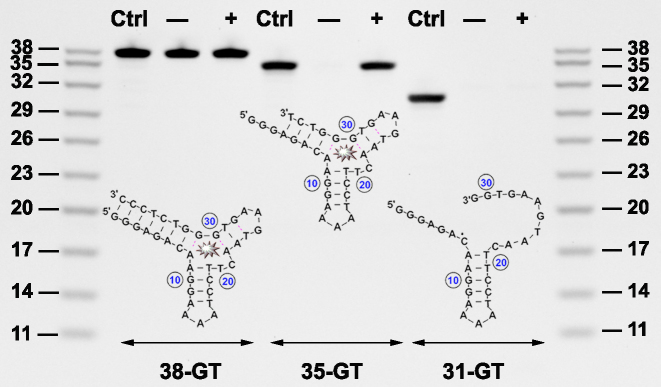
Exo III digestion of 38-GT was inhibited four nucleotides upstream of the target-binding TWJ domain, and we predicted that the truncated 35-nt aptamer would be thermally destabilized and possess target-induced structure-switching functionality. To confirm this, we synthesized this 35-nt product (35-GT) and digested it with or without cocaine using Exo I, a 3′-to-5′ exonuclease that specifically digests single-stranded DNA but has no activity towards double-stranded DNA (29). 35-GT was completely digested in the absence of cocaine, indicating that the unbound aptamer is predominately single-stranded. However, digestion of cocaine-bound 35-GT was strongly inhibited, with only 12% of the aptamer digested within 30 min (Figure 5, 35-GT). These results showed that 35-GT can effectively fold into a double-stranded TWJ structure upon cocaine binding, suggesting that the aptamer has structure-switching functionality. Using ITC, we determined that 35-GT has a binding affinity of KD = 12 ± 1.0 μM, (Supplementary Figure S10), and thus retains strong target binding affinity. We also used Exo I to perform control experiments with 38-GT and another aptamer variant (31-GT) that was engineered by removing seven nucleotides from the 3′ end of 38-GT. Since 38-GT is folded with a blunt-ended stem, Exo I could not digest the duplexed aptamer regardless of the absence or presence of cocaine (Figure 5, 38-GT). On the other hand, 31-GT predominantly exists in a single-stranded state and was completely digested by Exo I regardless of the absence or presence of target (Figure 5, 31-GT).
Figure 5.
PAGE analysis of Exo I digestion of aptamers after 30 min in the absence (–) or presence (+) of cocaine. Structure and PAGE analysis of digestion products for (A) 38-GT, (B) 35-GT and (C) 31-GT.
Performance of an electrochemical aptamer-based (E-AB) sensor fabricated with an Exo III-truncated cocaine aptamer
E-AB sensors utilize target-induced conformational changes of structure-switching aptamers to achieve specific detection of various targets (30). To evaluate the structure-switching functionality of 35-GT, we used a modified version of the aptamer to fabricate an E-AB sensor. Specifically, we labeled 35-GT with a 5′ thiol and a 3′ methylene blue (MB) redox tag and conjugated the modified aptamer to a gold electrode surface via thiol-gold chemistry and tested the performance of this E-AB sensor (23). In the absence of target, the aptamer was primarily unfolded, prohibiting electron transfer from MB to the electrode. The addition of 1000 μM cocaine induced a conformational change of the aptamer that brought MB close to the electrode surface, facilitating efficient electron transfer and resulting in a 170% increase in current within 3 s (Supplementary Figure S11A). We then constructed a calibration curve for this E-AB sensor using various concentrations of cocaine, and obtained a linear range of 0–100 μM with a detection limit of 1 μM (Supplementary Figure S11B). We also challenged our E-AB sensor with common interferent drugs (DPH and SCP) and cutting agents (caffeine, levamisole, lidocaine, and sucrose) to test specificity. Notably, our sensor had low cross-reactivity to DPH (20%) and SCP (10%) and minimal cross-reactivity to all cutting agents (<5%) (Supplementary Figure S11C). These results confirm the structure-switching properties of the Exo III digestion product, and demonstrate the usefulness of such truncated aptamers for sensor applications based on target-induced folding.
Generation of a structure-switching aptamer from a hairpin-structured ATP-binding aptamer
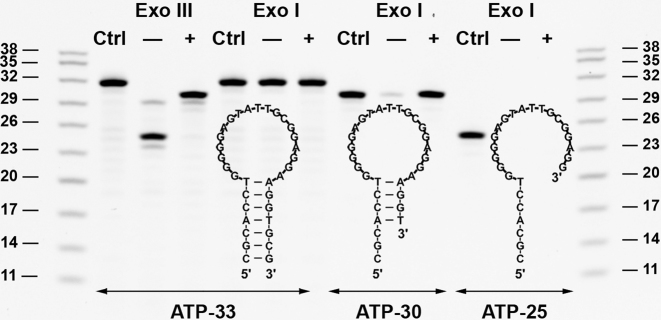
We then demonstrated the generality of target-binding-inhibited Exo III digestion with a non-TWJ-structured ATP-binding aptamer, ATP-33 (22). ATP-33 forms a hairpin structure with a 7-base-pair blunt-ended stem (Figure 6), which is a favorable substrate for Exo III but not Exo I. In the absence of ATP, Exo III removed eight nucleotides from the 3′-end of ATP-33, yielding a 25-nt major product. However, the presence of ATP caused Exo III digestion to halt four base-pairs prior to the ATP-binding domain, producing a 30-nt digestion product (Figure 6, ATP-33 with Exo III). We synthesized the 30-nt (ATP-30) and 25-nt (ATP-25) digestion products, and used ITC to measure their binding affinity. Given that ATP-binding aptamers bind to two ATP molecules (31), we determined the affinity of the first and second ATP binding events using a sequential binding model with two binding sites. We determined that the binding affinity of ATP-30 (KD1 = 0.8 ± 0.1 μM and KD2 = 10.1 ± 0.3 μM) was much higher than that of ATP-25 (KD = 595 ± 86 μM), and was essentially the same as its parent aptamer, ATP-33 (KD1 = 0.6 ± 0.1 μM and KD2 = 12.3 ± 0.6 μM) (Supplementary Figure S12). Negative cooperative binding was observed with ATP-33, which is consistent with a previous study (32). To confirm target-binding-induced inhibition, we then engineered a mutant of ATP-33 (ATP-33-M) by changing guanine to adenine at position 12 and adenine to thymine at position 23 (Supplementary Figure S13A). ATP-33-M maintains the same secondary structure as ATP-33, but possesses no ATP-binding affinity, as confirmed by ITC (Supplementary Figure S13B). We performed Exo III digestion with ATP-33-M and observed no inhibition in the presence of ATP (Supplementary Figure S14), which confirms that target binding is essential for the inhibition of Exo III digestion.
Figure 6.
PAGE analysis of Exo III and Exo I digestion of ATP-binding aptamers after 30 min, without (–) or with (+) ATP. Structure and gel analysis of digestion products for (A) ATP-33, (B) ATP-30 and (C) ATP-25.
To further determine whether Exo III digestion of other ATP-binding aptamers was inhibited upon binding to ATP, we synthesized a previously-described structure-switching ATP-binding aptamer (ATP-SW-34), which has been used for both fluorescence (33) and electrochemical (34) detection of ATP. We first used ITC to characterize the binding affinity of ATP-SW-34 under our experimental conditions (10 mM Tris, 10 mM MgCl2, pH 7.4, 23°C). The dissociation constants of the first (KD1) and second (KD2) binding events for ATP-SW-34 were 21.1 ± 1.3 μM and 7.2 ± 0.3 μM, respectively (Supplementary Figure S15A). ATP-SW-34 has a lower ATP-binding affinity relative to the pre-folded ATP-33 (KD1 = 0.6 ± 0.1 μM and KD2 = 12.3 ± 0.6 μM). This was expected, since the target-binding domain of ATP-SW-34 is purposely destabilized to achieve target-induced structure switching. We also observed slight positive cooperativity with ATP-SW-34, possibly because binding of the first ATP molecule induces a conformational change in the aptamer, which greatly facilitates binding of the second ATP molecule.
We then performed Exo III digestion of ATP-SW-34 in our experimental conditions (10 mM Tris buffer (pH 7.4) including 10 mM MgCl2, 1 μM DNA, 0.05 U/μl Exo III, 250 μM ATP, at 23°C). We found that ATP-SW-34 was completely digested to products shorter than 25 nt in length in the absence of ATP within 10 min. However, in the presence of 250 μM ATP, Exo III was halted four nucleotides prior to the target-binding domain to generate a 33-nt major product (Supplementary Figure S16, A and B). Even after 60 min of digestion, the 33-nt product was still present (Supplementary Figure S16C). As a comparison, we also performed Exo III digestion of ATP-SW-34 under the literature-reported experimental conditions (10 mM Tris buffer (pH 7.9) including 50 mM NaCl, 10 mM MgCl2, 2 μM DNA, 2 U/μl Exo III, 250 μM ATP, at 37°C). We observed that Exo III digests much faster under these conditions regardless of the presence or absence of ATP, most likely due to the higher concentration of Exo III and higher temperature. Specifically, ATP-SW-34 was completely digested into products around 25 nt in length within 10 s in the absence of ATP, whereas the 33-nt product was detectable for up to 60 s in the presence of ATP (Supplementary Figure S17A and B). The aptamer was completely digested into products less than 25 nt in length after 3 min regardless of the presence or absence of ATP (Supplementary Figure S17C). This weaker inhibition of Exo III in the literature-reported conditions is likely attributable to the lower ATP-binding affinity of the aptamer at elevated temperatures. Thus, we believe that this Exo III inhibition phenomenon is generalizable for other target-bound small-molecule-binding aptamers with different secondary structures, such as stem–loop.
To confirm that ATP-30 had structure-switching functionality, we digested ATP-33, ATP-30 and ATP-25 with Exo I in the presence and absence of ATP (Figure 6, Exo I). As expected, Exo I completely digested ATP-30 in the absence of target but was unable to digest the aptamer in the presence of target. This result indicated that ATP-30 has target-induced structure-switching functionality. On the other hand, no digestion of ATP-33 was observed regardless of the presence or absence of ATP due to its fully folded stem–loop structure. ATP-25 was completely digested by Exo I regardless of the presence or absence of target, confirming that this aptamer exists in a single-stranded state. We then exploited the structure-switching functionality of ATP-30 to fabricate a fluorophore-quencher ATP sensor. We labeled ATP-30 with a 3′ Cy5 fluorophore and a 5′ Iowa Black quencher (ATP-30-FQ). In the absence of target, the aptamer is primarily unfolded and the fluorophore is separated from the quencher, producing a large fluorescence background (Supplementary Figure S18A). The addition of 100 μM ATP induces a conformational change in the aptamer that brings the fluorophore in close proximity to the quencher, yielding a strong reduction in fluorescence within seconds (Supplementary Figure S18B). We challenged this sensor with various concentrations of ATP, and obtained a linear range of 0–10 μM with a measurable detection limit of 2.5 μM (Supplementary Figure S18C). This suggests that Exo III digestion of target-bound aptamers offers an apparently generalizable means for introducing structure-switching functionality.
CONCLUSION
In this work, we demonstrated a broadly applicable strategy for introducing structure-switching functionality into small-molecule-binding aptamers, based on the finding that the binding of a small-molecule target to its aptamer inhibits nuclease digestion a few nucleotides prior to the target-binding domain. As proof of concept, we used Exo III to digest a cocaine-binding aptamer, 38-GT, which folds into a TWJ structure with a blunt-ended terminus. Exo III initiated 3′-to-5′ digestion of 38-GT to generate single-stranded 15-nt and 16-nt products in the absence of cocaine. The binding of cocaine stabilizes the TWJ structure at the aptamer-binding domain, which inhibits Exo III digestion and results in accumulation of a 35-nt major digestion product. To understand the mechanism of this inhibition, we engineered a set of 38-GT mutants with reduced binding affinity to cocaine. We performed Exo III digestion of these mutants and determined that binding of cocaine to its aptamer is essential for the inhibition of Exo III digestion. We exploited this finding to develop an Exo III-based, SYBR Green I-reported fluorescence assay that could detect cocaine at concentrations as low as 1 μM in 10% saliva within 20 min. There are two reasons for this high sensitivity. First, 38-GT binds to cocaine with high affinity even at low concentrations of target, efficiently forming duplexed structures that are sensitively reported by SYBR Green I. Second, our assay yields minimal background signal, because unbound aptamers are completely digested by Exo III into short single-stranded products that are not labeled by SYBR Green I. Our assay is simple and label-free, and enhances the inherent specificity of the cocaine aptamer, because Exo III is highly sensitive to minor target-induced conformational changes within the aptamer.
Exo III digestion of target-bound 38-GT was inhibited four nucleotides prior to the aptamer's target binding domain. We found that the resulting 35-nt product has structure-switching functionality, based on the fact that it was completely digested by the single-strand exonuclease Exo I in the absence of cocaine while remaining intact in the presence of cocaine. This truncated aptamer retained high target-binding affinity, and demonstrated excellent performance in an E-AB sensor based on its structure-switching functionality. We subsequently demonstrated that our Exo III-based method can be more generally applied to introduce structure-switching functionality into other small-molecule binding aptamers. Exo III digestion of a stem–loop-structured ATP-binding aptamer produced a 25-nt single-stranded product in the absence of ATP, whereas digestion in the presence of ATP was halted four nucleotides prior to the target-binding domain, generating a double-stranded 30-nt product. As with the truncated cocaine aptamer, this 30-nt product exhibited structure-switching capability, as confirmed by Exo I digestion in the absence and presence of target. We further evaluated the structure-switching functionality of the Exo III-truncated ATP aptamer by modifying it with a fluorophore-quencher pair and employing it for the detection of low concentrations of ATP. We believe that Exo III can more generally be used for the selective truncation of other TWJ- or stem–loop-structured aptamers in order to home in on their target-binding domains and to generate structure-switching aptamers for the detection of small-molecule targets. Since Exo III is highly sensitive to subtle target-induced aptamer conformational changes, we see great potential in developing adaptations of SELEX that can enable the generation of aptamers with superior specificity and discrimination against structurally-similar target analogs.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health—National Institute on Drug Abuse [R15DA036821]; National Institute of Justice, Office of Justice Programs, U.S. Department of Justice [2013-DN-BX-K032]; National Natural Science Foundation of China [21505020]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Tuerk C., Gold L.. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990; 249:505–510. [DOI] [PubMed] [Google Scholar]
- 2. Ellington A.D., Szostak J.W.. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990; 346:818–822. [DOI] [PubMed] [Google Scholar]
- 3. Stojanovic M.N., de Prada P., Landry D.W.. Aptamer-based folding fluorescent sensor for cocaine. J. Am. Chem. Soc. 2001; 123:4928–4931. [DOI] [PubMed] [Google Scholar]
- 4. Sun H.G., Zhu X., Lu P.Y., Rosato R.R., Tan W., Zu Y.L.. Oligonucleotide aptamers: new tools for targeted cancer therapy. Mol. Ther. Nucleic Acids. 2014; 3:e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nimjee S.M., Rusconi C.P., Sullenger B.A.. Aptamers: an emerging class of therapeutics. Annu. Rev. Med. 2005; 56:555–583. [DOI] [PubMed] [Google Scholar]
- 6. Hayat A., Marty J.L.. Aptamer based electrochemical sensors for emerging environmental pollutants. Front. Chem. 2014; 2:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jayasena S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999; 45:1628–1650. [PubMed] [Google Scholar]
- 8. Dunn M.R., Jimenez R.M., Chaput J.C.. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017; 1:0076. [Google Scholar]
- 9. Neves M.A.D., Reinstein O., Saad M., Johnson P.E.. Defining the secondary structural requirements of a cocaine-binding aptamer by a thermodynamic and mutation study. Biophys. Chem. 2010; 153:9–16. [DOI] [PubMed] [Google Scholar]
- 10. Roncancio D., Yu H.X., Xu X.W., Wu S., Liu R., Debord J., Lou X.H., Xiao Y.. A label-free aptamer-fluorophore assembly for rapid and specific detection of cocaine in biofluids. Anal. Chem. 2014; 86:11100–11106. [DOI] [PubMed] [Google Scholar]
- 11. Stojanovic M.N., de Prada P., Landry D.W.. Fluorescent sensors based on aptamer self-assembly. J. Am. Chem. Soc. 2000; 122:11547–11548. [DOI] [PubMed] [Google Scholar]
- 12. Simon A.J., Vallée-Bélisle A., Ricci F., Plaxco K.W.. Intrinsic disorder as a generalizable strategy for the rational design of highly responsive, allosterically cooperative receptors. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15048–15053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stojanovic M.N., Kolpashchikov D.M.. Modular aptameric sensors. J. Am. Chem. Soc. 2004; 126:9266–9270. [DOI] [PubMed] [Google Scholar]
- 14. Gao S.X., Zheng X., Jiao B.H., Wang L.H.. Post-SELEX optimization of aptamers. Anal. Bioanal. Chem. 2016; 408:4567–4573. [DOI] [PubMed] [Google Scholar]
- 15. Fischer N.O., Tok J.B.-H, Tarasow T.M.. Massively parallel interrogation of aptamer sequence, structure and function. PLoS One. 2008; 3:e2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shangguan D.H., Tang Z.W., Mallikaratchy P., Xiao Z.Y., Tan W.H.. Optimization and modifications of aptamers selected from live cancer cell lines. ChemBioChem. 2007; 8:603–606. [DOI] [PubMed] [Google Scholar]
- 17. Neves M.A., Reinstein O., Johnson P.E.. Defining a stem length-dependent binding mechanism for the cocaine-binding aptamer. A combined NMR and calorimetry study. Biochemistry. 2010; 49:8478–8487. [DOI] [PubMed] [Google Scholar]
- 18. Xiao Y., Piorek B.D., Plaxco K.W., Heeger A.J.. A reagentless signal-on architecture for electronic, aptamer-based sensors via target-induced strand displacement. J. Am. Chem. Soc. 2005; 127:17990–17991. [DOI] [PubMed] [Google Scholar]
- 19. Rogers S.G., Weiss B.. Exonuclease III of Escherichia coli K-12, an AP endonuclease. Methods Enzymol. 1980; 65:201–211. [DOI] [PubMed] [Google Scholar]
- 20. Rao K.E., Lown J.W.. DNA sequence selectivities in the covalent bonding of antibiotic saframycins Mx1, Mx3, A, and S deduced from MPE•Fe(II) footprinting and exonuclease III stop assays. Biochemistry. 1992; 31:12076–12082. [DOI] [PubMed] [Google Scholar]
- 21. Albert F.G., Eckdahl T.T., Fitzgerald D.J., Anderson J.N.. Heterogeneity in the actions of drugs that bind in the DNA minor groove. Biochemistry. 1999; 38:10135–10146. [DOI] [PubMed] [Google Scholar]
- 22. Huizenga D.E., Szostak J.W.. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995; 34:656–665. [DOI] [PubMed] [Google Scholar]
- 23. Xiao Y., Lai R.Y., Plaxco K.W.. Preparation of electrode-immobilized, redox-modified oligonucleotides for electrochemical DNA and aptamer-based sensing. Nat. Protoc. 2007; 2:2875–2880. [DOI] [PubMed] [Google Scholar]
- 24. Mol C.D., Kuo C.F., Thayer M.M., Cunningham R.P., Tainer J.A.. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995; 374:381–386. [DOI] [PubMed] [Google Scholar]
- 25. Hoheisel J.D. On the activities of escherichia coli exonuclease III. Anal. Biochem. 1993; 209:238–246. [DOI] [PubMed] [Google Scholar]
- 26. Shida T., Noda M., Sekiguchi J.. Cleavage of single- and double-stranded DNAs containing an abasic residue by Escherichia coli exonuclease III (AP endonuclease VI). Nucleic Acids Res. 1996; 24:4572–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zipper H., Brunner H., Bernhagen J., Vitzthum F.. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucleic Acids Res. 2004; 32:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng D.M., Zou R.X., Lou X.H.. Label-free fluorescent detection of ions, proteins, and small molecules using structure-switching aptamers, SYBR Gold, and exonuclease I. Anal. Chem. 2012; 84:3554–3560. [DOI] [PubMed] [Google Scholar]
- 29. Lehman I.R., Nussbaum A.L.. The deoxyribonucleases of Escherichia coli V. On the specificity of exonuclease I (phosphodiesterase). J. Biol. Chem. 1964; 239:2628–2636. [PubMed] [Google Scholar]
- 30. Xiao Y., Lubin A.A., Heeger A.J., Plaxco K.W.. Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew. Chem., Int. Ed. 2005; 44:5592–5595. [DOI] [PubMed] [Google Scholar]
- 31. Lin C.H., Patei D.J.. Structural basis of DNA folding and recognition in an AMP-DNA aptamer complex: distinct architectures but common recognition motifs for DNA and RNA aptamers complexed to AMP. Chem. Biol. 1997; 4:817–832. [DOI] [PubMed] [Google Scholar]
- 32. Armstrong R.E., Strouse G.F.. Rationally manipulating aptamer binding affinities in a stem–loop molecular beacon. Bioconjugate. Chem. 2014; 25:1769–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu X.Q., Freeman R., Willner I.. Amplified fluorescence aptamer-based sensors using exonuclease III for the regeneration of the analyte. Chem. - Eur. J. 2012; 18:2207–2211. [DOI] [PubMed] [Google Scholar]
- 34. Liu S.F., Wang Y., Zhang C.X., Lin Y., Li F.. Homogeneous electrochemical aptamer-based ATP assay with signal amplification by exonuclease III assisted target recycling. Chem. Commun. 2013; 49:2335–2337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.