Abstract
RNase P is a ubiquitous site-specific endoribonuclease primarily responsible for the maturation of tRNA. Throughout the three domains of life, the canonical form of RNase P is a ribonucleoprotein (RNP) built around a catalytic RNA. The core RNA is well conserved from bacteria to eukaryotes, whereas the protein parts vary significantly. The most complex and the least understood form of RNase P is found in eukaryotes, where multiple essential proteins playing largely unknown roles constitute the bulk of the enzyme. Eukaryotic RNase P was considered intractable to in vitro reconstitution, mostly due to insolubility of its protein components, which hindered its studies. We have developed a robust approach to the in vitro reconstitution of Saccharomyces cerevisiae RNase P RNPs and used it to analyze the interplay and roles of RNase P components. The results eliminate the major obstacle to biochemical and structural studies of eukaryotic RNase P, identify components required for the activation of the catalytic RNA, reveal roles of proteins in the enzyme stability, localize proteins on RNase P RNA, and demonstrate the interdependence of the binding of RNase P protein modules to the core RNA.
INTRODUCTION
RNase P is an essential enzyme universally responsible for the site-specific cleavage of precursor tRNA leading to the formation of mature 5′-ends of tRNA (1). While in some cases the role of RNase P is played by regular protein enzymes (PRORPs), the canonical RNase P found in all three domains of life is a catalytic RNP that uses its conserved RNA moiety for catalysis (2–5). For reasons that are not well understood, the complexity and the apparent degree of the reliance on the protein components grew progressively from bacterial to archaeal to eukaryotic RNases P (6).
Bacterial RNase P holoenzyme is composed of a large (typically, ∼400 nucleotides (nt) long) RNA and a small (∼14 kDa) basic protein and can be reconstituted in vitro (7). The RNA component of bacterial RNase P is catalytically active in vitro under conditions of elevated ionic strength (2). The protein component of the holoenzyme interacts with the 5′-leader region of the precursor tRNA substrate, reducing the electrostatic repulsion between the enzyme and the substrate (8–10).
The protein part of a typical archaeal RNase P has five different components; none of them are homologous to the bacterial RNase P protein (3,11). Catalytically active archaeal RNase P can be reconstituted in vitro from proteins and RNA, which has enabled detailed studies of the interplay of its components (3,11). In archaeal RNase P, four of the proteins were shown to function as interacting pairs: Rpp30/aPop5 (yeast homologues are Rpp1/Pop5, Figure 1B) and Rpp21/Rpp29 (yeast homologues are Rpr2/Pop4, Figure 1B) (11,12). The Rpp30/aPop5 pair was shown to promote substrate cleavage and was suggested to have a functional overlap with the bacterial RNase P protein (15,16). The Rpp21/Rpp29 pair was shown to contribute to the pre-tRNA substrate affinity (15). Additionally, ribosomal protein L7Ae is a part of archaeal RNase P and was shown to bind to kink-turn motifs in RNase P RNA and contribute to the activity and temperature stability of the archaeal RNP (17,18).
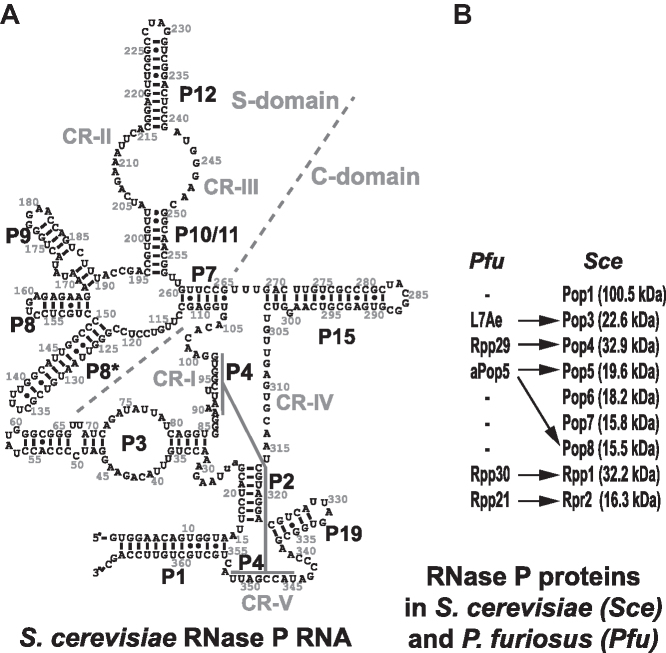
Figure 1.
(A) Secondary structure diagram of S. cerevisiae RNase P RNA (4). The nomenclature of structural elements is based on (13). (B) Protein components of archaeal RNase P from P. furiosus (Pfu) (12) and eukaryotic RNase P from S. cerevisiae (14). Arrows indicate homology between archaeal and yeast proteins.
Eukaryotic RNase P is considerably more complex than its bacterial and archaeal counterparts. The RNA component of eukaryotic RNase P (Figure 1A) has lost peripheral parts found in bacterial and most of the archaeal RNase P RNAs, but retains the core elements, and remains the catalytic moiety of the RNP (4,19). Drawing parallels to the better studied bacterial enzyme, the phylogenetically conserved regions CR-I, CR-IV, CR-V and the loop connecting stem P4 with P19 (Figure 1A) are expected to form the catalytic core of eukaryotic RNase P and are a part of the Catalytic domain (C-domain, Figure 1A), whereas the CR-II/CR-III area is expected to be involved in the recognition of the characteristic ‘elbow’ region of the pre-tRNA substrate (10), forming the core of the Specificity domain (S-domain, Figure 1A).
The protein part of eukaryotic RNase P constitutes the bulk of the RNP. S. cerevisiae RNase P has nine protein components (Figure 1B) (14); six of them have homologues in archaeal RNase P, and none are homologous to the sole bacterial RNase P protein. In S. cerevisiae, all RNase P proteins are essential for the cell viability and their depletion affects pre-tRNA processing (14). Human RNase P has a composition similar to that in yeast with 10 known protein subunits (3,20). In addition to its canonical role in the maturation of tRNA, eukaryotic RNase P and its components have been implicated in processes not directly related to tRNA metabolism ((21) and references therein).
The more complex eukaryotic RNase P is less studied than its bacterial and archaeal counterparts, in large extent due to the intractability of its reconstitution in vitro. In vitro reconstitution of the enzyme from components proved to be very productive for studies of bacterial and archaeal RNases P (3,10,11); however, previous attempts to reconstitute structurally homogeneous eukaryotic RNase P were not successful, which severely hampered the progress in our understanding of this enigmatic enzyme. Here, we report results of the analysis of the interplay of components of yeast RNase P revealed by a stepwise reconstitution of the RNP.
MATERIALS AND METHODS
Protein expression and purification
All procedures were performed at 4°C unless stated otherwise; all buffers were prepared to be RNase-free. Protein concentrations were determined using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific) based on absorbance at 280 nm; the extinction coefficients were calculated using ExPASy ProtParam tool (http://www.expasy.org (22)). The identities of the proteins were verified using mass spectrometry. For the detailed expression and purification protocols see Supplementary Data.
Pop1
Co-expression with Pop4 was necessary to produce soluble Pop1 as previously described (23); during Pop1 purification the co-expressed Pop4 was separated and not purified further. The reading frames of both proteins did not include any purification tags.
Pop4, Pop3, Rpr2
Expression and purification of these three proteins followed essentially the same protocol. Proteins were fused to the C-terminus of maltose-binding protein (MBP) via a linker that contained a tobacco etch virus (TEV) protease cleavage site. Purified proteins had a single additional glycine fused to their N-termini.
Rpp1/Pop5/Pop8 complex
Rpp1, Pop5 and Pop8 were co-expressed from a single codon-optimized polycistronic construct. The reading frames of the proteins did not include any purification tags.
Rpp1/Pop5 complex
The purification procedure was similar to that used for the Rpp1/Pop5/Pop8 complex, with minor modifications.
Pop6/Pop7 complex
The Pop6/Pop7 complex was expressed and purified as previously described (24).
RNA production and formation of ribonucleoprotein complexes
Saccharomyces cerevisiae RNase P RNA was produced by run-off in vitro transcription with T7 RNA polymerase using linearized pYRP (24), pYRP2 (23) or pYRP2-HDV (Supplementary Data) plasmids as templates. Plasmids pYRP2 and pYRP2-HDV carried A24, U25 deletions to facilitate proper folding (23). Plasmid pYRP2-HDV had a self-cleaving ribozyme construct fused to its 3′-termini to ensure the homogeneity of the 3′-ends of the final RNase P RNA (25). All plasmids had 5′- and 3′-terminal mutations to facilitate transcription and ribozyme cleavage (Supplementary Data).
Saccharomyces cerevisiae tRNAThr(AGT) (chromosomal location chrIII:295469–295565) was produced using in vitro T7 RNA polymerase transcription with a synthetic oligonucleotide as a template; the resulting tRNA construct included a 15-nucleotide-long leader and a 10-nucleotide-long trailer, as well as a GG added to the 5′-end to facilitate transcription (Supplementary Figure S1).
The products of transcription were purified using denaturing polyacrylamide gels as previously described (24). RNA concentration was determined with a NanoDrop ND-1000 spectrophotometer (Thermo Scientific) based on the absorbance at 260 nm and using the absorbance coefficient 0.025 (μg/ml)−1/cm−1.
Immediately before use, RNase P RNA was refolded as follows. RNA was incubated 10 min at 75°C in 20 mM HEPES–NaOH pH 7.4, followed by 15 min incubation at 30°C, then MgCl2 was added to 0.5 mM. Following 30 min incubation at 23°C, KCl (to 200 mM) and SUPERase-In RNase inhibitor (Ambion) were added. It should be noted that high magnesium concentrations interfered with the subsequent binding of the Rpp1/Pop5/Pop8 subcomplex, similar to the effect reported for the binding of human Pop5 to human RNase P RNA (20).
To form RNA–protein complexes, RNase P RNA was folded as described above, mixed with equimolar amounts of the protein(s) of interest, and incubated at 30°C for 15 min, followed by 15 min incubation at room temperature; when multiple proteins were added in a succession, the incubations were repeated for each protein added.
Gel mobility shift and activity assays
RNA–protein complexes of interest were formed as described above. The resulting complexes were loaded on 5% native polyacrylamide 1× TBE gels (3 μg of RNA and the corresponding amounts of proteins per lane), and fractionated at 4°C. RNA in the gel was visualized with Toluidine Blue stain.
For the activity assays, the complexes of interest were formed as described above, mixed with 5′-end 32P-labeled tRNAThr precursor, and incubated in a reaction buffer containing 50 mM HEPES–NaOH pH 7.8, 100 mM ammonium acetate, 10 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 0.5% glycerol, 0.5 μg/ml BSA at 30°C, unless stated otherwise. The reactions were stopped by the addition of an equal volume of loading buffer containing 8 M urea and 25 mM Na-EDTA pH 8.0. The products of the reactions were analyzed on 8% denaturing (8 M urea) polyacrylamide gels; the radioactive bands were visualized and quantified using a PhosphorImager (Molecular Dynamics). Cleavage performed by the endogenous RNase P holoenzyme was used as a control; the control reactions were performed in the same reaction buffer. RNasin RNase inhibitor (Promega) was added to all reactions at the recommended concentration. Kinetic assays were performed using the procedure described above. In the kinetic assays, the enzyme concentrations varied from 0.5 to 10 nM, the pre-tRNA substrate concentrations exceeded the enzyme concentration at least 10-fold and varied from 12.5 to 8000 nM, time points (between 2 and 120 min) were selected so that no more than 25% of the substrate was cleaved. For the estimation of the kinetic constants kcat and Km, the kinetic data were fitted to the Michaelis–Menten equation.
Footprinting assays
Fe(II)-EDTA footprinting analysis was performed as previously described (26) with modifications. In the experiments involving 5′-end 32P-labeled RNase P RNA, linearized plasmid pYRP2 (23) was used as the template for transcription. To control the heterogeneity of the 3′-ends of in vitro transcribed RNA, in the experiments involving 3′-end 32P-labeled RNase P RNA, linearized plasmid pYRP2-HDV was used as the template. The use of DEPC-treated solutions was avoided; glycerol was largely removed from samples using dialysis; the use of non-stabilized H2O2 was essential.
Proteins and RNA were taken at a 1:1 molar ratio. RNase P RNA was refolded and the RNP complexes of interest were formed as described above; the reaction buffer contained 0.5 mM MgCl2 (it should be noted that changing Mg2+concentrtation in the 0–5 mM range does not lead to appreciable changes of yeast RNase P holoenzyme footprinting results (27)). The Fenton's reactions were initiated by the addition of 1 mM (NH4)2Fe(SO4)2, 2 mM Na2EDTA (pH 8.0), 5 mM sodium ascorbate and 0.24% H2O2 and run for 15 min on ice. The reactions were stopped by the addition of 10 volumes of a stop buffer containing 10 mM thiourea, 5% ethanol (v/v), 300 mM sodium acetate (pH 5.2), 200 mM DTT, 0.5% SDS and 1/100 (v/v) of polyacryl carrier (MRC), followed by three rounds of phenol/chloroform extraction, ethanol precipitation, and electrophoretic analysis using 5–6% denaturing (8 M urea) polyacrylamide gels. To extend RNA coverage, electrophoresis was run twice for each reaction: a standard run and a prolonged run to analyze areas >100 nt away from the labeled terminus. The dried gels were scanned using a PhosphorImager (Molecular Dynamics). To quantify the results of Fe(II)-EDTA footprinting analysis, the intensity of each band was determined using ImageQuant software (Molecular Dynamics). The intensities of bands in individual lanes were normalized to account for variations in sample loading. The degree of protection in the presence of proteins was estimated from the intensities of the bands as Protection = [Intensity(RNA-only band) – Intensity(RNA with proteins band)]/Intensity(RNA-only band) for each sequence position, and the results were plotted for each gel.
Size-exclusion chromatography
RNP complexes were assembled as described above and run through a Superdex 200 10/300GL size exclusion column (GE Healthcare) in a buffer containing 50 mM HEPES–NaOH pH 7.5, 100 mM ammonium acetate, 50 mM KCl, 50 mM NaCl, 10 mM Mg-acetate, 10% (v/v) glycerol, 5 mM DTT, 0.1 mM PMSF, 0.1 mM EDTA, 0.1% (v/v) Tween 20. Fractions of interest were concentrated using Amicon Ultra-0.5 concentrator (10 000 MWCO, Millipore) and analyzed on a 15% SDS-polyacrylamide gel.
Isolation of the RNase P holoenzyme from yeast
Active RNase P holoenzyme was isolated from yeast strain OE101 (Supplementary Data) using a tandem affinity tag fused to the carboxyl terminus of RNase P protein component Rpr2. The affinity tag was based on the canonical TAP-tag (28), but had the calmodulin-binding domain substituted with a His8 tag. The isolation procedure followed the one previously used for the isolation of a closely related RNase MRP holoenzyme (29).
RESULTS AND DISCUSSION
Reconstitution of yeast RNase P RNPs
Insolubility of multiple individually expressed RNase P proteins was the major problem that prevented the reconstitution of structurally homogeneous eukaryotic RNase P in the past (14,20). We have overcome this hurdle by co-expressing problematic proteins and working with multi-subunit RNase P protein subcomplexes as well as with individual proteins. Of the nine S. cerevisiae RNase P proteins (Figure 1B), five were produced as parts of two multi-subunit protein subcomplexes (Pop6/Pop7 and Rpp1/Pop5/Pop8, Supplementary Figure S2).
The first of the protein subcomplexes used in our S. cerevisiae RNase P reconstitution is formed by Pop6 and Pop7, homologous proteins adopting the Alba protein fold (30,31). Co-expressed proteins form a heterodimer; co-expression with Pop6 is required for solubility of yeast Pop7, whereas the formation of the heterodimer is necessary for the proteins’ binding to RNase P RNA (24).
The second protein subcomplex comprises proteins Rpp1, Pop5 and Pop8. Co-expression of both archaeal and yeast Rpp1 and Pop5 was shown to result in essentially irreversible formation of a complex between the two proteins (15,32); this co-expression is required for solubility of the yeast proteins (32). Further screening has revealed that co-expression of yeast Rpp1, Pop5, and Pop8 (a homologue of Pop5) results in the formation of a complex that includes all three proteins (Materials and Methods). We did not observe any dissociation of the components of the Rpp1/Pop5/Pop8 complex produced by the co-expression of all three proteins together. The existence of the Rpp1/Pop5/Pop8 subcomplex in RNase P was previously suggested as a low-resolution electron microscopy study of yeast RNase P holoenzyme has positioned labeled carboxyl termini of Rpp1 and Pop8 in close proximity to each other (33).
Individually expressed protein Pop1 is not soluble (14,34). UV-crosslinking experiments performed on RNase P isolated from yeast (35) suggested that parts of Pop1 were in contact with Pop4, prompting us to attempt co-expression of Pop1 and Pop4. Indeed, co-expression of Pop1 and Pop4 allowed us to produce and isolate soluble Pop1 (23). Unexpectedly, co-expression of the two proteins did not yield a Pop1/Pop4 complex that could be purified: Pop4 was required for Pop1 solubility in cell lysate, but the two proteins (now both soluble) separated early in the purification process (23). Accordingly, in this work, Pop1 was co-expressed with Pop4, but isolated separately (Materials and Methods). While the exact mechanism of the observed effect of Pop4 on Pop1 folding is not clear, one can speculate that Pop4 plays a chaperoning role due to transient interactions between the two proteins during co-expression in E. coli, and it cannot be excluded that a similar interplay may potentially take place in yeast cells as well.
The remaining RNase P proteins, Pop3, Pop4, and Rpr2, were expressed individually (Materials and Methods).
We attempted reconstitution of RNase P RNP by the incubation of equimolar amounts of RNase P proteins added in a stepwise fashion to the equimolar amount of refolded in vitro transcribed RNase P RNA with essentially a wild-type sequence (Materials and Methods). The formation of the RNA–protein complexes was monitored using electrophoretic mobility shift assays (EMSA). EMSA indicated the formation of RNP complexes that were not homogeneous (Supplementary Figure S3). The partial heterogeneity of the multicomponent complexes was traced to partial RNA misfolding as described previously (23): a fraction in vitro transcribed S. cerevisiae RNase P RNA misfolds as nucleotides A24, U25 and U26 (parts of a non-conserved loop bridging stems P2 and P3 (13), Figure 1A) base pair with complementary nucleotides A314, A315, U316 of the CR-IV region, thus overextending stem P2 and sequestering a phylogenetically conserved part of the CR-IV region in vitro, and affecting the binding of the Rpp1/Pop5/Pop8 subcomplex (Supplementary Figure S3, lane 4). To facilitate the proper RNase P RNA folding, the non-conserved nucleotides A24 and U25 (shown in lower case in Figure 1A) were deleted as previously described (23), and the resulting RNA was used in lieu of the wild-type RNase P RNA in this work; for brevity, this RNA will be referred to as RNase P RNA; the effects of the deletion on the activity of the reconstituted RNP will be discussed later.
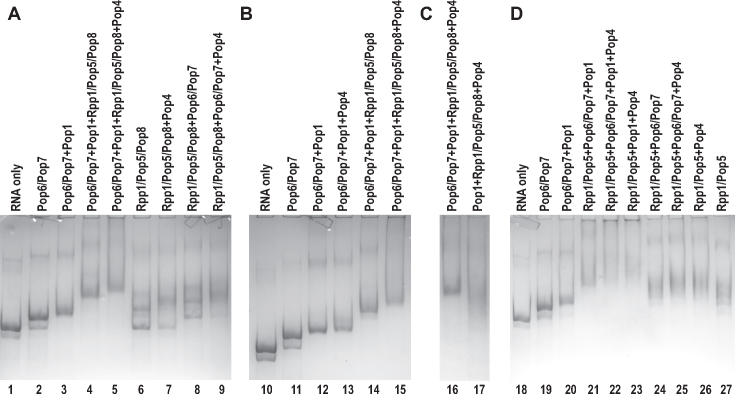
Reconstitution of a structurally homogeneous 8-component RNase P RNP (RNA plus proteins Pop1, Pop4, Pop5, Pop6, Pop7, Pop8, Rpp1) was achieved by the incubation of equimolar amounts of proteins Pop6/Pop7 (as a subcomplex), Rpp1/Pop5/Pop8 (as a subcomplex), Pop1 and Pop4 added, in a stepwise fashion (RNA→Pop6/Pop7→Pop1→Rpp1/Pop5/Pop8→Pop4) to the equimolar amount of refolded in vitro transcribed RNase P RNA that carried A24, U25 deletions. The resultant 8-component RNP migrates as a single band on a native polyacrylamide gel (Figure 2A, lanes 1–5), and a size-exclusion chromatography analysis of the assembled RNP further confirms that it contains all of the expected protein components (Supplementary Figure S4)
Figure 2.
(A) Stepwise assembly of the reconstituted RNase P RNP (lanes 1–5) and binding of RNase P proteins to RNase P RNA (lanes 6–9). (B) The addition of Pop4 does not result in a mobility shift unless the Rpp1/Pop5/Pop8 protein subcomplex is already present. (C) The presence of Pop6/Pop7 is required for the structural stability of the reconstituted RNP. (D) RNP assembly with the Rpp1/Pop5 complex substituting for Rpp1/Pop5/Pop8. Lanes 1, 10, 18: RNase P RNA alone; other lanes: electrophoretic mobility shifts upon the addition of protein components as indicated above the gel. Protein components are added to RNase P RNA at a 1:1 molar ratio; the resulting RNP complexes are resolved on a native polyacrylamide gel. RNA is stained with Toluidine Blue. Pop6 and Pop7, as well as Rpp1, Pop5, Pop8 or Rpp1, Pop5 formed subcomplexes and were co-expressed and co-purified together.
Proteins/protein subcomplexes Pop1, Pop6/Pop7, Rpp1/Pop5/Pop8 did not require the presence of other components to bind to RNase P RNA ((23,24) and Figure 2A), whereas the binding of Pop4 was sensitive to the presence of other proteins (Figure 2B and D), as discussed later. The order in which proteins have been added to RNA to form RNPs was chosen taking this observation into account.
We did not observe the binding of the remaining two RNase P proteins, Pop3 and Rpr2, to the reconstituted RNP, its assembly intermediates, or RNase P RNA using the same approach as applied for the incorporation of other components into the RNPs. This suggests that the addition of these proteins may require a chaperoning activity that is present in the cell, but not in vitro. It is important to note that yeast RNase P RNP assembly involves the formation of a stable precursor RNP form, which has all RNase P RNP proteins, except for Pop3 and Rpr2. This precursor form constitutes 10–20% of all cellular RNase P, can be isolated from yeast, and is catalytically active (36). The protein composition of the 8-component in vitro assembled RNase P RNP reported here (RNase P RNA plus proteins Pop1, Pop4, Pop5, Pop6, Pop7, Pop8, Rpp1) matches that of the stable catalytically active RNase P RNP precursor observed in yeast cells (36).
Catalytic activity and footprinting analysis of the reconstituted RNase P RNP
To validate the relevance of the in vitro assembled RNase P RNP, we assayed the activity of the RNP containing, simultaneously, RNase P RNA and RNase P proteins Pop1, Pop4, Pop5, Pop6, Pop7, Pop8, Rpp1, as well as of RNPs containing subsets of these proteins (Figure 3). The activity was assayed under conditions of moderate ionic strength and magnesium concentration using a yeast tRNAThr precursor (pre-tRNA) (Supplementary Figure S1) as a substrate. As a positive control, we used the digestion of the same substrate by the RNase P holoenzyme isolated from S. cerevisiae (Figure 3, lane 4). To ensure that the observed activity of the reconstituted RNPs was not a contamination artifact, as negative controls, we used RNPs that contained the same proteins in complexes with a mutated RNase P RNA that had the two phylogenetically conserved catalytic core nucleotides that bulge out of the P4 stem (A90 and U93 (27,37), Figure 1A) deleted (Figure 3, lanes 7, 11, 16, 18, 25). This deletion did not affect protein binding (Supplementary Figure S5).
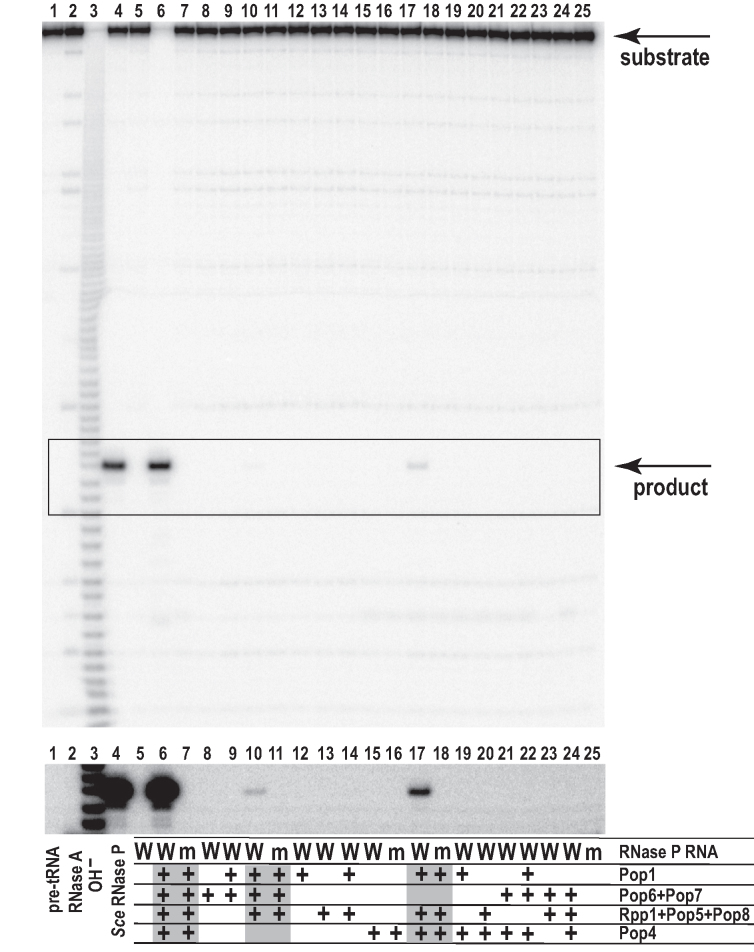
Figure 3.
Catalytic activity of reconstituted RNPs assayed under conditions of moderate ionic strength. The insert below the main gel shows the overexposed boxed section of the gel. 4 pmol (0.36 μM) of RNase P RNA was used to form RNPs; proteins were added to RNA at a 1:1 molar ratio. 40 pmol (3.6 μM) of 5′-end 32P-labeled S. cerevisiae pre-tRNAThr(AGT) substrate was incubated with the reconstituted RNPs for 30 min at 30°C in 50 mM HEPES-NaOH pH 7.8, 100 mM ammonium acetate, 10 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 0.5% glycerol, 0.5 μg/ml BSA. The protein combinations that resulted in activity (highlighted in gray below the gel) were assayed with the ‘wild type’ RNase P RNA (labeled as ‘W’ in the ‘RNase P RNA’ description lane, bottom part of the figure), as well as with a mutated RNA that lacked catalytic core nucleotides A90, U93 essential for catalysis (labeled as ‘m’ in the ‘RNase P RNA’ description lane), which served as a negative control. Lane 1: pre-tRNA substrate before cleavage. Lane 2: pre-tRNA substrate partially digested with RNase A. Lane 3: pre-tRNA substrate digested in alkali. Lane 4: pre-tRNA substrate digested with the endogenous RNase P (control). Lanes 5: incubation with RNase P RNA alone. Lanes 6–25: activities of the reconstituted RNPs with compositions as indicated below the gel.
Activity assays performed for the in vitro assembled RNP containing RNase P RNA in complex with proteins Pop1, Pop4, Pop5, Pop6, Pop7, Pop8, Rpp1 revealed robust multi-turnover cleavage of the substrate pre-tRNA (Figure 3, lane 6). The position of the cleavage site matched that produced by RNase P isolated from S. cerevisiae (Figure 3, lane 4). The control A90, U93 deletion in RNA (above) completely eliminated observed specific RNase P activity (Figure 3, lane 7). Kinetic constants under the assay conditions at 30°C were estimated to be kcat = 2.0 ± 1.0 min−1, Km = 3.5 ± 1.5 μM (compared to kcat = 40 ± 20 min−1, Km = 50 ± 20 nM estimated for RNase P isolated from yeast, assayed under the same conditions). The A24, U25 deletions (above) were not detrimental for the RNP activity; moreover, RNase P RNP assembled on the RNA with the wild-type sequence demonstrated 1.5 ± 0.2-fold lower level of the apparent catalytic activity compared to the RNP assembled on the RNase P RNA carrying the A24, U25 deletions (Supplementary Figure S6), consistent with the reduced yield of the fully assembled RNP of the former type (Supplementary Figure S3). The in vitro assembled RNase P RNP is catalytically active in a broad range of ionic strength and magnesium concentrations (Supplementary Figure S7).
The difference between kinetic parameters observed for the reconstituted RNase P RNP and the fully assembled endogenous enzyme is most likely largely attributable to the difference in the protein compositions. Indeed, the activity of both archaeal and human RNases P is affected by the presence of Pop3 homologues (17,38). Human RNase P purification fractions depleted of Rpp38 (a homologue of yeast Pop3 (36)) have been reported to have only low levels of RNase P activity (38). Moreover, the effects of the addition of L7Ae (archaeal homologue of Pop3) to the in vitro reconstituted archaeal RNase P RNP from Methanococcus maripaludis (Mma) (an archaeal RNase P of the M-type (39), which has RNA most similar to eukaryotic RNase P RNA) are in a good quantitative agreement with the difference between the activities of our in vitro assembled RNase P RNP and the endogenous RNase P. Specifically, Mma RNase P RNP lacking L7Ae (Pop3) demonstrated kcat = 10 ± 1 min−1, Km = 2.6 ± 0.3 μM assessed at 37°C under optimal buffer conditions (17). This level of activity is similar to what we observe for the reconstituted yeast RNase P missing Pop3 and Rpr2 (kcat = 2.0 ± 1.0 min−1, Km = 3.5 ± 1.5 μM, assessed at 30°C). Importantly, the addition of the constituent Pop3 homologue to the reconstituted Mma RNase P increased to kcat to 63 ± 7 min−1 and decreased Km to 44 ± 7 nM at 37°C (17), the values that are very similar to those observed for yeast RNase P isolated from cells (here and (40)). At the same time, under conditions of high substrate concentrations (5 μM, compared to mature RNase P Km ∼ 0.05 μM ((40) and this work)), the precursor form of RNase P isolated from S. cerevisiae that was missing Pop3 and Rpr2 demonstrated a pre-tRNA substrate cleavage rate similar to that of the fully assembled enzyme (36). It should be noted that at this substrate concentration, the effects of differences in Km on the observed cleavage rates would be largely suppressed, partly masking the potential difference in the catalytic efficiencies of the precursor and the mature forms. Indeed, the addition of a constituent Pop3 homologue to Mma RNase P RNP (above) resulted in a ∼6-fold increase of kcat and ∼60-fold drop in Km (17), but the 60-fold difference in Km would not be prominent in assays performed at the 5 μM concentration of the substrate. Additionally, it is possible that differences in activities of the cellular RNase P precursor (36) and the in vitro assembled RNP of the same protein composition are caused by a chaperoning activity that takes place in the cell, but is missing in vitro. Finally, the effects of potential posttranslational modifications that are not present in the reconstituted RNPs, but increase the activity of the precursor form assembled in vivo cannot be excluded.
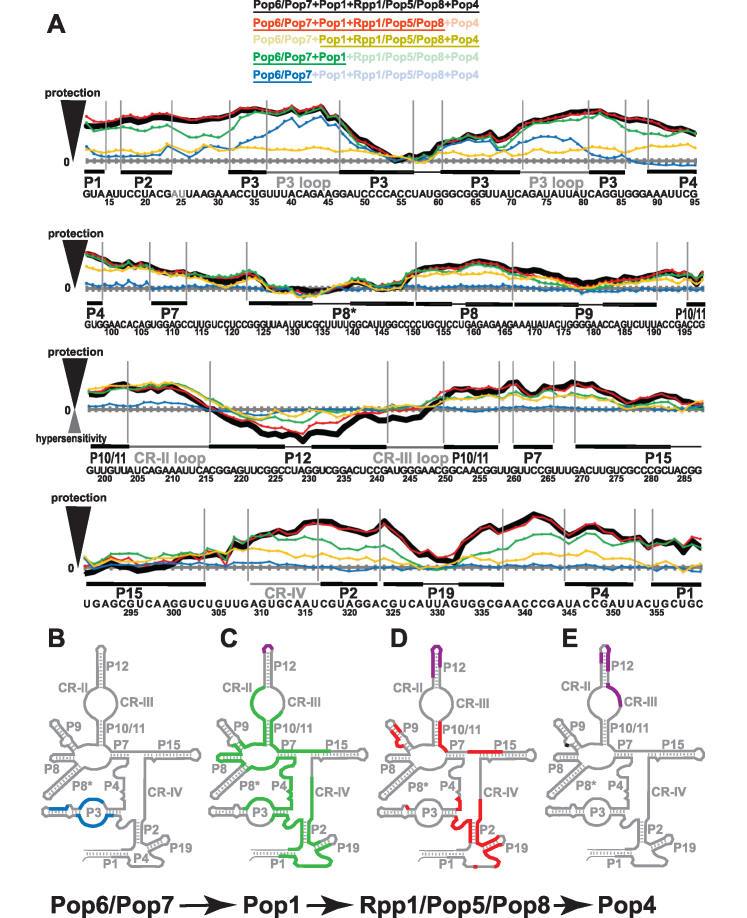
To further validate the affinity of the assembly of the 8-component RNase P RNP (RNase P RNA with Pop1, Pop4, Pop5, Pop6, Pop7, Pop8 and Rpp1), the reconstituted complex, as well as its assembly intermediates, were subjected to Fe(II)-EDTA footprinting analysis (Figure 4A and Supplementary Figure S8). The results of the footprinting analysis of the catalytically active 8-component RNase P RNP (Figure 4A in black) are similar to those previously obtained for the fully assembled RNase P holoenzyme isolated from yeast (27,41). Most parts of the RNase P RNA gain protection in the context of the in vitro assembled RNP, notable exceptions being distal parts of stems P3, P8*, P9, P12, P15 and the corresponding terminal loops, as well as the apical loop of P19. The differences between the footprinting data obtained for the in vitro assembled RNase P RNP and the fully assembled enzyme are localized to the element CR-III, which is predominantly protected in the presence of the proteins in the holoenzyme (27,41), but exposed to the solvent in the in vitro assembled RNP (Figure 4A in black). This difference in the protection of the CR-III element is consistent with the elevated Km observed for the in vitro assembled RNP as this phylogenetically conserved element participates in the pre-tRNA substrate recognition in bacterial RNase P (10,42). It should be noted that protein Rpr2 that is missing in the in vitro assembled RNase P RNP has been approximately localized to this general area in both archaea (11,43) and yeast (33), and it is likely that the observed difference in the protection of the CR-III element is caused by the absence of Rpr2.
Figure 4.
Footprinting data for RNase P RNP assemblies. (A) Quantification of hydroxyl radicals (Fe(II)-EDTA) footprinting assays. The grey horizontal axes correspond to no additional protection (same reactivity as the RNA-only reference); the elevation above an axis reflects the degree of the protection (compared to RNA-only reference) in the presence of proteins. Protein compositions of the assayed RNP complexes are color-coded as shown on top of the figure; the proteins present in the RNP are underscored. Thick black line: proteins Pop6/Pop7, Pop1, Rpp1/Pop5/Pop8, Pop4. Red line: Pop6/Pop7, Pop1, Rpp1/Pop5/Pop8. Sand line: Pop1, Rpp1/Pop5/Pop8, Pop4. Green line: Pop6/Pop7, Pop1. Blue line: Pop6/Pop7. Sequence and secondary structure elements of the RNase P RNA (Figure 1A) are shown below the graphs. Helical stems are shown by thick solid lines; terminal loops are shown by thin solid lines; large internal loops and conserved elements are shown by dotted lines. See Supplementary Figure S8 for gels. (B–E) Changes in RNase P RNA reactivity induced by protein binding during RNase P RNP assembly. (B) Protection upon the binding of Pop6/Pop7 to RNA (blue). (C) Additional protection (green) and hypersensitivity (violet) upon the binding of Pop1 to the RNA-Pop6/Pop7 RNP. (D) Additional protection (red) and hypersensitivity (violet) upon the binding of Rpp1/Pop5/Pop8 to the RNA-Pop6/Pop7/Pop1 RNP. (E) Additional protection (black) and hypersensitivity (violet) upon the binding of Pop4 to the RNA-Pop6/Pop7/Pop1/Rpp1/Pop5/Pop8 RNP.
Binding of RNase P proteins to RNA
To help better understand the interplay of the RNase P components, we assayed RNase P proteins for their binding to RNase P RNA (Figure 2). Consistent with the previous reports (23,24), protein Pop1 and the Pop6/Pop7 subcomplex do not require other proteins to bind to RNase P RNA. The Rpp1/Pop5/Pop8 subcomplex also binds to RNase P RNA in the absence of other RNase P proteins; however, when this protein subcomplex and RNA are taken at a 1:1 molar ratio, only ∼50% of RNA appears to be bound to the proteins (Figure 2A, lane 6). In the presence of Pop1 plus Pop6/Pop7, all RNase P RNA appears to be bound to Rpp1/Pop5/Pop8 (Figure 2A, lane 4); note that the presence of Pop6/Pop7 alone does not improve the binding of RNA to Rpp1/Pop5/Pop8 (Figure 2A, lane 8 vs lane 6). The observed effect of Pop1 on the binding of Rpp1/Pop5/Pop8 is consistent with the proximity of the UV-induced Pop1-RNA and Pop5-RNA crosslinks observed in the closely related RNase MRP holoenzyme (35), indicative of interactions between the two proteins.
Studies of the RNase P holoenzyme isolated from yeast showed that protein Pop4 formed UV-induced crosslinks with nucleotides A171, A173, and C192, C193 of RNase P RNA (35). The formation of these crosslinks indicates that Pop4 is involved in direct interactions with RNA, consistent with the results of yeast three-hybrid assays (44). Unexpectedly, in our assembly assays, Pop4 does not stably bind to RNase P RNA unless this RNA is decorated with specific protein components: Pop4 does not bind to the RNP containing Pop1 plus Pop6/Pop7, but binds to the RNP that has simultaneously Pop1, Pop6/Pop7, plus Rpp1/Pop5/Pop8 (Figure 2B, lanes 12, 13 versus 14, 15). The requirements for the Pop4 recruitment can be narrowed down further: the presence of Pop1, Pop6/Pop7 is not necessary for Pop4 binding as this binding is observed in the presence of only Rpp1/Pop5/Pop8; additionally, the presence of Pop4 raises the completeness of the Rpp1/Pop5/Pop8 binding to RNA (Figure 2A, lanes 6, 7). Furthermore, the presence of the Rpp1/Pop5 subcomplex appears to be sufficient for the recruitment of Pop4 (Figure 2D, lanes 26, 27). It should be noted that results of two-hybrid assays (44) are consistent with robust interactions between Pop4 and Rpp1.
Archaeal homologues of Rpp1/Pop5 can form a heterotetramer containing two copies of each protein (3,11). It was hypothesized that in eukaryotes one of the Pop5 copies was replaced with a homologous protein Pop8 (33). The formation of the Rpp1/Pop5/Pop8 subcomplex reported here is consistent with this hypothesis. To shed light on the role of Pop8, we performed assays where the Rpp1/Pop5/Pop8 subcomplex was substituted with Rpp1/Pop5 (Figure 2D). The Rpp1/Pop5 subcomplex is able to bind to RNase P RNA in the absence of other proteins, in keeping with the results previously reported for the closely related RNase MRP (32), although the binding products run as multiple bands in EMSA (Figure 2D, lane 27). The addition of Rpp1/Pop5 to the RNPs containing Pop1 plus Pop6/Pop7 (but not Pop6/Pop7 alone) leads to a noticeable and continuous precipitation of the resulting complex. Overall, it appears that Pop8, the only acidic RNase P protein (pI 4.6), is required for the proper engagement of Pop1 when Pop5 and Rpp1 are present. It should be noted that both Pop1 and Pop8 are absent in archaeal RNase P and appear only in eukaryotic enzymes.
Effects of the protein composition on RNP activity
To clarify the protein requirements for RNase P activity, we assayed pre-tRNA substrate cleavage by RNPs containing various combinations of RNase P proteins (Figure 3).
RNase P RNPs with protein combinations that did not include simultaneously Pop1, Rpp1, and Pop5 failed to demonstrate RNase P activity at detectable levels (Figure 3).
Compared to the 8-component RNP (RNase P RNA with Pop1, Pop4, Pop5, Pop6, Pop7, Pop8, Rpp1), the exclusion of the Pop6/Pop7 subcomplex or Pop4 did not affect the location of the cleavage site, but resulted in a considerable reduction in the observed catalytic activity (Figure 3, lanes 10, 17). Additionally, the exclusion of the Pop6/Pop7 subcomplex led to the instability of the resultant 6-component RNP, which was prone to aggregation and precipitation. We did not pursue kinetic measurements for these complexes due to low activity levels and precipitation. The simultaneous exclusion of both Pop6/Pop7 and Pop4 resulted in a complex that demonstrated no detectable activity in our assays (Figure 3, lane 14).
The substitution of the Rpp1/Pop5/Pop8 subcomplex with the Rpp1/Pop5 subcomplex did not affect the location of the cleavage site, but resulted in a ∼20-fold reduction of the observed rate of substrate cleavage (Supplementary Figure S9). Additionally, RNP complexes containing Rpp1/Pop5 instead of Rpp1/Pop5/Pop8 were prone to aggregation and continuous precipitation (above); the observed reduction in the cleavage rate can be attributed to a combination of a reduction of RNP activity and a reduction of the RNP concentration due to its partial precipitation. Given the difficulty of controlling the actual concentrations of RNPs containing Rpp1/Pop5 instead of Rpp1/Pop5/Pop8, we did not pursue detailed kinetic measurements for complexes that were missing Pop8; however, it is apparent that, as opposed to Rpp1/Pop5, Pop8 is not absolutely required for the catalytic activity of yeast RNase P and likely plays a structural role to properly accommodate the binding of Pop1 (above).
Localization of RNase P proteins using footprinting assays
To shed light on interactions between RNA and proteins in RNase P, we used footprinting assays to monitor the effects of protein binding on RNA accessibility in the context of partially assembled RNase P RNPs, generally following the assembly order used to reconstitute the catalytically active 8-component RNase P RNP (RNA→Pop6/Pop7→Pop1→Rpp1/Pop5/Pop8→Pop4, Figure 4).
The Pop6/Pop7 heterodimer was previously shown to interact with the P3 region of yeast RNase P and the related RNase MRP, as well as with a structurally similar CS2a/TeSS element of the telomerase RNA (31–33,45). Human homologues of Pop6 and Pop7 (Rpp25 and Rpp20, correspondingly) have also been reported to bind to the general P3 area of human RNase P and RNase MRP (20,46). Our footprinting assays show that the binding of Pop6/Pop7 to RNase P RNA results in a strong protection of the bottom part of the P3 internal loop (the U37-G46 region, Figure 4A, B, in blue) and the adjacent P3 helices, consistent with the crystal structure of Pop6/Pop7 in a complex with the P3 region of RNase MRP (31). The observed protection of the distal part of the P3 stem (G61-C71) extends further than the RNA–protein interactions observed in the crystal structure. The distal part of RNase P P3 stem has an internal bulge (C50, U67, U68, Figure 1A), which is not present in RNase MRP. The stem flexibility allowed by that bulge can explain this discrepancy between RNase MRP data and the observed protection in RNase P. The binding of Pop6/Pop7 does not cause any noticeable changes in RNA sensitivity beyond the P3 region (Figure 4A, B, in blue).
The Pop6/Pop7 heterodimer bound to the P3 stem/internal loop was proposed to form an interface that facilitates the binding of Pop1 to the surrounding areas (stems P1-P4) (23,47). The RNA protection differences between the 8-component RNase P RNP (RNase P RNA plus proteins Pop1, Pop4, Pop5, Pop6, Pop7, Pop8, Rpp1) and an RNP that is missing the Pop6/Pop7 heterodimer (Figure 4A, black versus yellow) are consistent with this Pop6/Pop7 role. It should be noted that the area affected by the removal of Pop6/Pop7 from the 8-component complex is much larger than the area affected by the addition of Pop6/Pop7 to the RNA alone. Both this difference and the drop in the RNase P activity observed when Pop6/Pop7 are excluded from the RNP (Figure 3, lane 6 versus lane 17) are attributable to the destabilization of Pop1 interactions with the P1-P4 region. The results of EMSA are also consistent with the structural destabilization of the RNase P RNP in the absence of Pop6/Pop7 as the corresponding RNP runs on a native polyacrylamide gel as a smear, as opposed to a well-defined band (Figure 2C). These observations provide support to the hypothesis that Pop6/Pop7 is a structural protein complex serving to facilitate the binding of Pop1 to the P1-P4 area of RNase P RNA.
The next step in the assembly of the catalytically active RNase P RNP, the addition of Pop1 (the largest (100.5 kDa) RNase P protein, Figure 1B) results in RNA sensitivity changes that dwarf the changes caused by other proteins (Figure 4A, C, in green). In the presence of Pop1 and Pop6/Pop7, the overall pattern of RNA protection is approaching that observed for the endogenous holoenzyme (23,27,41) and for the fully assembled 8-component RNP (Figure 4A, compare traces in green and in black). Studies of the RNase P holoenzyme isolated from yeast (35) showed that Pop1 formed a UV-induced crosslink to phylogenetically conserved nucleotide A309, which is involved in the formation of the catalytic core in bacterial RNase P RNA (10,37). Consistent with the location of that crosslink, our footprinting data (Figure 4A) indicate protection of the RNA region adjacent to A309 in the presence of Pop1. Moreover, Pop1 binding dominates the protection of the P4 stem and most of the CR-IV region (Figure 4C), key phylogenetically conserved regions of the catalytic core in bacterial RNase P RNA (10,37).
The protection of another essential part of RNase P RNA, the CR-II loop, is also dominated by Pop1 binding. In bacteria, the conserved CR-II loop forms a part of a structure responsible for the recognition of the pre-tRNA substrate (10,42,48), and it is most likely that it plays the same role in eukaryotic RNase P. The effects of Pop1 binding on the protection of CR-II loop are consistent with results of a low-resolution electron microscopy study of yeast RNase P holoenzyme (33) that positioned the labeled N-terminus of Pop1 in the vicinity of the P10/11-CR-II/III-P12 region of RNA.
Taken together, these results indicate that Pop1 spans across the RNase P RNP, from the parts forming the catalytic center, to the parts involved in pre-tRNA substrate recognition, and is positioned to (at least partially) replace the network of RNA-RNA interactions that compacts the RNA structure in bacterial RNase P RNA, but have been lost in eukaryotic enzymes (23). It is interesting to note that Pop1 binding leads to an anomalously small change in the electrophoretic mobility of the RNP (Figure 2A, lanes 2, 3), disproportionate to Pop1 size. This observed anomaly is consistent with the proposed compactization of RNA upon Pop1 binding. Further studies are required to discern the details of the roles played by Pop1 in RNase P, as it remains to be seen if its involvement in the catalytic core and the substrate recognition element is limited to facilitating RNA folding, or it plays more direct roles in interactions with substrates and in catalysis.
The subsequent addition of the Rpp1/Pop5/Pop8 protein subcomplex activates RNase P RNA (Figure 3). Both in archaeal RNase P and in yeast RNase MRP, the Rpp1/Pop5 (Rpp30/aPop5 in archaea) subcomplex was shown to bind to RNA in the regions that generally corresponded to the area of the binding of the sole protein in bacterial RNase P (10,32,43). The observed effects of the Rpp1/Pop5/Pop8 binding (Figure 4D) are the strongest in the geometric vicinity of the catalytic center of RNase P RNA (Figure 4A in red versus green). Studies of the closely related RNase MRP show that, in the holoenzyme isolated from yeast, Pop5 forms a UV-induced crosslink to a phylogenetically conserved nucleotide located at a position equivalent to A315 in RNase P (32,35). In keeping with that, our footprinting data show increased protection of the area surrounding A315 when the Rpp1/Pop5/Pop8 subcomplex is introduced (Figure 4A, in red versus green); it is interesting to note that the protection of the area near A309 (where Pop1 crosslinks to RNase P RNA, above) is not affected. The area that is most affected by Rpp1/Pop5/Pop8 binding (A314-G343) includes stem P19 (except for its apical loop), one strand of P2 stem (C317-A323), and phylogenetically conserved areas involved in the formation of the catalytic center A314-U316, A338-A344. The other strand of the P2 stem (U17-G23) is already well-protected in the presence of Pop1, Pop6/Pop7, and the addition of Rpp1/Pop5/Pop8 results in a minimal change of the RNA backbone sensitivity; however, the loop A24-A32 becomes considerably better protected in the presence of Rpp1/Pop5/Pop8. It should be noted that this loop can be expected to face the same direction as the strand of P2 that is subjected to additional protection in the presence of Rpp1/Pop5/Pop8 (C317-A323). Both the protection of RNA in the vicinity of the catalytic center, and the observation that Rpp1/Pop5 are required for RNase P RNA activation (Figure 3 and Supplementary Figure S9) are consistent with the notion of the functional overlap between eukaryotic Pop5 and the sole bacterial protein (16,32). However, the role of Rpp1/Pop5/Pop8 is evidently broader than mostly local and non-structural role played by the protein in bacterial RNase P (10), as the binding of Rpp1/Pop5/Pop8 also affects RNA regions away from the catalytic core (Figure 4D), suggesting a role of this subcomplex in the stabilization of the global RNA structure.
The addition of Pop4, the last step in the assembly of the catalytically active 8-component RNase P RNP, results a drastic improvement of the catalytic activity, while the position of the cleavage site is not affected (Figure 3). Interestingly, in spite of the strong effect on RNase P RNP activity, Pop4 binding has only a minimal effect on the protection of the RNA backbone in our footprinting assays (Figure 4A, red vs black): the addition of Pop4 resulted in only marginal (if any) increase of the protection near the location of the previously reported A171, A173 UV-induced crosslinks in the P9 stem (Figure 4E), and no change near the C192, C193 crosslinks (35). It is likely that Pop4 predominantly interacts with other proteins and RNase P RNA nucleobases, while the RNA backbone (which is probed in the Fe(II)-EDTA assays) remains mostly unprotected. It should be noted that Rpp1/Pop5/Pop8 binding to RNA results in an additional protection of the P9 stem in the immediate vicinity of Pop4 crosslinking sites (35) (Figure 4D), consistent with the observed requirement to have Rpp1/Pop5 for the Pop4 binding (above) and the results of two-hybrid assays (44) (as the latter suggest interactions between Pop4 and Rpp1). The interactions of Pop4 with, simultaneously, Rpp1/Pop5/Pop8 and the S-domain of RNA (35), position Pop4 to play a role in the global RNase P structure. It should be noted that human homologue of Pop4 (Rpp29) was reported to activate human RNase P RNA (20,49).
Pop4 binding results in RNA hypersensitivity localized to the conserved CR-III element directly involved in the recognition of pre-tRNA substrates in bacteria (10,48) (Figure 4E) and the adjacent P12 stem. The hypersensitivity of the P12 stem observed upon the addition of RNase P proteins (Figure 4C–E) is consistent with the proteins-associated hypersensitivity of this region previously observed for the endogenous yeast RNase P (27).
The potential role of Pop4 in the substrate binding will need further investigation. In any case, the result that active RNase P RNPs cleave pre-tRNA substrate at the proper location regardless of their protein composition suggests that yeast RNase P recognition of its pre-tRNA substrates relies predominantly on the RNA component. The roles of proteins in the recognition of alternative RNase P substrates will require further analysis.
Archaeal homologue of Pop4 functions as a part of a Rpr2/Pop4 (Rpp21/Rpp29) subcomplex (11,15); however, all our attempts to produce yeast Rpr2/Pop4 complexes (including co-expression of the proteins, as well as attempts to form complexes by mixing individually purified Pop4 and Rpr2) did not yield positive results, suggesting that, as opposed to archaeal proteins (11,15), the Rpr2/Pop4 subcomplex either does not form, or, more likely, is formed late in the RNase P RNP assembly and requires the presence of other RNase P components or chaperons. The latter is supported by the previously reported observation that yeast RNase P precursor RNP has Pop4, but not Rpr2 (36).
SUMMARY
Here, we report a stepwise reconstitution of structurally homogeneous and catalytically active eukaryotic RNase P RNPs, and an analysis of the interplay of RNase P components. Yeast RNase P has lost RNA elements that serve as structural braces in its bacterial and, by inference, archaeal counterparts, but has gained proteins Pop1, Pop6, Pop7 and Pop8 that are not found in archaea. We show that Pop1 is required for the catalytic RNA activation and is positioned to provide a major contribution to the its global fold, and, simultaneously, to potentially contribute to both substrate binding and the organization of the catalytic core. Proteins Pop6, Pop7 appear to be structural subunits that, together with the specialized RNA domain P3, form an interface for the Pop1 binding, while Pop8 is required for the proper interactions between Pop1 and proteins shared with the archaeal enzymes, Rpp1/Pop5. Proteins Pop6, Pop7, and Pop8 do not affect the position of the pre-tRNA substrate cleavage site, but increase the activity and stability of the RNP. Proteins Rpp1, Pop5 are required for RNA activation, and bind in the immediate vicinity of the RNA-based catalytic core, similar to what was observed in archaeal RNase P. In addition, Rpp1, Pop5 affect the Specificity domain of yeast RNase P RNA, and are required for the engagement of another protein shared with the archaeal enzymes, Pop4. Pop4 binding affects a phylogenetically conserved part of RNase P RNA that is directly involved in substrate recognition in bacteria, and dramatically increases the level of RNase P activity; however, Pop4 does not affect the location of the cleavage site in the pre-tRNA substrate and is not absolutely required for the activation of the catalytic RNA. While eukaryotic Pop4 appears to play a structural role, its potential involvement in the recognition of alternative RNase P substrates cannot be ruled out.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Phil Bevilacqua (Penn State University) and Venkat Gopalan (Ohio State University) for their valuable suggestions. We thank the staff of the Proteomics Core Facility at the Huck Institutes of Life Sciences (PSU) for their help with mass spectrometry analysis, and Chao Quan, Olga Esakova, and Robert Fagerlund for their help at the initial stages of the project.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [GM085149 to A.S.K.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Altman S. A view of RNase P. Mol. Biosyst. 2007; 3:604–607. [DOI] [PubMed] [Google Scholar]
- 2. Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S.. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983; 35:849–857. [DOI] [PubMed] [Google Scholar]
- 3. Jarrous N., Gopalan V.. Archaeal/eukaryal RNase P: subunits, functions and RNA diversification. Nucleic Acids Res. 2010; 38:7885–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Esakova O., Krasilnikov A.S.. Of proteins and RNA: the RNase P/MRP family. RNA. 2010; 16:1725–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lechner M., Rossmanith W., Hartmann R.K., Tholken C., Gutmann B., Giege P., Gobert A.. Distribution of ribonucleoprotein and protein-only RNase P in eukarya. Mol. Biol. Evol. 2015; 32:3186–3193. [DOI] [PubMed] [Google Scholar]
- 6. Gopalan V., Jarrous N., Krasilnikov A.S.. Chance and necessity in the evolution of RNase P. RNA. 2018; 24:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stark B.C., Kole R., Bowman E.J., Altman S.. Ribonuclease P: An enzyme with an essential RNA component. Proc. Natl. Acad. Sci. U.S.A. 1978; 75:3717–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reich C., Olsen G.J., Pace B., Pace N.R.. Role of the protein moiety of Ribonuclease P, a ribonucleoprotein enzyme. Science. 1988; 239:178–181. [DOI] [PubMed] [Google Scholar]
- 9. Crary S.M., Niranjanakumari S., Fierke C.A.. The protein component of Bacillus subtilis Ribonuclease P increases catalytic efficiency by enhancing interactions with the 5′ leader sequence of pre-tRNAAsp. Biochemistry. 1998; 37:9409–9416. [DOI] [PubMed] [Google Scholar]
- 10. Reiter N.J., Osterman A., Torres-Larios A., Swinger K.K., Pan T., Mondragón A.. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010; 468:784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kimura M. Structural basis for activation of an archaeal ribonuclease P RNA by protein cofactors. Biosci. Biotechnol. Biochem. 2017; 81:1670–1680. [DOI] [PubMed] [Google Scholar]
- 12. Samanta M.P., Lai S.M., Daniels C.J., Gopalan V.. Sequence analysis and comparative study of the protein subunits of archaeal RNase P. Biomolecules. 2016; 6:E22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frank D.N., Adamidi C., Ehringer M.A., Pitulle C., Pace N.R.. Phylogenetic-comparative analysis of the eukaryal ribonuclease P RNA. RNA. 2000; 6:1895–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chamberlain J.R., Lee Y., Lane W.S., Engelke D.R.. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998; 12:1678–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsai H.Y., Pulukkunat D.K., Woznick W.K., Gopalan V.. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:16147–16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen W.Y., Pulukkunat D.K., Cho I.M., Tsai H.Y., Gopalan V.. Dissecting functional cooperation among protein subunits in archaeal RNase P, a catalytic ribonucleoprotein complex. Nucleic Acids Res. 2010; 38:8316–8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cho I.M., Lai L.B., Susanti D., Mukhopadhyay B., Gopalan V.. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:14573–14578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lai L.B., Tanimoto A., Lai S.M., Chen W.Y., Marathe I.A., Westhof E., Wysocki V.H., Gopalan V.. A novel double kink-turn module in euryarchaeal RNase P RNAs. Nucleic Acids Res. 2017; 45:7432–7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kikovska E., Svard S.G., Kirsebom L.A.. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:2062–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reiner R., Alfiya-Mor N., Berrebi-Demma M., Wesolowski D., Altman S., Jarrous N.. RNA binding properties of conserved protein subunits of human RNase P. Nucleic Acids Res. 2011; 39:5704–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jarrous N. Roles of RNase P and its subunits. Trends Genet. 2017; 33:594–603. [DOI] [PubMed] [Google Scholar]
- 22. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. Walker JM. Protein identification and analysis tools on the ExPASy server. The Proteomics Protocols Handbook. 2005; Humana Press; 571–607. [Google Scholar]
- 23. Fagerlund R.D., Perederina A., Berezin I., Krasilnikov A.S.. Footprinting analysis of interactions between the largest eukaryotic RNase P/MRP protein Pop1 and RNase P/MRP RNA components. RNA. 2015; 21:1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perederina A., Esakova O., Koc H., Schmitt M.E., Krasilnikov A.S.. Specific binding of a Pop6/Pop7 heterodimer to the P3 stem of the yeast RNase MRP and RNase P RNAs. RNA. 2007; 13:1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Obayashi E., Oubridge C., Pomeranz Krummel D., Nagai K.. Crystallization of RNA–protein complexes. Methods Mol. Biol. 2007; 363:259–276. [DOI] [PubMed] [Google Scholar]
- 26. Celander D.W., Cech T.R.. Iron(II)-ethylenediaminetetraacetic acid catalyzed cleavage of RNA and DNA oligonucleotides: similar reactivity toward single- and double-stranded forms. Biochemistry. 1990; 29:1355–1361. [DOI] [PubMed] [Google Scholar]
- 27. Esakova O., Perederina A., Quan C., Schmitt M.E., Krasilnikov A.S.. Footprinting analysis demonstrates extensive similarity between eukaryotic RNase P and RNase MRP holoenzymes. RNA. 2008; 14:1558–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B.. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999; 17:1030–1032. [DOI] [PubMed] [Google Scholar]
- 29. Esakova O., Perederina A., Quan C., Berezin I., Krasilnikov A.S.. Substrate recognition by ribonucleoprotein Ribonuclease MRP. RNA. 2011; 17:356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aravind L., Lakshminarayan M.I., Anantharaman V.. The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 2003; 4:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perederina A., Esakova O., Quan C., Khanova E., Krasilnikov A.S.. Eukaryotic ribonucleases P/MRP: the crystal structure of the P3 domain. EMBO J. 2010; 29:761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perederina A., Khanova E., Quan C., Berezin I., Esakova O., Krasilnikov A.S.. Interactions of a Pop5/Rpp1 heterodimer with the catalytic domain of RNase MRP. RNA. 2011; 17:1922–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hipp K., Galani K., Batisse C., Prinz S., Bottcher B.. Modular architecture of eukaryotic RNase P and RNase MRP revealed by electron microscopy. Nucleic Acids Res. 2012; 40:3275–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xiao S., Hsien J., Nugent R.L., Coughlin D.J., Fierke C.A., Engelke D.R.. Functional characterization of the conserved amino acids in Pop1p, the largest common protein subunit of yeast RNases P and MRP. RNA. 2006; 12:1023–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Khanova E., Esakova O., Perederina A., Berezin I., Krasilnikov A.S.. Structural organizations of yeast RNase P and RNase MRP holoenzymes as revealed by UV-crosslinking studies of RNA–protein interactions. RNA. 2012; 18:720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Srisawat C., Houser-Scott F., Bertrand E., Xiao S., Singer R.H., Engelke D.R.. An active precursor in assembly of yeast nuclear ribonuclease P. RNA. 2002; 8:1348–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kazantsev A.V., Krivenko A.A., Harrington D.J., Holbrook S.R., Adams P.D., Pace N.R.. Crystal structure of a bacterial ribonuclease P RNA. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:13392–13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jarrous N., Eder P.S., Guerrier-Takada C., Hoog C., Altman S.. Autoantigenic properties of some protein subunits of catalytically active complexes of human ribonuclease P. RNA. 1998; 4:407–417. [PMC free article] [PubMed] [Google Scholar]
- 39. Harris J.K., Haas E.S., Williams D., Frank D.N., Brown J.W.. New insight into RNase P RNA structure from comparative analysis of the archaeal RNA. RNA. 2001; 7:220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hsieh J., Walker S.C., Fierke C.A., Engelke D.R.. Pre-tRNA turnover catalyzed by the yeast nuclear RNase P holoenzyme is limited by product release. RNA. 2009; 15:224–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tranguch A.J., Kindelberger D.W., Rohlman C.E., Lee J.Y., Engelke D.R.. Structure-sensitive RNA footprinting of yeast nuclear ribonuclease P. Biochemistry. 1994; 33:1778–1787. [DOI] [PubMed] [Google Scholar]
- 42. Krasilnikov A.S., Xiao Y., Pan T., Mondragón A.. Basis for structural diversity in homologous RNAs. Science. 2004; 306:104–107. [DOI] [PubMed] [Google Scholar]
- 43. Xu Y., Amero C.D., Pulukkunat D.K., Gopalan V., Foster M.P.. Solution structure of an archaeal RNase P binary protein complex: formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-RNA interactions. J. Mol. Biol. 2009; 393:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Houser-Scott F., Xiao S., Millikin C.E., Zengel J.M., Lindahl L., Engelke D.R.. Interactions among the protein and RNA subunits of Saccharomyces cerevisiae nuclear RNase P. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:2684–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lemieux B., Laterreur N., Perederina A., Noël J.F., Dubois M.L., Krasilnikov A.S., Wellinger R.J.. Active yeast telomerase shares subunits with ribonucleoproteins RNase P and RNase MRP. Cell. 2016; 165:1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hands-Taylor K.L., Martino L., Tata R., Babon J.J., Bui T.T., Drake A.F., Beavil R.L., Pruijn G.J., Brown P.R., Conte M.R.. Heterodimerization of the human RNase P/MRP subunits Rpp20 and Rpp25 is a prerequisite for interaction with the P3 arm of RNase MRP RNA. Nucleic Acids Res. 2010; 38:4052–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Perederina A., Krasilnikov A.S.. The P3 domain of eukaryotic RNases P/MRP: Making a protein-rich RNA-based enzyme. RNA Biol. 2010; 7:534–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krasilnikov A.S., Yang X., Pan T., Mondragón A.. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003; 421:760–764. [DOI] [PubMed] [Google Scholar]
- 49. Mann H., Ben-Asouli Y., Schein A., Moussa S., Jarrous N.. Eukaryotic RNase P: role of RNA and protein subunits of a primordial catalytic ribonucleoprotein in RNA-based catalysis. Mol. Cell. 2003; 12:925–935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.