Abstract
In February and September each year the World Health Organisation (WHO) recommends influenza viruses to be included in influenza vaccines for the forthcoming winters in the Northern and Southern Hemispheres respectively. These recommendations are based on data collected by National Influenza Centres (NIC) through the Global Influenza Surveillance and Response System (GISRS) and a more detailed analysis of representative and potential antigenically variant influenza viruses from the WHO Collaborating Centres for Influenza (WHO CCs) and Essential Regulatory Laboratories (ERLs). This article provides a detailed summary of the antigenic and genetic properties of viruses and additional background data used by WHO experts during development of the recommendations for the 2012 Southern Hemisphere influenza vaccine composition.
1. Introduction
In contrast to many other vaccines, influenza vaccines are frequently updated so as to be most effective against newly evolving human influenza viruses that are likely to circulate in the following influenza season. WHO convenes technical consultations (vaccine composition meetings (VCM)) twice a year to provide guidance to national public health authorities and vaccine manufacturers on the viruses to be included in trivalent influenza vaccines for the following influenza seasons in the Northern and Southern Hemispheres. The committee assembled by WHO comprises representatives from six WHO Collaborating Centres (Melbourne, Australia; Beijing, China; Tokyo, Japan; London, United Kingdom; Atlanta, USA; Memphis, USA) and four Essential Regulatory Laboratories (ERL) (Therapeutic Goods Administration (TGA), Australia; National Institute of Infectious Diseases (NIID), Japan; National Institute for Biological Standards and Control (NIBSC), UK; Food and Drug Administration (FDA), USA), with observers from several H5 Reference Laboratories, WHO National Influenza Centres (NICs) and other expert groups. In a previous publication [1], the main responsibilities of the WHO committee were described. The committee focuses on the geographic spread and epidemiological, antigenic and genetic characteristics of the most recently circulating influenza viruses in order to assess which are likely to predominate in the forthcoming season. Additionally, sera panels from individuals who received seasonal trivalent inactivated vaccines are tested to measure the presence of antibodies to recent influenza viruses. National and international regulatory agencies make the final decision about which influenza viruses are to be used in influenza vaccines to be licensed in their country.
In the present report we describe the basis for the selection of vaccine viruses recommended by the WHO for use in the 2012 Southern Hemisphere influenza season. This report describes only those data that were available at the time of the WHO VCM held September 26–28, 2011, in Geneva, Switzerland. The recommended viruses for use in the 2012 Southern Hemisphere influenza season were:
-
–
An A/California/7/2009 (H1N1)pdm09-like virus
-
–
An A/Perth/16/2009 (A(H3N2)-like virus
-
–
A B/Brisbane/60/2008-like virus
2. Influenza Activity, February – September 2011
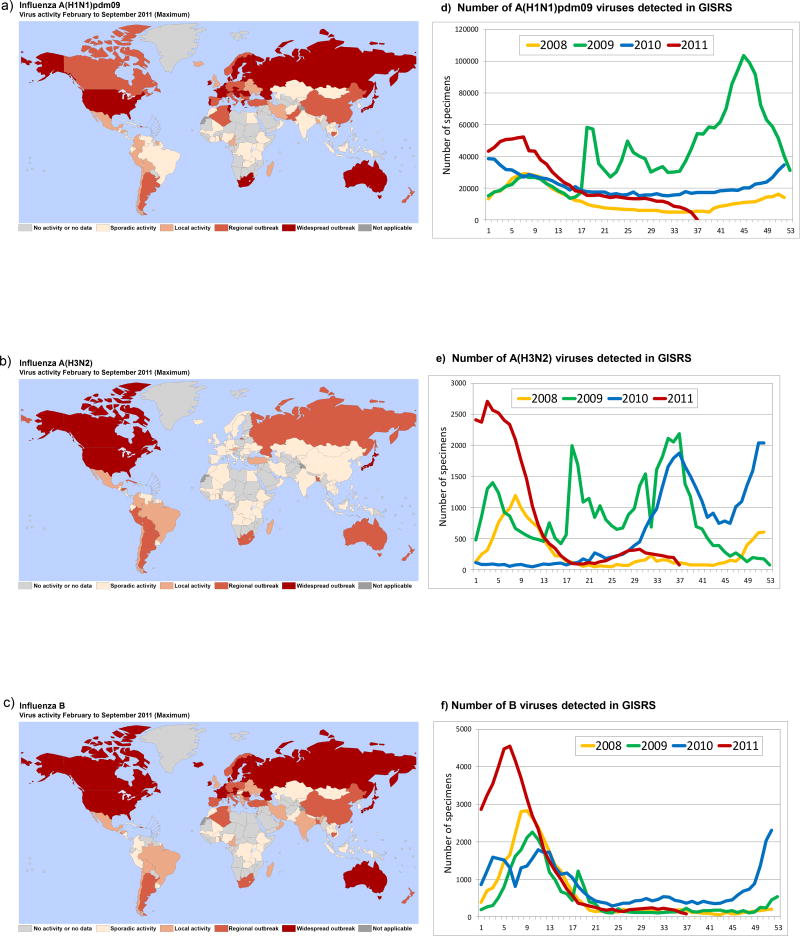
Influenza activity between the previous WHO Consultation on the Composition of Influenza Vaccines in February 2011 [2] and September 2011 [3] was reported by NICs and collated in the WHO FluNet database (see http://www.who.int/flunet). During this period, influenza was active worldwide and reported in Africa, the Americas, Asia, Europe and Oceania. Co-circulation of influenza A(H1N1) 2009 pandemic (subsequently referred to as A(H1N1)pdm09 as recommended by WHO [4]), A(H3N2) and influenza B viruses continued, to infect humans with the predominant circulating virus type and sub-type varying between countries and regions. In general, influenza activity was low or moderate compared with recent years. Influenza activity maps for the period February to September 2011 along with graphs showing the number of influenza viruses detected, typed and subtyped by the GISRS laboratories are presented in Fig. 1. During this time the majority of global influenza activity and detection occurred in the Northern Hemispherein February and March. At the time of the consultation, data collected from the GISRS laboratory network showed that of the influenza viruses collected from February to September 2011, approximately 40% were A(H1N1)pdm09, 16% were A(H3N2), 14% were not subtyped influenza A and 30% were influenza B.
Fig. 1.
Maps showing peak levels of laboratory confirmed influenza (reported to FluNet) in each country with an NIC, for the period February to September 2011: a) A(H1N1)pdm09 viruses; b) A(H3N2) viruses; c) B viruses. The total number of seasonal and pandemic viruses detected by GISRS from February to September 2011 for each type/subtype is shown in panels: d) A(H1N1)pdm09; e) A(H3N2); f) B viruses.
3. Overview of antigenic and genetic characteristics of the viruses
For the Consultation, WHO CCs performed detailed antigenic analyses on approximately 4400 influenza viruses (Table 1). Viruses were collected from March 2011 to the end of August 2011 and recovered from either clinical specimens or virus isolates provided by over 70 NICs and other laboratories within and outside of GISRS. Antigenic characterisation was carried out predominantly in haemagglutination inhibition (HI) assays using viruses isolated and propagated in either mammalian tissue culture cells (most frequently Madin-Darby canine kidney cells (MDCK), or MDCK-SIAT-1 cells engineered to express increased levels of α2–6 sialyl transferase [5]) or in embryonated hens’ eggs. HI assays using turkey or guinea pig red blood cells (RBC) were performed to compare the reactivity of cultured viruses with post-infection ferret antisera raised against egg or cell grown reference, vaccine and recent representative viruses [6]. A subset of viruses also underwent genetic characterisation. Genetic analyses routinely focused on the sequencing of the haemagglutinin (HA) (the complete HA or the HA1 coding region) and neuraminidase (NA) genes with full genome sequencing performed on a smaller subset of viruses. Gene sequencing provides the molecular basis for observed antigenic variation and phylogenetic analysis of HA and NA genes defines the genetic relatedness of antigenic variants. The initial phylogenetic tree of the HA1 domain nucleotide sequences of each type or subtype was constructed with the PhyML software package version 3.0 [7] using GTR+I+Γ4. For this analysis the general time-reversible model with the proportion of invariant sites and the gamma distribution of among-site rate variation with four categories was estimated from the empirical data, determined by ModelTest [8] as the evolutionary model. GARLI v0.961 [9] was run on the best tree from PhyML for 2 million generations to optimise tree topology and branch lengths for each virus type or sub-type. The list of virus gene sequence accession numbers and their originating laboratories used in this report are listed in Tables S8a–d.
Table 1.
Influenza isolates characterized by hemagglutination inhibition assay by WHO Collaborating Centres (samples collected in March – August, 2011)
| A(H1N1)pdm09a | A(H3N2)b | B/Victoriac | B/Yamagatad | |
|---|---|---|---|---|
| Total tested | 1232 | 776 | 1770 | 598 |
| Low reactors (% of total) | 160 (3%) | 40 (5%) | 53 (3%) | 0 |
Compared to the A/California/7/2009 vaccine virus
Compared to the A/Perth/16/2009 vaccine virus
Compared to the B/Brisbane/60/2008 vaccine virus
Compared to reference viruses B/Wisconsin/01/2010, B/Hubei-Wujiagang/158/2009 or B/Bangladesh/3333/2007
The combination of antigenic and genetic data is used routinely to identify emergent antigenic variants. Antigenic cartography [10] was used to visualize the HI data. These results are used to identify virus variants with future epidemic potential, and to be considered as potential vaccine candidates.
As discussed previously, the behaviour of A(H3N2) viruses in HA and HI assays has changed in recent years and their antigenic analyses have become more complex [1]. In particular, guinea pig RBC are now preferred for antigenic characterisation of current A(H3N2) viruses in HA and HI assays. To control for the possible participation of the virus NA in the agglutination of RBC, HI assays can also be performed in the presence of oseltamivir [11]. Virus neutralisation (plaque reduction and micro-neutralisation) assays were performed in addition to HI tests for a subset of A(H3N2) viruses.
In addition to antigenic studies using post-infection ferret antisera, human serum panels obtained pre- and post- vaccination with seasonal influenza vaccine formulations were used to assess current vaccine coverage against representative recently circulating viruses. . Serum panels for adults), older adults) and paediatric populations received from Australia, China, Japan, the UK and the USA were tested where available.
4. A(H1N1)pdm09 viruses
The vast majority of influenza A(H1N1)pdm09 viruses collected February - August 2011 remained antigenically closely related to the vaccine virus A/California/7/2009. The results of a typical HI assay are presented in Table 2 (see also supplementary Table S1). Only two viruses in this table (A/Argentina/656/2011 and A/Sapporo/163/2011) demonstrated more than a 4-fold reduction in HI titre compared with the homologous titre obtained using post-infection ferret antiserum raised against the A/California/7/2009 vaccine virus. It should be noted that the A/Argentina/656/2011 and A/Sapporo/163/2011 viruses have amino acid (AA) substitutions G156E and K154E, respectively in their HA molecules. AA substitutions in the 153–157 region of the HA were identified in many cell or egg grown A(H1N1)pdm09 viruses that had low reactivity to the ferret antisera raised against A/California/7/2009 (see below) and nucleotide polymorphism is frequently seen within samples in the HA gene encoding these amino acids. Notably, more recent A(H1N1)pdm09 viruses from Australia demonstrated reduced reactivity with ferret antisera raised against A/California/7/2009 (Table S1), with many having HA AA substitutions or polymorphisms at positions 153–157. A summary of HI test results for A(H1N1)pdm09 viruses from all WHO CCs is shown in Table 1.
Table 2.
Hemagglutination inhibition test of influenza A(H1N1)pdm09 viruses (WHO CC Atlanta) a
| REFERENCE VIRUSES | Collection date |
Reference Ferret Antisera
|
Passage history |
|||||
|---|---|---|---|---|---|---|---|---|
| A/Cal/7/09 | X-179A | A/Minn/3/11 | A/Vor/1/11 | A/ST.P/100/11 | A/HK/3934/11 | |||
| A/California/7/2009 | 2009-04-09 | 2560 | 1280 | 2560 | 1280 | 1280 | 2560 | E3 |
| A/California/7/2009 X-179A | reassortant | 5120 | 2560 | 5120 | 1280 | 2560 | 2560 | EX/E1 |
| A/Minnesota/03/2011 | 2011-02-18 | 2560 | 1280 | 2560 | 1280 | 1280 | 2560 | C3 |
| A/Voronezh/01/2011 | 2011-03-14 | 5120 | 2560 | 5120 | 2560 | 5120 | 2560 | E1/E2 |
| A/St Petersburg/100/2011 | 2011-03-14 | 2560 | 1280 | 5120 | 1280 | 1280 | 2560 | E1/E2 |
| A/Hong Kong/3934/2011 | unknown | 2560 | 640 | 2560 | 640 | 1280 | 1280 | C2C2/C1 |
|
| ||||||||
| TEST VIRUSES | ||||||||
|
| ||||||||
| A/Argentina/779/2011 | 2011-07-05 | 5120 | 1280 | 2560 | 640 | 5120 | 1280 | S1/C1 |
| A/Bolivia/802/2011 | 2011-08-29 | 5120 | 1280 | 5120 | 1280 | 2560 | 2560 | C1 |
| A/Brazil/568/2011 | 2011-07-02 | 5120 | 1280 | 5120 | 1280 | 1280 | 2560 | C1 |
| A/Concepcion/16695/2011 | 2011-07-30 | 5120 | 2560 | 5120 | 2560 | 5120 | 5120 | C2/C1 |
| A/Guangdong-Xiangzhou/1623/2011 | 2011-07-07 | 5120 | 2560 | 5120 | 1280 | 2560 | 5120 | C2/C1 |
| A/Santiago/17529/2011 | 2011-08-16 | 5120 | 1280 | 5120 | 1280 | 5120 | 1280 | C2/C1 |
| A/Santiago/18553/2011 | 2011-08-27 | 5120 | 640 | 2560 | 640 | 2560 | 640 | C1 |
| A/St. Petersburg/27/2011 | unknown | 5120 | 2560 | 5120 | 2560 | 2560 | 2560 | E1E1/E1 |
| A/Tasmania/14/2011 | 2011-05-07 | 5120 | 1280 | 2560 | 640 | 2560 | 1280 | E3/E2 |
| A/Uruguay/863/2011 | 2011-07-06 | 5120 | 1280 | 2560 | 1280 | 1280 | 2560 | C1 |
| A/Astrakhan/01/2011 | unknown | 2560 | 2560 | 5120 | 1280 | 2560 | 2560 | C1C2/C1 |
| A/Bangladesh/9751/2011 | 2011-07-28 | 2560 | 1280 | 5120 | 1280 | 2560 | 2560 | C1/C1 |
| A/Brisbane/70/2011 | 2011-02-18 | 2560 | 1280 | 2560 | 640 | 1280 | 1280 | E4/E1 |
| A/Colombia/416/2011 | 2011-05-10 | 2560 | 1280 | 5120 | 1280 | 2560 | 1280 | C1 |
| A/Guadeloupe/104/2011 | 2011-04-16 | 2560 | 640 | 1280 | 320 | 1280 | 1280 | C2 |
| A/Guyane/103/2011 | 2011-04-24 | 2560 | 1280 | 2560 | 640 | 2560 | 1280 | C2 |
| A/Honduras/8083/2011 | 2011-07-19 | 2560 | 1280 | 5120 | 1280 | 2560 | 1280 | C1/C1 |
| A/Miyagi/32/2011 | 2011-02-22 | 2560 | 640 | 2560 | 640 | 1280 | 1280 | C4/C1 |
| A/Valparaiso/17275/2011 | 2011-08-10 | 2560 | 1280 | 5120 | 1280 | 1280 | 2560 | E2/E1 |
| A/Venezuela/06/2011 | 2011-05-28 | 2560 | 1280 | 2560 | 1280 | 1280 | 2560 | C1/C1 |
| A/Wellington/01/2011 | 2011-05-11 | 2560 | 1280 | 2560 | 640 | 1280 | 640 | E3/E1 |
| A/Martinique/67/2011 | 2011-02-09 | 1280 | 640 | 2560 | 320 | 1280 | 1280 | C1 |
| A/Victoria/502/2010 IVR-159 | 2011-01-24 | 1280 | 640 | 1280 | 640 | 2560 | 320 | E3D7/E1 |
| A/Philippines/477/2011 | 2011-03-09 | 640 | 320 | 320 | 160 | 640 | 320 | C3/C1 |
| A/Argentina/656/2011 (G155Eb) | 2011-08-05 | 320 | 160 | 160 | 80 | 160 | 160 | S1/C1 |
| A/Sapporo/163/2011 (K154Eb) | 2011-03-04 | 160 | 80 | 160 | 80 | 80 | 160 | C3/C1 |
Shaded column: A/California/7/2009 current vaccine virus
Amino acid changes in the HA molecule
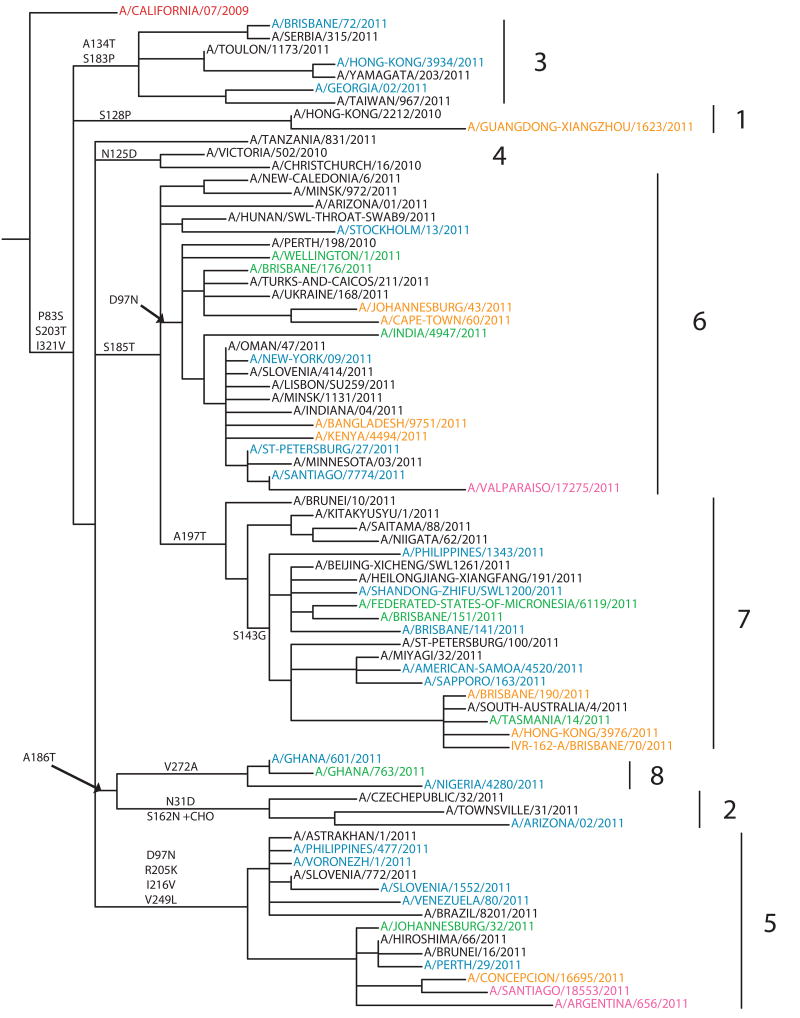
Genetic analyses of the HA genes of A(H1N1)pdm09 viruses are shown in Fig. 2 (see also Fig. S1 that presents a high resolution tree constructed from sequences of >1,600 A(H1N1)pdm09 isolates collected through GISRS since 2010-01-01). Currently the phylogenetic tree of the A(H1N1)pdm09 HA gene can be divided into eight major genetic subgroups. At the time of the VCM the majority of recent isolates belonged to subgroups 6 and 7 with a signature AA substitution of S185T in the HA1. Subgroup 6 viruses, which had been circulating worldwide, carry AA substitution D97N. Subgroup 7 viruses, mainly seen in Europe, Asia and Oceania, carry the AA substitution A197T and many also carry the AA substitution S143G. Subgroup 5 viruses were also found worldwide with signature AA substitutions of D97N, R205K, I216V and V249L. Viruses of HA subgroup 4 have not circulated widely since early 2011 carry the AA substitution N125D. Subgroup 3 includes viruses with AA substitutions A134T and S183P in the HA. Subgroup 2 and 8 share the AA substitution A186T. Subgroup 2 includes viruses that had been collected globally, with AA substitutions N31D and S162N (which results in the gain of a potential glycosylation site at residue 162 of the HA) while subgroup 8 includes viruses collected from Africa with an AA substitution of V272A. Lastly genetic subgroup 1 includes early 2011 isolates from Asia and North America that carry the AA substitution S128P. Sequences of isolates with substitutions at positions 153–157 in the HA do not form a monophyletic group; instead they are distributed throughout the phylogenetic tree and appear in nearly all circulating genetic groups (Fig. 2). Full genome sequencing has been carried out on viruses from several geographic regions and no evidence of reassortment with co-circulating A(H3N2) viruses was observed (data not shown).
Fig. 2.
Phylogenetic trees of representative A(H1N1)pdm09 HA1 domain nucleotide sequences were constructed with the PhyML software package 3.0 [7] using GTR+I+Γ4 as determined by ModelTest [8]. GARLI v0.961 [9] was run on the best tree from PhyML for 2 million generations to optimise tree topology and branch lengths. Amino acid substitutions are marked on major nodes. Viruses are colour-coded by month of collection as follows: March-April 2011 in Blue, May 2011 in Green, June – July 2011 in Orange, August 2011 in Pink. The current vaccine strain is in Red. Genetic subgroups are labelled 1–8.
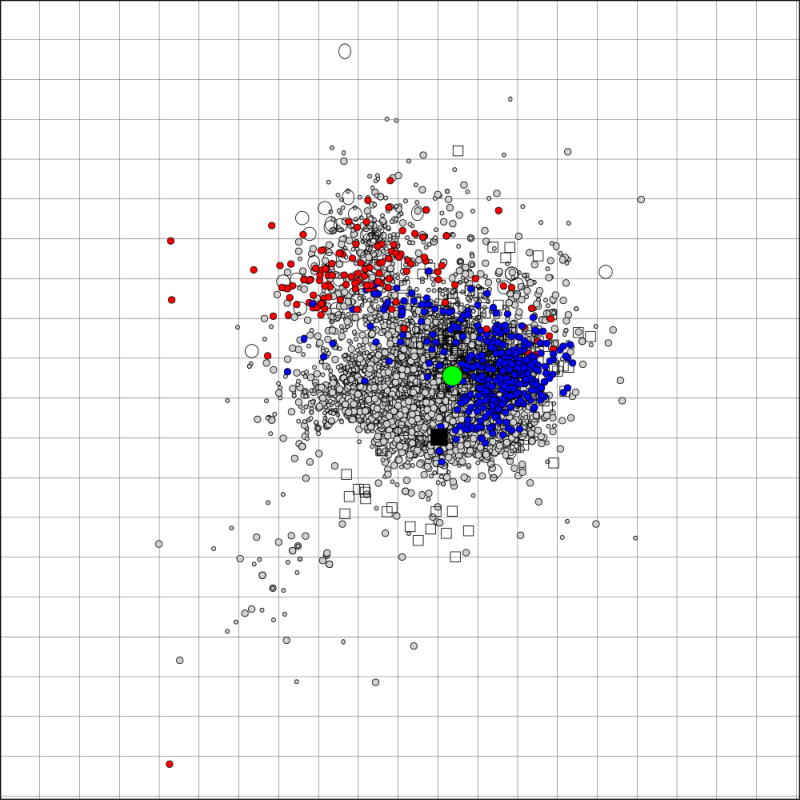
Antigenic cartography illustrates that the majority of A(H1N1)pdm09 isolates are antigenically similar to A/California/7/2009 and cluster together demonstrating little antigenic diversity during this period or since 2009 (Fig. 3). Although viruses with 8-fold or greater reductions in HI with the A/California/7/2009 ferret antisera cluster together somewhat apart from antigenically similar antigens in the antigenic map, as mentioned above they do not form a separate genetic group and are spread throughout the HA phylogenic tree (Fig. 3).
Fig. 3.
An antigenic map [10] generated from A(H1N1)pdm09 HI data from WHO CCs in Atlanta, London, Melbourne and Tokyo from April 2009 – August 2011. Viruses are represented as circles, antisera as squares. The current vaccine strain A/California/7/2009 is represented by the large green dot, 2011 viruses with 8-fold or greater reductions in HI titres to the A/California/7/2009 ferret antiserum are in red, the remaining 2011 viruses in blue. The Grid indicates a 2-fold dilution in HI titre, 1 unit of antigenic distance [10].
Vaccines containing the A/California/7/2009 (H1N1) antigen stimulated anti-HA antibodies of similar geometric mean HI titres to the vaccine virus and the majority of representative A(H1N1)pdm09 isolates. Fig. S2 summarises human serology data obtained by all WHO CCs and ERLs that perform human serology studies. Only two viruses used in human serology tests - A/Sapporo/163/2011 and A/South Australia/4/2011 – demonstrated a 50% or greater reduction in geometric mean titres (GMT) in HI tests with human sera from vaccinees who received the A/California/7/2009 antigen. Both of these viruses also showed reduced HI reactivity (4- to 8-fold) with post-infection ferret antisera raised against A/California/7/2009. Additionally, A/Sapporo/163/2011 is representative of isolates with AA substitutions seen in positions 153 through 157 of the HA.
Based on analyses of data presented at the VCM, it was concluded that the observed genetic diversity of A(H1N1)pdm09 viruses did not result in changes in their antigenic properties and that A/California/7/2009 remained appropriate for the 2012 Southern Hemisphere vaccine composition [12].
5. A(H3N2) viruses
Combined data from all WHO CCs using a variety of antigenic assays identified a low proportion of February - August 2011 isolates (5%) with greater than 8-fold reduction in titre (as compared to the titres of the homologous antigen) to ferret antisera raised against the A/Perth/16/2009 vaccine virus (Table 1). Table 3 presents an example of an HI assay using guinea pig RBC in the presence of 20 nM oseltamivir. Based on results of both HI (see also Table S2) and virus neutralisation (Table S3) assays, it was concluded that the majority of A(H3N2) viruses that circulated from February to August 2011 in different parts of the world were antigenically closely related to the A/Perth/16/2009 vaccine virus.
Table 3.
Hemagglutination inhibition test of A(H3N2) viruses using Guinea Pig RBC in the presence of 20nM Oseltamivir (WHO CC London) a
| REFERENCE VIRUSES | Collection date |
Reference Ferret Antisera
|
Passage history |
|||||
|---|---|---|---|---|---|---|---|---|
| A/Bris/10/07 | A/Per/16/09 | A/Vic/208/09 | A/Vic/210/09 | A/Perth/10/10 | A/S Aus/3/11 | |||
| A/Brisbane/10/2007 | 2007-02-06 | 1280 | 80 | 80 | 80 | 80 | 160 | E2/E1 |
| A/Perth/16/2009 | 2009-07-04 | 40 | 1280 | 80 | 320 | 320 | 640 | E3/E1 |
| A/Victoria/208/2009 | 2009-06-02 | 320 | 640 | 1280 | 2560 | 2560 | 2560 | E3/E1 |
| A/Victoria/210/2009 | 2009-06-02 | 320 | 1280 | 1280 | 2560 | 1280 | 1280 | E2/E2 |
| A/Perth/10/2010 | 2010-05-25 | 160 | 1280 | 1280 | 1280 | 1280 | 1280 | E2/E1 |
| A/South Australia/3/2011 | 2011-06-10 | 40 | 320 | 80 | 160 | 320 | 640 | C2/S1 |
|
| ||||||||
| TEST VIRUSES | ||||||||
|
| ||||||||
| A/Ghana/FS-11-1014/2011 | 2011-06-21 | 40 | 640 | 160 | 160 | 640 | nt | S2 |
| A/Johannesburg/113/2011 | 2011-05-17 | 80 | 640 | 320 | 320 | 640 | 1280 | C1/S1 |
| A/Johannesburg/114/2011 | 2011-05-26 | < | 640 | 160 | 160 | 640 | 1280 | C1/S1 |
| A/La Plata/65507/2011 | 2011-05-23 | 40 | 640 | 160 | 320 | 640 | nt | SI/S1 |
| A/Norway/1789/2011 | 2011-08-02 | 40 | 640 | 160 | 160 | 320 | 1280 | C1/S1 |
| A/Buenos Aires/10140261/2011 | 2011-06-13 | < | 320 | 160 | 160 | 320 | nt | SI/S1 |
| A/Buenos Aires/66090/11 | 2011-06-21 | 40 | 320 | 160 | 160 | 320 | nt | SI/S1 |
| A/Buenos Aires/R230/2011 | 2011-07-08 | < | 320 | 160 | 160 | 320 | nt | SI/S1 |
| A/Durban/92/2011 | 2011-06-15 | 40 | 320 | 160 | 160 | 320 | 320 | C1/S1 |
| A/Entre Ríos/755282/2011 | 2011-07-05 | < | 320 | 160 | 160 | 320 | nt | S2/S1 |
| A/Ghana/731/2011 | 2011-05-10 | 40 | 320 | 160 | 160 | 320 | nt | S1/C1 |
| A/Ghana/FS1037/2011 | 2011-06-22 | 40 | 320 | 160 | 80 | 320 | nt | S3 |
| A/Ghana/FS1045/2011 | 2011-06-27 | 40 | 320 | 160 | 160 | 640 | nt | S3 |
| A/Ghana/FS591/2011 | 2011-04-07 | 40 | 320 | 160 | 160 | 320 | nt | S2 |
| A/Ghana/FS648/2011 | 2011-04-15 | 40 | 320 | 80 | 80 | 320 | nt | S2 |
| A/Hong Kong/3968/2011 | 2011-05-17 | 40 | 320 | 80 | 80 | 320 | nt | C1/S1 |
| A/Hong Kong/3969/2011 | 2011-05-19 | 80 | 320 | 80 | 80 | 160 | nt | C1/S1 |
| A/Johannesburg/107/2011 | 2011-06-29 | < | 320 | 80 | 160 | 320 | 640 | C1/S1 |
| A/Johannesburg/153/2011 | 2011-07-05 | < | 320 | 80 | 160 | 320 | 640 | C1/S1 |
| A/Johannesburg/27/2011 | 2011-05-06 | 40 | 320 | 80 | 80 | 320 | nt | C1/S1 |
| A/Johannesburg/94/2011 | 2011-06-24 | 40 | 320 | 160 | 160 | 640 | 1280 | C1/S1 |
| A/Johannesburg/99/2011 | 2011-06-29 | < | 320 | 80 | 160 | 320 | 640 | C1/S1 |
| A/Norway/1775/2011 | 2011-07-29 | 40 | 320 | 80 | 160 | 320 | 640 | C1/S1 |
| A/Santa Fé/1157-04/2011 | 2011-06-15 | < | 320 | 160 | 160 | 640 | nt | SI/S1 |
| A/Santa Fé/1431/2011 | 2011-07-04 | 40 | 320 | 160 | 160 | 640 | nt | SI/S1 |
| A/Bangladesh/5071/2011 | 2011-04-15 | < | 160 | 40 | 80 | 160 | 320 | CX/S1 |
| A/Johannesburg/79/2011 | 2011-06-21 | 40 | 160 | 80 | 160 | 320 | 640 | C1/S1 |
| A/Norway/1762/2011 | 2011-07-21 | < | 160 | 40 | 80 | 160 | 320 | C2/S1 |
| A/Sakai/20/2011 | 2011-02-15 | < | 160 | 40 | 80 | 160 | 320 | C4/S1 |
Shaded column: A/Perth/16/2009 current vaccine virus
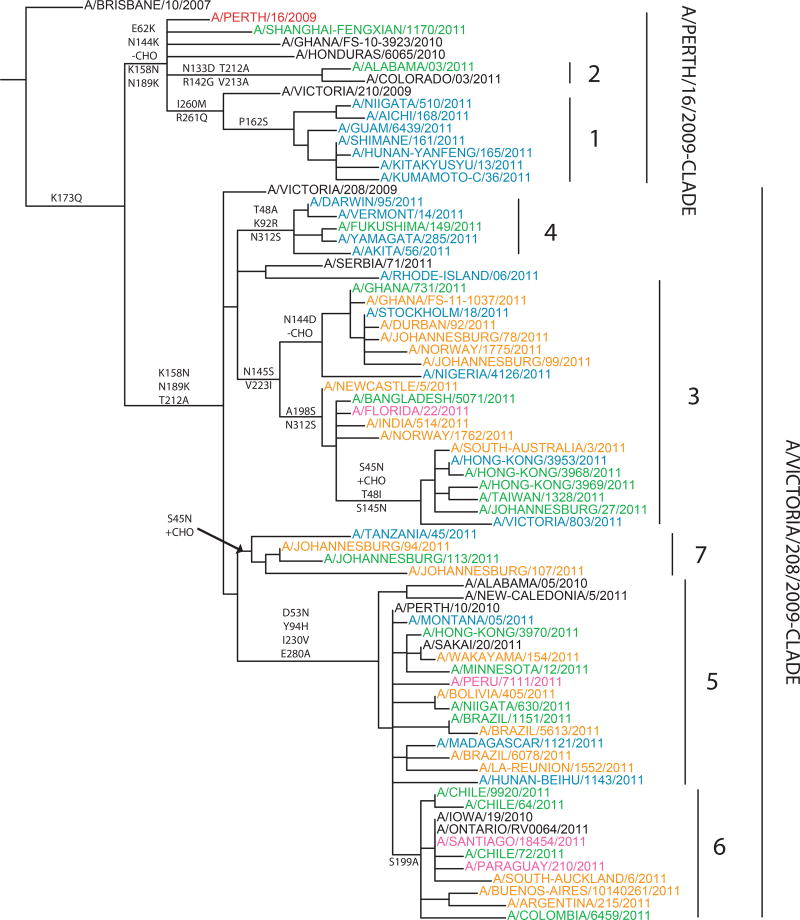
A phylogenetic tree for the HA1 of A(H3N2) viruses is presented in Fig. 4 (see also Fig. S3 that presents a high resolution tree constructed from sequences of >1,000 A(H3N2) isolates collected through GISRS since 2010-01-01). A(H3N2) viruses continued to fall into two major genetic clades represented by A/Victoria/208/2009 and A/Perth/16/2009. Viruses with reduced HI titres to post-infection ferret antisera raised against A/Perth/16/2009 were scattered throughout the HA tree and did not form monophyletic groups or share common AA substitutions.
Fig. 4.
Phylogenetic trees of representative A(H3N2) HA1 domain nucleotide sequences were constructed and annotated as described for Fig. 2. Genetic subgroups are labelled 1–7.
The A(H3N2) HA tree can be further divided into seven genetic subgroups. Isolates in the smaller A/Perth/16/2009 genetic clade carry signature AA substitutions E62K and N144K (leading to the loss of a potential glycosylation site). Within the A/Perth/16/2009 group there are subgroups 1 and 2 with several AA substitutions. Subgroup 1 consists mainly of isolates from Japan, China and other parts of Asia; many viruses within this subgroup also have signature AA substitutions P162S, I260M and R261Q. Subgroup 2 includes early 2011 U.S. isolates with AA substitutions N133D (leading to the loss of a potential glycosylation site), R142G, T212A and V213A in the HA1.
The majority of A(H3N2) isolates collected worldwide belong to the A/Victoria/208/2009 genetic clade, but remain antigenically related to A/Perth/16/2009. Within the A/Victoria/208/2009 clade, the most commonly circulating subgroups were subgroups 5 and 6, which have signature AA substitutions D53N, Y94H, I230V and E280A in HA1. Subgroup 6 isolates carrying an additional AA substitution, S199A. The remaining isolates in the Victoria/208/2009 clade that were in circulation between January 2011 and September 2011 group formed three smaller subgroups, 3, 4 and 7. Subgroup 3 isolates have a signature AA substitution V223I. Within subgroup 3, one cluster of viruses collected from Europe and Africa shared substitutions N144D (leading to the loss of a potential glycosylation site) and N145S. Another cluster within subgroup 3 isolates collected from Asia and Oceania also carry additional AA substitutions A198S and N312S. Subgroup 5 isolates have three AA substitutions, T48A, K92R and N312S and subgroup 7 isolates share AA substitution N45S (resulting in the gain of a potential glycosylation site).
Vaccines containing influenza A/Perth/16/2009 (H3N2)-like antigen stimulated anti-HA antibodies of similar geometric mean HI titres to the vaccine virus and the majority of representative A(H3N2) isolates, consistent with results obtained in HI assays with ferret antisera (Fig. S4). Among 12 tested A(H3N2) antigens, three (A/Sakai/20/2011 from genetic subgroup 5; A/South Australia/3/2011 and A/Sydney/27/2011, both from genetic subgroup 3) consistently demonstrated a 50% or greater reduction in post vaccination GMT compared to A/Perth/16/2009-like vaccine viruses and an 8-fold or greater reduction in HI titre with ferret antisera (data not shown).
Based on surveillance data available in September 2011, it was concluded that the A(H3N2) component of the 2012 Southern Hemisphere influenza vaccine should continue to be an A/Perth/16/2009-like virus.
6. Influenza B viruses
There are two lineages of influenza B viruses that show little antigenic cross reactivity, the B/Victoria/2/87- and B/Yamagata/16/88 [13]. Viruses from both of these lineages have been observed in various proportions in different countries since 1988–89. Only one of these two lineages of influenza B virus has been included in trivalent seasonal influenza vaccines.
From January to April 2011 most B viruses reported to WHO belonged to the B/Victoria lineage. The analyses of influenza B viruses by HI assays continue to demonstrate that sera raised in ferrets infected with egg-grown B viruses may react poorly with cell-grown B viruses, prompting the use of cell-grown viruses for serum production in ferrets [1]. In addition, influenza B viruses often generate antisera with lower titres than those raised against influenza A viruses, some CCs use additional boosting of ferrets potentially broadening the cross-reactivity of the antibody response.
Combines HI data from all WHO CCs found only 3% of February - August 2011 isolates had reduced HI titres to post-infection ferret antiserum raised against A/Brisbane/60/2008, the current vaccine virus (Table 1) using data combined from all WHO CCs. Table 4 represents results of a typical HI assay performed using turkey RBC with recent influenza B/Victoria lineage viruses. These and other data showed that the majority of 2011 influenza B viruses were antigenically closely related to the cell grown equivalents of the B/Brisbane/60/2008 vaccine virus .
Table 4.
Hemagglutination inhibition test of B Victoria lineage viruses (WHO CC Melbourne) a
| REFERENCE VIRUSES | Date collected |
Reference Ferret Antiserab
|
Passage history |
||||||
|---|---|---|---|---|---|---|---|---|---|
| B/Mal/2506/04 | B/Br/60/08 | B/Br/33/08 | B/HK/90/08 | B/HK/259/10 | B/Syd/508/10 | B/Phil/6363/09 | |||
| B/Malaysia/2506/2004 | 2004-06-12 | 640 | 20 | 640 | 160 | 640 | 320 | 1280 | E4 |
| B/Brisbane/60/2008 | 2008-08-04 | <20 | 160 | 640 | 160 | 80 | 160 | 160 | CX/C4 |
| B/Brisbane/33/2008 | 2008-07-13 | 160 | 160 | 2560 | 640 | 1280 | 1280 | 640 | E4 |
| B/Hong Kong/90/2008 | 2008-02-04 | 160 | 80 | 1280 | 640 | 640 | 640 | 640 | E5 |
| B/Hong Kong/259/2010 | 2010-03-02 | 320 | 80 | 1280 | 320 | 640 | 640 | 320 | E4 |
| B/Sydney/508/2010 | 2010-10-11 | 80 | 80 | 2560 | 640 | 640 | 1280 | 320 | E2 |
| B/Philippines/6363/2009 | 2009-12-03 | 320 | 20 | 320 | 160 | 160 | 160 | 640 | C4 |
|
| |||||||||
| TEST VIRUSES | |||||||||
|
| |||||||||
| B/Christchurch/21/2011 | 2011-08-03 | <20 | 640 | >2560 | 640 | 640 | 640 | 320 | R-MIX1/C1 |
| B/Christchurch/22/2011 | 2011-08-03 | <20 | 640 | >2560 | 640 | 640 | 640 | 640 | R-MIX1/C1 |
| B/Newcastle/42/2011 | 2011-08-10 | <20 | 320 | 1280 | 160 | 160 | 160 | 160 | C1 |
| B/Newcastle/48/2011 | 2011-08-11 | <20 | 320 | 1280 | 160 | 160 | 160 | 160 | C1 |
| B/Newcastle/49/2011 | 2011-08-12 | <20 | 320 | 1280 | 160 | 160 | 160 | 160 | C1 |
| B/South Australia/294/2011 | 2011-08-19 | <20 | 320 | 1280 | 320 | 160 | 320 | 160 | C1 |
| B/South Australia/297/2011 | 2011-08-22 | <20 | 320 | 1280 | 320 | 160 | 320 | 160 | C1 |
| B/South Australia/298/2011 | 2011-08-20 | <20 | 320 | 1280 | 160 | 160 | 160 | 160 | C1 |
| B/Victoria/829/2011 | 2011-08-20 | <20 | 320 | 1280 | 160 | 160 | 320 | 160 | C1 |
| B/Victoria/831/2011 | 2011-08-24 | <20 | 320 | 1280 | 160 | 160 | 160 | 160 | C1 |
| B/Brisbane/67/2011 | 2011-08-03 | <20 | 160 | 1280 | 160 | 160 | 160 | 160 | C1 |
| B/Newcastle/50/2011 | 2011-08-15 | <20 | 160 | 640 | 160 | 160 | 160 | 160 | C1 |
| B/Newcastle/53/2011 | 2011-08-16 | <20 | 160 | 640 | 160 | 80 | 160 | 160 | C1 |
| B/Newcastle/55/2011 | 2011-08-15 | <20 | 160 | 640 | 160 | 80 | 160 | 80 | C1 |
| B/Newcastle/63/2011 | 2011-08-10 | <20 | 160 | 640 | 160 | 160 | 160 | 80 | C1 |
| B/Sydney/17/2011 | 2011-07-16 | <20 | 160 | 640 | 160 | 160 | 160 | 80 | CX/C1 |
| B/Sydney/18/2011 | 2011-07-23 | <20 | 160 | 1280 | 160 | 160 | 160 | 160 | CX/C1 |
| B/Sydney/19/2011 | 2011-07-24 | <20 | 160 | 640 | 160 | 160 | 160 | 160 | CX/C1 |
| B/Sydney/20/2011 | 2011-08-11 | <20 | 160 | 640 | 160 | 160 | 160 | 160 | CX/C1 |
| B/Sydney/21/2011 | 2011-07-21 | <20 | 160 | 1280 | 160 | 160 | 160 | 80 | CX/C1 |
| B/Sydney/22/2011 | 2011-06-20 | <20 | 160 | 640 | 160 | 160 | 160 | 80 | CX/C1 |
| B/Sydney/26/2011 | 2011-07-13 | <20 | 160 | 640 | 160 | 160 | 160 | 160 | CX/C1 |
| B/Cambodia/20/2011 | 2011-06-16 | 40 | <20 | 80 | 20 | 20 | 20 | 640 | X1/C1 |
| B/Cambodia/22/2011 | 2011-06-17 | 20 | <20 | 80 | 20 | 20 | 20 | 640 | X1/C1 |
Shaded column: B/Brisbane/60/2008 current vaccine virus
Reference sera from ferrets after boost with homologous antigen
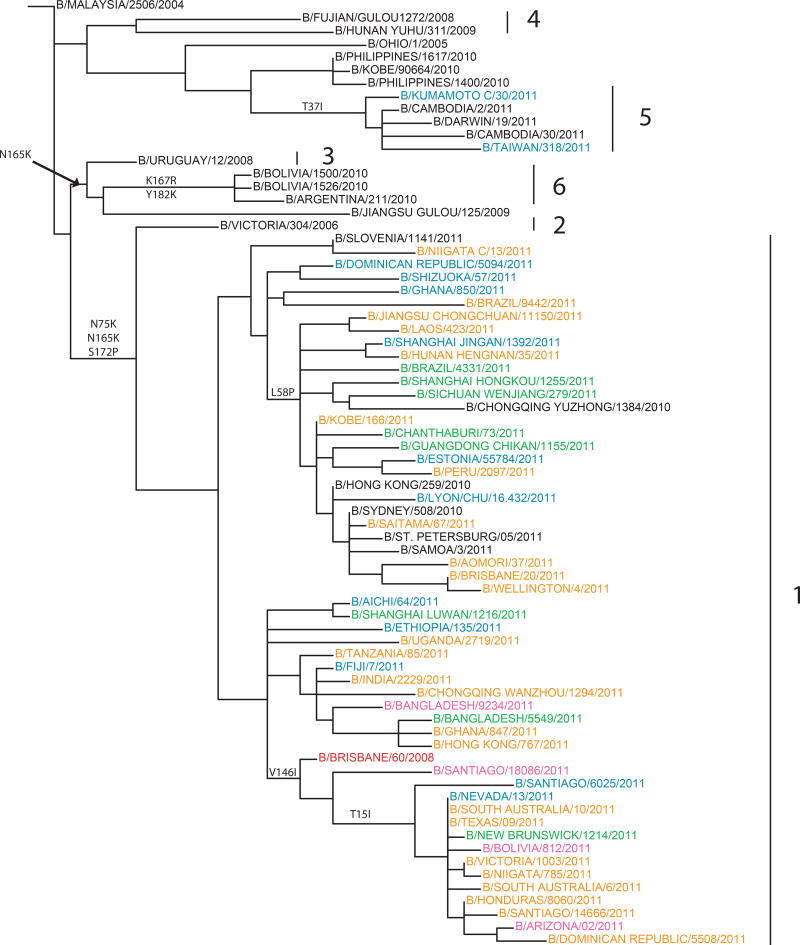
The HA genes of recent B/Victoria-lineage viruses fell into six main phylogenetic groups, with the vast majority of them falling in group 1 represented by B/Brisbane/60/2008 with signature AA substitutions of N75K, N165K and S172P (Fig. 5). (A high resolution tree constructed from sequences of >1,000 B/Victoria lineage isolates collected through GISRS since 2010-01-01 is shown in Fig. S5). Of the B/Victoria-lineage genetic groups, 2–6, only isolates from group 5 have been detected recently; most of these group 5 viruses were isolated at the beginning of 2011 in South East Asia and Oceania (Figs 5 and S5). All group 1 viruses remained antigenically similar to B/Brisbane/60/2008. Small genetic subgroups within group 1 were identified including a subgroup with no AA substitutions in HA that clustered in the nucleotide tree with contemporary isolates from Africa, Asia and the U.S. and a second small subgroup that included isolates collected recently in Asia and South America with the HA AA substitution L58P. Many of the recent isolates from China in this second subgroup had HA and NA genes from different groups of the B/Victoria-lineage; HA genes from the B/Victoria-lineage group 1 and NA genes from HA group 4; the NA of these intra-lineage reassortant viruses had additional AA substitutions D384N and A465T (leading to the gain of a potential glycosylation site) in the NA compared with viruses that carried both the HA and NA gene of group 4. Viruses in the third small subgroup within group 1 have AA substitution V146I in the HA1. An additional subgroup within group 1 has undergone intra-Victoria lineage reassortment inheriting the NA gene from isolates similar to those in HA group 3 (represented by B/Uruguay/12/2008), but with additional AA substitutions of L73F, S397R and A389T in the NA. The majority of isolates within this subgroup have AA substitution V15I in the HA1 and circulated in North and South America and Oceania.
Fig. 5.
Phylogenetic trees of representative B/Victoria lineage HA1 domain nucleotide sequences were constructed and annotated as described for Fig. 2. Genetic groups are labelled 1–7.
B/Yamagata-lineage viruses were detected at low levels in all countries with the exception of China where B/Yamagata-lineage viruses comprised over 45% of B viruses detected (data not shown). Regional differences in the distribution of B/Yamagata- and B/Victoria-lineages were seen between the north and south of China. While B/Yamagata-lineage viruses were isolated throughout the country, the majority of B/Victoria-lineage isolates were isolated in the south of China.
Most recent B/Yamagata isolates were antigenically distinct from the previous vaccine virus, B/Florida/4/2006, and were more closely related to the B/Wisconsin/1/2010 or B/Hubei-Wujiagang/158/2009 reference viruses (Table 5) and the B/Bangladesh/3333/207 reference virus (data not shown). The HA genes of recent B/Yamagata-lineage viruses clustered phylogenetically into group 3 with signature AA substitutions S150I, N166Y and G230D (see Fig. S6 for a high resolution tree constructed from sequences of over 250 B/Yamagata lineage isolates collected since 2010-01-01).
Table 5.
Hemagglutination inhibition test of B Yamagata lineage viruses (WHO CC Beijing)
| REFERENCE VIRUSES | Collection date |
Reference Ferret Antisera a
|
Passage history |
|||||
|---|---|---|---|---|---|---|---|---|
| B/Fl/4/06 | B/Hub/158/09 | B/Guan/1512/10 | B/Fuj/1790/10 | B/WI/1/10 | B/Sic/139/11 | |||
| B/Florida/4/2006 | 2006-11-01 | 10240 | 2560 | 320 | 640 | 10240 | 5120 | unknown |
| B/Hubei-Wujiagang/158/2009 | unknown | 320 | 2560 | 80 | 80 | 2560 | 1280 | unknown |
| B/Guangdong-Luohu/1512/2010 | 2010-06-21 | 1280 | 1280 | 320 | 640 | 5120 | 2560 | unknown |
| B/Fujian-Gulou/1790/2010 | 2010-04-13 | 160 | 1280 | 160 | 160 | 2560 | 640 | unknown |
| B/Wisconsin/01/2010 | 2010-02-20 | 320 | 1280 | 160 | 160 | 5120 | 1280 | unknown |
| B/Sichuan-Anyue/139/2011 | 2011-03-02 | 640 | 1280 | 160 | 320 | 2560 | 1280 | unknown |
|
| ||||||||
|
TEST VIRUSES
| ||||||||
| B/Shanghai-Pudongxin/1941/2010 | 2010-12-13 | 320 | 2560 | 320 | 320 | 2560 | 1280 | C1+C1 |
| B/Hunan-Beihu/113/2011 | 2011-01-09 | 640 | 5120 | 320 | 320 | 5120 | 1280 | C2 |
| B/Shanghai-Pudongxin/13/2011 | 2011-01-04 | 640 | 5120 | 640 | 640 | 5120 | 2560 | C2 |
| B/Sichuan-Qingyang/17/2011 | 2011-01-07 | 160 | 1280 | 160 | 160 | 5120 | 640 | C2 |
| B/Hunan-Yuhu/134/2011 | 2011-03-02 | 2560 | 10240 | 1280 | 1280 | 10240 | 5120 | C1+C1 |
| B/Hunan-Lusong/1126/2011 | 2011-02-16 | 160 | 1280 | 160 | 80 | 2560 | 320 | C1+C1 |
| B/Shanghai-Luwan/187/2011 | 2011-02-15 | 320 | 2560 | 320 | 320 | 5120 | 1280 | C1+C1 |
| B/Heilongjiang-Gongnong/182/2011 | 2011-02-16 | 320 | 2560 | 320 | 320 | 5120 | 1280 | C1+C1 |
| B/Shaanxi-Beilin/1165/2011 | 2011-02-15 | 320 | 2560 | 320 | 320 | 5120 | 1280 | C1+C1 |
| B/Beijing-Huairou/1338/2011 | 2011-03-10 | 320 | 2560 | 320 | 320 | 5120 | 1280 | C1+C1 |
| B/Shanghai-Songjiang/1108/2011 | 2011-03-02 | 320 | 2560 | 320 | 320 | 5120 | 1280 | C1+C1 |
| B/Jiangsu-Xinpu/192/2011 | 2011-03-02 | 640 | 2560 | 320 | 320 | 10240 | 1280 | C2 |
| B/Shanghai-Changning/1143/2011 | 2011-03-01 | 640 | 5120 | 320 | 320 | 5120 | 5120 | C2 |
| B/Beijing-Xicheng/1291/2011 | 2011-03-09 | 640 | 5120 | 320 | 320 | 5120 | 1280 | C2 |
| B/Gansu-Chengguan/572/2011 | 2011-03-10 | 320 | 2560 | 320 | 320 | 5120 | 2560 | C1+C1 |
| B/Sichuan-Anyue/139/2011 | 2011-03-02 | 160 | 2560 | 160 | 160 | 5120 | 1280 | C1+C1 |
| B/Chongqing-Wanzhou/1152/2011 | 2011-03-08 | 320 | 2560 | 160 | 160 | 5120 | 1280 | C1+C1 |
| B/Beijing-Xicheng/1267/2011 | 2011-02-27 | 320 | 2560 | 160 | 160 | 5120 | 1280 | C2 |
| B/Shanghai-Songjiang/1100/2011 | 2011-02-23 | 320 | 2560 | 320 | 320 | 5120 | 1280 | E1+E1 |
| B/Shanghai-Songjiang/190/2011 | 2011-02-21 | 1280 | 5120 | 640 | 640 | 5120 | 2560 | E1+E1 |
| B/Shanghai-Songhui/1104/2011 | 2011-03-08 | 640 | 5120 | 320 | 320 | 10240 | 2560 | E2 |
| B/Jiangsu-Xinpu/183/2011 | 2011-02-28 | 320 | 2560 | 160 | 160 | 5120 | 1280 | E2 |
| B/Hubei-Wuchang/1108/2011 | 2011-04-03 | 320 | 2560 | 320 | 320 | 5120 | 1280 | C1+C1 |
| B/Jiangsu-Beitang/1135/2011 | 2011-04-05 | 320 | 2560 | 320 | 320 | 2560 | 1280 | C1+C1 |
| B/Shanghai-Luwan/1188/2011 | 2011-04-12 | 320 | 1280 | 160 | 160 | 2560 | 1280 | C1+C1 |
| B/Beijing-Xicheng/1677/2011 | 2011-04-06 | 160 | 1280 | 160 | 160 | 2560 | 1280 | C1+C1 |
| B/Sichuan-Yucheng/1212/2011 | 2011-04-03 | 160 | 1280 | 160 | 160 | 2560 | 640 | C2 |
| B/Anhui-Huashan/1216/2011 | 2011-05-10 | 160 | 2560 | 160 | 160 | 5120 | 1280 | C4 |
| B/Jiangsu-Quanshan/1329/2011 | 2011-04-27 | 160 | 2560 | 160 | 160 | 10240 | 640 | C2 |
| B/Tianjin-Hedong/1174/2011 | 2011-05-15 | 160 | 10240 | 160 | 160 | 20480 | 1280 | C1+C1 |
| B/Hunan-Beihu/1300/2011 | 2011-05-26 | 160 | 1280 | 160 | 160 | 2560 | 640 | C2 |
| B/Shanghai-Pudongxin/1238/2011 | 2011-05-13 | 160 | 1280 | 160 | 160 | 2560 | 640 | C2 |
| B/Beijing-Xicheng/1962/2011 | 2011-05-23 | 160 | 2560 | 80 | 80 | 10240 | 640 | C1+C1 |
| B/Shanghai-Hongkou/1299/2011 | 2011-06-10 | 1280 | 5120 | 640 | 1280 | 5120 | 2560 | C2 |
| B/Shandong-Boshan/1163/2011 | 2011-05-11 | 320 | 2560 | 160 | 160 | 5120 | 1280 | C2 |
| B/Hunan-Beihu/1374/2011 | 2011-07-05 | 320 | 2560 | 320 | 320 | 5120 | 1280 | C2 |
| B/Tianjin-Hexi/180/2011 | 2011-02-28 | 160 | 1280 | 160 | 80 | 2560 | 640 | E1+E1 |
| B/Shanghai-Changning/1133/2011 | 2011-02-24 | 320 | 1280 | 80 | 80 | 1280 | 640 | E2 |
| B/Tianjin-Hexi/156/2011 | 2011-02-15 | 320 | 2560 | 320 | 320 | 5120 | 2560 | E1+E1 |
| B/Shanghai-Songhui/1104/2011 | 2011-03-08 | 320 | 2560 | 160 | 160 | 5120 | 1280 | E2 |
| B/Shanghai-Luwan/1217/2011 | 2011-05-04 | 160 | 640 | 40 | 40 | 1280 | 320 | E2 |
Reference sera from ferrets after boost with homologous antigen
Data generated by WHO CCs and ERLs showed that the post-vaccination sera obtained from people immunized with vaccines containing B/Brisbane/60/2008-like viruses reacted well with recent influenza B viruses from the B/Victoria lineage, but less well with B viruses from the B/Yamagata lineage (Fig. S7). There was on average only a 23% reduction in post-vaccination GMTs to the recent B/Victoria lineage viruses compared with those to the vaccine virus, whereas the equivalent reduction in post-vaccination GMT to B/Yamagata lineage viruses was on average 67%.
Based on the predominance of B/Victoria-lineage viruses in many parts of the world throughout the surveillance period covered here, and their antigenic similarity to B/Brisbane/60/2008, it was concluded that vaccines containing a B/Brisbane/60/2008-like virus remained appropriate for the Southern Hemisphere for 2012.
7. Antiviral resistance
The two classes of antiviral drugs currently licensed for the prevention and treatment of influenza are the adamantanes or M2 ion channel inhibitors (amantadine and rimantadine) and the neuraminidase inhibitors (oseltamivir and zanamivir). More than 99% of A(H1N1)pdm09 viruses tested carry the AA substitution S31N in the M2 protein associated with resistance to amantadine and rimantadine. There were sporadic detections of oseltamivir resistance in A(H1N1)pdm09 viruses due to an H275Y substitution in NA. Although the level of resistance to oseltamivir remains low worldwide (~2%, Table 6), available data indicate that the proportion of resistant A(H1N1)pdm09 viruses isolated from patients not exposed to the drug increased in Australia, Japan, the UK and U.S. compared to previous seasons (data not shown). All A(H1N1)pdm09 viruses were sensitive to zanamivir (data not shown). More than 99% of A(H3N2) viruses characterised were resistant to adamantanes (based on the presence of the M2 protein AA substitution S31N) but all were sensitive to neuraminidase inhibitors, oseltamivir (Table 6) and zanamivir (data not shown). Over 99% of all B viruses characterized were sensitive to neuraminidase inhibitors oseltamivir (Table 6) and zanamivir (data not shown).
Table 6.
Frequency of resistance to oseltamivir in isolates or clinical materials collected from March – August, 2011
H275Y mutation, also confers resistance to Peramivir
B/Lyon/CHU/16.432/2011 (B/Victoria lineage): also resistant to Zamamivir and Peramivir
B/Kochi/61/2011 (B/Victoria lineage): also resistant to Zanamivir and Peramivir
Supplementary Material
Supplementary Fig. 1. Phylogenetic trees of the A(H1N1)pdm09 HA1 domain nucleotide sequences were constructed with the PhyML software package version 3.0 [7] using GTR+I+Γ4 as determined by ModelTest [8]. GARLI v0.961 [9] was run on the best tree from PhyML for 2 million generations to optimise tree topology and branch lengths. The eight genetic subgroups are boxed and labelled and key amino acid substitutions are marked on major nodes. Viruses are colour-coded by region of origin as follows: Dark Blue-North America, Light Blue-South America, Green-Europe, Orange-Africa, Purple-Middle East, Maroon-Russia, Red-E-SE Asia, Pink-Oceania, unknown-grey. The month in which the strains in the tree were isolated is indicated by horizontal bars to the right of the tree drawn at the same vertical position as the position of the strain in the tree. The horizontal bars are also colour-coded by region. This tree is presented in high resolution the strain names and dates of virus collection can be read upon magnification.
Supplementary Fig. 2. Human post vaccination serology analysis comparing mean geometric mean titres of HI antibody responses to representative A(H1N1)pdm09 isolates relative to A/California/7/2009.
Supplementary Fig. 3. Phylogenetic trees of the A(H3N2) HA1 domain nucleotide sequences were constructed and annotated as described fro Supplementary Fig. 1.
This tree is presented in high resolution the strain names and dates of virus collection can be read upon magnification.
Supplementary Fig. 4. Human post vaccination serology analysis comparing mean geometric mean titres of HI antibody responses to representative A(H3N2) isolates relative to A/Perth/16/2009 or A/Victoria/210/2009 (A/Perth/16/2009-like viruses).
Supplementary Fig. 5. Phylogenetic trees of the B Victoria Lineage HA1 domain nucleotide sequences were constructed and annotated as described for Supplementary Fig. 1.
This tree is presented in high resolution the strain names and dates of virus collection can be read upon magnification.
Supplementary Fig. 6. Phylogenetic trees of the B/Yamagata HA1 domain nucleotide sequences were constructed and annotated as described for Supplementary Fig. 1.
This tree is presented in high resolution the strain names and dates of virus collection can be read upon magnification.
Supplementary Fig. 7. Human post vaccination serology analysis comparing mean geometric mean titres of HI antibody responses to representative B isolates relative to B/Brisbane/60/2008 or B/Brisbane/33/2008 (A/Brisbane/60/2008-like viruses).
Acknowledgments
The writing committee would like to thank all of their colleagues in their institutes, the WHO NICs and other laboratories and organisations for their efforts in supplying, testing and analysing the influenza viruses contained in this report.
Appendix A
The writing committee’s affiliations are as follows: WHO Collaborating Centre for Surveillance, Epidemiology and Control of Influenza, Centers for Disease Control and Prevention, Atlanta, USA, (AIK, RG, XX, NJC); WHO Collaborating Centre for Reference and Research on Influenza, Victoria Infectious Diseases Reference Laboratory (VIDRL), Melbourne, Australia, (IGB, AK); WHO Collaborating Centre for Reference and Research on Influenza, National Institute for Medical Research (NIMR), London UK, (JM, RD); WHO Collaborating Centre for Reference and Research on Influenza, National Institute of Infectious Diseases (NIID), Tokyo, Japan, (TO, MT, SI); St Jude Children’s Research Hospital, Memphis, USA (RW); WHO Collaborating Center for Reference and Research on Influenza, Chinese National Influenza Center (CNIC), Beijing, PRC China (YS, DW); WHO Global Influenza Programme (GIP), Geneva, Switzerland, (WZ, TB); National Institute for Biological Standards and Control (NIBSC), Health Protection Agency (HPA), Potters Bar, UK, (OGE); Food and Drug Administration (FDA), Center for Biologics Evaluation and Research (CBER), Bethesda, MD, USA, (ZY); Therapeutic Goods Administration (TGA), Canberra, Australia (GG); University of Cambridge, UK and Fogarty International Centre, National Institutes of Health (NIH), USA, (DS, CR).
Appendix B. Supplementary data
Supplementary data associated with this article can be found in the online version,
Footnotes
The members of the writing committee (Alexander I. Klimov, Rebecca Garten, Colin Russell, Ian G. Barr, Terry G. Besselaar, Rod Daniels, Othmar G. Engelhardt, Gary Grohmann, Shigeyuki Itamura, Anne Kelso, John McCauley, Takato Odagiri, Derek Smith, Masato Tashiro, Xiyan Xu, Richard Webby, Dayan Wang, Zhiping Ye, Shu Yuelong, Wenqing Zhang, Nancy Cox) assume responsibility for the overall content and integrity of the article.
The boundaries and names shown and the designations used in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement.
References
- 1.Barr IG, McCauley J, Cox N, Daniels R, Engelhardt OG, Fukuda K, et al. Epidemiological, antigenic and genetic characteristics of seasonal influenza A(H1N1), A(H3N2) and B influenza viruses: basis for the WHO recommendation on the composition of influenza vaccines for use in the 2009–2010 Northern Hemisphere season. Vaccine. 2010 Feb 3;28(5):1156–67. doi: 10.1016/j.vaccine.2009.11.043. [DOI] [PubMed] [Google Scholar]
- 2.Organization WH. Recommended composition of influenza virus vaccines for use in the 2011–2012 northern hemisphere influenza season. Weekly epidemiological record. 2011;86(10):81–92. [PubMed] [Google Scholar]
- 3.Organization WH. Recommended composition of influenza vaccines for use in the 2012 southern hemisphere influenza season. Weekly epidemiological record. 2011;86:457–68. [PubMed] [Google Scholar]
- 4.Organization WH. Standardization of terminology of the pandemic A(H1N1)2009 virus. 2011 10/18/2011 [cited 2011 11/01/2011]; Available from: http://www.who.int/influenza/gisrs_laboratory/terminology_ah1n1pdm09/en/ [PubMed]
- 5.Matrosovich M, Matrosovich T, Carr J, Roberts NA, Klenk HD. Overexpression of the alpha-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J Virol. 2003 Aug;77(15):8418–25. doi: 10.1128/JVI.77.15.8418-8425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Press W, editor. Network WGIS. Manual for the laboratory diagnosis and virological surveillance of influenza. Geneva: World Health Organization; 2011. [Google Scholar]
- 7.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic biology. 2003 Oct;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 8.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–8. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 9.Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion.: Ph.D. 2006 [Google Scholar]
- 10.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004 Jul 16;305(5682):371–6. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 11.Lin YP, Gregory V, Collins P, Kloess J, Wharton S, Cattle N, et al. Neuraminidase receptor binding variants of human influenza A(H3N2) viruses resulting from substitution of aspartic acid 151 in the catalytic site: a role in virus attachment? J Virol. 2010 Jul;84(13):6769–81. doi: 10.1128/JVI.00458-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Organization WH. [cited; 09/29/2011];Recommended composition of influenza virus vaccines for use in the 2012 southern hemisphere influenza season. 2011 Available from: http://www.who.int/influenza/vaccines/virus/recommendations/2012south/en/index.html.
- 13.Rota PA, Wallis TR, Harmon MW, Rota JS, Kendal AP, Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990 Mar;175(1):59–68. doi: 10.1016/0042-6822(90)90186-u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Phylogenetic trees of the A(H1N1)pdm09 HA1 domain nucleotide sequences were constructed with the PhyML software package version 3.0 [7] using GTR+I+Γ4 as determined by ModelTest [8]. GARLI v0.961 [9] was run on the best tree from PhyML for 2 million generations to optimise tree topology and branch lengths. The eight genetic subgroups are boxed and labelled and key amino acid substitutions are marked on major nodes. Viruses are colour-coded by region of origin as follows: Dark Blue-North America, Light Blue-South America, Green-Europe, Orange-Africa, Purple-Middle East, Maroon-Russia, Red-E-SE Asia, Pink-Oceania, unknown-grey. The month in which the strains in the tree were isolated is indicated by horizontal bars to the right of the tree drawn at the same vertical position as the position of the strain in the tree. The horizontal bars are also colour-coded by region. This tree is presented in high resolution the strain names and dates of virus collection can be read upon magnification.
Supplementary Fig. 2. Human post vaccination serology analysis comparing mean geometric mean titres of HI antibody responses to representative A(H1N1)pdm09 isolates relative to A/California/7/2009.
Supplementary Fig. 3. Phylogenetic trees of the A(H3N2) HA1 domain nucleotide sequences were constructed and annotated as described fro Supplementary Fig. 1.
This tree is presented in high resolution the strain names and dates of virus collection can be read upon magnification.
Supplementary Fig. 4. Human post vaccination serology analysis comparing mean geometric mean titres of HI antibody responses to representative A(H3N2) isolates relative to A/Perth/16/2009 or A/Victoria/210/2009 (A/Perth/16/2009-like viruses).
Supplementary Fig. 5. Phylogenetic trees of the B Victoria Lineage HA1 domain nucleotide sequences were constructed and annotated as described for Supplementary Fig. 1.
This tree is presented in high resolution the strain names and dates of virus collection can be read upon magnification.
Supplementary Fig. 6. Phylogenetic trees of the B/Yamagata HA1 domain nucleotide sequences were constructed and annotated as described for Supplementary Fig. 1.
This tree is presented in high resolution the strain names and dates of virus collection can be read upon magnification.
Supplementary Fig. 7. Human post vaccination serology analysis comparing mean geometric mean titres of HI antibody responses to representative B isolates relative to B/Brisbane/60/2008 or B/Brisbane/33/2008 (A/Brisbane/60/2008-like viruses).