Abstract
Purpose of review
Erythropoiesis, in which hematopoietic stem cells (HSCs) generate lineage-committed progenitors that mature into erythrocytes, is regulated by numerous chromatin modifying and remodeling proteins. We will focus on how epigenetic and genetic mechanisms mesh to establish the erythroid transcriptome and how studying erythropoiesis can yield genomic principles.
Recent findings
Trans-acting factor binding to small DNA motifs (cis-elements) underlies regulatory complex assembly at specific chromatin sites, and therefore unique transcriptomes. As cis-elements are often very small, thousands or millions of copies of a given element reside in a genome. Chromatin restricts factor access in a context-dependent manner, and cis-element-binding factors recruit chromatin regulators that mediate functional outputs. Technologies to map chromatin attributes of loci in vivo, to edit genomes and to sequence whole genomes have been transformative in discovering critical cis-elements linked to human disease.
Summary
Cis-elements mediate chromatin-targeting specificity, and chromatin regulators dictate cis-element accessibility/function, illustrating an amalgamation of genetic and epigenetic mechanisms. Cis-elements often function ectopically when studied outside of their endogenous loci, and complex strategies to identify nonredundant cis-elements require further development. Facile genome-editing technologies provide a new approach to address this problem. Extending genetic analyses beyond exons and promoters will yield a rich pipeline of cis-element alterations with importance for red cell biology and disease.
Keywords: chromatin, cis-element, epigenetics, erythropoiesis
INTRODUCTION
The progressive transition of a nucleated erythroblast into an enucleated erythrocyte requires profound morphological and functional changes to ensure the generation of billions of erythrocytes daily [1,2]. As this process involves massive chromatin structure reconfiguration, there is considerable interest in identifying the regulators, how they are integrated to yield a vital network, and factors/signals that control the regulators. The proteins mediating chromatin transitions are often referred to as ‘epigenetic regulators’. Although the semantics of equating ‘epigenetics’ with chromatin mechanisms is hotly debated, these factors may generate memory that dictates the daughter cell transcriptome and phenotypes. Moreover, the DNA methylation epigenetic mark 5-methylcytosine is inextricably linked to chromatin mechanisms [3]. We shall focus on the role of chromatin regulators in generating the erythroid transcriptome. As cis-elements recognized by activators and repressors underlie chromatin-targeting specificity, their genetic integrity is crucial to ensure normal chromatin landscapes. On the basis of the amalgamation of genetic and epigenetic mechanisms, ascribing genetic or epigenetic components to a biological process can be murky.
Large numbers of erythroid cells representing distinct maturation states can be isolated, and powerful models exist for studying erythropoiesis and erythroid cell function [1,2]. As such, erythroid cells are ideal for investigating chromatin control of genome function, development and homeostasis. Erythropoiesis is controlled by a restricted cohort of lineage-specific master transcriptional regulators functioning in concert with broadly expressed factors. The founding member of the GATA-binding protein (GATA) transcription factor family GATA-1 [4,5] and Kruppel-like factor 1 (KLF1) [6] exemplify core determinants of the erythroid transcriptome that regulate both shared and unique target genes. GATA-2 [7,8], runt-related transcription factor 1 (RUNX1) [9,10] and T-cell acute lymphocytic leukemia protein 1 (TAL1) [11] have pivotal roles to promote the genesis and/or function of hematopoietic stem/progenitor cells (HSPCs), although TAL1 is also expressed and functions in erythroid cells [12].
Massive efforts utilizing genome-wide technologies have mapped transcription factor, coregulator and chromatin landscapes in living cells [13,14,15▪▪,16,17], including erythroid cells [18–22,23▪,24,25,26▪]. An ongoing challenge is to leverage these datasets into innovative discoveries to explain epigenetic mechanisms underlying erythroid precursor cell development into erythrocyte, and how these mechanisms are impacted by stress and pathophysiological states.
TRANSCRIPTIONAL CONTROL OF ERYTHROID CELL DEVELOPMENT AND FUNCTION
Although the dual zinc finger transcription factors GATA-1 and GATA-2 share a similar DNA binding domain [27], they differ in significant ways. They are differentially expressed during hematopoiesis, with GATA-2 expressed predominantly in HSPCs and GATA-1 expressed in erythroid, megakaryocytic, mast and eosinophil cells [5,7,8,28]. They have distinct functions to mediate generation, proliferation and/or survival of hematopoietic cell types [7,8,29,30,31▪,32▪]. GATA-2 induces hematopoietic stem cell (HSC) generation from hemogenic endothelium in the aorta gonad mesonephros region of the embryo and regulates HSPC function [31▪–34▪]. By contrast, hemogenic endothelium and HSCs express little to no GATA-1. GATA-1 functions in erythroid precursors to promote erythroid cell development and maturation [29]. GATA-1 and GATA-2 have distinct biochemical attributes, for example, GATA-2 is less stable than GATA-1 [35], and GATA-1 selectively requires the coregulator friend of GATA-1 (FOG-1) for many of its actions [36,37]. Finally, GATA-1 and GATA-2 differentially regulate target gene transcription [38,39].
Despite their differences, GATA-1 and GATA-2 actions are intricately linked. GATA-1 is upregulated at an early stage of erythropoiesis and directly represses Gata2 expression [38,40]. In erythroid precursors, GATA-2 occupies five sites at the active Gata2 locus, indicative of positive autoregulation [40–42]. As GATA-1 levels/activity rise, GATA-1 displaces GATA-2 from genomic sites – a process termed GATA switching [38–40]. GATA-1 upregulation in erythropoiesis occurs during the early transition (‘S0 to S1’) involving commitment to terminal differentiation and dramatic chromatin reconfiguration [43]. Given the GATA-2 function in hemogenic endothelium and HSPCs, and GATA-1 promotion of erythropoiesis, GATA switches control erythroid precursor cell differentiation into erythrocytes. Not all GATA-1/GATA-2 chromatin occupancy sites are GATA switch sites, as certain sites are occupied preferentially or exclusively by GATA-1 or GATA-2 [18,26▪,44].
As GATA-1 upregulation induces GATA switches, it is instructive to consider the factors/signals that control GATA-1 expression and activity. Coregulators mediate GATA-1 activity in a context-dependent manner [20,37,45▪,46–48]. GATA-1-mediated activation and repression is facilitated by FOG-1 at many target loci [37,49]. This multi-zinc finger protein binds the GATA-1 N-terminal zinc finger and appears to lack DNA binding activity [37,50]. The GATA-1 C-terminal zinc finger binds to the GATA motif (A/TGATAA) [51,52], which in a chromatin context has the consensus (C/G)(A/T)-GATAA(G/A/C)(G/A/C) [18]. FOG-1 copurifies with the NuRD chromatin remodeling complex, and NuRD mediates certain GATA-1 functions [53,54]. FOG-1 facilitates GATA-1 chromatin occupancy at select sites [55,56], precludes GATA-1 occupancy at others [23▪] and facilitates GATA switches [55]. GATA-1 occupies several thousand genomic sites, with the highest frequency at introns and sites distal to promoters [18–21]. GATA-1 and GATA-2 form a multimeric transcriptional regulatory complex with TAL1, LIM domain only protein 2 (LMO2) and LIM domain-binding protein 1 (LDB1) [57]. GATA-1-occupied and GATA-2-occupied loci are commonly associated with one or more of these factors [20,58–62,63▪▪], and enrichments in histone H3K4me1 [21]. LDB1 and LMO2 can contribute to GATA-1-mediated activation and repression [47]. The Brahma-related gene 1 (BRG1) component of the switch/sucrose non-fermentable (SWI/SNF) chromatin remodeling complex induces chromatin accessibility at GATA-1-occupied enhancers [64,65]. Interferon regulatory factors colocalize with GATA-1 at certain sites and confer stage-specific transcription [66]. E-twenty-six (ETS) transcription factors can cooccupy chromatin with GATA-1/GATA-2, influencing their activities [44,67,68]. Given the numerous ETS factors, it will be instructive to consider how multiple ETS factors in the same cell influence GATA factor function.
Although it is in vogue to study factor colocalization at endogenous sites, extrapolating chromatin immunoprecipitation (ChIP) data to function can be precarious, as formaldehyde cross-linking might reflect nonredundant or redundant function, or an interaction with no functional significance. Knockdown studies in a genetic complementation assay in GATA-1-null G1E cells represent a powerful approach to identify functional determinants of GATA-1-mediated transcriptional regulation. G1E cells, which resemble normal proerythroblasts, were derived from mouse embryonic stem cells via disruption of the Gata1 locus [69]. Expression of an estrogen receptor ligand binding domain fusion to GATA-1 allows one to rapidly activate GATA-1, which induces a normal erythroid program [40,70] that recapitulates a physiologically relevant window of erythroid maturation [70].
Studies in G1E cells indicate that the mechanistic requirements for GATA-1-mediated activation and repression differ in distinct contexts. GATA-1-regulated loci can be sensitive or insensitive to FOG-1, the NuRD component Mi2β and the histone H3K20 monomethyltransferase SetD8 [45▪]. The differential locus sensitivity to partial factor knockdowns (50–80%) may reflect overt qualitative differences or locus-specific concentration requirements for the factors. The context-dependent FOG-1 requirement for GATA-1 activity was first demonstrated using GATA-1 mutants (V205G or V205M) impaired in FOG-1 binding [37]. As these mutants do not eliminate FOG-1 binding, alternative strategies have been devised, including genetic complementation in FOG-1-null hematopoietic precursor cells [71]. However, these cells lack certain factors required for erythroid gene expression. Multiple lines of evidence using these strategies provide evidence for differential FOG-1 requirements at distinct loci.
Of high relevance to GATA-1 function is its capacity to regulate local chromatin structure [72–74] and to induce long-range chromatin looping [75]. GATA-1 induces a chromatin loop between the β-globin locus control region (LCR) and the downstream βmajor promoter [75]. FOG-1 is required for GATA-1-dependent looping, at least at the limited number of loci examined. This might relate to its activity to facilitate GATA-1 occupancy or an unidentified mechanism. LDB1 [76] and BRG1 [77] also promote this chromatin loop. GATA-1 expels the β-globin locus from the nuclear periphery concomitant with looping [78,79], and expulsion characterizes primary erythroid cell maturation [80]. As the nuclear periphery can create an inhospitable environment for transcription [81], expulsion may relocate the locus into a favorable environment to generate high levels of β-globin. Tiling the β-globin locus and neighboring sequences with bacterial artificial chromosome probes indicated that expulsion is restricted to the β-globin locus and does not involve a considerably broader region [79]. KLF1 promotes the LCR–βmajor promoter loop [82], contributes to expulsion [79] and has a broader role in conferring subnuclear positioning of genes [83]. Recent evidence indicates that GATA-1 functions as a mitotic ‘bookmark’ to ensure the stable maintenance of lineage-specific gene expression throughout development [84▪▪].
Given the instrumental role of GATA-1 to establish/maintain the erythroid cell transcriptome and to promote erythropoiesis, defining mechanisms that control GATA-1 expression/activity are very important. Gata1 expression in erythroid cells is regulated by an upstream GATA motif-containing enhancer [85]. Targeted deletion of this cis-element, which contains GATA and CACCC motifs, yields an erythroid maturation defect only when the NeoR gene remains at the targeted site; NeoR excision is associated with normal erythropoiesis [86]. In vivo footprinting and overexpression analysis in zebra-fish suggest that GATA-1 autoregulates its own expression [87,88]. PBX and MEIS1 act upstream of GATA-1 in erythropoiesis [89], and Biklf [90] and ZBP-89 [91] promote Gata1 expression in zebrafish. In an embryonic stem cell differentiation system, bone morphogenetic protein signaling induces Gata1 [92]. The sumo ligase PIAS1 represses Gata1 in HSCs [93]. Despite this entourage of factors, considerable work is required to achieve a coherent model for how their activities are integrated through Gata1 cis-elements.
The myeloid transcription factor PU.1 is expressed reciprocally with GATA-1 and represses GATA-1 activity [94]. PU.1 downregulation enhances GATA-1 activity concomitant with, or as a prelude to, Gata2 repression, and GATA-1 represses PU.1 expression [95]. Phosphorylation, sumoylation and acetylation regulate GATA-1 activity. Although GATA-1 is multisite phosphorylated [96], with Ser302 phosphorylated by Akt [97], how phosphorylation influences GATA-1 activity is unclear. GATA-1 sumoylation at Lys137 facilitates its regulation of FOG-1-dependent target genes and expulsion of the loci from the nuclear periphery [78]. As FOG-1 [55] and GATA-1 multisite acetylation [98] promote GATA-1 chromatin occupancy, mechanisms regulating FOG-1 levels/activity and the acetylation pathway dictate GATA-1 activity, and therefore GATA switches. FOG-1 is multisite sumoylated [99] and presumably regulated by diverse post-translational modifications.
Even though GATA-1 and GATA-2 control distinct processes during hematopoiesis, they confer primitive erythroblast survival redundantly [100]. GATA-1 and GATA-2 can colocalize with a cohort of factors [TAL1, lympoblastomic leukemia 1 (LYL1), LMO2, RUNX1, ETS-related gene (ERG) and friend leukemia integration 1 (FLI1)] that have important functions to control hematopoiesis [62,101–104]. Coregulators including p300 [105], MED1 [106] and HDAC3/4 [105,107] have been reported to mediate GATA-2 function. Although GATA-2 is acetylated [105,108] and phosphorylated [109], and MAP kinase and Akt phosphorylate GATA-2 [109,110], the modification sites and consequences are unclear.
In addition to established GATA-1 and GATA-2 coregulators, other chromatin modifying enzymes are implicated in erythropoiesis and/or erythroblast function. The histone methyltransferases Mll, Dot1L, Ezh2 and SetD8 regulate hematopoiesis and/or hematopoietic cell function [45▪,111–113]. The lysine-specific demethylase LSD1 [114] regulates erythroid progenitor differentiation by repressing genes associated with HSPCs [115,116]. As histone marks and DNA methylation dramatically change upon erythroid maturation [117,118], elucidating molecular determinants of these patterns will yield important mechanistic insights. Attempting to unravel how factors control specific loci without knowing the requisite cis-elements is analogous to embarking on the construction of a sophisticated architectural structure devoid of a blueprint.
IDENTIFYING AND ANALYZING NONREDUNDANT CIS-ELEMENTS IN COMPLEX GENOMES
ChIP-seq commonly reveals several thousand factor occupancy sites with only a small fraction of the cis-elements occupied. Although thousands of GATA motifs exist in a genome, each of which would bind GATA-1/GATA-2 with high affinity as naked DNA, less than 1% are occupied in chromatin [18,19]. The mechanisms that endow motifs with the capacity to bind GATA factors in chromatin and why only certain occupied cis-elements confer nonredundant activity in vivo remain enigmatic.
Genomic maps of transcription factor occupancy and chromatin landscape are used to infer function. H3K4me1 and H3K27ac enrichments are interpreted to demarcate active enhancers [119▪▪]. Large chromosomal segments enriched in H3K27me3 and H3K4me3 (bivalent domains) are thought to poise loci for rapid activation during development [119▪▪]. p300 occupancy [120] is considered to pinpoint enhancers, whereas DNaseI hypersensitivity [121] and formaldehyde-assisted isolation of regulatory elements [122] score for accessibility. Although these approaches can lead to the discovery of putative cis-elements, predictions are tenuous without accompanying functional analysis at the endogenous locus. Another limitation is that mapping studies do not always utilize systems that recapitulate the relevant biology.
Traditional assays to evaluate cis-element function commonly rely on reporter gene measurements in transfected cells or transgene activity at ectopic chromatin sites. Cis-elements can elicit substantial activities within plasmids and at ectopic chromatin sites, which are irrelevant to endogenous locus function. The β-globin LCR consists of four DNaseI hypersensitivity sites approximately 15–50 kilo-bases upstream of the embryonic ε-globin and adult βmajor promoters, respectively [123,124], and is a pivotal determinant of β-like globin gene transcription at all developmental stages. The LCR confers position-independent and copy number-dependent expression of transgenes in transgenic mice, and targeted deletion of the hypersensitivity sites collectively in mice strongly reduces β-like globin expression at all developmental stages [125]. Many reports have documented impressive enhancer activities of individual hypersensitivity sites and subfragments thereof in transfection assays and transgenic mice. However, removing entire hypersensitivity sites of the LCR, which contain multiple cis-elements, only modestly (~10–30%) influence β-like globin expression [126]. Comparison of deletions of one or more of the hypersensitivity sites revealed that the individual hypersensitivity sites function additively to yield the powerful enhancer activity. This work, combined with numerous other studies, indicates that in vitro models of enhancer activity often do not recapitulate physiological function. The limitations of assessing cis-element function using historically accepted strategies are considerable.
The low frequency of GATA-1/GATA-2 occupancy of GATA motifs in chromatin, different permutations of GATA motifs (sequence variants, proximity to neighboring cis-elements, distinct chromatin attributes etc.), biologically critical functions of GATA-1 and GATA-2, unanswered questions about GATA switching and unresolved issues in epigenetics constitute a strong rationale for addressing how GATA motifs function at endogenous loci. An ideal system to explore this problem is the Gata2 locus, given its five GATA switch sites, essential function to control HSPC generation/function, and links to hematologic malignancies and vascular disorders.
Three GATA switch sites were deleted individually in mouse embryonic stem cells, and mutant mouse strains were generated. Deletion of the –1.8 site containing a conserved palindromic GATA motif revealed little to no role in activating Gata2 expression, but it is essential for maintaining Gata2 repression during erythroid maturation [127]. However, hematopoiesis is largely normal. Deletion of the –2.8 site, which contains multiple conserved GATA motifs, modestly reduces maximal Gata2 expression, but does not influence Gata2 repression upon erythroid maturation, nor does it significantly affect hematopoiesis [128]. Knockout of the intronic +9.5 site, which contains a conserved E-box–GATA composite element, is embryonic lethal at E14.5. +9.5−/− embryos have very few fetal liver HSPCs in E12.5 embryos, and Gata2 expression is strongly reduced, consistent with the loss of Gata2-expressing cells [31▪,33▪] (Fig. 1). Although these sites share comparable GATA-1/GATA-2 occupancy, certain chromatin attributes and enhancer activity in vitro, their deletions yielded gross qualitative differences in activity.
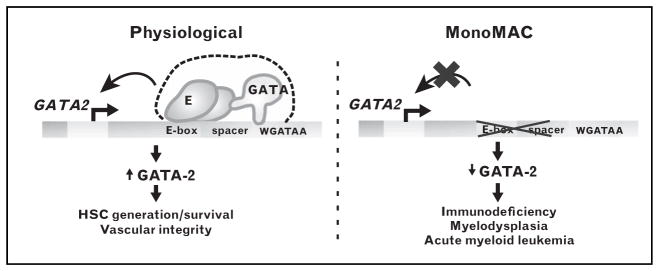
FIGURE 1.
Gata2 +9.5 cis-element. The Gata2 intronic +9.5 GATA switch site, which contains an E-box–GATA motif composite element, functions nonredundantly to confer hematopoietic stem cell (HSC) generation in the aorta gonad mesonephros (AGM) region, the fetal liver hematopoietic stem/progenitor cell (HSPC) compartment and vascular integrity. Heterozygous mutation of the +9.5 element leads to monocytopenia and mycobacteria infection (MonoMAC) syndrome, with a phenotype indistinguishable from MonoMAC patients with GATA2 zinc finger mutations. Adapted with permission from [33▪].
Recent genome-editing innovations allow one to analyze cis-elements at endogenous loci in essentially any system [129▪▪,130▪▪]. The utility of this approach was highlighted in an application of transcription activator-like effector nucleases (TALENs) to delete a potential enhancer in intron 2 of Bcl11a, which encodes a fetal γ-globin repressor [131]. The deletion demonstrated the importance of the element for conferring Bcl11a expression in erythroid cells. Loss of the element downregulated BCL11A, which induced the γ-globin genes [132▪▪]. As γ-globin gene upregulation counteracts toxic effects of mutant β-globin in sickle cell disease, this study highlighted a potential therapeutic approach. The use of TALENs and zinc finger–nuclease fusions, along with the clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9 system, to generate site-directed deletions is revolutionizing analyses of cis-element function at endogenous loci and their contribution to epigenetic mechanisms.
CIS-ELEMENT MECHANISMS UNDERLYING HEMATOPOIETIC PATHOPHYSIOLOGIES
Inherited and acquired hematologic disorders are often caused by mutations at loci essential for normal hematopoiesis. Although many mutations and polymorphisms in protein-coding regions have been described, less is known about variation in noncoding DNA motifs that influence disease susceptibility. Regulatory single nucleotide polymorphisms (SNPs) have been identified within a GATA motif upstream of the α-globin gene that causes α-thalassemia by interfering with activation of α-like globin genes [133] (Fig. 2). An intron 1 mutation disrupts a GATA motif in ALAS2, yielding X-linked sideroblastic anemia [134▪] (Fig. 2). SNPs within the BCL11A intronic enhancer impact hemoglobin levels and chromatin occupancy [132▪▪] (Fig. 2). Evidence for the role of cis-elements in suppressing malignant hematopoiesis emerged from analysis of patients with monocytopenia and mycobacteria infection (MonoMAC), an immunodeficiency associated with a predisposition for myelodysplastic syndrome and acute myeloid leukemia [135–138]. MonoMAC is caused by heterozygous mutations in the DNA binding zinc finger of GATA-2 and appears to involve haplo-insufficiency [139]. One MonoMAC patient lacking coding region mutations harbors a heterozygous deletion of the E-box and five base pairs of the spacer of the +9.5 composite element (Fig. 1), and the phenotype of this patient is indistinguishable from those with GATA-2 zinc finger mutations [33▪]. Additional patients harbor point mutations in an ETS motif residing near the +9.5 composite element [139]. In a transfection context, the ETS motif was important for composite element enhancer activity [139]. It seems likely that further studies will reveal disruptions in many cis-elements that function nonredundantly to control hematopoiesis and/or hematopoietic cell function, and these alterations will underlie malignant and nonmalignant hematologic disorders.
FIGURE 2.
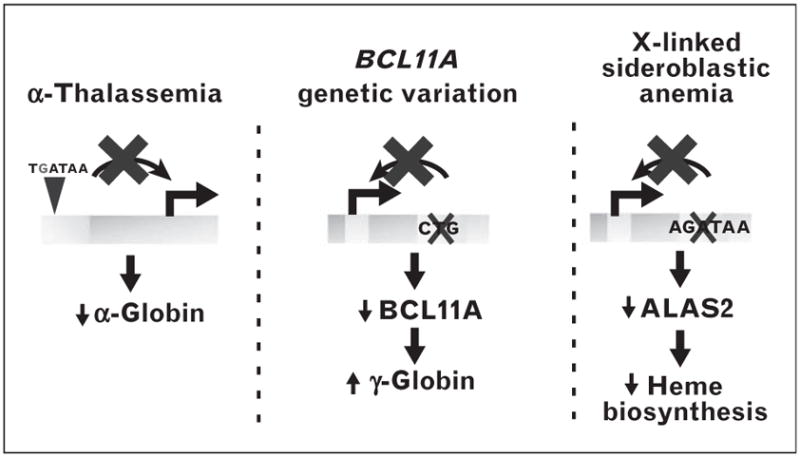
Cis-element variation in normal and disease states. Mutational generation of a GATA motif interferes with α-globin transcription leading to α-thalassemia [133]. Natural variation in a GATA-1-binding region of the BCL11A locus as a determinant of γ-globin expression [132▪▪]. Mutational disruption of a GATA-1 motif reduces expression of ALAS2, which encodes a critical heme biosynthetic enzyme [134▪].
Genome-wide association studies (GWASs) are increasingly identifying putative noncoding sequence variants linked to a predisposition for specific pathophysiologies. SNPs linked to predisposition for development of Hodgkin’s lymphoma occur in genomic regions lacking known functions [140]. Seventy-five independent loci containing SNPs correlate with red cell phenotypes [141]. Although the majority of SNPs reside in noncoding regions, most GWASs have been disproportionately biased toward tabulating coding variants. Expanding these studies to identify additional noncoding variants that predispose for pathophysiological conditions will reveal novel disease-relevant loci that can be functionally validated using genome-editing technologies.
CONCLUSION
‘Epigenetics’ is often used to refer to chromatin modifying/remodeling mechanisms. Others argue that epigenetics should be restricted to scenarios involving unequivocal heritable transmission of traits without altered genetic content, which might or might not involve chromatin mechanisms. As cis-elements underlie chromatin-targeting specificity, and chromatin controls cis-element accessibility/function, disentangling genetic versus epigenetic contributions can be daunting. Sifting through abundant prospective cis-elements to identify those with nonredundant function is challenging. How many cis-elements resemble the +9.5 in controlling hematopoiesis nonredundantly? Can the thousands of GATA-1/GATA-2-occupied elements be segregated into functional versus nonfunctional elements via bioinformatics alone (Fig. 3)? How do critical cis-elements function in a context-dependent manner, for example, in distinct developmental stages? How important are combinatorial mechanisms in which merged cis-elements constitute entities with activities that cannot be predicted based on how the individual cis-elements function and factor occupancy patterns? To what extent do cis-element mutations, natural variation and alterations in chromatin mechanisms that control cis-element function underlie pathophysiologies and interindividual variation? Addressing these types of questions will yield important insights into red cell biology, normal and malignant hematopoiesis, and more broadly biological and genomic principles.
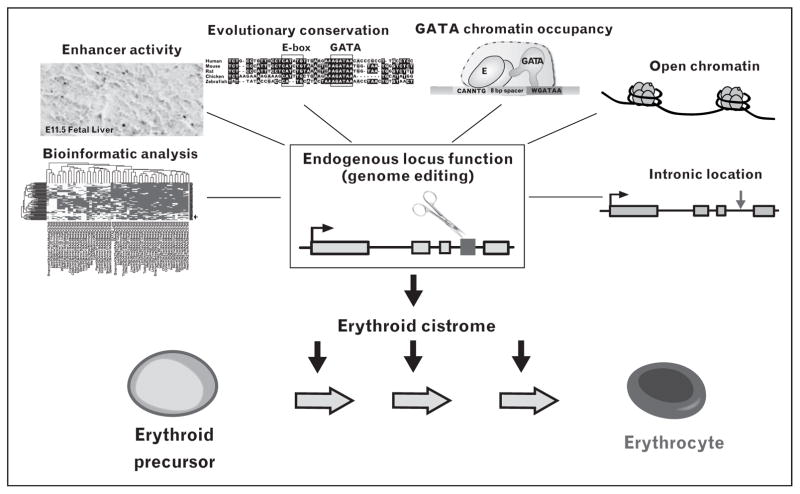
FIGURE 3.
Erythroid cistrome discovery strategy. Prospective cis-elements are prioritized based on multiple parameters and subjected to functional analysis by cis-element editing. GATA-1-occupied cis-elements functional at their endogenous loci are predicted to be important determinants of erythroid cell genesis and/or function.
KEY POINTS.
Cis-elements underlie the complex amalgamation of genetic and epigenetic mechanisms.
GATA-1 and GATA-2 occupy a small fraction of their abundant cis-elements in a genome, and occupancy does not predict the functional output.
Although predicting GATA motifs that function nonredundantly in vivo is not yet possible, multiple parameters collectively may have predictive value.
Validation of cis-element function at endogenous loci is crucial, and novel genome-editing tools are revolutionizing such analyses.
Mutational disruption of cis-elements underlies malignant and nonmalignant hematologic disorders, and single nucleotide polymorphisms within cis-elements can yield significant interindividual differences in hematologic parameters.
Acknowledgments
Funding sources: NIH R37DK50107 (E.H.B.), NIH R0168034 (E.H.B.), postdoctoral training grant T32 HL 007899 (K.J.H.).
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
- 1.Hattangadi SM, Wong P, Zhang L, et al. From stem cell to red cell: regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood. 2011;118:6258–6268. doi: 10.1182/blood-2011-07-356006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGrath K, Palis J. Ontogeny of erythropoiesis in the mammalian embryo. Curr Top Dev Biol. 2008;82:1–22. doi: 10.1016/S0070-2153(07)00001-4. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 4.Evans T, Felsenfeld G. The erythroid-specific transcription factor Eryf1: a new finger protein. Cell. 1989;58:877–885. doi: 10.1016/0092-8674(89)90940-9. [DOI] [PubMed] [Google Scholar]
- 5.Tsai SF, Martin DI, Zon LI, et al. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- 6.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai FY, Keller G, Kuo FC, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 8.Tsai FY, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89:3636–3643. [PubMed] [Google Scholar]
- 9.Wang Q, Stacy T, Binder M, et al. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okuda T, van Deursen J, Hiebert SW, et al. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 11.Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 12.Hall MA, Slater NJ, Salmon JM, et al. Functional but abnormal adult erythropoiesis in the absence of the stem cell leukemia gene. Mol Cell Biol. 2005;25:6355–6362. doi: 10.1128/MCB.25.15.6355-6362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stamatoyannopoulos JA, Snyder M, Hardison R, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15▪▪.Neph S, Vierstra J, Stergachis AB, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. Through a massive genome-wide analysis of DNaseI cleavage of chromatin, this study describes the genomic complement of ‘footprints’ reflecting, in part, protein–chromatin interactions. This uniquely powerful resource will impact upon many problems in genome biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Consortium EP. Dunham I, Kundaje A, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen Y, Yue F, McCleary DF, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara T, O’Geen H, Keles S, et al. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang YA, Sanalkumar R, O’Geen H, et al. Autophagy driven by a master regulator of hematopoiesis. Mol Cell Biol. 2012;32:226–239. doi: 10.1128/MCB.06166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu M, Riva L, Xie H, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y, Wu W, Kumar SA, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Wu W, Cheng Y, et al. Primary sequence and epigenetic determinants of in vivo occupancy of genomic DNA by GATA1. Nucleic Acids Res. 2009;37:7024–7038. doi: 10.1093/nar/gkp747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23▪.Chlon TM, Dore LC, Crispino JD. Cofactor-mediated restriction of GATA-1 chromatin occupancy coordinates lineage-specific gene expression. Mol Cell. 2012;47:608–621. doi: 10.1016/j.molcel.2012.05.051. By comparing GATA-1 and GATA-1(V205G) chromatin occupancy genome-wide, evidence is provided that FOG-1 binding to GATA-1 has a profound impact upon chromatin target site occupancy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tallack MR, Whitington T, Yuen WS, et al. A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res. 2010;20:1052–1063. doi: 10.1101/gr.106575.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pilon AM, Ajay SS, Kumar SA, et al. Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood. 2011;118:e139–e148. doi: 10.1182/blood-2011-05-355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26▪.May G, Soneji S, Tipping AJ, et al. Dynamic analysis of gene expression and genome-wide transcription factor binding during lineage specification of multipotent progenitors. Cell Stem Cell. 2013;13:754–768. doi: 10.1016/j.stem.2013.09.003. This study describes a rigorous hematopoietic factor ChIP-seq/network analysis with the factor-dependent cell progenitors mix model system, which provided evidence for GATA-2-mediated repression of PU.1 as an important step in lineage specification and the existence of a functionally critical ‘GATA-1–GATA-2–PU.1 kernal’. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gamsjaeger R, Liew CK, Loughlin FE, et al. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci. 2007;32:63–70. doi: 10.1016/j.tibs.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 28.Leonard M, Brice M, Engel JD, et al. Dynamics of GATA transcription factor expression during erythroid differentiation. Blood. 1993;82:1071–1079. [PubMed] [Google Scholar]
- 29.Pevny L, Simon MC, Robertson E, et al. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 30.Ling KW, Ottersbach K, van Hamburg JP, et al. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med. 2004;200:871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31▪.Gao X, Johnson KD, Chang Y-I, et al. Gata2 cis-element is required for hematopoietic stem cell generation in the mammalian embryo. J Exp Med. 2013;210:2833–2842. doi: 10.1084/jem.20130733. The study illustrates how deleting a single GATA factor-binding cis-element from a Gata2 intron abrogates the capacity of hemogenic endothelium to form long-term repopulating HSCs by severely disrupting the requisite genetic network. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32▪.de Pater E, Kaimakis P, Vink CS, et al. Gata2 is required for HSC generation and survival. J Exp Med. 2013;210:2843–2850. doi: 10.1084/jem.20130751. Through rigorous analysis of Gata2 conditional knockouts, this study describes the importance of GATA-2 for regulating the generation and survival of HSCs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33▪.Johnson KD, Hsu AP, Ryu MJ, et al. Cis-element mutated in GATA2-dependent immunodeficiency governs hematopoiesis and vascular integrity. J Clin Invest. 2012;122:3692–3704. doi: 10.1172/JCI61623. The article describes the discovery of a Gata2 intronic enhancer (+9.5) required for generation of the fetal liver hematopoietic stem/progenitor cell compartment, for conferring vascular integrity, and for embryogenesis, which was mutated in a patient with MonoMAC immunodeficiency syndrome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34▪.Lim KC, Hosoya T, Brandt W, et al. Conditional Gata2 inactivation results in HSC loss and lymphatic mispatterning. J Clin Invest. 2012;122:3705–3717. doi: 10.1172/JCI61619. This article describes the use of a Gata2 +9.5 enhancer-Cre mouse strain to generate a conditional Gata2 knockout, which revealed its importance for establishment of HSPCs in the fetal liver and bone marrow and for lymphatic development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lurie LJ, Boyer ME, Grass JA, et al. Differential GATA factor stabilities: implications for chromatin occupancy by structurally similar transcription factors. Biochemistry. 2008;47:859–869. doi: 10.1021/bi701692p. [DOI] [PubMed] [Google Scholar]
- 36.Tsang AP, Fujiwara Y, Hom DB, et al. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev. 1998;12:1176–1188. doi: 10.1101/gad.12.8.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crispino JD, Lodish MB, MacKay JP, et al. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol Cell. 1999;3:219–228. [Google Scholar]
- 38.Bresnick EH, Lee HY, Fujiwara T, et al. GATA switches as developmental drivers. J Biol Chem. 2010;285:31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bresnick EH, Katsumura KR, Lee HY, et al. Master regulatory GATA transcription factors: mechanistic principles and emerging links to hematologic malignancies. Nucleic Acids Res. 2012;40:5819–5831. doi: 10.1093/nar/gks281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grass JA, Boyer ME, Pal S, et al. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci U S A. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martowicz ML, Grass JA, Boyer ME, et al. Dynamic GATA factor interplay at a multicomponent regulatory region of the GATA-2 locus. J Biol Chem. 2005;280:1724–1732. doi: 10.1074/jbc.M406038200. [DOI] [PubMed] [Google Scholar]
- 42.Grass JA, Jing H, Kim SI, et al. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol Cell Biol. 2006;26:7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pop R, Shearstone JR, Shen Q, et al. A key commitment step in erythropoiesis is synchronized with the cell cycle clock through mutual inhibition between PU. 1 and S-phase progression. PLoS Biol. 2010;8:e1000484. doi: 10.1371/journal.pbio.1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dore LC, Chlon TM, Brown CD, et al. Chromatin occupancy analysis reveals genome-wide GATA factor switching during hematopoiesis. Blood. 2012;119:3724–3733. doi: 10.1182/blood-2011-09-380634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45▪.DeVilbiss AW, Boyer ME, Bresnick EH. Establishing a hematopoietic genetic network through locus-specific integration of chromatin regulators. Proc Natl Acad Sci U S A. 2013;110:E3398–E3407. doi: 10.1073/pnas.1302771110. This article demonstrates the importance of the histone methyltransferase SetD8 for GATA-1-mediated repression of a subset of its target genes, and how different combinations of coregulators mediate repression at distinct target genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pope NJ, Bresnick EH. Differential coregulator requirements for function of the hematopoietic transcription factor GATA-1 at endogenous loci. Nucleic Acids Res. 2010;38:2190–2200. doi: 10.1093/nar/gkp1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujiwara T, Lee HY, Sanalkumar R, et al. Building multifunctionality into a complex containing master regulators of hematopoiesis. Proc Natl Acad Sci U S A. 2010;107:20429–20434. doi: 10.1073/pnas.1007804107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim SI, Bultman SJ, Jing H, et al. Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation. Mol Cell Biol. 2007;27:4551–4565. doi: 10.1128/MCB.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson KD, Boyer ME, Kang JA, et al. Friend of GATA-1-independent transcriptional repression: a novel mode of GATA-1 function. Blood. 2007;109:5230–5233. doi: 10.1182/blood-2007-02-072983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fox AH, Liew C, Holmes M, et al. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 1999;18:2812–2822. doi: 10.1093/emboj/18.10.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin DI, Orkin SH. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 1990;4:1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- 52.Evans T, Reitman M, Felsenfeld G. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc Natl Acad Sci U S A. 1988;85:5976–5980. doi: 10.1073/pnas.85.16.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miccio A, Wang Y, Hong W, et al. NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J. 2010;29:442–456. doi: 10.1038/emboj.2009.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hong W, Nakazawa M, Chen YY, et al. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 2005;24:2367–2378. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pal S, Cantor AB, Johnson KD, et al. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc Natl Acad Sci U S A. 2004;101:980–985. doi: 10.1073/pnas.0307612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Letting DL, Chen YY, Rakowski C, et al. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc Natl Acad Sci U S A. 2004;101:476–481. doi: 10.1073/pnas.0306315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wadman IA, Osada H, Grutz GG, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tripic T, Deng W, Cheng Y, et al. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 2009;113:2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wozniak RJ, Keles S, Lugus JJ, et al. Molecular hallmarks of endogenous chromatin complexes containing master regulators of hematopoiesis. Mol Cell Biol. 2008;28:6681–6694. doi: 10.1128/MCB.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anguita E, Hughes J, Heyworth C, et al. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 2004;23:2841–2852. doi: 10.1038/sj.emboj.7600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soler E, Andrieu-Soler C, de Boer E, et al. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 2010;24:277–289. doi: 10.1101/gad.551810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li L, Freudenberg J, Cui K, et al. Ldb1-nucleated transcription complexes function as primary mediators of global erythroid gene activation. Blood. 2013;121:4575–4585. doi: 10.1182/blood-2013-01-479451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63▪▪.Love PE, Warzecha C, Li L. Ldb1 complexes: the new master regulators of erythroid gene transcription. Trends Genet. 2014;30:1–9. doi: 10.1016/j.tig.2013.10.001. A highly instructive survey of Ldb1 mechanisms and biology relevant to many systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bultman SJ, Gebuhr TC, Magnuson T. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in beta-globin expression and erythroid development. Genes Dev. 2005;19:2849–2861. doi: 10.1101/gad.1364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu G, Schones DE, Cui K, et al. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 2011;21:1650–1658. doi: 10.1101/gr.121145.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu J, Shao Z, Glass K, et al. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev Cell. 2012;23:796–811. doi: 10.1016/j.devcel.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Linnemann AK, O’Geen H, Keles S, et al. Genetic framework for GATA factor function in vascular biology. Proc Natl Acad Sci U S A. 2011;108:13641–13646. doi: 10.1073/pnas.1108440108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilson NK, Foster SD, Wang X, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 69.Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Welch JJ, Watts JA, Vakoc CR, et al. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- 71.Cantor AB, Katz SG, Orkin SH. Distinct domains of the GATA-1 cofactor FOG-1 differentially influence erythroid versus megakaryocytic maturation. Mol Cell Biol. 2002;22:4268–4279. doi: 10.1128/MCB.22.12.4268-4279.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kiekhaefer CM, Grass JA, Johnson KD, et al. Hematopoietic-specific activators establish an overlapping pattern of histone acetylation and methylation within a mammalian chromatin domain. Proc Natl Acad Sci U S A. 2002;99:14309–14314. doi: 10.1073/pnas.212389499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Letting DL, Rakowski C, Weiss MJ, et al. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol Cell Biol. 2003;23:1334–1340. doi: 10.1128/MCB.23.4.1334-1340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Im H, Grass JA, Johnson KD, et al. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc Natl Acad Sci U S A. 2005;102:17065–17070. doi: 10.1073/pnas.0506164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vakoc CR, Letting DL, Gheldof N, et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 76.Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim SI, Bultman SJ, Kiefer CM, et al. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc Natl Acad Sci U S A. 2009;106:2259–2264. doi: 10.1073/pnas.0806420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee HY, Johnson KD, Fujiwara T, et al. Controlling hematopoiesis through sumoylation-dependent regulation of a GATA factor. Mol Cell. 2009;36:984–995. doi: 10.1016/j.molcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee HY, Johnson KD, Boyer ME, et al. Relocalizing genetic loci into specific subnuclear neighborhoods. J Biol Chem. 2011;286:18834–18844. doi: 10.1074/jbc.M111.221481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ragoczy T, Bender MA, Telling A, et al. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Politz JC, Scalzo D, Groudine M. Something silent this way forms: the functional organization of the repressive nuclear compartment. Annu Rev Cell Dev Biol. 2013;29:241–270. doi: 10.1146/annurev-cellbio-101512-122317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Drissen R, Palstra RJ, Gillemans N, et al. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schoenfelder S, Sexton T, Chakalova L, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84▪▪.Kadauke S, Udugama MI, Pawlicki JM, et al. Tissue-specific mitotic book marking by hematopoietic transcription factor GATA1. Cell. 2012;150:725–737. doi: 10.1016/j.cell.2012.06.038. This extremely interesting study describes a new function for the master erythroid transcription factor GATA-1 – mitotic bookmarking. Further analysis of the underlying mechanisms will almost certainly yield important insights vis-á-vis GATA factor function and more broadly for genome biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McDevitt MA, Fujiwara Y, Shivdasani RA, et al. An upstream, DNaseI hypersensitive region of the hematopoietic-expressed transcription factor GATA-1 gene confers developmental specificity in transgenic mice. Proc Natl Acad Sci U S A. 1997;94:7976–7981. doi: 10.1073/pnas.94.15.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McDevitt MA, Shivdasani RA, Fujiwara Y, et al. A ‘knockdown’ mutation created by cis-element gene targeting reveals the dependence of erythroid cell maturation on the level of transcription factor GATA-1. Proc Natl Acad Sci U S A. 1997;94:6781–6785. doi: 10.1073/pnas.94.13.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tsai SF, Strauss E, Orkin SH. Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev. 1991;5:919–931. doi: 10.1101/gad.5.6.919. [DOI] [PubMed] [Google Scholar]
- 88.Kobayashi M, Nishikawa K, Yamamoto M. Hematopoietic regulatory domain of GATA-1 gene is positively regulated by GATA-1 protein in Zebrafish embryos. Development. 2001;128:2341–2350. doi: 10.1242/dev.128.12.2341. [DOI] [PubMed] [Google Scholar]
- 89.Pillay LM, Forrester AM, Erickson T, et al. The Hox cofactors Meis1 and Pbx act upstream of gata1 to regulate primitive hematopoiesis. Dev Biol. 2010;340:306–317. doi: 10.1016/j.ydbio.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 90.Kawahara A, Dawid IB. Critical role of biklf in erythroid cell differentiation in zebrafish. Curr Biol. 2001;11:1353–1357. doi: 10.1016/s0960-9822(01)00398-0. [DOI] [PubMed] [Google Scholar]
- 91.Li X, Xiong JW, Shelley CS, et al. The transcription factor ZBP-89 controls generation of the hematopoietic lineage in zebrafish and mouse embryonic stem cells. Development. 2006;133:3641–3650. doi: 10.1242/dev.02540. [DOI] [PubMed] [Google Scholar]
- 92.Adelman CA, Chattopadhyay S, Bieker JJ. The BMP/BMPR/Smad pathway directs expression of the erythroid-specific EKLF and GATA1 transcription factors during embryoid body differentiation in serum-free media. Development. 2002;129:539–549. doi: 10.1242/dev.129.2.539. [DOI] [PubMed] [Google Scholar]
- 93.Liu B, Yee KM, Tahk S, et al. PIAS1 SUMO ligase regulates the self-renewal and differentiation of hematopoietic stem cells. EMBO J. 2014;33:101–113. doi: 10.1002/embj.201283326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 95.Chou ST, Khandros E, Bailey LC, et al. Graded repression of PU. 1/Sfpi1 gene transcription by GATA factors regulates hematopoietic cell fate. Blood. 2009;114:983–994. doi: 10.1182/blood-2009-03-207944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Crossley M, Orkin SH. Phosphorylation of the erythroid transcription factor GATA-1. J Biol Chem. 1994;269:16589–16596. [PubMed] [Google Scholar]
- 97.Zhao W, Kitidis C, Fleming MD, et al. Erythropoietin stimulates phosphorylation and activation of GATA-1 via the PI3-kinase/AKT signaling pathway. Blood. 2006;107:907–915. doi: 10.1182/blood-2005-06-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lamonica JM, Vakoc CR, Blobel GA. Acetylation of GATA-1 is required for chromatin occupancy. Blood. 2006;108:3736–3738. doi: 10.1182/blood-2006-07-032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Snow JW, Kim J, Currie CR, et al. Sumoylation regulates interaction of FOG1 with C-terminal-binding protein (CTBP) J Biol Chem. 2010;285:28064–28075. doi: 10.1074/jbc.M109.096909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fujiwara Y, Chang AN, Williams AM, et al. Functional overlap of GATA-1 and GATA-2 in primitive hematopoietic development. Blood. 2004;103:583–585. doi: 10.1182/blood-2003-08-2870. [DOI] [PubMed] [Google Scholar]
- 101.Yamada Y, Warren AJ, Dobson C, et al. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci U S A. 1998;95:3890–3895. doi: 10.1073/pnas.95.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li L, Jothi R, Cui K, et al. Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat Immunol. 2011;12:129–136. doi: 10.1038/ni.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pimanda JE, Ottersbach K, Knezevic K, et al. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc Natl Acad Sci U S A. 2007;104:17692–17697. doi: 10.1073/pnas.0707045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Souroullas GP, Salmon JM, Sablitzky F, et al. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell Stem Cell. 2009;4:180–186. doi: 10.1016/j.stem.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hayakawa F, Towatari M, Ozawa Y, et al. Functional regulation of GATA-2 by acetylation. J Leukoc Biol. 2004;75:529–540. doi: 10.1189/jlb.0603389. [DOI] [PubMed] [Google Scholar]
- 106.Crawford SE, Qi C, Misra P, et al. Defects of the heart, eye, and mega-karyocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J Biol Chem. 2002;277:3585–3592. doi: 10.1074/jbc.M107995200. [DOI] [PubMed] [Google Scholar]
- 107.Ozawa Y, Towatari M, Tsuzuki S, et al. Histone deacetylase 3 associates with and represses the transcription factor GATA-2. Blood. 2001;98:2116–2123. doi: 10.1182/blood.v98.7.2116. [DOI] [PubMed] [Google Scholar]
- 108.Dalgin G, Goldman DC, Donley N, et al. GATA-2 functions downstream of BMPs and CaM KIV in ectodermal cells during primitive hematopoiesis. Dev Biol. 2007;310:454–469. doi: 10.1016/j.ydbio.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Towatari M, Ciro M, Ottolenghi S, et al. Involvement of mitogen-activated protein kinase in the cytokine-regulated phosphorylation of transcription factor GATA-1. Hematol J. 2004;5:262–272. doi: 10.1038/sj.thj.6200345. [DOI] [PubMed] [Google Scholar]
- 110.Menghini R, Marchetti V, Cardellini M, et al. Phosphorylation of GATA2 by Akt increases adipose tissue differentiation and reduces adipose tissue-related inflammation: a novel pathway linking obesity to atherosclerosis. Circulation. 2005;111:1946–1953. doi: 10.1161/01.CIR.0000161814.02942.B2. [DOI] [PubMed] [Google Scholar]
- 111.Jude CD, Climer L, Xu D, et al. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kamminga LM, Bystrykh LV, de Boer A, et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107:2170–2179. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Feng Y, Yang Y, Ortega MM, et al. Early mammalian erythropoiesis requires the Dot1L methyltransferase. Blood. 2010;116:4483–4491. doi: 10.1182/blood-2010-03-276501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 115.Sprussel A, Schulte JH, Weber S, et al. Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia. 2012;26:2039–2051. doi: 10.1038/leu.2012.157. [DOI] [PubMed] [Google Scholar]
- 116.Kerenyi MA, Shao Z, Hsu YJ, et al. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. Elife. 2013;2:e00633. doi: 10.7554/eLife.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shearstone JR, Pop R, Bock C, et al. Global DNA demethylation during mouse erythropoiesis in vivo. Science. 2011;334:799–802. doi: 10.1126/science.1207306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wong P, Hattangadi SM, Cheng AW, et al. Gene induction and repression during terminal erythropoiesis are mediated by distinct epigenetic changes. Blood. 2011;118:e128–e138. doi: 10.1182/blood-2011-03-341404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119▪▪.Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. A highly instructive review of state-of-the-art genomics/epigenomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Visel A, Blow MJ, Li Z, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stergachis AB, Neph S, Reynolds A, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Simon JM, Giresi PG, Davis IJ, et al. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc. 2012;7:256–267. doi: 10.1038/nprot.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Grosveld F, van Assendelft GB, Greaves DR, et al. Position-independent high-level expression of the human b-globin gene in trangsenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 124.Forrester WC, Takegawa S, Papayannopoulou T, et al. Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res. 1987;15:10159–10177. doi: 10.1093/nar/15.24.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bender MA, Bulger M, Close J, et al. B-globin gene switching and DNaseI sensitivity of the endogenous b-globin locus in mice do not require the locus control region. Mol Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- 126.Bender MA, Roach JN, Halow J, et al. Targeted deletion of 5′HS1 and 5′HS4 of the beta-globin locus control region reveals additive activity of the DNaseI hypersensitive sites. Blood. 2001;98:2022–2027. doi: 10.1182/blood.v98.7.2022. [DOI] [PubMed] [Google Scholar]
- 127.Snow JW, Trowbridge JJ, Fujiwara T, et al. A single cis element maintains repression of the key developmental regulator Gata2. PLoS Genet. 2010;6:e1001103. doi: 10.1371/journal.pgen.1001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Snow JW, Trowbridge JJ, Johnson KD, et al. Context-dependent function of ‘GATA switch’ sites in vivo. Blood. 2011;117:4769–4772. doi: 10.1182/blood-2010-10-313031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129▪▪.Kim Y, Kweon J, Kim A, et al. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol. 2013;31:251–258. doi: 10.1038/nbt.2517. This study describes transformative technology that allows for facile genome editing with TALENs. [DOI] [PubMed] [Google Scholar]
- 130▪▪.Cho SW, Kim S, Kim JM, et al. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. This study describes transformative technology that allows for facile genome editing with the CRISPR/Cas9 system. [DOI] [PubMed] [Google Scholar]
- 131.Sankaran VG, Menne TF, Xu J, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 132▪▪.Bauer DE, Kamran SC, Lessard S, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342:253–257. doi: 10.1126/science.1242088. This ground-breaking study describes the identification of a GATA-1-binding cis-element in intron 1 of BCL11A, which encodes a critical repressor of fetal hemoglobin expression, and how SNPs in this element influence BCL11A levels and fetal hemoglobin expression. TALENs were utilized to excise this cis-element from mouse cell lines, which demonstrated that the deletion reduces Bcl11a expression and increases fetal hemoglobin levels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.De Gobbi M, Viprakasit V, Hughes JR, et al. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312:1215–1217. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]
- 134▪.Kaneko K, Furuyama K, Fujiwara T, et al. Identification of the novel erythroid specific enhancer for ALAS2 gene and its loss of function mutation associated with congenital sideroblastic anemia. Hematologica. 2014;99:252–261. doi: 10.3324/haematol.2013.085449. This study describes a human disease mutation of a GATA-1-binding cis-element within an intron of the heme biosynthetic enzyme ALAS2, which underlies X-linked sideroblastic leukemia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hsu AP, Sampaio EP, Khan J, et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118:2653–2655. doi: 10.1182/blood-2011-05-356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ostergaard P, Simpson MA, Connell FC, et al. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome) Nat Genet. 2011;43:929–931. doi: 10.1038/ng.923. [DOI] [PubMed] [Google Scholar]
- 137.Hahn CH, Chong C-E, Carmichael CL, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome, acute myeloid leukemia. Nat Genet. 2011;43:1012–1017. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dickinson RE, Griffin H, Bigley V, et al. Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood. 2011;118:2656–2658. doi: 10.1182/blood-2011-06-360313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hsu AP, Johnson KD, Falcone EL, et al. GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. Blood. 2013;121:3830–3837. doi: 10.1182/blood-2012-08-452763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Frampton M, da Silva Filho MI, Broderick P, et al. Variation at 3p24.1 and 6q23.3 influences the risk of Hodgkin’s lymphoma. Nat Commun. 2013;4:2549. doi: 10.1038/ncomms3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ganesh SK, Zakai NA, van Rooij FJ, et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet. 2009;41:1191–1198. doi: 10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]