Abstract
The polyadenosine RNA-binding protein ZC3H14 is important in RNA processing. Although ZC3H14 is ubiquitously expressed, mutation of the ZC3H14 gene causes a non-syndromic form of intellectual disability. Here, we examine the function of ZC3H14 in the brain by identifying ZC3H14-interacting proteins using unbiased mass spectrometry. Through this analysis, we identified physical interactions between ZC3H14 and multiple RNA processing factors. Notably, proteins that comprise the THO complex were amongst the most enriched proteins. We demonstrate that ZC3H14 physically interacts with THO components and that these proteins are required for proper RNA processing, as loss of ZC3H14 or THO components leads to extended bulk poly(A) tail length. Furthermore, we identified the transcripts Atp5g1 and Psd95 as shared RNA targets of ZC3H14 and the THO complex. Our data suggest that ZC3H14 and the THO complex are important for proper processing of Atp5g1 and Psd95 RNA, as depletion of ZC3H14 or THO components leads to decreased steady-state levels of each mature transcript accompanied by accumulation of Atp5g1 and Psd95 pre-mRNA in the cytoplasm. Taken together, this work provides the first unbiased identification of nuclear ZC3H14-interacting proteins from the brain and links the functions of ZC3H14 and the THO complex in the processing of RNA.
INTRODUCTION
Control of eukaryotic gene expression involves a highly ordered regulatory network of cis-acting elements and trans-acting factors to ensure that a given gene product is produced in the correct amount at the proper time and location. RNA-binding proteins are critical regulators of the co- and posttranscriptional events that control gene expression (1). These events include processing steps such as the initial capping of the nascent mRNA emerging from RNA polymerase, the splicing of the primary transcript, and the formation of the 3′end of the mRNA through cleavage and polyadenylation (2,3). Upon completion of these steps, a mature mRNA is generated that is packaged with RNA-binding proteins to form a messenger ribonucleoprotein (mRNP) complex (4). This mRNP can then be exported from the nucleus and undergo further regulation in the cytoplasm.
The importance of RNA-binding proteins is evident in both their critical role in gene expression and the growing number of diseases that are associated with the mutation of genes encoding these proteins (5,6). Interestingly, an emerging subset of these diseases are tissue-specific diseases that result from mutation in genes encoding ubiquitously expressed proteins (6). Recently, mutations in the gene encoding the ubiquitously expressed RNA-binding protein ZC3H14 were linked to an inherited form of autosomal recessive intellectual disability (7). This finding suggests that ZC3H14 plays a critical role in the brain. However, the function of ZC3H14 has been largely studied in cultured cell lines and lower order model organisms (7–9).
Initial understanding of the function of ZC3H14 came from studies of the budding yeast orthologue Nab2 (10). ZC3H14 and Nab2 share conserved domain structures that consist of an N-terminal PWI-like domain and C-terminal tandem CCCH zinc fingers (10,11). This work showed that NAB2 is an essential yeast gene that is required for proper poly(A) tail length control and poly(A) RNA export from the nucleus (12–14). Like Nab2, ZC3H14 is primarily localized to the nucleus and both Nab2 and ZC3H14 bind with high affinity to polyadenosine RNA (15,16). Although Nab2 and ZC3H14 bind to polyadenosine RNA tracts (17) and therefore could bind to all mRNA transcripts, previous work has shown that loss of ZC3H14 only alters the steady-state levels of a small number of transcripts (9). This finding focuses the regulation of mRNA processing by ZC3H14 to a select group of transcripts.
As patients with mutations in the ZC3H14 gene that cause loss of the ZC3H14 protein display brain phenotypes, the role of ZC3H14 in the brain has begun to be studied (7). Studies of the ZC3H14 orthologue in Drosophila, dNab2, provide evidence that the function of dNab2 is critical in neurons (7). dNab2 mutant flies have compromised brain function and severe brain morphology defects (18). Importantly, these defects in the fly can be rescued by the expression of either dNab2 or the mammalian ZC3H14 protein exclusively in neurons (18). Furthermore, the generation of a Zc3h14 mutant mouse model revealed that global loss of ZC3H14 impairs higher order brain function with the mutant mice displaying defects in working memory (19). These findings coupled with the defects reported in the ZC3H14 patients (7) further emphasize the need to understand the function of ZC3H14 in the brain.
In this study, we identify proteins that interact with ZC3H14 in the brain. We employ a proteomic approach using fractionated mouse brain lysate to identify the nuclear factors that interact with ZC3H14. Among the most enriched factors is a group of proteins that belong to an RNA-processing complex termed the THO complex. The THO complex is a multi-subunit complex that is essential for RNA processing (20). Most studies of this complex have been performed in budding yeast and these studies have linked this complex to transcription elongation, mRNA splicing, and mRNA export (20–23). Interestingly, mutations in genes encoding components of this complex THOC2 and THOC6 are associated with brain disorders, similar to mutations in ZC3H14 (24–26).
Here, we demonstrate that ZC3H14 and the THO complex physically interact and coordinately regulate the processing of mRNA transcripts. As previously shown for ZC3H14, we demonstrate that THO components are required for proper control of bulk poly(A) tail length. As these factors do not affect all RNAs (9,27,28), we have identified two transcripts Atp5g1, which encodes a component of the ATP synthase machinery (29), and Psd95 (Dlg4), which encodes a component of the neuronal post synaptic density (30), as shared targets of ZC3H14 and the THO complex. This work reveals that ZC3H14 and the THO complex are required to coordinate nuclear processing of Atp5g1 and Psd95 mRNAs with export from the nucleus. Loss of these factors leads to a decrease in the steady-state levels of the mature Atp5g1 and Psd95 mRNAs and accumulation of the pre-mRNAs of these transcripts in the cytoplasm. Taken together, these results suggest coordination between ZC3H14 and the THO complex to ensure proper mRNA processing prior to export from the nucleus.
MATERIALS AND METHODS
Tissue fractionation and immunoprecipitations
Brain tissue was collected from wild-type C57BL/6 mice and fractionated as first described in Guillem et al. 2005 (31). Nuclear pellets were suspended and sonicated (5 times at 10% output) in immunoprecipitation (IP) buffer (50 mM Tris–HCl [pH 7.4], 150 mM NaCl and 0.5% NP-40) supplemented with 1 cOmplete mini protease inhibitor tablet (Roche). Lysates were spun at 13 000 rpm for 10 min (min) at 4°C and protein concentrations were determined using a standard bicinchoninic acid (BCA) assay (Pierce). Protein A-magnetic beads (Dynabeads; Invitrogen) were suspended in IP buffer and incubated for 30 min with preimmune rabbit serum or an equal volume of experimental antibody (4 μg/mg of protein lysate) at room temperature. Bead/antibody complexes were added to clarified cell lysates, followed by incubation at 4°C overnight while tumbling end over end (10% removed prior to overnight incubation for input samples). After incubation, the beads were magnetized and washed five times with ice-cold IP buffer. At this point, samples were prepared for mass spectrometry analysis or for immunoblotting. For immunoblotting, protein complexes were eluted with reducing sample buffer (250 mM Tris–HCl, 500 mM dithiothreitol [DTT], 10% SDS, 0.5% bromophenol blue, and 50% glycerol).
Mass spectrometry analysis
Mass spectrometry analysis was performed as previously described by Soucek et al. (8). Magnetic beads from control and ZC3H14 immunoprecipitates were suspended in 8 M urea–100 mM NaHPO4 (pH 8.5; 50 μl, final volume) and treated with 1 mM DTT at 25°C for 30 min, followed by 5 mM iodoacetamide at 25°C for 30 min in the dark. The samples were then diluted to 1 M urea with 50 mM ammonium bicarbonate (final volume, 400 μl) and digested with lysyl endopeptidase (Wako; 1.25 ng/l, final concentration) at 25°C for 4 h and further digested overnight with trypsin (Promega; 1.25 ng/l, final concentration) at 25°C. The resulting peptides were desalted with a Sep-Pak C18 column (Waters) and dried under vacuum. Each sample was suspended in loading buffer (0.1% formic acid, 0.03% trifluoroacetic acid, 1% acetonitrile) and analyzed independently by reverse-phase liquid chromatography (LC) coupled with tandem mass spectrometry (MS/MS) essentially as previously described with some minor modifications (32,33). Briefly, peptide mixtures were loaded onto a C18 nanoLC column (75 m [inner diameter], 15-cm long, 1.9-m resin [Maisch GmbH]) and eluted over a 5 to 0% gradient (buffer A: 0.1% formic acid; buffer B: 0.1% formic acid in 100% acetonitrile). Eluates were monitored in an MS survey scan, followed by 10 data-dependent MS/MS scans on a Q-Exactive plus Orbitrap mass spectrometer (Thermo Scientific). The acquired MS/MS spectra were searched against a concatenated target decoy mouse reference database (v.62) of the National Center for Biotechnology Information (downloaded 14 November 2013 with 30 267 target entries) using the SEQUEST Sorcerer algorithm (version 4.3.0; SAGE-N). The search parameters included the following: fully tryptic restriction, parent ion mass tolerance of 50 ppm, up to two missed trypsin cleavages, and dynamic modifications for oxidized Met (15.9949 Da). The peptides were classified by charge state and first filtered by mass accuracy (10 ppm for high-resolution MS) and then dynamically by increasing XCorr and Cn values to reduce the protein false discovery rate to 1%. If peptides were shared by multiple members of a protein family, the matched members were clustered into a single group in which each protein identified by a unique peptide represented a subgroup. The total peptide spectra match (PSM) counts are provided in Supplementary Table S1 in the Supplementary material.
Gene ontology (GO) enrichment and network
Functional enrichment of the modules was determined using the GO-Elite (v1.2.5) package (34). The set of total proteins (1,350) identified was used as the background. We analyzed the 62 proteins enriched in IP ([PSM] IP/bead fold change of 2). The Z-score determines the overrepresentation of ontologies in a module, and a permutation P-value was used to assess the significance of a Z-score cutoff of 4.5 and a P-value cutoff of 0.05, with a minimum of three proteins per category used as filters in pruning the ontologies. A horizontal bar graph was plotted in R. The networks were constructed using the Circlize package in R.
Immunoblotting
Protein lysates were boiled in reducing sample buffer and resolved on 4–20% Criterion TGX polyacrylamide gels (Bio Rad). Proteins were transferred to nitrocellulose membranes and incubated for at least 1 h in blocking buffer (5% non-fat dry milk in 0.1% TBS-Tween). This step was followed by an overnight incubation at 4°C in primary antibody diluted in blocking buffer. Primary antibodies were detected using species-specific horse radish peroxidase (HRP) conjugated secondary antibodies (Jackson ImmunoResearch) followed by incubation with enhanced chemiluminescence substrate (ECL, Sigma). Chemiluminescence was detected by exposing blots to autoradiography film (Daigger). Immunoblots were quantified using ImageJ software. Primary antibodies and dilutions employed as they appear: ZC3H14, (1:6000) (15), Nuclear Pore complex NUP93 and NUP62, (1:5000), (414, Abcam, ab24609), eIF5, (1:5000) (Santa Cruz, sc-282), THOC1, (1:1000), (Bethyl, A302-839A), THOC2, (1:2000), (Bethyl, A303-630A), ALYREF, (1:1000), (ALY, Santa Cruz, sc-32311), HuR, (1:1000), (Santa Cruz, sc-5261), NPM1, (1:1000), (B23, Santa Cruz, sc-6013), THOC5, (1:1000) (Bethyl, A302-119A), HSP90, (1:1000), (Santa Cruz, sc-13119), and NXF1, (1:1000), (TAP, Santa Cruz, sc-32319).
Glycerol density gradients
Nuclei from mouse brain tissue were collected and lysed as described for tissue fractionation and immunoprecipitation. Nuclear lysate was nuclease treated using Benzonase (Sigma). Approximately 1.5 mg of nuclear lysate in a 300-μl volume was layered on top of a 10–50% glycerol gradient (10 mM HEPES pH 7.5, 2 mM MgCl2, 10 mM KCl, 0.5 mM EDTA and 150 mM NaCl) in 14 × 89 mm tubes (Beckman Coulter), and spun in a SW41Ti rotor (Beckman Coulter) at 30 000 × RPM for 16 h at 4°C. Gradients were fractionated top to bottom into 500-μl fractions using the Gradient Master automated fraction collector (BioComp).
Cell culture and siRNA transfections
Neuro2a (N2a) cells are a mouse neuroblastoma cell line (35). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and antibiotics and grown under standard environmental conditions. Individual siRNAs were transfected into cultured cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol in antibiotic free media. In experiments where cells were treated with two different siRNAs, cells were transfected simultaneously with double the amount of siRNA as the single transfections. siRNAs: Scrambled (Scr) control (Stealth siRNA Neg Control Medium GC, Invitrogen), Zc3h14 (Stealth siRNA, Invitrogen, GAAGAGCCTCGATACTGACTCCAAA), Thoc1 (MISSION esiRNA, Sigma EMU081331), Thoc5 (MISSION esiRNA, Sigma EMU003181).
RNA isolation and quantitative RT-PCR
Total RNA was isolated from N2a cells using the TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Isolated RNA was treated with Turbo DNase (Invitrogen) to degrade contaminating DNA. For qRT-PCR analyses, cDNA was generated using the MMLV Reverse transcriptase (Invitrogen) from 1 μg of total RNA. Relative mRNA levels were measured by quantitative PCR analysis of duplicate samples of 15 ng cDNA with QuantiTect SYBR Green Master Mix using an Applied Biosystems StepOne Plus real time machine (ABI). Results were analyzed using the ΔΔCT method and normalized to 18S rRNA. Oligonucleotide sequences are provided in Supplementary Table S2.
Bulk Poly(A) tail length assays
Bulk poly(A) tails were analyzed as described previously (8,36). Briefly, total RNA was 3′end labeled with [32P]-pCp (cytidine 3′,5′ bisphosphate) (Perkin Elmer) using T4 RNA ligase. RNA was then digested with RNase A and T1 and resolved on TBE-Urea (90 mM Tris-borate, 2 mM EDTA, 8 M urea) 7% polyacrylamide gels. Gel images were obtained using a Typhoon phosphorimager and quantified using ImageQuant software.
RNA immunoprecipitation
N2a cells were grown on 100-mm plates and UV crosslinked at 254 nm using a Stratalinker UV 2400 (Stratagene). Cells were then suspended in RNA-IP buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 10% RIPA-2 buffer in DEPC-treated water) supplemented with 1 cOmplete mini protease inhibitor tablet (Roche). Lysates were then sonicated on ice 5 times at 10% output and spun at 13,000 RPM for 10 min at 4°C. Protein concentration was determined with a standard BCA assay (Pierce) and standard immunoprecipitation protocol followed.
Cell fractionation
N2a cells were collected on ice, spun down, and suspended in ice-cold fractionation buffer (10 mM Tris–HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.5% (v/v) NP-40), supplemented with 1 mini cOmplete protease inhibitor tablet (Roche) for 10 min on ice. Cell lysates were centrifuged at 1000 × g for 5 min and the supernatant or cytoplasmic fraction was separated from the nuclear pellet. The nuclear pellet was then washed once with fractionation buffer and centrifuged at 1000 × g for 5 min. The resulting pellet was collected as the nuclear fraction. RNA was isolated from each fraction with TRIzol reagent (Invitrogen). Protein samples were suspended with RIPA-2 buffer and prepared for immunoblotting.
Statistical analysis
Comparisons between experimental groups were made using one-way analysis of variance (ANOVA), unless noted otherwise. All data are presented as means and standard error of the mean (SEM) (error bars) for at least three independent experiments. Asterisks (*) indicate statistical significance at *P-value < 0.05.
RESULTS
ZC3H14 interacts with the THO complex
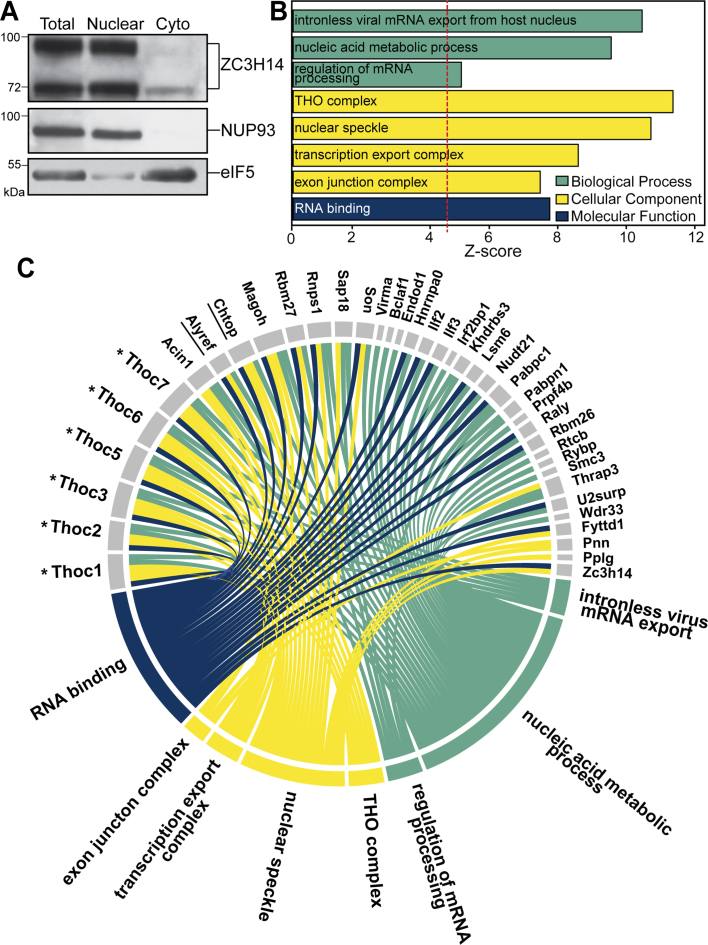
To examine the function of ZC3H14 in the brain, we first analyzed the nucleocytoplasmic distribution of the protein in the brain. We collected and fractionated mouse brain tissue as described in Materials and Methods. Figure 1A shows the distribution of the endogenous ZC3H14 protein. The polyclonal antibody raised against the N-terminal domain of ZC3H14 detects multiple ZC3H14 isoforms generated by alternative splicing, but does not detect a predicted cytoplasmic isoform that lacks the N-terminal domain (15). The nuclear fraction contains the bulk of ZC3H14 protein. However, ZC3H14 can be detected in the cytoplasmic (Cyto) fraction with enrichment for the ∼70 kDa isoforms of ZC3H14 as compared to the ∼100 kDa isoform.
Figure 1.
ZC3H14 interacts with multiple mRNA processing proteins in the brain. (A) Fractionation of mouse brain lysate. Equal amounts of protein lysate from Total, Nuclear, or Cytoplasmic (Cyto) fractions were analyzed by immunoblotting with ZC3H14 antibody. The ZC3H14 antibody recognizes multiple isoforms of ZC3H14 resulting from alternative splicing of the ZC3H14 gene (15). To ensure proper fractionation, a nuclear marker (NUP93) and a cytoplasmic marker (eIF5) are shown. Molecular weights are indicated in kDa. (B) The gene ontology (GO) terms for the proteins that are mostly highly enriched within the immunoprecipitation of ZC3H14 compared to IgG control as identified by tandem mass spectrometry. These proteins are grouped based on the three GO domains: Biological Process, Cellular Component, and Molecular Function. The inclusion criteria for this analysis were a Z-score of > 4.5, a P-value of < 0.05, and at least three gene terms per GO term. (C) The gene terms that cluster into the listed GO terms in (B) are depicted. Colored lines connect GO categories to GO terms and individual term members. All core members of the THO complex (THOC1, THOC2, THOC3, THOC5, THOC6, THOC7) are present (indicated by an asterisk) as well as the THO associated proteins CHTOP and ALYREF (underlined).
As the majority of ZC3H14 is nuclear, we sought to identify factors that interact with ZC3H14 in the nucleus. To identify ZC3H14-interacting proteins, we collected mouse brain tissue and isolated the nuclear fraction. We then immunoprecipitated endogenous ZC3H14 and associated proteins from the nuclear fraction using a polyclonal ZC3H14 antibody (15). With unbiased mass spectrometry, we identified ZC3H14-interacting proteins and compared results from our ZC3H14 immunoprecipitation to the results obtained with control pre-immune serum. These interacting proteins are presented in detail in Supplementary Table S1 as well as Figure 1B and C.
To explore the relationship between ZC3H14 and the interacting proteins identified, we used GO analysis. Figure 1B shows the GO analysis of the 62 ‘top’ interacting candidates identified by the mass spectrometry analysis. These ‘top’ candidates were selected based on the criteria that they were only identified in the ZC3H14 immunoprecipitation and not with the control pre-immune serum and they showed a ≥ 2 peptide spectra matched (PSM) quantification. Of the 62 proteins, 38 proteins grouped based on the three GO domains: Biological Process, Cellular Component, and Molecular Function. A Z-score cutoff of > 4.5, a P-value cutoff of < 0.05, and a three-gene per category minimum were used as filters to refine the ontologies. The 62 proteins selected for GO analysis are shaded in grey in Supplementary Table S1.
From our analysis, we found that all the identified GO terms relate to mRNA processing (Figure 1B). This result is consistent with previous reports on the proteins that associate with ZC3H14 and the role for ZC3H14 as a regulator of mRNA processing (8). Previous studies show that ZC3H14 localizes to nuclear speckles and interacts with components of the exon junction complex (EJC) (8,15,37). Consistent with these data, the GO terms for nuclear speckle and the EJC serve as positive controls for our analysis. Figure 1C presents a circle graph depicting the specific genes identified in each of the GO categories. The colored sections correspond to the GO domain, where each GO term is linked to the individual gene symbol. All of the identified factors that cluster into GO terms (38 total) are shown. Notably, among the most significant categories identified is the THO complex. Every member of the mammalian THO complex was identified in the ZC3H14-interacting proteome, as indicated in Figure 1C by the asterisks. In addition, two THO-associated factors were identified, ALYREF and CHTOP (38,39). These results suggest a potential physical link between ZC3H14 and the THO complex.
ZC3H14 physically associates with the THO complex
The mammalian THO complex is a six-member complex implicated in various RNA processing events that plays critical roles in dictating mRNA fate (40,41). Functional studies of various THO components in mammalian cells and budding yeast suggest roles in transcription elongation, mRNA splicing and nuclear mRNA export (20–23). Like ZC3H14, THO components are ubiquitously expressed and yet mutations in two THO component genes THOC2 and THOC6 are linked to brain disorders (24–26). This finding strengthens the rationale to explore the relationship between ZC3H14 and the individual THO components.
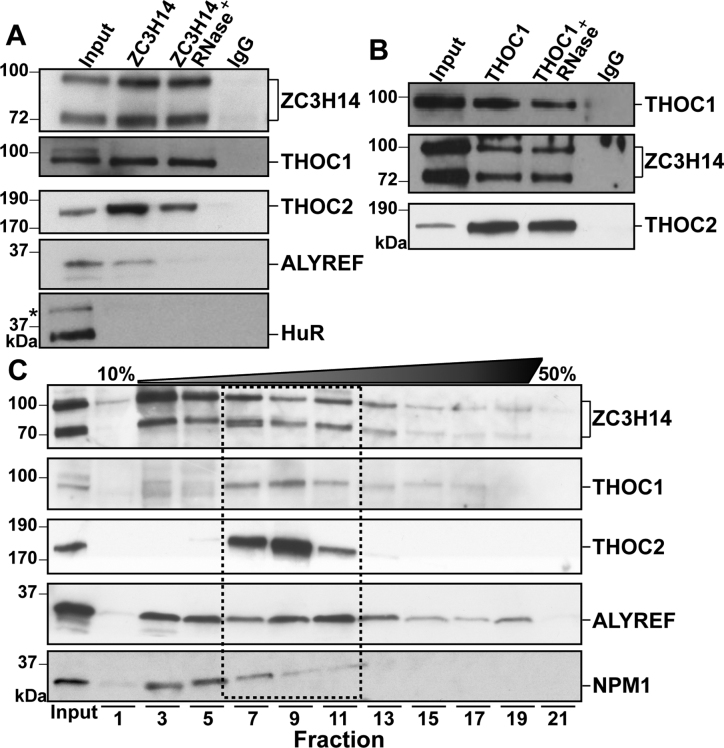
To validate the interaction between ZC3H14 and individual THO components, we immunoprecipitated ZC3H14 from mouse brain nuclear lysate as described for the mass spectrometry analysis. Utilizing SDS-PAGE and immunoblotting, we assessed the enrichment of THO components in the ZC3H14 immunoprecipitate. As shown in Figure 2A, the ZC3H14 protein is enriched in the immunoprecipitation compared to the input and the IgG control. The THO components THOC1 and THOC2 and the THO-associated protein ALYREF are all enriched in the bound fraction with ZC3H14 but not with the IgG control (Figure 2A). The interaction between ZC3H14 and THOC1 is not affected by the addition of RNase, however, the interaction between ZC3H14 and THOC2 is partially reduced upon addition of RNase and the interaction with ALYREF is completely abolished. As a negative control, we used the RNA-binding protein HuR, which was not detected as an interacting protein in our mass spectrometry experiments. Consistent with our analysis, HuR shows no co-enrichment with ZC3H14. To further examine the interaction between ZC3H14 and THO components, we performed a reciprocal immunoprecipitation with THOC1. As shown in Figure 2B, ZC3H14 co-immunoprecipitates with THOC1 in a manner that is insensitive to RNase treatment. THOC2 serves as a positive control for THOC1 binding, as these proteins interact within the THO complex (42–44).
Figure 2.
ZC3H14 interacts with the THO complex. (A) Co-immunoprecipitation of ZC3H14 and THO components from mouse brain lysate. The Input and immunoprecipitated samples for ZC3H14, ZC3H14 + RNase, and control IgG are shown. THOC1 and THOC2 serve as representative members of the THO complex, together with the THO-associated protein ALYREF. HuR serves as a negative control. Asterisks indicate non-specific protein bands detected by the commercial antibodies. (B) Reciprocal co-immunoprecipitation using anti-THOC1. The Input and immunoprecipitated samples for THOC1, THOC1 + RNase and control IgG immunoprecipitations were probed to detect THOC1, ZC3H14 and THOC2 proteins. (C) Sedimentation of ZC3H14 protein complexes from the nuclear fraction of mouse brain lysate through a 10–50% glycerol gradient. Gradient fractions from top to bottom were resolved on SDS-PAGE from left to right. Fractions were probed with antibodies to detect ZC3H14, THOC1, THOC2, ALYREF and Nucleophosmin (NPM1) as a control. The Input sample is shown on the left. Overlapping fractions are outlined. Molecular weights are shown in kDa and indicated to the left.
To test whether ZC3H14 and the THO complex exist in a macromolecular complex, we employed glycerol density gradients to sediment nuclear lysate. We purified nuclei from mouse brains and fractionated the nuclease-treated nuclear lysate through a 10–50% glycerol density gradient. Figure 2C shows the resulting immunoblot performed on the glycerol gradient fractions. The fractions increase in density from left to right, with the lightest protein complexes sedimenting towards the left and the heaviest complexes towards the right. ZC3H14 has a wide distribution throughout the gradient. The THO components THOC1 and THOC2 sediment together and are largely restricted to fractions 7–9. Fractions 7–9 also are enriched for ZC3H14 and the THO-associated protein ALYREF. However, these THO-containing fractions do not show enrichment for all nuclear RNA-binding proteins as evidenced by the distribution of the Nucleophosmin (NPM1) protein. These data show a clear overlap between ZC3H14, ALYREF and THO component containing fractions suggesting that these proteins can exist in a complex.
Loss of ZC3H14 and THO components affect processing of mRNAs
THO components are required for proper bulk poly(A) tail length control
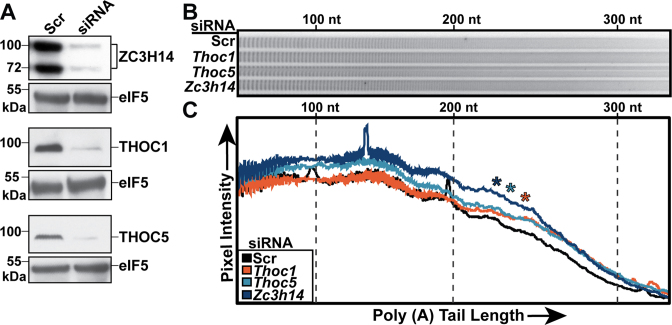
Previous studies have shown that ZC3H14 plays an evolutionarily conserved role in poly(A) tail length control (14,18,19,45). We tested whether THO components are also required for proper poly(A) tail length control. For these studies, we used a mouse neuronal cell line, Neuro2a (N2a) cells, as our experimental model (35). We treated these cells with siRNA to knockdown Zc3h14, Thoc1 or Thoc5 and compared to cells treated with a scrambled (Scr) control siRNA. Total RNA from these cells was then isolated and subjected to a poly(A) tail length assay as described in Materials and Methods. Figure 3A shows an immunoblot demonstrating efficient siRNA-mediated depletion of THOC1, THOC5 and ZC3H14. The protein eIF5 serves as a loading control. Figure 3B shows detection of end-labeled poly(A) tracts that were resolved on a polyacrylamide gel with the number of adenosines increasing from left to right. As shown previously for loss of ZC3H14 (45), depletion of the THO components THOC1 and THOC5 causes extended bulk poly(A) tails when compared to the control. This result is quantified by a line scan analysis of the samples (Figure 3C). The signal from each lane is normalized to the beginning of the gel to correct for loading. This analysis demonstrates that loss of ZC3H14, THOC1 or THOC5 causes a significant increase in the length of bulk poly(A) tail revealing that these factors are required for proper control of poly(A) tail length.
Figure 3.
Knockdown of ZC3H14 and THO components cause extended bulk poly(A) tails. N2a cells were treated with scrambled (Scr), Thoc1, Thoc5 or Zc3h14 siRNA to deplete the corresponding proteins. (A) Immunoblot of protein samples from siRNA-treated N2a cells. Antibodies against THOC1, THOC5, and ZC3H14 were used to confirm depletion of each protein. eIF5 serves as a loading control for each sample. Molecular weights are given in kDa and indicated to the left. (B) Representative image of 3′-end labeled poly(A) tracts from bulk poly(A) tails of the siRNA-treated cells that have been resolved on a denaturing polyacrylamide gel and detected by autoradiography. (C) Line scan analysis of bulk poly(A) tail length. Poly(A) tails were quantified using ImageQuant software by plotting Pixel Intensity versus poly(A) tail length and normalizing to the start of the gel. Size markers are shown for 100 nt, 200 nt, and 300 nt. Line scan data are presented as means for three independent experiments. Statistical analysis was performed using a non-parametric, one-way ANOVA Kruskal-Wallis test. Asterisks (*) indicate results that are statistically significant at *P-value < 0.05.
ZC3H14 and the THO complex regulate the processing of specific transcripts
To begin to define the function(s) shared by ZC3H14 and the THO complex in RNA processing, specific target transcripts needs to be defined. However, the complete set of transcripts that is regulated by ZC3H14 and/or THO components in the brain is not known. To identify common transcripts regulated by ZC3H14 and the THO complex, we performed RNA immunoprecipitations of endogenous ZC3H14, THOC5 and ALYREF and analyzed candidate target RNA transcripts. While the RNA-binding mechanism for the THO complex is not yet understood, THOC5 and ALYREF were chosen based on their suggested role as the mRNA adapters for the THO complex (39,46).
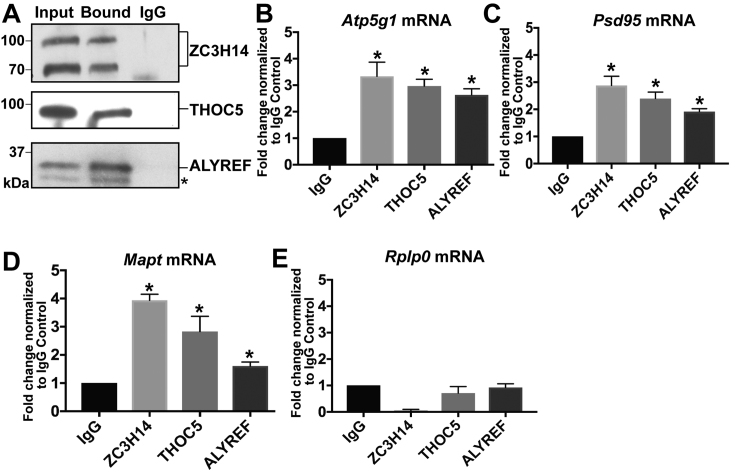
For RNA immunoprecipitations, endogenous ZC3H14, THOC5 and ALYREF were immunoprecipitated from UV crosslinked N2a cells (Figure 4A). Associated RNA was isolated, converted to cDNA and then subjected to qPCR for analysis (Figure 4B–E). Transcript levels were normalized to input and then to IgG control. Using a candidate-based approach, we tested for enrichment of RNA targets using a previously identified ZC3H14 target transcript Atp5g1 (9) and other important neuronal transcripts with potential links to ZC3H14 (unpublished results), Psd95 (Dlg4) and Mapt. The Atp5g1, Psd95 and Mapt mRNA are all significantly enriched in ZC3H14, THOC5 and ALYREF precipitations as compared to the IgG control (Figure 4B–D). The Rplp0 transcript is not enriched in these precipitations, showing that not all transcripts associate with these factors (Figure 4E).
Figure 4.
ZC3H14 and THO components bind to common mRNA targets. (A) Endogenous ZC3H14, THOC5 and ALYREF RNA immunoprecipitation from N2a cells. The Input fractions and the Bound immunoprecipitated fractions were compared to the IgG immunoprecipitated control to confirm efficient and specific immunoprecipitation of ZC3H14, THOC5, and ALYREF. Molecular weights are indicated in kDa to the left. Asterisk indicates a non-specific band recognized by the commercial ALYREF antibody. (B–E) Quantification of RNA IP enrichment in (A). RNA isolated from the RNA-IPs was subjected to RT-qPCR analyses to detect the (B) Atp5g1, (C) Psd95, (D) Mapt and (E) Rplp0 transcripts. RNA levels in the bound fractions are normalized to input levels and are presented as fold change normalized to IgG control, with the value of the IgG control set to 1.0. Values represent the mean ± SEM for n = 3 independent experiments. Asterisks (*) indicate results that are statistical significance at *P-value < 0.05.
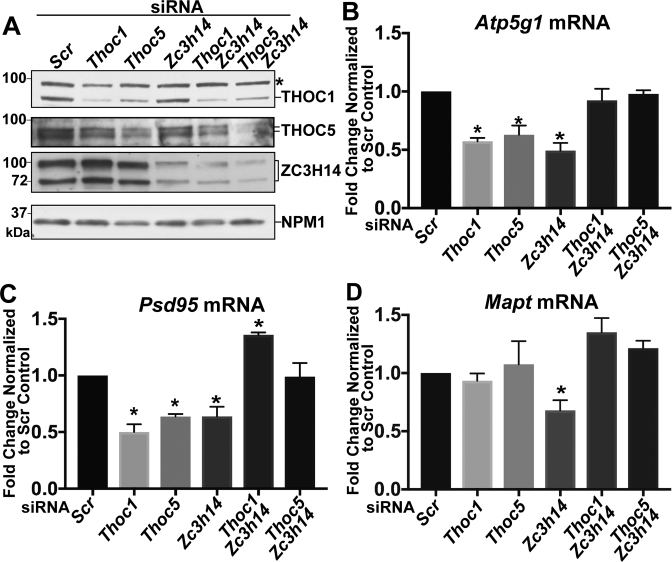
We next tested whether loss of ZC3H14 or THO components affects the steady-state levels of the Atp5g1, Psd95 or Mapt transcripts. We treated N2a cells with siRNA against Thoc1, Thoc5, Zc3h14, or a scrambled control siRNA. We isolated total protein and total RNA samples from these cells. Protein samples were separated by SDS-PAGE and analyzed by immunoblotting to confirm efficient siRNA-mediated depletion (Figure 5A). RNA from each siRNA-treated sample was analyzed by qRT-PCR to assess steady-state levels of the Atp5g1, Psd95, and Mapt transcripts. Depletion of ZC3H14, THOC1, and THOC5 all cause a statistically significant decrease (∼50%) in the steady-state levels of both the Atp5g1 and Psd95 transcripts (Figure 5B and C). In contrast, Mapt mRNA is only affected by depletion of ZC3H14 with an ∼50% decrease in steady-state levels (Figure 5C). Depletion of the THO components THOC1 or THOC5 does not alter the steady-state levels of Mapt mRNA. These results suggest that Atp5g1 and Psd95 are shared transcripts regulated by ZC3H14 and the THO complex but not all ZC3H14 target transcripts are shared with the THO complex.
Figure 5.
Depletion of ZC3H14 or THO components decreases steady-state mRNA levels. N2a cells were treated with siRNA to deplete THOC1, THOC5, or ZC3H14. Cells where either treated with individual siRNA or siRNA used in combination to deplete both THOC1 and ZC3H14 or both THOC5 and ZC3H14. Protein samples were analyzed by immunoblotting in (A) and RNA was analyzed by qRT-PCR in (B-D). (A) Antibodies against THOC1, THOC5, and ZC3H14 were used to confirm depletion of each protein with NPM1 serving as a loading control. Molecular weights are given in kDa and indicated to the left. Asterisk indicates a non-specific band recognized by the commercial THOC1 antibody. (B-D) RNA levels for (B) Atp5g1, (C) Psd95, and (D) Mapt mRNA are normalized to 18S transcript levels and are presented as fold change normalized to Scr control, with the value for the Scr control set to 1.0. Values represent the mean ± SEM for n = 3 independent experiments. All primers span exon-exon boundaries to specifically detect mature mRNA. Asterisks (*) indicate results that are statistically significant at *P < 0.05.
One potential explanation for the shared phenotypes of ZC3H14 and THO depletion on poly(A) tail length and steady-state levels of RNA targets could be that depletion of ZC3H14 decreases the levels of THO components or vice versa. To address this possibility, we treated N2a cells with siRNA against Zc3H14 or a scrambled control. We collected protein samples from the transfected cells and used immunoblotting to assess the effect of depletion of ZC3H14 on the protein level of THO components (Supplementary Figure S1A). We quantified protein levels comparing ZC3H14 depleted samples and control samples using Image J software. Depletion of ZC3H14 did not cause any significant changes in THO protein levels (Supplementary Figure S1B–E). However, depletion of ZC3H14 does cause a slight decrease in THOC5 levels that is not statistically significant (Supplementary Figure S1D). We also assessed whether depletion of THO components alters ZC3H14 proteins levels. Results of this analysis confirm that depletion of THO components does not decrease ZC3H14 protein levels (Supplementary Figure S1F and G). These data show that the shared phenotypes detected upon depletion of ZC3H14 and THO components are not due to changes in the levels of the associated proteins.
Previous work has explored genetic interactions between the THO complex genes in budding yeast and the gene that encodes the ZC3H14 orthologue, Nab2 (47–49). These studies identified complex genetic interactions suggesting that these factors coordinate proper mRNA export in yeast. To extend this analysis to mammalian cells, we used siRNA to simultaneously deplete either ZC3H14 and THOC1 or ZC3H14 and THOC5. As shown in Figure 5A, we confirmed depletion of the targeted proteins. Interestingly, in cells simultaneously treated with siRNA against Thoc1 and Zc3h14 or Thoc5 and Zc3H14, we found that the decrease in steady-state mRNA levels detected upon depletion of each of these individual components is rescued. In contrast to depletion of any single protein, co-depletion of ZC3H14 and either THOC1 or THOC5 causes no statistically significant change in the steady-state levels of Atp5g1, Psd95 or Mapt (Figure 5B–D). In fact, simultaneous depletion of ZC3H14 and THOC1 causes an increase in the steady-state levels of Psd95 compared to the scrambled control (Figure 5C) and depletion of ZC3H14 together with THOC1 or THOC5 causes a slight, but not statistically significant, increase in the steady-state levels of Mapt mRNA (Figure 5D). These results show that simultaneous depletion of both ZC3H14 and THO components ameliorates the individual effect of depletion of either one of these RNA processing factors.
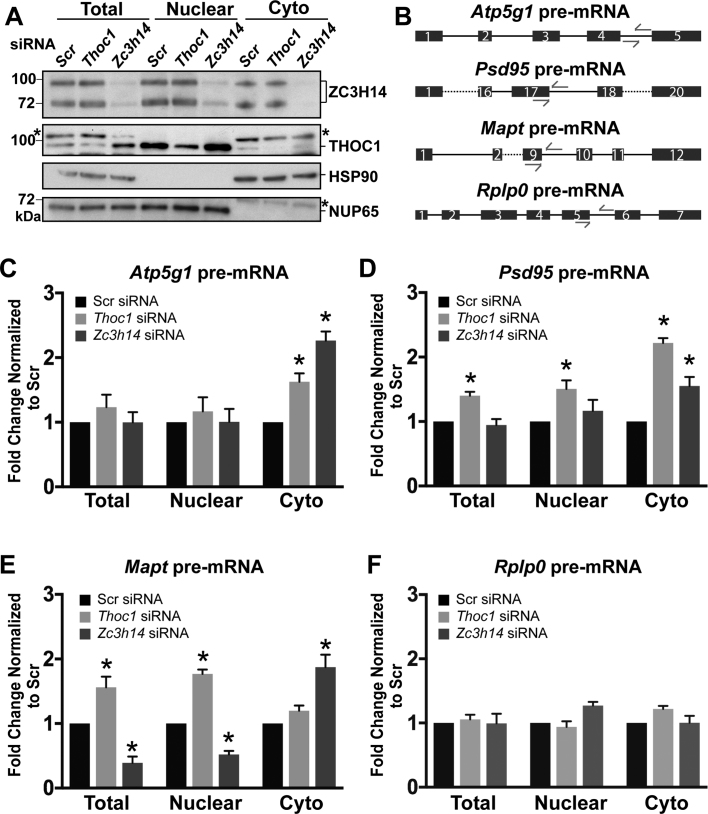
Previous work demonstrated that ZC3H14 is critical to prevent the escape of premature or intron-containing Atp5g1 transcript to the cytoplasm in cultured breast cancer cells (9). To investigate the effect of ZC3H14 and the THO complex on pre-mRNA, we depleted either ZC3H14 or THOC1 using siRNA in N2a cells and fractionated the cells into nuclear and cytoplasmic fractions. We confirmed successful knockdown and fractionation by immunoblotting (Figure 6A). As markers for fractionation, we used NUP62 (nuclear) and HSP90 (cytoplasmic) proteins. As illustrated in Figure 6B, we designed primers that specifically detect pre-mRNA of Atp5g1, Psd95, Mapt and a control transcript Rplp0. The location of the qPCR primers is indicated by arrows above their targeted regions in the schematic corresponding to each pre-mRNA. We then assessed whether there is an increase in cytoplasmic pre-mRNA upon siRNA-mediated depletion of ZC3H14 or THOC1 by measuring total, nuclear, and cytoplasmic levels of Atp5g1, Psd95, Mapt, and Rplp0 pre-mRNA (Figure 6C–F). RNA levels were normalized first to 18S rRNA and then to the Scr control for each fraction. After normalization, the fold changes normalized to the Scr control (which were set to 1.0) were compared within each fraction. The amount of both Atp5g1 and Psd95 pre-mRNA detected in the cytoplasm significantly increases upon depletion of either ZC3H14 or THOC1 (Figure 6C and D). In addition, the total and nuclear levels of Psd95 pre-mRNA significantly increase with depletion of THOC1 (Figure 6D). Consistent with previous data suggesting that Mapt is an unshared target of ZC3H14, Mapt pre-mRNA levels in the cytoplasm significantly increase upon depletion of ZC3H14 but not THOC1 depletion (Figure 6E). However, the total and nuclear levels of Mapt pre-mRNA do significantly increase upon depletion of THOC1 (Figure 6E). The pre-mRNA of the control transcript Rplp0 shows no statistically significant change in any of the fractions analyzed (Figure 6F). These data demonstrate that depletion of both ZC3H14 and THOC1 can allow escape of specific pre-mRNAs to the cytoplasm.
Figure 6.
Depletion of ZC3H14 or THOC1 causes accumulation of target pre-mRNA in the cytoplasm. N2a cells were treated with scrambled (Scr), Thoc1, or Zc3h14 siRNA and fractionated into Total, Nuclear, and Cytoplasmic (Cyto) fractions. Protein and RNA from these fractions were analyzed by immunoblotting (A) and qRT-PCR (C-F). (A) Fractions were probed with antibodies to detect ZC3H14 and THOC1 to confirm efficient depletion. Antibodies against the nuclear protein NUP62 and the cytoplasmic protein HSP90 were used to normalize loading and confirm proper fractionation. Molecular weights are given in kDa and indicated to the left. Asterisks indicate non-specific bands recognized by the commercial THOC1 and NUP62 antibody. (B) Schematic of pre-mRNA primers. The locations of the qPCR primers are indicated by arrows in the schematics above the targeted regions. Exons are represented as boxes and introns are represented as lines for each pre-mRNA. Dashed lines indicate regions of the transcript that are not shown. (C–F) Quantification of the total, nuclear, and cytoplasmic (Cyto) pre-mRNA levels in ZC3H14 or THOC1 depleted samples compared to Scr control treated samples. Pre-mRNA levels for (C) Atp5g1, (D) Psd95, (E) Mapt and (F) Rplp0 pre-mRNA were normalized to 18S transcript levels and are presented as fold change normalized to Scr control. Experimental groups for each fraction (total, nuclear, and cytoplasmic) were compared to the Scr control (which was set to 1.0) within each fraction to assess statistically significant differences. Values represent the mean ± SEM for n = 3 independent experiments. Asterisks (*) indicate results that are statistically significant at *P-value < 0.05.
As the distribution of specific pre-mRNAs changes upon loss of ZC3H14 and THOC1, we examined whether the distribution of the mature mRNAs is affected upon loss of these factors. Using qRT-PCR and primers specific to mature mRNAs, we found that there are no statistically significant changes in the nucleocytoplasmic distribution of mature Atp5g1, Psd95 and Mapt mRNA upon depletion of ZC3H14 or THOC1 (Supplementary Figure S2A–C).
The accumulation of pre-mRNA in the cytoplasm upon loss of ZC3H14 or THOC1 led us to investigate how these pre-mRNAs could prematurely exit the nucleus. One possibility is that ZC3H14 and THOC1 modulate the interaction of transcripts with the mRNA export machinery. The ZC3H14 orthologue Nab2 mediates interactions between mRNA export factors and the nuclear pore (10,50,51). To test whether ZC3H14 modulates interactions between RNA and mRNA export components, we tested whether pre-mRNA aberrantly associates with the mRNA export factor NXF1 upon depletion of ZC3H14 or THOC1. Normally, NXF1 binds to processed mRNA and mediates the rapid export of these mature mRNAs through the nuclear pore (52). Thus, NXF1 should not associate with pre-mRNA that is not competent for export.
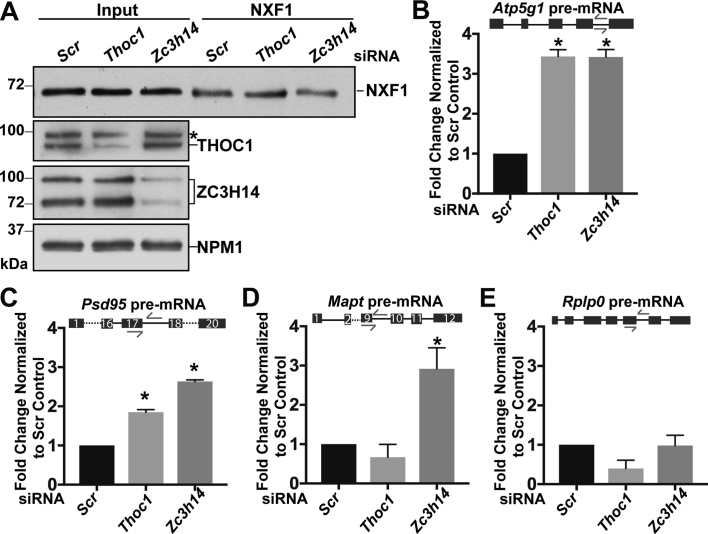
To assess whether loss of ZC3H14 or THOC1 allows premature association of NXF1 with transcripts that are not yet ready for export, we used siRNA to deplete either ZC3H14 or THOC1 in N2a cells, immunoprecipitated endogenous NXF1, and analyzed the NXF1-bound RNAs by qRT-PCR. Figure 7A shows an immunoblot of the input levels of NXF1, THOC1 and ZC3H14 and the NXF1 immunoprecipitation from the siRNA-treated cells. Depletion of ZC3H14 slightly decreases the steady-state level of NXF1 as compared to the control, while depletion of THOC1 has no significant effect on the levels of NXF1.
Figure 7.
Loss of ZC3H14 affects NXF1 target mRNA selection. Endogenous NXF1 was immunoprecipitated from N2a cells treated with scrambled (Scr), Thoc1 or Zc3h14 siRNA. (A) Immunoblot of NXF1 immunoprecipitation. Antibodies against THOC1 and ZC3H14 were used to confirm knockdown of ZC3H14 and THOC1. NPM1 serves as a loading control. Molecular weights are given in kDa and indicated on the left. Asterisk indicates a non-specific band recognized by the commercial THOC1 antibody. (B–E) Quantification of RNA-IP enrichment. RNA isolated from the NXF1 RNA-IP was subjected to qRT-PCR analyzed to detect (B) Atp5g1, (C) Psd95, (D) Mapt and (E) Rplp0 pre-mRNA. RNA levels in the NXF1 bound fractions are normalized to respective input levels and are presented as fold change normalized to Scr control, which was set to 1.0. The locations of the qPCR primers are indicated by arrows in the schematic above the graph. Exons are represented as boxes and introns are represented as lines for each pre-mRNA. Dashed lines indicate regions of the transcript that are not shown. Values represent the mean ± SEM for n = 3 independent experiments. Asterisks (*) indicate that the data are statistically significant at *P-value < 0.05.
To test for the association of NXF1 with pre-mRNA, RNA was isolated from immunoprecipitated NXF1 samples from siRNA-treated cells and subjected to qRT-PCR. RNA was normalized to the input of each condition and then compared to the scrambled control. As shown in Figure 7B and C, depletion of ZC3H14 or THOC1 causes a significant increase in the association of NXF1 with both Atp5g1 (Figure 7B) and Psd95 (Figure 7C) pre-mRNA compared to the siRNA control. Depletion of ZC3H14 but not THOC1 causes a significant increase in the association of NXF1 with Mapt pre-mRNA compared to scrambled control (Figure 7D). As a control, we measured NXF1 enrichment with RPLP0 pre-mRNA because this transcript was unchanged upon depletion of ZC3H14 nor THOC1. Neither depletion of ZC3H14 or THOC1 increases the association of NXF1 with Rplp0 pre-mRNA (Figure 7E).
DISCUSSION
In this study, we employed an unbiased proteomic approach to identify factors that interact with ZC3H14 in the murine brain. From this analysis, we identified a link between ZC3H14 and proteins that comprise the THO mRNA processing complex. Detailed analysis reveals that ZC3H14 interacts with THO components in the nucleus. We present evidence that ZC3H14 and the THO complex can cooperate to ensure proper processing of mRNA. In fact, ZC3H14 and the THO components THOC1 and THOC5 are required for proper control of bulk poly(A) tail length. In addition, we identified the transcripts Atp5g1, a nuclear encoded component of the mitochondria ATP synthase machinery (29), and the neuronal post synaptic density component, Psd95 (30), as shared targets of ZC3H14 and the THO complex. Both ZC3H14 and the THO complex are required for the proper processing of the Atp5g1 and Psd95 transcripts, as loss of either of these factors leads to a decrease in the steady-state levels of the mature transcripts. Furthermore, we provide evidence that the decrease in the steady-state level of the mature transcripts could be due to the improper export of the unprocessed pre-mRNAs from the nucleus via association with RNA export factors. Thus, these data suggest a model where ZC3H14 and the THO complex are critical RNA regulatory factors that cooperate to coordinate nuclear mRNA processing events with export from the nucleus.
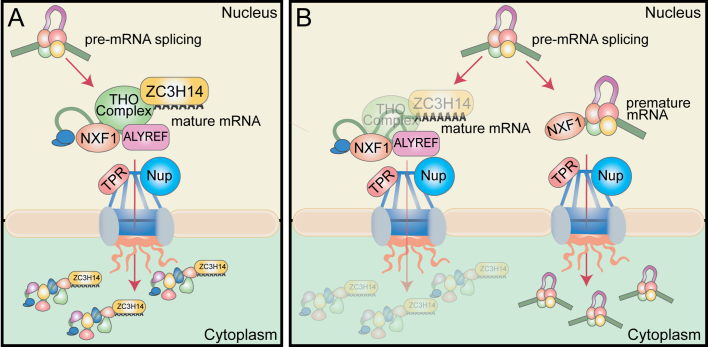
This work highlights the requirement for ZC3H14 and the THO complex in mRNA processing and establishes ZC3H14 and the THO complex as surveillance factors for nuclear mRNA processing in vertebrate cells. These data support the model presented in Figure 8A, where ZC3H14 and the THO complex are recruited to the transcript as early as splicing to ensure that the proper processing of the transcript is coupled with nuclear export. Once the transcript is checked for quality, ZC3H14 and the THO complex would then facilitate subsequent association with export factors such as NXF1.
Figure 8.
Model. Nuclear surveillance model for ZC3H14 and the THO complex. (A) In our speculative model, ZC3H14 and the THO complex are recruited to mRNA either concomitant with or after splicing. Following binding to the mRNA, ZC3H14 and the THO complex facilitate the subsequent interaction of the mRNA with export factors such as NXF1 and nuclear pore proteins (Nups), such as the nuclear basket Nup, TPR (73). (B) Loss of ZC3H14 or the THO complex causes the uncoupling of mRNA processing from nuclear export. This impairment decreases the amount of mature mRNA exported to the cytoplasm and allows an increase in the export of intron-containing premature mRNA. Loss of ZC3H14 or the THO complex allows the pre-mRNA to bypass further processing steps and improperly gain access to the mRNA export machinery.
Consistent with this model, loss of ZC3H14 or the THO complex causes defects in nuclear mRNA processing, possibly by uncoupling processing events and mRNA export (Figure 8B). This compromised quality control pathway, resulting from loss of ZC3H14 and the THO complex, could underlie the increase in the bulk poly(A) tail length of RNA and a decrease in the steady-state levels of mature target transcripts. These defects could occur because the improperly processed transcripts can bypass further processing steps and gain access to the export machinery. This defect would then cause an increase in the amount of pre-mRNA in the cytoplasm and possibly lead to downstream effects on gene expression.
Although our studies provide evidence that ZC3H14 and the THO complex cooperate to regulate mRNA processing, additional studies are required to define the precise nature of the interaction. Our mass spectrometry data reveal that all six members of the THO complex as well as the THO-associated proteins ALYREF (39) and CHTOP (38) co-immunoprecipitate with ZC3H14. We confirmed that ZC3H14 interacts with the THO components THOC1, THOC2 and the THO-associated protein ALYREF and that these proteins can exist in a macromolecular complex. The interaction between ZC3H14 and THOC1 is RNase insensitive, but the interaction between ZC3H14 and THOC2 is decreased by RNase treatment suggesting an RNA-independent interaction between ZC3H14 and THOC1. While the composition of the evolutionarily-conserved THO complex has been studied extensively in budding yeast (44,53–55), there is little information about the organization of the vertebrate THO complex or how the complex binds RNA. The THO complex could interface with various RNA-binding proteins to dictate the fate of different target transcripts. As our data reveal that ZC3H14 interacts with the THO complex, ZC3H14 could be an auxiliary component of the THO complex that targets the complex to certain mRNAs for specific functions. Thus, the THO complex could cooperate with distinct RNA-binding proteins to modulate different RNA processing events. Additional studies will be required to determine if ZC3H14 interacts directly with the THO complex, however; more knowledge of the overall organization of the THO complex must be acquired to lay the groundwork for these studies.
Both ZC3H14 and components of the THO complex are localized to nuclear speckles (15,20), which are nuclear domains enriched for RNA processing factors such as those required for pre-mRNA splicing (56,57). The location of ZC3H14 and the THO complex within these nuclear speckles appropriately poises these factors to surveille mRNA processing. While a direct role for ZC3H14 in mRNA quality control has not been fully established, our proteomic data provide additional evidence for a link between ZC3H14 and proteins that comprise the exon junction complex (EJC). The EJC is deposited 20–24 nucleotides upstream of the 5′ end of splice junctions and then dictates further processing events including mRNA export (58,59). Together with the EJC, ZC3H14 and the THO complex could ensure that the proper processing of mRNA occurs prior to nuclear export.
Both our analysis of combined depletion of ZC3H14 and THO components and genetic studies in budding yeast (49,60) provide interesting insight into how these factors cooperate. Studies in budding yeast have shown that growth defects of nab2 mutants can be suppressed by other mutants in the mRNA export pathway including tho1Δ and pml39Δ (49,60). Deletion of THO1 results in restoration of nuclear export and improved growth of nab2 mutants (49), while pml39Δ mutants suppress the growth phenotypes of nab2 mutants (60). In agreement with these data, our work shows that the combined loss of ZC3H14 and THO components rescues the steady-state levels of target mRNAs that are decreased upon depletion of either individual component. We suggest that this rescue could be the result of an increase in the export of defective RNA. As quality control factors, ZC3H14 and the THO complex would normally prevent export of defective or unprocessed mRNAs from the nucleus. The loss of one factor would cause an increase in mRNA processing defects as both ZC3H14 and the THO complex contribute to proper mRNA processing. The defective RNAs would then be retained in the nucleus and presumably degraded, resulting in the decreased steady-state levels of the mature mRNA seen with individual depletion of one component. However, in our model with the simultaneous loss of both ZC3H14 and the THO complex, the defective RNAs are not retained and degraded in the nucleus but instead can escape to the cytoplasm, which accounts for the restored steady-state levels of the target mRNA. Further characterization of the defects observed upon simultaneous depletion of ZC3H14 and THO components is needed to test this model and elucidate the exact role these factors play in the mRNA quality control pathway. Nonetheless, the confluence of the results obtained with genetic studies in budding yeast and siRNA in cultured cells strongly supports functional interplay between these mRNA processing factors.
A future challenge remains in understanding precisely how ZC3H14 and the THO complex cooperate to dictate the fate of a given mRNA. The mechanisms by which ZC3H14 and the THO complex are recruited to transcripts and the exact roles they play in mRNA processing are still not understood. With ZC3H14 and the THO complex localized to nuclear speckles, one possible model is that these factors are recruited to the transcript concomitant with the processing of the pre-mRNA. As the pre-mRNA is undergoing processing, ZC3H14 and the THO complex would govern proper polyadenylation of the 3′end and then the eventual export of the mRNA. Furthermore, recent reports have found that ZC3H14 and some THO components are detected in the cytoplasm (61–63), suggesting these proteins could accompany the mature mRNP to the cytoplasm for further regulation. Thus, these components potentially influence gene expression both through mediating and/or monitoring processing steps in the nucleus and/or through direct modulation of mRNA fate in the cytoplasm. For example, ZC3H14, which binds with high affinity to polyadenosine RNA (16) could compete with the cytoplasmic poly(A) binding protein, PABPC1, for binding to poly(A) tails to modulate translation.
As ZC3H14 and members of the THO complex are ubiquitously expressed (15,19,43), how the loss of these factors can cause brain-specific pathology is unclear. If ZC3H14 and the THO complex influence the fate of mRNA transcripts in the cytoplasm either by ensuring proper completion of nuclear processing events or by accompanying specific RNAs to the cytoplasm, this function could be more critical in neurons than in other cell types (64,65). Alternatively, ZC3H14 and the THO complex could be critical for the regulation of a specific subset of key neuronal transcripts. The loss of ZC3H14 and THO components does not affect all transcripts (9,27) raising the possibility that these factors have specificity for certain neuronal transcripts. Indeed, the Atp5g1 and Psd95 transcripts studied here are critical for proper neuronal function and are also trafficked to neurite extensions to modulate local mitochondrial function in the case of Atp5g1 (66) or to maintain the integrity of the post synaptic density in the case of Psd95 (30). Loss of either ZC3H14 or THO components would impair the processing of these transcripts, potentially interfering with proper temporal and spatial control of gene expression.
While we identified the interaction between ZC3H14 and the THO complex in the brain, this interaction is likely conserved in other tissues and cell types as these factors are all ubiquitously expressed. Thus, interplay between ZC3H14 and the THO complex could provide a fundamental mechanism to couple nuclear mRNA processing with export from the nucleus. We speculate that ZC3H14 and THO may be critical for a subset of transcripts. Indeed, the THO complex has been implicated in the resolution of R loops (67–70). Transcripts prone to form R loops may be dependent on THO and ZC3H14 for efficient processing. In fact, recent studies report that the splicing out of introns prevents the formation of R-loops (67) and that retained introns with high GC contents are prone to the formation of R-loops leading to DNA damage (71). While the introns examined in this study have not been classified as retained introns, they do have high GC contents (> 50%). Interestingly, in addition to high GC contents, R-loops are enriched for polyadenosine tracts (72). As ZC3H14 binds to polyadenosine RNA, ZC3H14 and the THO complex could work alongside the splicing machinery to aid in the resolution of R-loops during the splicing out of introns with high GC content prior to nuclear export. ZC3H14 would bind to the A-rich regions of these introns, preventing them from forming R-loops. Depletion of ZC3H14 or THO components could lead to an increase in R-loops and impact mRNA processing such that pre-mRNA accumulates, allowing improperly processed mRNA to gain access to the export machinery. Studies are underway to define the spectrum of direct targets for these factors in a common cell type. If ZC3H14 and the THO complex are needed to prevent R-loop formation, transcripts that are more prone to R-loop formation could represent a class of RNAs that are co-regulated by ZC3H14 and the THO complex. Future studies are needed to assess whether ZC3H14 and the THO complex are required for the proper processing of transcripts that are prone to R-loop formation.
In summary, our work identifies the THO complex as a novel protein interactor of ZC3H14. Using detailed biochemical and molecular approaches, we characterized the interaction between ZC3H14 and the THO complex as well as their regulation of target transcripts. Our work supports a model where ZC3H14 and the THO complex work together as quality control factors for nuclear mRNA processing and this work highlights the importance of both ZC3H14 and the THO complex in the mRNA quality control pathway.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported in part by the Emory Proteomics Core, which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [5R01GM058728, 1R21AG054206 to A.H.C., 5F31GM112418 to K.J.M.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Siomi H., Dreyfuss G.. RNA-binding proteins as regulators of gene expression. Curr. Opin. Genet. Dev. 1997; 7:345–353. [DOI] [PubMed] [Google Scholar]
- 2. Moore M.J. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005; 309:1514–1518. [DOI] [PubMed] [Google Scholar]
- 3. Singh G., Pratt G., Yeo G.W., Moore M.J.. The clothes make the mRNA: past and present trends in mRNP fashion. Annu. Rev. Biochem. 2015; 84:325–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dreyfuss G., Kim V.N., Kataoka N.. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 2002; 3:195–205. [DOI] [PubMed] [Google Scholar]
- 5. Lukong K.E., Chang K.W., Khandjian E.W., Richard S.. RNA-binding proteins in human genetic disease. Trends Genet.: TIG. 2008; 24:416–425. [DOI] [PubMed] [Google Scholar]
- 6. Zhou H., Mangelsdorf M., Liu J., Zhu L., Wu J.Y.. RNA-binding proteins in neurological diseases. Sci. China Life Sci. 2014; 57:432–444. [DOI] [PubMed] [Google Scholar]
- 7. Pak C., Garshasbi M., Kahrizi K., Gross C., Apponi L.H., Noto J.J., Kelly S.M., Leung S.W., Tzschach A., Behjati F. et al. Mutation of the conserved polyadenosine RNA binding protein, ZC3H14/dNab2, impairs neural function in Drosophila and humans. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:12390–12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Soucek S., Zeng Y., Bellur D.L., Bergkessel M., Morris K.J., Deng Q., Duong D., Seyfried N.T., Guthrie C., Staley J.P. et al. The evolutionarily-conserved polyadenosine RNA binding protein, Nab2, cooperates with splicing machinery to regulate the fate of pre-mRNA. Mol. Cell. Biol. 2016; 36:2714–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wigington C.P., Morris K.J., Newman L.E., Corbett A.H.. The polyadenosine RNA-binding protein, zinc finger Cys3His protein 14 (ZC3H14), regulates the pre-mRNA processing of a key ATP synthase subunit mRNA. J. Biol. Chem. 2016; 291:22442–22459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grant R.P., Marshall N.J., Yang J.C., Fasken M.B., Kelly S.M., Harreman M.T., Neuhaus D., Corbett A.H., Stewart M.. Structure of the N-terminal Mlp1-binding domain of the Saccharomyces cerevisiae mRNA-binding protein, Nab2. J. Mol. Biol. 2008; 376:1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marfatia K.A., Crafton E.B., Green D.M., Corbett A.H.. Domain analysis of the Saccharomyces cerevisiae heterogeneous nuclear ribonucleoprotein, Nab2p. Dissecting the requirements for Nab2p-facilitated poly(A) RNA export. J. Biol. Chem. 2003; 278:6731–6740. [DOI] [PubMed] [Google Scholar]
- 12. Anderson J.T., Wilson S.M., Datar K.V., Swanson M.S.. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol. Cell. Biol. 1993; 13:2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Green D.M., Marfatia K.A., Crafton E.B., Zhang X., Cheng X., Corbett A.H.. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 2002; 277:7752–7760. [DOI] [PubMed] [Google Scholar]
- 14. Hector R.E., Nykamp K.R., Dheur S., Anderson J.T., Non P.J., Urbinati C.R., Wilson S.M., Minvielle-Sebastia L., Swanson M.S.. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 2002; 21:1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leung S.W., Apponi L.H., Cornejo O.E., Kitchen C.M., Valentini S.R., Pavlath G.K., Dunham C.M., Corbett A.H.. Splice variants of the human ZC3H14 gene generate multiple isoforms of a zinc finger polyadenosine RNA binding protein. Gene. 2009; 439:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kelly S.M., Pabit S.A., Kitchen C.M., Guo P., Marfatia K.A., Murphy T.J., Corbett A.H., Berland K.M.. Recognition of polyadenosine RNA by zinc finger proteins. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:12306–12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kelly S.M., Leung S.W., Apponi L.H., Bramley A.M., Tran E.J., Chekanova J.A., Wente S.R., Corbett A.H.. Recognition of polyadenosine RNA by the zinc finger domain of nuclear poly(A) RNA-binding protein 2 (Nab2) is required for correct mRNA 3′-end formation. J. Biol. Chem. 2010; 285:26022–26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kelly S.M., Bienkowski R., Banerjee A., Melicharek D.J., Brewer Z.A., Marenda D.R., Corbett A.H., Moberg K.H.. The Drosophila ortholog of the Zc3h14 RNA binding protein acts within neurons to pattern axon projection in the developing brain. Dev. Neurobiol. 2016; 76:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rha J., Jones S.K., Fidler J., Banerjee A., Leung S.W., Morris K.J., Wong J.C., Inglis G.A.S., Shapiro L., Deng Q. et al. The RNA-binding protein, ZC3H14, is required for proper Poly(A) tail length control, expression of synaptic proteins, and brain function in mice. Hum. Mol. Genet. 2017; 26:3663–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Masuda S., Das R., Cheng H., Hurt E., Dorman N., Reed R.. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005; 19:1512–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huertas P., Garcia-Rubio M.L., Wellinger R.E., Luna R., Aguilera A.. An hpr1 point mutation that impairs transcription and mRNP biogenesis without increasing recombination. Mol. Cell. Biol. 2006; 26:7451–7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rougemaille M., Dieppois G., Kisseleva-Romanova E., Gudipati R.K., Lemoine S., Blugeon C., Boulay J., Jensen T.H., Stutz F., Devaux F. et al. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell. 2008; 135:308–321. [DOI] [PubMed] [Google Scholar]
- 23. Zenklusen D., Vinciguerra P., Wyss J.C., Stutz F.. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell Biol. 2002; 22:8241–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kumar R., Corbett M.A., van Bon B.W., Woenig J.A., Weir L., Douglas E., Friend K.L., Gardner A., Shaw M., Jolly L.A. et al. THOC2 mutations implicate mRNA-export pathway in X-linked intellectual disability. Am. J. Hum. Genet. 2015; 97:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Di Gregorio E., Bianchi F.T., Schiavi A., Chiotto A.M., Rolando M., Verdun di Cantogno L., Grosso E., Cavalieri S., Calcia A., Lacerenza D. et al. A de novo X;8 translocation creates a PTK2-THOC2 gene fusion with THOC2 expression knockdown in a patient with psychomotor retardation and congenital cerebellar hypoplasia. J. Med. Genet. 2013; 50:543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beaulieu C.L., Huang L., Innes A.M., Akimenko M.A., Puffenberger E.G., Schwartz C., Jerry P., Ober C., Hegele R.A., McLeod D.R. et al. Intellectual disability associated with a homozygous missense mutation in THOC6. Orphanet. J. Rare Dis. 2013; 8:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guria A., Tran D.D., Ramachandran S., Koch A., El Bounkari O., Dutta P., Hauser H., Tamura T.. Identification of mRNAs that are spliced but not exported to the cytoplasm in the absence of THOC5 in mouse embryo fibroblasts. RNA. 2011; 17:1048–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang L., Miao Y.L., Zheng X., Lackford B., Zhou B., Han L., Yao C., Ward J.M., Burkholder A., Lipchina I. et al. The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self-renewal and somatic cell reprogramming. Cell Stem Cell. 2013; 13:676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bonora M., Bononi A., De Marchi E., Giorgi C., Lebiedzinska M., Marchi S., Patergnani S., Rimessi A., Suski J.M., Wojtala A. et al. Role of the c subunit of the FO ATP synthase in mitochondrial permeability transition. Cell Cycle. 2013; 12:674–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kornau H.C., Schenker L.T., Kennedy M.B., Seeburg P.H.. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995; 269:1737–1740. [DOI] [PubMed] [Google Scholar]
- 31. Guillemin I., Becker M., Ociepka K., Friauf E., Nothwang H.G.. A subcellular prefractionation protocol for minute amounts of mammalian cell cultures and tissue. Proteomics. 2005; 5:35–45. [DOI] [PubMed] [Google Scholar]
- 32. Donovan L.E., Dammer E.B., Duong D.M., Hanfelt J.J., Levey A.I., Seyfried N.T., Lah J.J.. Exploring the potential of the platelet membrane proteome as a source of peripheral biomarkers for Alzheimer's disease. Alzheimers Res. Ther. 2013; 5:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Herskowitz J.H., Seyfried N.T., Gearing M., Kahn R.A., Peng J., Levey A.I., Lah J.J.. Rho kinase II phosphorylation of the lipoprotein receptor LR11/SORLA alters amyloid-beta production. J. Biol. Chem. 2011; 286:6117–6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zambon A.C., Gaj S., Ho I., Hanspers K., Vranizan K., Evelo C.T., Conklin B.R., Pico A.R., Salomonis N.. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 2012; 28:2209–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mao A.J., Bechberger J., Lidington D., Galipeau J., Laird D.W., Naus C.C.. Neuronal differentiation and growth control of neuro-2a cells after retroviral gene delivery of connexin43. J. Biol. Chem. 2000; 275:34407–34414. [DOI] [PubMed] [Google Scholar]
- 36. Apponi L.H., Leung S.W., Williams K.R., Valentini S.R., Corbett A.H., Pavlath G.K.. Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Hum. Mol. Genet. 2010; 19:1058–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Singh G., Kucukural A., Cenik C., Leszyk J.D., Shaffer S.A., Weng Z., Moore M.J.. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012; 151:750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chang C.T., Hautbergue G.M., Walsh M.J., Viphakone N., van Dijk T.B., Philipsen S., Wilson S.A.. Chtop is a component of the dynamic TREX mRNA export complex. EMBO J. 2013; 32:473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou Z., Luo M.J., Straesser K., Katahira J., Hurt E., Reed R.. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000; 407:401–405. [DOI] [PubMed] [Google Scholar]
- 40. Luna R., Rondon A.G., Aguilera A.. New clues to understand the role of THO and other functionally related factors in mRNP biogenesis. Biochim. Biophys. Acta. 2012; 1819:514–520. [DOI] [PubMed] [Google Scholar]
- 41. Rondon A.G., Jimeno S., Aguilera A.. The interface between transcription and mRNP export: from THO to THSC/TREX-2. Biochim. Biophys. Acta. 2010; 1799:533–538. [DOI] [PubMed] [Google Scholar]
- 42. Rehwinkel J., Herold A., Gari K., Kocher T., Rode M., Ciccarelli F.L., Wilm M., Izaurralde E.. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 2004; 11:558–566. [DOI] [PubMed] [Google Scholar]
- 43. Strasser K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondon A.G., Aguilera A., Struhl K., Reed R. et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002; 417:304–308. [DOI] [PubMed] [Google Scholar]
- 44. Ren Y., Schmiege P., Blobel G.. Structural and biochemical analyses of the DEAD-box ATPase Sub2 in association with THO or Yra1. Elife. 2017; 6:e20070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kelly S.M., Leung S.W., Pak C., Banerjee A., Moberg K.H., Corbett A.H.. A conserved role for the zinc finger polyadenosine RNA binding protein, ZC3H14, in control of poly(A) tail length. RNA. 2014; 20:681–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Katahira J., Inoue H., Hurt E., Yoneda Y.. Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J. 2009; 28:556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Piruat J.I., Aguilera A.. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 1998; 17:4859–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jimeno S., Rondon A.G., Luna R., Aguilera A.. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002; 21:3526–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jimeno S., Luna R., Garcia-Rubio M., Aguilera A.. Tho1, a novel hnRNP, and Sub2 provide alternative pathways for mRNP biogenesis in yeast THO mutants. Mol. Cell. Biol. 2006; 26:4387–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Iglesias N., Tutucci E., Gwizdek C., Vinciguerra P., Von Dach E., Corbett A.H., Dargemont C., Stutz F.. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 2010; 24:1927–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Saroufim M.A., Bensidoun P., Raymond P., Rahman S., Krause M.R., Oeffinger M., Zenklusen D.. The nuclear basket mediates perinuclear mRNA scanning in budding yeast. J. Cell. Biol. 2015; 211:1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Viphakone N., Hautbergue G.M., Walsh M., Chang C.T., Holland A., Folco E.G., Reed R., Wilson S.A.. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat. Commun. 2012; 3:1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gewartowski K., Cuellar J., Dziembowski A., Valpuesta J.M.. The yeast THO complex forms a 5-subunit assembly that directly interacts with active chromatin. Bioarchitecture. 2012; 2:134–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Poulsen J.B., Sanderson L.E., Agerschou E.D., Dedic E., Boesen T., Brodersen D.E.. Structural characterization of the Saccharomyces cerevisiae THO complex by small-angle X-ray scattering. PLoS One. 2014; 9:e103470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pena A., Gewartowski K., Mroczek S., Cuellar J., Szykowska A., Prokop A., Czarnocki-Cieciura M., Piwowarski J., Tous C., Aguilera A. et al. Architecture and nucleic acids recognition mechanism of the THO complex, an mRNP assembly factor. EMBO J. 2012; 31:1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wansink D.G., Schul W., van der Kraan I., van Steensel B., van Driel R., de Jong L.. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J. Cell Biol. 1993; 122:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Phair R.D., Misteli T.. High mobility of proteins in the mammalian cell nucleus. Nature. 2000; 404:604–609. [DOI] [PubMed] [Google Scholar]
- 58. Le Hir H., Gatfield D., Izaurralde E., Moore M.J.. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001; 20:4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Le Hir H., Moore M.J., Maquat L.E.. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000; 14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 60. Palancade B., Zuccolo M., Loeillet S., Nicolas A., Doye V.. Pml39, a novel protein of the nuclear periphery required for nuclear retention of improper messenger ribonucleoparticles. Mol. Biol. Cell. 2005; 16:5258–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bienkowski R.S., Banerjee A., Rounds J.C., Rha J., Omotade O.F., Gross C., Morris K.J., Leung S.W., Pak C., Jones S.K. et al. The conserved, disease-associated RNA binding protein dNab2 interacts with the fragile X protein ortholog in Drosophila neurons. Cell Rep. 2017; 20:1372–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tascou S., Kang T.W., Trappe R., Engel W., Burfeind P.. Identification and characterization of NIF3L1 BP1, a novel cytoplasmic interaction partner of the NIF3L1 protein. Biochem. Biophys. Res. Commun. 2003; 309:440–448. [DOI] [PubMed] [Google Scholar]
- 63. El Bounkari O., Guria A., Klebba-Faerber S., Claussen M., Pieler T., Griffiths J.R., Whetton A.D., Koch A., Tamura T.. Nuclear localization of the pre-mRNA associating protein THOC7 depends upon its direct interaction with Fms tyrosine kinase interacting protein (FMIP). FEBS Lett. 2009; 583:13–18. [DOI] [PubMed] [Google Scholar]
- 64. Holt C.E., Schuman E.M.. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron. 2013; 80:648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Davis H.P., Squire L.R.. Protein synthesis and memory: a review. Psychol. Bull. 1984; 96:518–559. [PubMed] [Google Scholar]
- 66. Natera-Naranjo O., Kar A.N., Aschrafi A., Gervasi N.M., Macgibeny M.A., Gioio A.E., Kaplan B.B.. Local translation of ATP synthase subunit 9 mRNA alters ATP levels and the production of ROS in the axon. Mol. Cell. Neurosci. 2012; 49:263–270. [DOI] [PubMed] [Google Scholar]
- 67. Bonnet A., Grosso A.R., Elkaoutari A., Coleno E., Presle A., Sridhara S.C., Janbon G., Geli V., de Almeida S.F., Palancade B.. Introns protect eukaryotic genomes from transcription-associated genetic instability. Mol. Cell. 2017; 67:608–621. [DOI] [PubMed] [Google Scholar]
- 68. Gomez-Gonzalez B., Garcia-Rubio M., Bermejo R., Gaillard H., Shirahige K., Marin A., Foiani M., Aguilera A.. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 2011; 30:3106–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pfeiffer V., Crittin J., Grolimund L., Lingner J.. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J. 2013; 32:2861–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dominguez-Sanchez M.S., Barroso S., Gomez-Gonzalez B., Luna R., Aguilera A.. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 2011; 7:e1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jangi M., Fleet C., Cullen P., Gupta S.V., Mekhoubad S., Chiao E., Allaire N., Bennett C.F., Rigo F., Krainer A.R. et al. SMN deficiency in severe models of spinal muscular atrophy causes widespread intron retention and DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E2347–E2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wahba L., Costantino L., Tan F.J., Zimmer A., Koshland D.. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016; 30:1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cordes V.C., Reidenbach S., Rackwitz H.-R., Franke W.W.. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex–attached intranuclear filaments. J. Cell Biol. 1997; 136:515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.