Abstract
The design of polymeric nanoparticles for gene therapy requires engineering of polymer structure to overcome multiple barriers, including prolonged colloidal stability during formulation and application. Poly(β-amino ester)s (PBAEs) have been shown effective as polymeric vectors for intracellular DNA delivery, but limited studies have focused on polymer modifications to enhance the stability of PBAE/DNA polyplexes. We developed block copolymers consisting of PBAE oligomer center units and poly(ethylene glycol) (PEG) end units. We fabricated a library of PEG-PBAE polyplexes by blending PEGylated PBAEs of different PEG molecular weights and non-PEGylated PBAEs of different structures at various mass ratios of cationic polymer to anionic DNA. Non-PEGylated PBAE polyplexes aggregated following a 24 h incubation in acidic and physiological buffers, presenting a challenge for therapeutic use. In contrast, among 36 PEG-PBAE polyplex formulations evaluated, certain polyplexes maintained a small size under these conditions. These selected polyplexes were further evaluated for transfection in human small cell lung cancer cells (H446) in the presence of serum, and the best formulation transfected ~ 40% of these hard-to-transfect cells while preventing polymer-mediated cytotoxicity. When PEG-PBAE polyplex delivered Herpes simplex virus thymidine kinase plasmid in combination with the prodrug ganciclovir, the polyplexes killed significantly more H446 cancer cells (35%) compared to healthy human lung fibroblasts (IMR-90) (15%). These findings indicate that PEG-PBAE polyplexes can maintain particle stability without compromising their therapeutic function for intracellular delivery to human small cell lung cancer cells, demonstrate potential cancer specificity, and have potential as safe materials for small cell lung cancer gene therapy.
Keywords: gene therapy, lung cancer, polymeric nanoparticle, polyethylene glycol, surface modification
1. Introduction
Small cell lung cancer (SCLC) is a neuroendocrine subtype of lung cancer that accounts for 15% of all lung cancer cases [1]. SCLC is initially sensitive to chemotherapy and radiation, most often involving a combination of cisplatin-etoposide chemotherapy with chest radiation, prophylactic cranial irradiation, or hyperfractionated thoracic radiation [2]. However, SCLC still has one of the highest fatality rates among cancers due to its high recurrence and metastasis [3, 4]. New therapies are needed to improve the survival of patients with SCLC.
Gene therapy is a promising technology due to its tremendous potential as a selective and potent therapeutic for genetic diseases including cancer. Many approaches to DNA-based therapeutics have been identified and validated, such as tumor suppressor genes including TNF-related apoptosis-inducing ligand (TRAIL) and p53 [5, 6]. Another method is suicidal gene therapy, which induces apoptosis of tumor cells by delivering exogenous DNA, such as that encoding Herpes simplex virus thymidine kinase (HSV-tk) that converts prodrugs in situ to an active form [7].
There has been parallel effort to develop efficient, safe, and stable gene delivery vectors. Although viral vectors have the advantage of high transduction efficacy, limitations in cargo capacity, difficulty of production, and safety concerns due to immunogenic and mutagenic factors have led to the emergence of non-viral approaches as alternatives [8, 9]. Poly(β-amino ester)s (PBAEs), a class of biodegradable cationic polymers, have been shown to exhibit low levels of toxicity and high rates of both DNA and siRNA transfection in various types of cells [10–14]. These cationic polymers are able to bind with negatively charged nucleotides and form polyplexes by electrostatic interactions. Previous studies have shown that the biophysical properties of these PBAE polyplexes allow them to overcome critical barriers to gene delivery at the cellular level, including cellular uptake and endosomal escape via pH buffering [15, 16]. However, there has been limited effort to modify PBAE polyplexes to promote biological stability at the systemic and tissue levels, which is a critical property to facilitate efficient in vivo utilization and crossing of extracellular barriers [17–19].
Poly(ethylene glycol) (PEG), a water-soluble molecule with low toxicity, is widely used with a variety of biomaterials to minimize unwanted interactions with biomolecules. Its neutral and hydrophilic structure not only reduces surface charge of particles but also provides steric hindrance to reduce non-specific adsorption and aggregation. These properties have been shown to significantly enhance stability and increase half-life of biologics and particles in systemic circulation [20, 21].
PEGylation has the potential to reduce non-specific interactions between polyplexes and biological molecules and off-target cells. However, PEGylating polyplexes also generally has a negative effect on cellular uptake and transfection to target cells, which has been referred to as the “PEG dilemma” [22]. Due to the shielding of positive surface charge by neutral PEG molecules, polyplexes are not only preventing non-specific protein adsorption but also become less associated with the plasma membrane of target cells. Decreased polyplex-cell interaction has been correlated to reduced cellular uptake and transfection [23, 24].
The present study introduces a synthesis method to conjugate PEG to PBAE polymers and a combinatorial method was used to formulate polyplexes from a blend of PEGylated PBAEs (PEG-PBAEs) and end-capped PBAEs (ePBAEs) developed in our lab [13] in order to overcome the PEG dilemma. The resulting PEG-PBAE polyplexes not only maintain particle stability and efficacy over time, but also efficiently deliver suicidal gene HSV-tk in vitro and activate ganciclovir to kill SCLC cells.
2. Materials and methods
2.1. Materials
1,4-butanediol diacrylate (B4), 4-amino-1-pentanol (S4), 5-amino-1-pentanol (S5), 1-(3-aminopropyl)-4-methylpiperazine (E7) (Alfa Aesar), 1,5-pentanediol diacrylate (B5) (Monomer Polymer & Dajac Labs), 2-methylpentane-1,5-diamine (E4) (TCI America), 2-(3-aminopropylamino)ethanol (E6) (Fluka), poly(ethylene glycol) methyl ether thiol (800 Da), branched 25 kDa poly(ethylenimine) (PEI) (Sigma-Aldrich), α-Mercaptoethyl-ω-methoxy polyoxyethylene (5000 Da) (NOF America Corporation), and cell culture media components were purchased and used as received. pEGFP-N1 (EGFP) DNA (purchased from Elim Biopharmaceuticals and amplified by Aldevron, Fargo, ND), ganciclovir (Invivogen, San Diego, CA), Label IT-Tracker Cy3 kit (Mirus Bio LLC), and CellTiter 96 AQueous One MTS assay (Promega, Fitchburg, WI) were obtained from commercial vendors and used per manufacturer’s instructions. HSV-tk gene was cloned into the pcDNA3.1 vector (Life Technologies) and amplified (Aldevron, Fargo, ND).
2.2. Polymer synthesis and characterization
ePBAEs and PEG-PBAEs were synthesized in a two-step reaction using small commercially-available molecules as described in Fig. 1. As an example, acrylate-terminated base polymer poly(1,4-butanediol diacrylate-co-4-amino-1-butanol) (B4S4) was first synthesized by mixing a backbone monomer (B4) and a side-chain monomer (S4) at 1.2:1 or 1.05:1 B4:S4 monomer molar ratios in DMSO as a 500mg/mL solution and stirring at 90°C for 24 h. The base polymer was purified in cold diethyl ether, dried under vacuum with desiccant for 24 h, and the molecular weight and chemical structure of the base polymer were confirmed by Bruker Avance III 500 MHz NMR spectrometer in CDCl3. Base polymers were end-capped with a small molecule (E4, E6, or E7) by dissolving base polymer and end-capping molecule at 1:30 molar ratio in THF as a 100 mg/mL solution and shaking the mixture at room temperature for 3 h. Final ePBAE polymers were purified in cold diethyl ether, dried under vacuum with desiccant for 24 h, confirmed with 1H NMR for complete conjugation, and then stored with desiccant at −20°C as 100 mg/mL solutions in DMSO.
Figure 1.
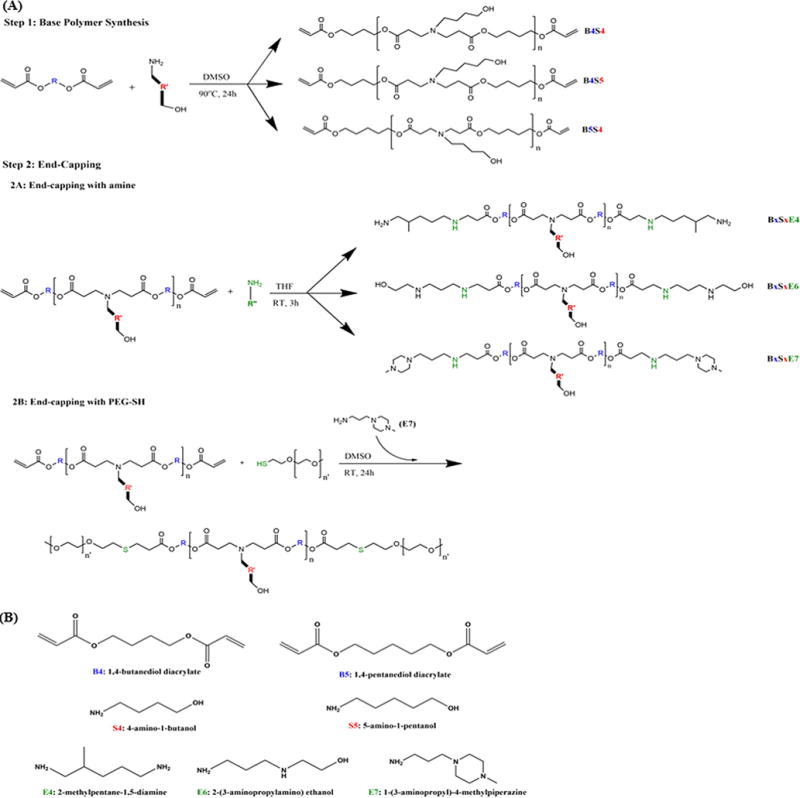
(A) Synthesis scheme of conventionally end-capped poly(β-amino ester)s (ePBAEs) and poly(ethylene glycol)-co-poly(β-amino ester)s (PEG-PBAEs). (B) Chemical structures of monomers used.
Amine-catalyzed, thiol-ene Michael addition reaction was used to conjugate PEG to the base polymer B4S4 [25]. Briefly, B4S4, methoxy PEG-thiol, and E7 molecules were mixed at 1:2.5:0.2 molar ratios as a 100 mg/mL solution in DMSO and stirred at room temperature for 24 h at 1000 rpm. Final block copolymers were precipitated in diethyl ether at room temperature without centrifugation, confirmed with 1H NMR for complete conjugation, and stored with desiccant at −20°C as 100 mg/mL solutions in DMSO. The nomenclature of different PEG-PBAEs used in this study is listed in Table 1.
Table 1.
Nomenclature of different PEG-PBAE polymers. The PBAE used was 1-4-butanediol diacrylate-co-1,4-aminobutanol (B4S4).
| PEG-PBAE | Name |
|---|---|
| PEG0.8k-B4S44k-PEG0.8k | 0.8k-4k |
| PEG0.8k-B4S413k-PEG0.8k | 0.8k-13k |
| PEG5k-B4S44k-PEG5k | 5k-4k |
| PEG5k-B4S413k-PEG5k | 5k-13k |
2.3. Particle formulation and characterization
PBAE polyplexes were made at 60 and 75 w/w mass ratios of ePBAE to DNA in 25 mM sodium acetate buffer (pH = 5). For example, diluted polymer solution at 3.6 mg/mL was mixed into diluted DNA solution at 0.06 mg/mL at equal volume to form 60 w/w polyplexes, and the mixture was incubated for 10 minutes to allow complexation. 75 w/w was tested to check for cytotoxicity of ePBAE at higher polymer concentration.
PEG-PBAE polyplexes were made at 30, 60, 90 w/w ratios of total polymer to DNA in 25 mM sodium acetate buffer (pH = 5). Polymer used to condense DNA was a mixture of ePBAE and PEG-PBAE at three different mass ratios of 1:2, 1:1, and 2:1. For example, 50 μg of ePBAE 447 and 100 μg of PEG-PBAE 5k-4k were diluted to 3.6 mg/mL total polymer concentration with 25 mM sodium acetate buffer (pH = 5), and the polymer solution was mixed with diluted DNA solution at 0.06 mg/mL at equal volume to form polyplexes with 447:5k-4k 1:2 w/w and polymer:DNA 60 w/w ratios. These polyplexes were incubated for 10 minutes to allow complexation.
The polyplex size was determined by nanoparticle tracking analysis (NTA) using Nanosight NS500 (Malvern Instruments, 532 nm laser) and dynamic light scattering (DLS) using Malvern Zetasizer Nano ZS (Malvern Instruments, detection angle 173°, 633 nm laser). The polyplexes prepared at DNA concentration of 0.1 mg/mL were diluted 1000-fold and 2-fold into 25 mM sodium acetate buffer or 2× PBS to a total volume of 400 μL for Nanosight and Zetasizer, respectively. To determine 24 h stability of the polyplexes, cuvettes with the polyplex solution were stored in dark at room temperature for 24 h, then the polyplex size was re-measured following a brief resuspension. Only number-weighted measurements with particle concentrations above 15 particles/frame by NTA and intensity-weighted Z-average measurements passing the quality control expert advice criteria by DLS are reported. Zeta potential was determined using Malvern a Zetasizer Nano ZS (Malvern Instruments) with samples prepared at DNA concentration of 0.03 mg/mL diluted 2-fold into 25 mM sodium acetate buffer (pH=5.0) for a total volume of 800 μL. The mean and standard deviation were calculated.
2.4. Cell culture
H446 small cell lung cancer cells (ATCC) were cultured at 37°C and 5% CO2 in ATCC-modified RPMI 1640 media (Life Technologies A10491-01), supplemented with 10% FBS and 1% penicillin/streptomycin. IMR-90 human lung fibroblast cells (ATCC) were cultured at 37°C and 5% CO2 in Eagle’s minimum essential media (Cellgro 10-009-CV), supplemented with 10% FBS.
2.5.DNA delivery assays
2.5.1. Polyplex delivery
Cells were plated at a density of 15,000 cells/well (100 μL/well) in 96-well tissue culture plates and were incubated for 24 h. pEGFP labeled with Cy3 per manufacturer’s instructions (Label IT Tracker kit) and unlabeled pEGFP were used for uptake and transfection experiments, respectively. Polyplexes were prepared as described above to a final DNA concentration of 0.03 mg/mL. Then, 20 μL of polyplexes was added to 100 μL of serum-containing medium in each well. For PEI polyplexes, pEGFP-Cy3 diluted into 150 mM NaCl to 60 μg/mL was mixed with equal volume of PEI diluted into 150 mM NaCl to 120 μg/mL (2 w/w) from a stock solution of 1 mg/mL in dH2O. PEI polyplexes were also incubated for 10 min to complex, and 20 μL of polyplex solution was added to 100 μL of medium in each well. For uptake experiments, cells were incubated with polyplexes for 4 h, washed twice with heparin-containing PBS (50 μg/mL), and prepared for flow cytometry. For transfection experiments, cells were incubated with polyplexes for 4 h, washed twice with heparin-containing PBS, and incubated with 100 μL fresh media for an additional 48 h, and analyzed qualitatively with fluorescent microscope and quantitatively with FACS analysis.
2.5.2. Cell viability
Cells were treated following the same protocol as transfection. Following 4 hours of incubation with polyplexes, cells were washed twice with heparin-containing PBS, added with 100 μL of fresh media, and incubated for an additional 24 h at 37°C. 20 μL of CellTiter 96 AqueousOne MTS reagent were added per well, cells were incubated with reagent at 37°C, and absorbance was measured at 490 nm using a Synergy 2 plate reader (Biotek) every 30 min until the highest absorbance signal reached 1.2. Absorbance signal was normalized to that of untreated cells after subtracting the background signal. All conditions were prepared in quadruplicates.
2.5.3. Flow Cytometry
To prepare for flow cytometry (Accuri C6 with HyperCyt high-throughput adaptor), cells were detached using 30 μL of 0.05% trypsin, resuspended with 170 μL of FACS buffer (PBS containing 2% v/v FBS), transferred to a round-bottom 96-well plate and centrifuged at 800 rpm at 40C for 5 min. 170 μL of supernatant was removed, and the remaining 30 μL was triturated to resuspend the cells. Propidium iodide (PI) (Invitrogen, Carlsbad, CA) was added to FACS buffer at 1:200 to detect cells in the process of apoptosis for transfection assay.
For uptake, % positive is the percentage of total cells that are Cy3+ as measured by two-dimensional gating of FL1 vs. FL2 using FlowJo 7.6.5 software. For transfection, % positive is the percentage of total cells that are EGFP+ as measured by sequential two-dimensional gating of PI- by FSC-H vs. FL2 and EGFP+ by FL1 vs. FL2. At least 500 cell counts were analyzed for each measurement. All conditions were prepared in quadruplicates.
2.6 Delivery of pHSV-tk and ganciclovir
Cells were treated following the same protocol as transfection using pHSV-tk DNA. Following 4 h of incubation with polyplexes, cells were washed twice with heparin-containing PBS and incubated with 100 μL of fresh media for 24 h at 37°C. The media was then replaced with fresh media containing 10 or 20 μg/mL of ganciclovir. Following additional 48 h incubation at 37°C, the media was replaced with fresh media containing 10 or 20 μg/mL of ganciclovir. Cell death was measured 24 h after the second ganciclovir treatment with CellTiter 96 AQueous CellTiter reagent as described above. All conditions were prepared in quadruplicates. Stock ganciclovir solution at 5 mg/mL was prepared by dissolving it in 2% 1M NaOH, and then neutralizing the pH with 1% 1M HCl, 40% dH2O, and 57% 1× PBS by volume.
2.7. Statistics
All statistical analysis was performed with GraphPad Prism 5 software package. One-way ANOVA with post-hoc Dunnett test was used to test statistical significance of multiple conditions against the control group (p < 0.05). A Student’s t-test was used to test statistical significance of cell death from the same HSV-tk and ganciclovir treatment between H446 and IMR-90 cells (p < 0.01).
3. Results
3.1 Synthesis and characterization of PBAE and PEG-PBAE polymer
We first sought to synthesize and confirm the molecular weight as well as the completion of synthesis of ePBAEs and PEG-PBAEs. Both types of PBAEs share the same base polymer, with the molecular weight controlled by the molar ratio of backbone (B) to side-chain (S) monomers in the Step 1 reaction (Fig. 1). Two molar ratios, 1.2:1 and 1.05:1, as well as two (B) and (S) monomer types each were used to synthesize four different acrylate-terminated base polymers with molecular weight distribution as shown in Fig. S1; the closer the monomer ratio is to unity, the greater the degree of polymerization.
A total of 9 ePBAEs were synthesized with three base polymers, B4S4, B4S5, and B5S5 of approximately 10 kDa, and three end-capping (E) molecules, E4, E6, and E7, by Step 2A end-capping reaction (Fig. 1). Similar molecular weights were selected for each polymer structure. An example of an ePBAE nomenclature is 457, which is base polymer B4S5 end-capped with E7. A total of 4 PEG-PBAEs were synthesized with two base polymers (B4S4 at 4 and 13 kDa), and two methoxy PEG-thiol molecules (0.8 and 5 kDa). E7 was selected as the amine catalyst in this Step 2B PEGylation reaction because of its use in our lab for end-capping of PBAEs and its non-toxicity in our studies [26]. A trace amount of E7 (5 % mol) was used to ensure PEGylation occurred and not E7-endcapping.
Because end-capping reactions involve a nucleophilic addition to acrylates, the completion of end-capping can be confirmed using1H NMR. Once the diacrylates on the base polymer (Fig. S2A/B) reacted with (E) molecules to yield ePBAE or with methoxy PEG-thiol molecules to yield PEG-PBAE (Fig. S2C), the signature peaks for protons on acrylates disappeared, verifying that every base polymer in the reaction was completely end-capped.
3.2 Preparation and characterization of PEG-PBAE polyplexes
Polyplexes are formulated via electrostatic interaction between cationic polymer and negatively charged DNA. Thus, N/P ratio, the ratio of amines in the polymer (N, positively charged) to phosphates in the DNA (P, negatively charged), is an important parameter for polyplex formulation. N/P ratio can also be converted to the total or the effective weight-to-weight ratio (w/w) between the polymer and the DNA, or vice versa, as long as the amine density and molecular weight are known. The total polymer:DNA w/w ratios that were used are converted to the effective PBAE:DNA w/w ratios and N/P ratios in Table S1. For PEG-PBAE polyplexes, ePBAE was also blended in with PEG-PBAE, and their weight to weight ratio was added as another parameter.
Polyplex stability over time was investigated with both nanoparticle tracking analysis (NTA) and dynamic light scattering (DLS) by measuring the increase in particle size over 24 h. While NTA directly measures the number-averaged hydrodynamic diameter, DLS reports an intensity-weighted average that is skewed towards larger or aggregated particles [27, 28]. Polyplex formulations that yielded significant aggregation or decomplexation as indicated by low particle concentration on NTA or large particle size over the detection limit on DLS were eliminated from further consideration as candidate gene delivery formulations (Fig. 2A). PBAE polyplex size of 447 60 w/w was similar to that reported in previous literature [29]. Four formulation conditions, indicated by red arrows, showed an initial particle size of 90-110 nm and minimal aggregation over time by NTA in 25 mM sodium acetate buffer. These small polyplex formulations were ePBAE blended with 5k-4k 1:2 30 w/w, 5k-4k 1:1 30 w/w, 5k-13k 1:2 30 w/w, and 5k-13k 1:1 30 w/w, and they were selected for subsequent transfection evaluation. A similar trend was observed when PEG-PBAE polyplexes are formulated with another ePBAE, 457 (Figure S3). While polyplexes with PEG-PBAE polymer synthesized from 4 kDa PBAE base polymer significantly aggregated over time in PBS, polyplexes with PEG-PBAE polymer synthesized from 13 kDa PBAE base polymer remained nanosized (~ 300 nm) after 24 h incubation in PBS. All four formulations showed a slight decrease in surface charge to +7 mV in NaAc, although not statistically significant, in comparison to 447 60 w/w PBAE polyplexes (Fig. 2B).
Figure 2.
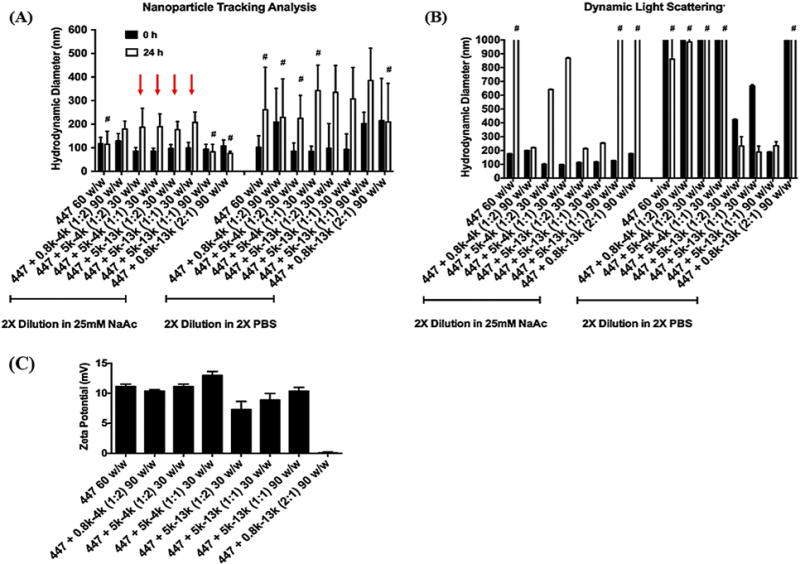
The size of polyplexes formed by self-assembly of enhanced green fluorescent protein (pEGFP) DNA with 447 alone or in combination with PEG-PBAE at various polymer:DNA and 447:PEG-PBAE w/w ratios was measured by (A) Nanosight (NTA) or (B) Zetasizer (DLS). The stability of the polyplexes was tested by sizing them after a 24-h incubation in either sodium acetate or PBS at room temperature. #: Indicates formulation conditions where polyplex aggregation is occurring, leading to unreliable size measurements (low particle concentration by NTA or greater than a micron in size by DLS). (C) The zeta potential of polyplexes. Data are mean ± SD of particle population for NTA and mean ± SD of 3 independent measurements for DLS. Red arrows indicate the four formulations that maintained particle stability and were selected for transfection evaluation.
3.3 High-throughput evaluation of uptake, transfection and cytotoxicity
High-throughput evaluation was sequentially performed at two levels to select the most optimized polyplex formulation based on uptake and transfection of polyplexes in H446 cells. Initially, PBAE polyplexes formed with 9 different ePBAEs were tested to select the best ePBAE polymer that would be blended with PEG-PBAE polymers in the subsequent screening. As shown in Fig. 3B, ePBAEs with more hydrophobic base polymer generally formed polyplexes with higher cytotoxicity, evidenced by B5S5 polyplexes leaving no viable cells 48 h after transfection. Higher uptake of ePBAE polyplexes with B4S5 base polymer did not result in a higher transfection rate than those with B4S4 base polymer, possibly due to different endocytosis pathways or rate-limiting downstream steps (Fig. 3A/C, S4) [16]. Top performing ePBAEs 444, 446, 447, and 457 with cell viability over 80% and transfection efficacies of 60-75% were blended into four selected PEG-PBAE polyplex formulations for subsequent evaluation.
Figure 3.
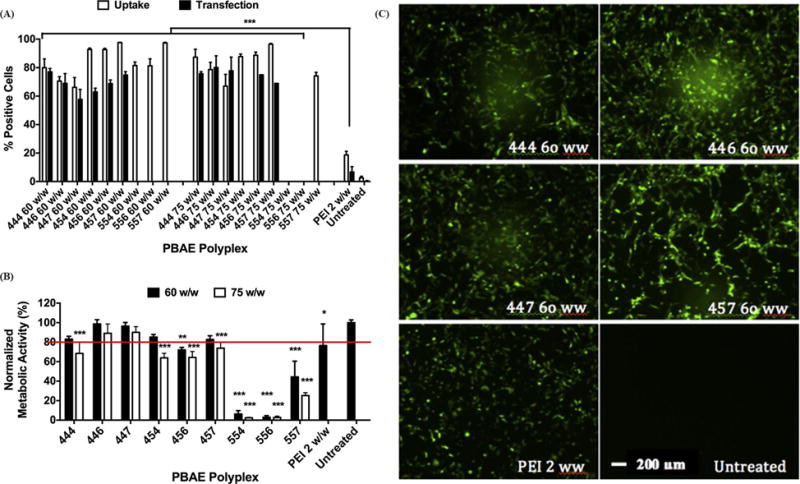
Flow cytometry data showing uptake at 4 hrs and transfection efficacy at 2 days post treatment of H446 cells with nine PBAE polyplexes. The efficiency is in terms of percentage of live H446 cells positive for Cy3 (uptake) or EGFP (transfection). Efficacy of ePBAEs is compared to that of polyethylenimine (PEI) 2 w/w. Data are mean ± SD (n=4) (*** p < 0.001 compared to untreated). (B) Cytotoxicity of PBAE polyplexes, quantified by normalizing metabolic activity to untreated cells. Data are mean ± SD (n=3) (*** p < 0.001, ** p < 0.01, * P < 0.05 compared to untreated, red line marks 80% viability). (C) Representative fluorescence microscope images (10×) at 48 h post-treatment of H446 cells transfected with 4 different PBAE polyplexes at 60 w/w and controls. Scale bar is 200 μm for all panels.
As anticipated due to the shielding properties of PEG, the cellular uptake and transfection efficacy of PEG-PBAE polyplexes significantly decreased in comparison to unPEGylated PBAE polyplexes (Fig. 4A/C, S4). Among PEG-PBAE polyplexes, formulation with 5k-4k polymer generally resulted in higher uptake efficacy than that with 5k-13k polymer, which is consistent with the enhanced particle stability of 5k-13k polyplexes (Figure 2A/B), potentially from greater PEG shielding, limiting the polyplexes’ interaction with the cell membrane. However, transfection efficacies of PEG-PBAE polyplexes formed with 5k-4k and 5k-13k polymer at 1:1 30 w/w condition were similar, indicating polyplexes with 5k-13k polymer more efficiently deliver the DNA cargo to the nucleus to be transcribed following endocytosis. Also, PEG-PBAE polyplexes blended with ePBAE 457 resulted in the highest uptake and transfection overall, which is comparable to the results from PBAE polyplex screening. This may be due to hydrophobicity of 457 that allows for stronger condensation and more stable particles. Specifically, PEG-PBAE polyplex formed from 457 blended with 5k-13k at 1:1 30 w/w condition was internalized in 30% of H446 cells, and transfected 40%. The higher measured transfection rate compared to the measured uptake rate is likely due to the lower sensitivity of measuring successful cellular uptake compared to successful gene expression (expressed plasmid leads to an amplified GFP florescence signal compared to the fluorescence signal from the labeled plasmid itself). This formulation also showed second highest geometric mean GFP fluorescence intensity, which is an indicator of the amount of protein expressed by the transgene per cell (Fig. S5).
Figure 4.
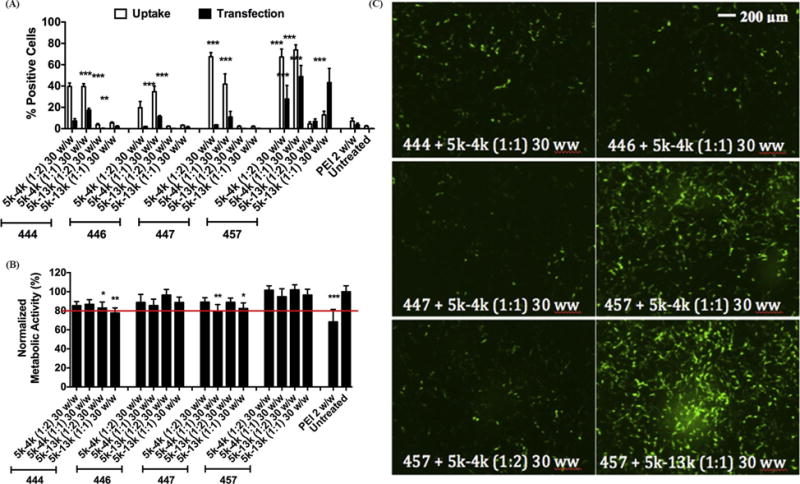
Flow cytometry data showing uptake efficacy at 4 hrs and transfection efficacy at 2 days post treatment of H446 cells with 16 different formulations of PEG-PBAE polyplexes. The efficiency is in terms of percentage of live H446 cells positive for Cy3 (uptake) or EGFP (transfection). Data are mean ± SD (n=4)(*** p < 0.001, ** p < 0.01 compared to untreated). (B) Cytotoxicity of PEG-PBAE polyplexes, quantified by normalizing metabolic activity to untreated cells. Data are mean ± SD (n=3)(*** p < 0.001, ** p < 0.01, * P < 0.05 compared to untreated, red line marks 80% viability). (C) Representative fluorescence microscope images (10×) at 48 h post-treatment of H446 cells transfected with 6 different PEG-PBAE polyplex formulations. Scale bar is 200 μm for all panels.
3.4 Therapeutic activity against small cell lung cancer with PEG-PBAE/pHSV-tk polyplexes and ganciclovir
Ganciclovir is a widely investigated prodrug of interest for suicide gene therapy for different types of cancer [26, 30, 31]. The nontoxic ganciclovir prodrug is phosphorylated into ganciclovir triphosphate by the HSV-tk gene product, which then disrupts DNA replication and causes cell death [32, 33]. We examined PEG-PBAE polyplexes as a functional vehicle for small cell lung cancer gene therapy by delivering PEG-PBAE/pHSV-tk polyplexes followed by ganciclovir treatment. The optimal PEG-PBAE polyplex formulation 457 + 5k-13k 1:1 30 w/w was chosen and was compared to 457 60 w/w PBAE polyplexes for transfection of both H446 human small cell lung cancer cells and IMR-90 human lung fibroblasts as a healthy control cell type. PBAE and PEG-PBAE polyplexes were able to kill 60% and 35% of cancer cells, respectively (Fig. 5A). Interestingly, the level of cell death induced by two types of polyplexes correlated closely with their EGFP transfection efficacies of 73% and 43% (Fig. 3A and 4A), but not with their EGFP geometric mean intensities of 170000 and 4000 RFUs (Fig. S5). The expression of HSV-tk, followed by treatment with ganciclovir, is expected to cause death of the transfected cell. A two-fold increase of the ganciclovir dosage had negligible effect on cell death, demonstrating that the exogenous gene expression of HSV-tk was the limiting factor determining cell killing.
Figure 5.
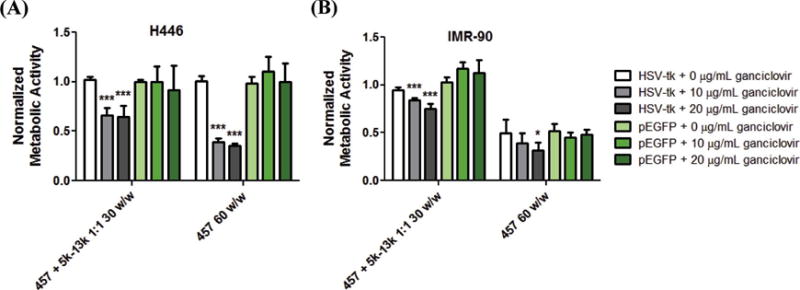
Cell death measured by MTS assay and reported as normalized metabolic activity to untreated group (not shown). (A) H446 and (B) IMR-90 cells are transfected with the optimized PEG-PBAE polyplex formulation (457 + 5k-13k 1:1 30 w/w) and PBAE polyplexes (457 60 w/w) delivering pHSV-tk, followed by two sequential ganciclovir treatments at either 10 or 20 g/mL dosage. Data are mean ± SD (n=4) (* p < 0.05, *** p < 0.001 compared to pEGFP control of each group).
The same PEG-PBAE and PBAE polyplexes showed different outcomes with IMR-90 human lung fibroblasts. First, polyplexes formed with 457 ePBAE complexed with pEGFP at 60 w/w had significant inherent cytotoxicity of 50% (Fig. 5B). This demonstrates the potential fragility of healthy human cells and the need for biocompatible, non-cytotoxic formulations. This concern with potential PBAE polyplex cytotoxicity is resolved when 457 ePBAE is blended with 5k-13k at 1:1 ratio, as evidenced by near 100% viability from pEGFP as well as pHSV-tk + 0 μg/mL ganciclovir controls. This reduced cytotoxicity is likely due to a combination of less 457 ePBAE being used to form PEG-PBAE polyplexes in comparison to non-PEGylated PBAE polyplexes of the same total w/w and due to PEG molecules shielding potentially unfavorable interaction between surface-exposed positively charged 457 and cellular components. In addition, the same PEG-PBAE/HSV-tk DNA polyplexes + ganciclovir system is more specific in promoting killing of human lung cancer H446 cells than healthy human lung IMR-90 fibroblasts with statistical significance (p < 0.01); 35% of H446 cells and 15% of IMR-90 cells are killed at the 10 μg/mL ganciclovir dosage. Overall, these results show the potential of stable and effective PEG-PBAE polyplexes for lung cancer gene therapy.
4. Discussion
Nanoparticles, including polyplexes that are formed by electrostatic interaction between cationic polymer and negatively charged nucleic acids, that are intended to be used for systemic administration, need to overcome challenges of destabilization in physiological saline, adsorption of serum proteins, and aggregation post-administration, which all can lead to rapid clearance from the blood. Furthermore, colloidal stability at sub-400 nm diameter is critical for nanoparticles in cancer therapy to utilize passive targeting to tumors and their leaky vasculature via the enhanced permeation and retention (EPR) effect [22, 34]. A new copolymer synthesized by conjugating the hydrophilic molecule PEG to selected PBAE base polymers provided steric hindrance to the resulting PEG-PBAE polyplexes that minimized particle aggregation and maintained an effective size for the EPR effect.
When formulating both PBAE and PEG-PBAE polyplexes, the N/P ratios used are relatively high in comparison to polyplexes of different polymers, such as PEI (Table S1). Non-degradable PEI, with its high charge density, becomes cytotoxic at higher N/P ratios unless it is modified with degradable moieties [35, 36]. Two features of the PBAE chemical structure allows for polyplexes with much higher N/P ratios. Firstly, PBAE has repeated ester bonds along its backbone and hence is hydrolytically degradable into small bioeliminable units and thus much higher w/w ratios can be utilized [15]. Secondly, most of PBAEs’ positive charge is from tertiary amines, some of which are not protonated in the physiological range of pH 5.1-7.4 [15]. Thus, N/P ratio is a function of pH and not the necessarily the same as the ratio of positive charges to negative charges within the polyplexes. This pH dependence of the PBAE polyplexes’ charge is an important feature as it provides pH buffering capacity, protecting DNA in endosomes and promoting endosomal escape consistent with the proton sponge hypothesis, enabling successful transfection [37].
PEG-PBAE polyplexes have ePBAE blended in at different mass ratios of ePBAE to PEG-PBAE. Although PEG-PBAE polymer has tertiary amines along the backbone that can be protonated and associate with the DNA, the end-group structure of ePBAE has been implicated to serve important and complimentary functions. For example, different end-groups were found to regulate specific uptake mechanisms and downstream steps leading to successful transfection [15, 16]. PEGylating polyplexes of various polymers has been shown to affect cellular uptake and intracellular trafficking significantly [38], often reducing cellular uptake and gene delivery efficacy in vitro. While residual positive charge on the surface can contribute to particle-cell interaction, the presence and exposure of select ePBAE in PEG-PBAE polyplexes can promote cellular uptake via specific pathways that leads to greater transfection. 457 ePBAE, which was selected from high-throughput screening to be blended into PEG-PBAE polyplexes, yielded results that were in agreement to previous literature, which showed high in vivo efficacy of 457 PBAE polyplexes in a subcutaneous H446 xenograft mice model [39].
Interestingly, the PEG-PBAE polyplexes used in this study were able to kill H446 small cell lung cancer cells more than IMR-90 lung fibroblasts through HSV-tk/ganciclovir treatment. This cancer cell selectivity in efficacy is possibly due to a higher doubling rate of cancer cells than fibroblasts, since ganciclovir phosphorylated by HSV-tk kills cells by disrupting DNA replication in actively dividing cells [32]. However, another potential explanation is cancer specificity of the ePBAE polymer. Our group has previously shown that specific ePBAE structure (including the (3-Aminopropyl)-4-methylpiperazine (E7) end-group) leads to increased transfection in various tumor cells in comparison to the healthy cells in the same tissue [40, 41]. Intriguingly, this ePBAE polyplex cancer cell transfection specificity with E7 is evident in corresponding tumor and non-tumor primary cell samples that show the same cell doubling time and have the same percentage of polyplex cellular uptake [40].
Since the current PEG-PBAE polyplexes do not have an active targeting functionality, further modification for cancer targeting, such as conjugation of a targeting ligand to the polymer and/or insertion of a cancer-specific promoter in the plasmid DNA, can further enhance their therapeutic efficiency in cancer therapy [42]. This work demonstrates an important step in the design of non-viral vectors that utilize the PBAE platform. Through the synthesis of new PEGylated PBAE polymers and new PEG-PBAE/ePBAE formulations via combinatorial approach, stability was enhanced, non-specific cytotoxicity was prevented, and selective killing of small cell lung cancer cells was enabled. Our PEG-PBAE polyplexes are anticipated to show enhanced in vivo results due to two main effects: bioavailability and cellular transfection. PEGylation of PBAE polyplexes may enhance pharmacokinetics in the systemic circulation and diffusion in the tissue, as reported with other polymeric systems. Also, a blend of best performing ePBAE in PEG-PBAE polyplex may also allow for cell specificity, efficient cellular uptake, and transfection, thereby overcoming the “PEG dilemma.”
5. Conclusions
PBAEs are a class of cationic polymers that has been shown to transfect a wide range of cell types with high efficiency. In an effort to make more stable PBAE polyplexes, we synthesized PEG-PBAEs using thiol-ene Michael addition reaction and fabricated new polyplexes with blends of PEG-PBAEs and ePBAEs. After selecting the best performing ePBAEs through screening against cytotoxicity and transfection in small cell lung cancer cells (H446), PEG-PBAE polyplexes of varying conditions, including PEG-PBAE molecular weight, mass ratios of ePBAE to PEG-PBAE, and total polymer to DNA mass ratios, were further evaluated for nanoparticle size, stability, cytotoxicity and transfection efficacy. The most effective formulation consisted of ePBAE 457 blended with PEG-PBAE 5k-13k at 1:1 w/w ratio, and the total polymer mixed with DNA at a 30 w/w ratio for polyplex self-assembly. This PEG-PBAE formulation maintained its size under 300 nm over 24 h in physiological PBS and transfected 40% of H446 cells. When human lung cancer cells were transfected with HSV-tk using the optimized PEG-PBAE polyplex and subsequently treated with ganciclovir, 35% of the cells were killed in contrast to 15% cell death to healthy human lung fibroblasts (IMR-90). The present study used a novel method to synthesize PEGylated PBAE polymer and to formulate stable polyplexes that do not exhibit biomaterial-based cytotoxicity, can successfully transfect human lung cancer cells, and can induce their death via HSV-tk/ganciclovir prodrug gene therapy.
Supplementary Material
Acknowledgments
H446 and IMR-90 cells were provided by Dr. Christine Lee Hann and Dr. Linzhao Cheng. Ron Shmueli is thanked for assistance with preliminary studies in the synthesis of PEG-PBAE. This work was in part supported by the NIH(R01EB016721).The authors also thank the Imaging and Microscopy Core Module of the Wilmer Core Grant, P30-EY001865. JK thanks Samsung Scholarship for fellowship support.
Appendix
Footnotes
Disclosure
The authors do not have any conflicts of interest to disclose.
References
- 1.Rodriguez E, Lilenbaum RC. Small cell lung cancer: past, present, and future. Curr Oncol Rep. 2010;12:327–334. doi: 10.1007/s11912-010-0120-5. [DOI] [PubMed] [Google Scholar]
- 2.Paumier A, Le Pechoux C. Radiotherapy in small-cell lung cancer: where should it go? Lung Cancer. 2010;69:133–140. doi: 10.1016/j.lungcan.2010.04.019. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 4.Brade AM, Tannock IF. Scheduling of radiation and chemotherapy for limited-stage small-cell lung cancer: repopulation as a cause of treatment failure? J Clin Oncol. 2006;24:1020–1022. doi: 10.1200/JCO.2005.04.9676. [DOI] [PubMed] [Google Scholar]
- 5.Lang FF, Bruner JM, Fuller GN, Aldape K, Prados MD, Chang S, Berger MS, McDermott MW, Kunwar SM, Junck LR, Chandler W, Zwiebel JA, Kaplan RS, Yung WK. Phase I trial of adenovirus-mediated p53 gene therapy for recurrent glioma: biological and clinical results. J Clin Oncol. 2003;21:2508–2518. doi: 10.1200/JCO.2003.21.13.2508. [DOI] [PubMed] [Google Scholar]
- 6.Wang S, El-Deiry WS. TRAIL and apoptosis induction by TNF-family death receptors. Oncogene. 2003;22:8628–8633. doi: 10.1038/sj.onc.1207232. [DOI] [PubMed] [Google Scholar]
- 7.Moolten FL. Tumor chemosensitivity conferred by inserted herpes thymidine kinase genes: paradigm for a prospective cancer control strategy. Cancer Res. 1986;46:5276–5281. [PubMed] [Google Scholar]
- 8.Hollon T. Researchers and regulators reflect on first gene therapy death. Nat Med. 2000;6:6–6. doi: 10.1038/71545. [DOI] [PubMed] [Google Scholar]
- 9.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 10.Anderson DG, Akinc A, Hossain N, Langer R. Structure/property studies of polymeric gene delivery using a library of poly(beta-amino esters) Mol Ther. 2005;11:426–434. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Bhise NS, Wahlin KJ, Zack DJ, Green JJ. Evaluating the potential of poly(beta-amino ester) nanoparticles for reprogramming human fibroblasts to become induced pluripotent stem cells. Int J Nanomed. 2013;8:4641–4658. doi: 10.2147/IJN.S53830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho SW, Yang F, Son SM, Park HJ, Green JJ, Bogatyrev S, Mei Y, Park S, Langer R, Anderson DG. Therapeutic angiogenesis using genetically engineered human endothelial cells. J Control Release. 2012;160:515–524. doi: 10.1016/j.jconrel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzeng SY, Green JJ. Subtle changes to polymer structure and degradation mechanism enable highly effective nanoparticles for siRNA and DNA delivery to human brain cancer. Adv Healthc Mater. 2013;2:468–480. doi: 10.1002/adhm.201200257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandenbroucke RE, De Geest BG, Bonne S, Vinken M, Van Haecke T, Heimberg H, Wagner E, Rogiers V, De Smedt SC, Demeester J, Sanders NN. Prolonged gene silencing in hepatoma cells and primary hepatocytes after small interfering RNA delivery with biodegradable poly(beta-amino esters) J Gene Med. 2008;10:783–794. doi: 10.1002/jgm.1202. [DOI] [PubMed] [Google Scholar]
- 15.Sunshine JC, Peng DY, Green JJ. Uptake and transfection with polymeric nanoparticles are dependent on polymer end-group structure, but largely independent of nanoparticle physical and chemical properties. Mol Pharm. 2012;9:3375–3383. doi: 10.1021/mp3004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J, Sunshine JC, Green JJ. Differential polymer structure tunes mechanism of cellular uptake and transfection routes of poly(beta-amino ester) polyplexes in human breast cancer cells. Bioconjugate Chem. 2014;25:43–51. doi: 10.1021/bc4002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris TJ, Green JJ, Fung PW, Langer R, Anderson DG, Bhatia SN. Tissue-specific gene delivery via nanoparticle coating. Biomaterials. 2010;31:998–1006. doi: 10.1016/j.biomaterials.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shenoy D, Little S, Langer R, Amiji M. Poly(ethylene oxide)-modified poly(beta-amino ester) nanoparticles as a pH-sensitive system for tumor-targeted delivery of hydrophobic drugs. 1. In vitro evaluations. Mol Pharm. 2005;2:357–366. doi: 10.1021/mp0500420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Y, Tang H, Zhan Y, Van Kirk EA, Murdoch WJ. Degradable poly(beta-amino ester) nanoparticles for cancer cytoplasmic drug delivery. Nanomedicine. 2009;5:192–201. doi: 10.1016/j.nano.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Harris JM, Chess RB. Effect of pegylation on pharmaceuticals. Nat Rev Drug Discov. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 21.Owens DE, 3rd, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 22.Hatakeyama H, Akita H, Harashima H. A multifunctional envelope type nano device (MEND) for gene delivery to tumours based on the EPR effect: a strategy for overcoming the PEG dilemma. Adv Drug Deliv Rev. 2011;63:152–160. doi: 10.1016/j.addr.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Ge Z, Chen Q, Osada K, Liu X, Tockary TA, Uchida S, Dirisala A, Ishii T, Nomoto T, Toh K, Matsumoto Y, Oba M, Kano MR, Itaka K, Kataoka K. Targeted gene delivery by polyplex micelles with crowded PEG palisade and cRGD moiety for systemic treatment of pancreatic tumors. Biomaterials. 2014;35:3416–3426. doi: 10.1016/j.biomaterials.2013.12.086. [DOI] [PubMed] [Google Scholar]
- 24.Williford JM, Archang MM, Minn I, Ren Y, Wo M, Vandermark J, Fisher PB, Pomper MG, Mao HQ. Critical Length of PEG Grafts on lPEI/DNA Nanoparticles for Efficient in Vivo Delivery. ACS Biomater Sci Eng. 2016;2:567–578. doi: 10.1021/acsbiomaterials.5b00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vandenbergh J, Ranieri K, Junkers T. Synthesis of (Bio)-Degradable Poly(β-thioester)s via Amine Catalyzed Thiol−Ene Click Polymerization. Macromol Chem Physic. 2012;213:2611–2617. [Google Scholar]
- 26.Mangraviti A, Tzeng SY, Kozielski KL, Wang Y, Jin Y, Gullotti D, Pedone M, Buaron N, Liu A, Wilson DR, Hansen SK, Rodriguez FJ, Gao GD, DiMeco F, Brem H, Olivi A, Tyler B, Green JJ. Polymeric nanoparticles for nonviral gene therapy extend brain tumor survival in vivo. ACS Nano. 2015;9:1236–1249. doi: 10.1021/nn504905q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filipe V, Hawe A, Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res. 2010;27:796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nobbmann U, Morfesis A. Light scattering and nanoparticles. Mater Today. 2009;12:52–54. [Google Scholar]
- 29.Bhise NS, Gray RS, Sunshine JC, Htet S, Ewald AJ, Green JJ. The relationship between terminal functionalization and molecular weight of a gene delivery polymer and transfection efficacy in mammary epithelial 2-D cultures and 3-D organotypic cultures. Biomaterials. 2010;31:8088–8096. doi: 10.1016/j.biomaterials.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boucher PD, Ruch RJ, Shewach DS. Differential ganciclovir-mediated cytotoxicity and bystander killing in human colon carcinoma cell lines expressing herpes simplex virus thymidine kinase. Hum Gene Ther. 1998;9:801–814. doi: 10.1089/hum.1998.9.6-801. [DOI] [PubMed] [Google Scholar]
- 31.Shalev M, Kadmon D, Teh BS, Butler EB, Aguilar-Cordova E, Thompson TC, Herman JR, Adler HL, Scardino PT, Miles BJ. Suicide gene therapy toxicity after multiple and repeat injections in patients with localized prostate cancer. J Urol. 2000;163:1747–1750. [PubMed] [Google Scholar]
- 32.Rubsam LZ, Boucher PD, Murphy PJ, KuKuruga M, Shewach DS. Cytotoxicity and accumulation of ganciclovir triphosphate in bystander cells cocultured with herpes simplex virus type 1 thymidine kinase-expressing human glioblastoma cells. Cancer Res. 1999;59:669–675. [PubMed] [Google Scholar]
- 33.Tomicic MT, Thust R, Kaina B. Ganciclovir-induced apoptosis in HSV-1 thymidine kinase expressing cells: critical role of DNA breaks, Bcl-2 decline and caspase-9 activation. Oncogene. 2002;21:2141–2153. doi: 10.1038/sj.onc.1205280. [DOI] [PubMed] [Google Scholar]
- 34.Steichen SD, Caldorera-Moore M, Peppas NA. A review of current nanoparticle and targeting moieties for the delivery of cancer therapeutics. Eur J Pharm Sci. 2013;48:416–427. doi: 10.1016/j.ejps.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E, Ogris M. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J Gene Med. 2004;6:1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 36.Park MR, Han KO, Han IK, Cho MH, Nah JW, Choi YJ, Cho CS. Degradable polyethylenimine-alt-poly(ethylene glycol) copolymers as novel gene carriers. J Control Release. 2005;105:367–380. doi: 10.1016/j.jconrel.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 37.Green JJ, Langer R, Anderson DG. A combinatorial polymer library approach yields insight into nonviral gene delivery. Acc Chem Res. 2008;41:749–759. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mishra S, Webster P, Davis ME. PEGylation significantly affects cellular uptake and intracellular trafficking of non-viral gene delivery particles. Eur J Cell Biol. 2004;83:97–111. doi: 10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- 39.Kamat CD, Shmueli RB, Connis N, Rudin CM, Green JJ, Hann CL. Poly(beta-amino ester) nanoparticle delivery of TP53 has activity against small cell lung cancer in vitro and in vivo. Mol Cancer Ther. 2013;12:405–415. doi: 10.1158/1535-7163.MCT-12-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guerrero-Cazares H, Tzeng SY, Young NP, Abutaleb AO, Quinones-Hinojosa A, Green JJ. Biodegradable polymeric nanoparticles show high efficacy and specificity at DNA delivery to human glioblastoma in vitro and in vivo. ACS Nano. 2014;8:5141–5153. doi: 10.1021/nn501197v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tzeng SY, Higgins LJ, Pomper MG, Green JJ. Student award winner in the Ph.D. category for the 2013 society for biomaterials annual meeting and exposition, april 10-13, 2013, Boston, Massachusetts: biomaterial-mediated cancer-specific DNA delivery to liver cell cultures using synthetic poly(beta-amino ester)s. J Biomed Mater Res A. 2013;101:1837–1845. doi: 10.1002/jbm.a.34616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J, Wilson DR, Zamboni CG, Green JJ. Targeted polymeric nanoparticles for cancer gene therapy. J Drug Target. 2015;23:627–641. doi: 10.3109/1061186X.2015.1048519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


