TOC GRAPHICS
Keywords: Receptor clustering, intermembrane junction, T-cell, artificial antigen presenting cell, molecular tension probe, immunological synapse
Abstract
Short-range communication between cells is required for the survival of multicellular organisms. One mechanism of chemical signaling between adjacent cells employs surface displayed ligands and receptors that only bind when two cells make physical contact. Ligand-receptor complexes that form at the cell-cell junction and physically bridge two cells likely experience mechanical forces. A fundamental challenge in this area pertains to mapping the mechanical forces experienced by ligand-receptor complexes within such a fluid intermembrane junction. Herein, we describe the development of ratiometric tension probes for direct imaging of receptor tension, clustering and lateral transport within a model cell-cell junction. These probes employ two fluorescent reporters that quantify both the ligand density and the ligand tension, and thus generate a tension signal independent of clustering. As a proof-of-concept, we applied the ratiometric tension probes to map the forces experienced by the T-cell receptor (TCR) during activation and showed the first direct evidence that the TCR-ligand complex experiences sustained pN forces within a fluid membrane junction. We envision that the ratiometric tension probes will be broadly useful for investigating mechanotransduction in juxtacrine signaling pathways.
Membrane receptors are ubiquitous in Nature and play a central role in transferring chemical information across the cell membrane.1 The first step in the majority of signal transduction cascades, ranging from growth factor signaling2,3 to T-cell activation,4–6 is ligand-induced dimerization and oligomerization of receptors. These higher-order clusters of receptors often function as a signaling complex for signal amplification, diversification, and in some cases serve to facilitate receptor internalization and signal degradation.7 Interestingly, receptor oligomers are typically coupled to the cytoskeleton which offers an active scaffold for receptor translocation and organization, thus further fine-tuning signaling circuits.8 Given the role of the cytoskeleton in force transmission and generation,9 it seems intuitive to conclude that ligand-induced receptor clustering is intimately linked with mechanotransduction but evidence for this connection is lacking. This is due to the absence of methods to measure the mechanical forces experienced by ligand-receptor complexes during active signaling at the cell membrane.
The archetypal example highlighting the complex interplay between ligand-induced binding, receptor clustering and mechanical coupling is illustrated by the T cell receptor (TCR).10 A polarized and crawling T cell constantly scans the surfaces of antigen-presenting cells (APCs) in search of foreign peptides that are bound to the major histocompatibility complex (pMHC).11,12 Upon TCR engagement and activation by their cognate pMHC ligand, the receptors form signaling microclusters that initiate T-cell activation cascades leading to Ca2+ flux and cytokine production. During activation, lymphocyte function-associated antigen 1 (LFA-1) receptors bind intercellular adhesion molecule 1 (ICAM-1) on the APC, thus facilitating the formation of a stable intercellular junction.8 Within this specialized junction, the T cell cytoskeleton associates with TCR and LFA-1 and sorts these receptors into distinct concentric zones. TCR microclusters undergo continuous translocation and become spatially organized in a structure known as the central supramolecular activation cluster (cSMAC), while LFA-1/ICAM-1 are re-organized into a ring-like structure surrounding the cSMAC.4,13
The migratory nature of T cells and the central role of the cytoskeleton in TCR activation14,15 suggest a fundamental connection between receptor oligomerization, signaling, and mechanotransduction. Several lines of evidence support an important role of mechanical force in T-cell activation and signaling.16,17 First, Reinherz, Lang and co-workers showed that TCR is a mechanosensor that responds to externally applied pN forces from an optical tweezer.18 This concept is further supported by Li and co-workers who demonstrated micropipette induced shear forces can activate T-cells,19 and recently by and recently by our optomechanical actuator (OMA) nanoparticles which, upon stimulation of NIR light, are able to mechanically activate T-cells.20 In addition, TCR undergoes distinct conformational transitions upon experiencing pN forces transmitted through the pMHC antigens.21 Second, bio-membrane force probe experiments by Zhu, Evavold and colleagues showed that TCR-pMHC bond lifetime (1/koff) is best correlated to antigen potency when 10–15 pN forces are applied to the TCR-pMHC complex.22 Third, T-cells apply nano-newton (nN) traction forces to deform micron-sized PDMS pillars within ~5 min of activation.23 Lastly, restriction of TCR transport with diffusion barriers upregulates TCR signaling, possibly due the increased forces on the TCR-pMHC complex and its altered spatial organization.24 Still, whether the interplay of mechanics, clustering and chemical signaling influences T-cell function remains a longstanding question due to the lack of tools to quantify nanoscale mechanics of T-cells.
To investigate how physical inputs regulate or couple to signal transduction in living cells, we invented molecular tension fluorescence-based microscopy (MTFM), which maps pN receptor forces generated by cells using fluorescence imaging with high spatial (~200 nm) and temporal (~ms) resolutions.25,26 Typically, MTFM probes consist of an extendable “spring” flanked by a fluorophore and quencher and immobilized on a surface. The probe is decorated with a ligand of interest, and is highly quenched in the resting conformation (>90% quenching efficiency). When cell surface receptors engage their ligand on the MTFM probe and apply sufficient mechanical load to extend the “spring” then the dye is de-quenched, which leads to significant enhancement in fluorescence. Second generation MTFM probes have further improved the dynamic force range,27 sensitivity28,29 and stability.30–33 We recently developed a gold nanoparticle (AuNP)-based DNA tension probe to directly image the TCR-pMHC forces on immobilized ligands and revealed that T-cells may harness mechanics for enhanced antigen sampling and discrimination.34
One remaining limitation for MTFM is that probes are immobilized onto a solid substrate, which is used for several reasons. First, it limits the lateral mobility of the ligand and thus the probe reports the integrated forces transmitted in the lateral and perpendicular directions through the ligand-receptor complex. Immobilized MTFM probes provide mimics of the cell–extracellular matrix interactions, where ligands are physically affixed onto an immobile scaffold.35 Second, by maintaining a constant probe density, the donor fluorescence intensity can be directly used to determine the quenching efficiency (QE) using the following relation:
| QE = 1 – (IDA/ID) |
where IDA is the donor fluorescence under zero force condition and ID is the donor fluorescence in the presence of applied forces.
In this way, a non-fluorescent acceptor (quencher) can be used, which reduces bleed-through, improves signal-to-noise ratio and also frees up additional fluorescence channels for live cell imaging of protein translocation and signaling activity. An alternate immobilization strategy would be to tether ligands onto laterally fluid surfaces mimicking the plasma membrane. In this scenario, it remains unknown whether MTFM probes on these surfaces would experience sufficient pN tension to generate detectable signal. In order to study mechanical forces at cell–cell junctions where ligands and receptors are allowed to diffuse laterally, it is necessary to introduce new methods that effectively integrate MTFM probes with fluid membranes.
One approach to mimic the cell–cell junction is the hybrid cell–supported lipid bilayer platform. The supported lipid bilayer (SLB) is comprised of phospholipids that self-assemble onto a solid substrate. These lipids form a fluid bilayer that can be chemically decorated with a variety of biomolecules, and thus the SLB displays chemical and physical properties that resemble those of the cellular plasma membrane.36,37 Importantly, when a living cell encounters an SLB displaying appropriate ligands, the cell surface receptors bind, oligomerize, and laterally translocate across the cell–SLB junction. Therefore, this platform provides a powerful tool to reconstitute juxtacrine signaling pathways such as immunological synapse formation during T-cell signaling,38–42 receptor clustering in E-cadherin,43 EphA2–EphrinA1,44,45 Notch–delta46 and integrin–RGD47–49 systems. Taking advantages of the cell–SLB platform, we aimed to tether MTFM tension probes onto an SLB to enable the study of receptor mechanics during active signaling at model cell-cell junctions.
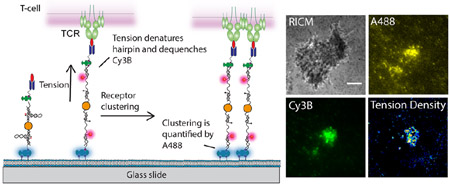
To generate SLB-tethered tension probes, we first designed a DNA hairpin structure that places a fluorophore-quencher pair (Cy3B-BHQ2) in close proximity. DNA hairpins are immobilized onto a AuNP that further quenches the Cy3B fluorescence. The AuNP-tension probe is in turn immobilized onto a SLB using biotin-streptavidin and is further functionalized with a ligand. DNA hairpins unfold in response to mechanical strain, which separates the fluorophore from the molecular quencher and the AuNP surface. The magnitude of force needed to unfold the hairpin can be estimated from the F1/2, which is the force that leads to 50% probability of unfolding at equilibrium.50 Note that the loading rate and the duration of forces are unknown and thus the F1/2 serves as a lower bound estimate of the applied force. Due to the dual quenching of the dye by a molecular quencher (FRET)51 and the AuNP via Nanometal Surface Energy Transfer (NSET),52 this construct provides a 10-fold improvement in sensitivity over the existing MTFM probes.27,28 To distinguish fluorescence enhancement due to clustering from that of tension, we introduced a second fluorophore (Alexa Fluor® 488, abbreviated A488 thereafter) to report on relative probe density (Scheme 1). The size of AuNP-tension probes is estimated to be 47.4±1.8 nm from dynamic light scattering (DLS) (Figure S1). Each particle is labeled with 38±1 hairpins as determined by fluorescence calibration (Figure S2).
Scheme 1.
Schematic representation of gold nanoparticle-based ratiometric tension probes
To demonstrate that the ratiometric tension probe can reliably report on clustering and mechanics, we employed a model system to mimic the cell–SLB junction. In this system, tension probes (F1/2 = 4.7 pN, see Table S1 for DNA sequences) were immobilized onto a SLB and decorated with streptavidin that engaged biotinylated microparticles (diameter = 5 μm) (Figure 1A). It has been shown that the junction between a spherical particle and planar SLB generates interfacial tension across the ligand-receptor complexes (biotin-streptavidin) bridging the particle to the SLB.53 This is due to the geometric mismatch between the particle and planar surface. We anticipated accumulation of streptavidin as well as the extension of a subset of DNA hairpins bridging the particle and the surface within the contact zone. Incubation of biotinylated microparticles onto the tension probe-SLB surface generated a strong interference pattern in reflection interference contrast microscopy (RICM), indicating close nanoscale contact between the particle and surface (Figure 1B). This interaction led to a drastic increase in fluorescence in both the Cy3B and the A488 signals (Figure 1B). Line scan analysis across the microparticles showed a maximum of ~14 fold increase in Cy3B fluorescence, which reports on probe clustering and hairpin unfolding, while only a ~2.5-fold enhancement was observed in the A488 channel, which exclusively reports on clustering (Figure 1C). Fluorescence from both channels colocalized with the RICM interference pattern and were most pronounced at the center of the microparticle-SLB contact zone, confirming microparticle-driven probe clustering (Figure 1D).
Figure 1.
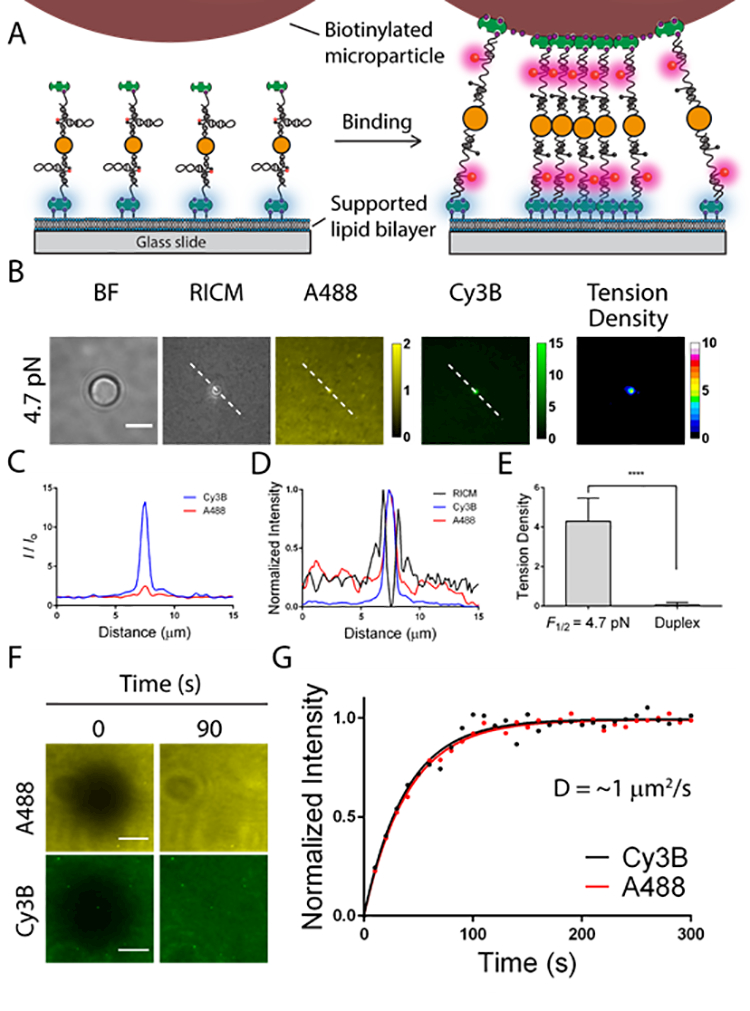
A) Illustration of the contact zone between a biotinylated microparticle and a DNA tension probe surface anchored to a supported lipid membrane. B) Representative brightfield, RICM, Cy3B, A488, and tension density images showing the contact zone of a microparticle (diameter = 5 μm) that binds to SLB tension probes (F1/2 = 4.7 pN). Scale bar = 5 μm. C) Plot displaying line scan of Cy3B and A488 channels for the microparticle shown in B. The intensity is normalized (I) to the background regions lacking the microparticle (I0). D) Plot overlaying the line scan profile of fluorescence and RICM channels, and demonstrating their spatial colocalization. E) Bar graph showing the tension density of microparticles engaged to tension probe SLBs (F1/2 = 4.7 pN) and control SLB surfaces decorated using DNA duplexes (n = 20 for each sample, error bar represents S.D. of the data) within the microparticle-SLB contact zone. F) Representative FRAP images showing recovery after 90 s. Scale bars = 10 μm. G) Representative FRAP recovery plots for Cy3B and A488 channels. Solid lines represent the fit made using the following equation I(t) = A(1−e-tτ). The Lateral diffusion coefficient (D) is calculated by: D = w2/4t1/2, where w is the radius of the Gaussian bleaching area; t1/2 is the time for 50% recovery obtained from the fit. The values used for the calculation were: w = 10.4 μm (for both channels); t1/2 = 26.5 s (Cy3B) and 27.9 s (A488).
To estimate the relative fraction of open hairpins induced by microparticle binding, we created normalized Cy3B and A488 images (normalized to a region of interest lacking cells). The normalized Cy3B image was divided by the normalized A488 image, and then was subtracted by 1 to obtain a tension density map, such that a tension density signal of ~0 corresponds to the background (Scheme 1 and Figure S3 for detailed image processing procedures). Tension density signals that are greater than 0 indicate a relative fraction of hairpins in the open state. Molecular binding between microparticles and hairpin tension probes within the contact zone generated fluorescence enhancement greatest at the center and had an average tension density value of 4.29±1.16 (Figure 1E). As a control, we used AuNP-duplexes lacking a hairpin loop and for which the fluorescence increase is exclusively due to clustering rather than hairpin unfolding (see Table S1 for DNA sequences). As expected, tension density signal on these surfaces were significantly lower (0.05±0.13) compared to the surfaces decorated with hairpin tension probes (Figure S4 and Figure 1E). Collectively, these results indicate that the ratiometric response of the tension probe is largely due to mechanical unfolding of the hairpin stem-loop of the tension probes.
We further evaluated the interaction between the microparticles and tension probes by immobilizing the tension probes on glass surfaces, where probe clustering is prohibited (Scheme S2). On these surfaces, the Cy3B signal was localized at the edges of the microparticle-surface junction. Unlike SLB surfaces, line scan analysis across the microparticles revealed two local maxima in Cy3B channel, and the peaks encased the center of microparticle-surface contact zone (Figure S5). In contrast, no accumulation of fluorescence in the A488 channel was observed (Figure S5). Control experiments using DNA duplexes showed a small increase in fluorescence (25.5±26.4%) in the Cy3B channel, whereas tension probes immobilized on glass surfaces had an average of 4-fold enhancement in Cy3B fluorescence (400±115%) underneath the microparticle-surface contact zone (Figure S5). These results show that ratiometric tension probes can be used to distinguish signals due to tension from that of clustering.
To test whether the ratiometric tension probe is suitable for mapping TCR tension and lateral transport (Scheme 1), we tethered the F1/2 = 4.7 pN probes onto SLBs presenting biotin and Ni-NTA (0.1% biotin-DPPE, 4% Ni-NTA DOGS and 95.9% DOPC). In a one-pot incubation, we decorated the tension probes with anti-CD3 antibody that binds and activates the TCR and also introduced His6-ICAM-1 on the lipid membrane to support T-cell spreading (Scheme S1 and method). Given that each gold nanoparticle presents ~38 biotinylated DNA hairpins, it is possible for each particle to bind multiple streptavidin molecules on the SLB, thus reducing the probe mobility. To minimize multivalent binding, we tuned the biotin doping level in the SLB (from 0.001% to 1% biotin) and measured the stoichiometry between the gold particles and streptavidin. We identified that a concentration of 0.1% biotin-DPPE provides the highest density and optimal coverage of probes while maintaining their long-range fluidity (Figure S6). The mobility of the tension probes and A488-labelled streptavidin on this surface was confirmed by fluorescence recovery after photobleaching (FRAP) experiments. Both the Cy3B and A488 fluorescence channels showed ~90% recovery within 90s (Figure 1F). The estimated lateral diffusion coefficients (D) of the fluorescent streptavidin and tension probes are almost identical (~1 μm2/s), confirming that the tension probes are primarily bound to streptavidin tethered on the lipid membrane (Figure 1G).
With the fluid AuNP-tension probes in hand, we next plated CD4+ T-cells directly onto the surface. T cells rapidly spread upon initial engagement to the SLB (within the first 3 min). In a representative cell shown in Figure 2A, time-lapse imaging showed the accumulation of fluorescence in both Cy3B and A488 channels underneath the cell (Figure 2A, t = 3 min, white circle). The tension density (ratiometric) signal showed a gradual increase in intensity that colocalized with a subset of the accumulating Cy3B signal. This indicates that TCRs transmit mechanical forces exceeding F1/2 = 4.7 pN to a fraction of the clustering anti-CD3 ligands. The T-cell continuously translocated anti-CD3 probes throughout the 15 min duration of the video. Starting at t = 4 min, centripetal movement of clusters was accompanied by waves of inward tension density signal (Figure 2A and supporting movie 1). A larger fraction of tension probes were unfolded at the periphery of the accumulating clusters (Supporting movie 1). Kymographs of different region of interests (ROIs) showed that tension gradually developed across the cell surface over the 15 min duration of the video, and TCR tension and clustering are closely linked in space and time (Figure 2B). Note that the tension density signal was most pronounced for larger clusters at the micron-scale and this signal was highly dynamic. Smaller oligomers or monomers may also experience mechanical strain that is not reported by our probes because it is below the 4.7 pN threshold for DNA unfolding or possibly due to the low signal-to-noise ratio associated with smaller assemblies. The lateral translocation of TCR-ligand complexes and their accumulation at sites of diffusion barriers strongly suggests that these complexes experience mechanical strain.24 Nonetheless, our results provide the first direct evidence that the TCR transmits pN mechanical strain to its ligand within a fluid intermembrane junction.
Figure 2.
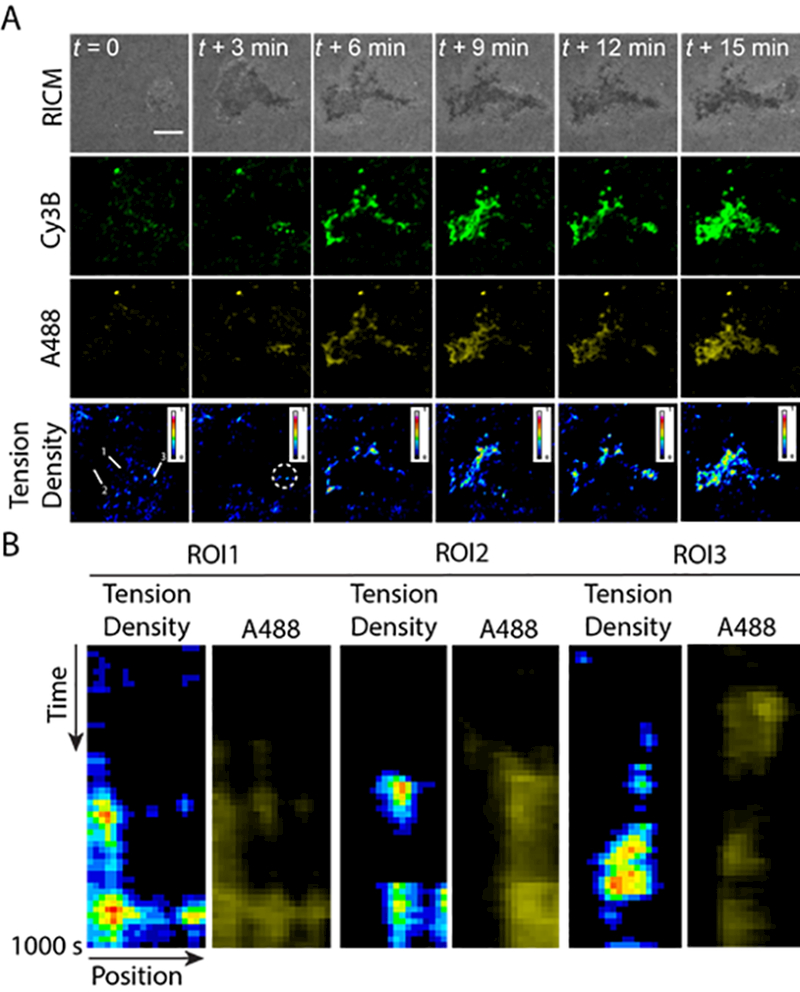
A) Representative time-lapse images (RICM, Cy3B and A488 and tension density) showing the first 15 min of CD4+ T-cell engagement with the CD3-tension probes anchored onto an SLB. B) The kymographs display tension density and the A488 intensity as a function of time within the three regions of interest (lines in the tension density channel from A). Scale bar = 5 μm.
We next aimed to investigate whether TCR-ligand complexes experience tension within the cSMAC, which forms at later points after formation of the immunological synapse and is associated with signal termination and receptor degradation.13,54 An important question pertains to the cSMAC structure is whether TCR-ligand complexes in this centralized assembly experience mechanical load during TCR recycling.55 To answer this question, T-cells were incubated with the tension probe surfaces for 30 min to allow for complete cSMAC formation. After 30 min of cell spreading, we observed Cy3B and A488 signal in a central region that is a hallmark feature of cSMAC formation (Figure 3A).56 The cSMAC displayed strong tension density signal with a maximum value of ~2, exceeding the values observed during initial TCR-ligand binding and clustering (Figure 3A). Also, the tension density within this structure was more homogeneous and less dynamic. Control experiments that used DNA duplexes showed the accumulation of anti-CD3 probes within a central cluster, but did not display significant tension density signal (Figure 3A). Scatter plot analysis revealed that an average tension density signal of 0.37±0.31 within the cSMAC while the control duplexes only had negligible signal (−0.02±0.08) (n = 25 cells for each group, Figure 3B). Immunostaining further confirmed that the Cy3B and A488 signals are strongly associated with TCR (Pearson correlation coefficients of ~0.8, Figure S7). Taken together these experiments demonstrate that the TCR-ligand complexes experience significant tension within the TCR recycling cSMAC structure.
Figure 3.
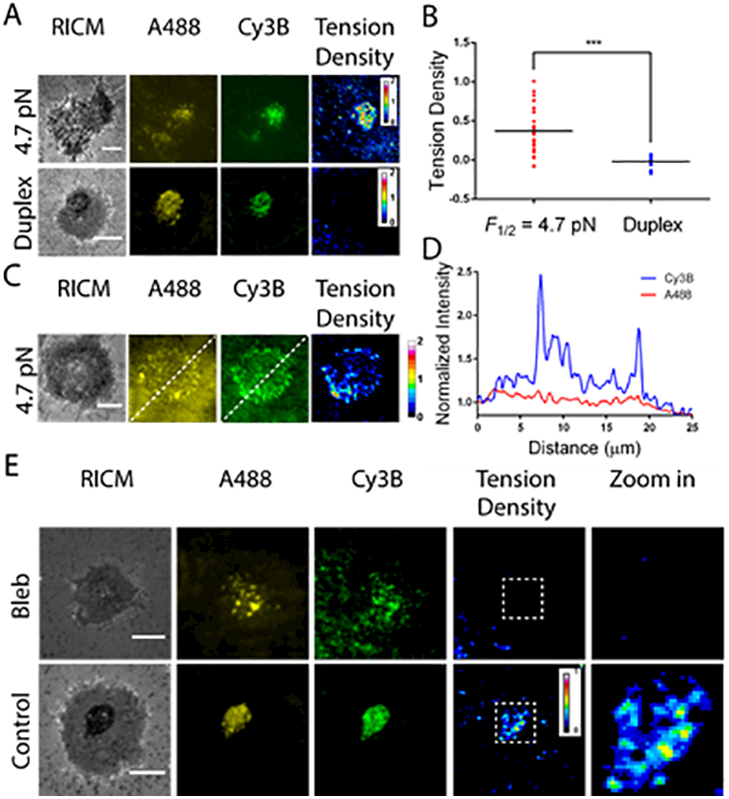
A) Representative images (RICM, Cy3B, A488 and tension density) of CD4+ T-cells plated on fluid SLBs containing tension probes (upper panel) or control duplexes (lower panel) for a duration of 30 min. B) Scatter plot showing the mean tension density signal generated on tension probes and control duplexes within the cSMAC structure (n = 25 cells). C) Representative images (RICM, Cy3B, A488 and tension density) of CD4+ T-cells plated on the hindered SLB displaying tension probes. The mobility of tension probes was limited due to the high density of streptavidin on the SLB. D) Line profile across the cell (dashed line in 3C) showing differential response in the Cy3B and A488 channels. E) Representative images (RICM, Cy3B, A488 tension density, and zoom in) of T-cells pre-treated with 50 μM blebbistatin (bleb) or without blebbistatin (control) and plated onto the 4.7 pN tension probe surface for a duration of 30 min. Scale bars = 5 μm.
To investigate the role of long-range lateral mobility of the ligand and how this influences TCR force transmission, we limited the mobility of SLBs by increasing streptavidin density (using 4% biotin-DPPE lipid composition). Lateral diffusion on these surfaces was significantly reduced as shown by FRAP measurements, in which the Cy3B signal from tension probes showed only ~40% recovery after 15 min (Figure S8). The reduced mobility could be due to local molecular crowding of the lipid bound streptavidin and the tension probes. T-cells plated on these surfaces showed reduced ligand translocation and accordingly, cSMAC formation was inhibited. Nonetheless, T cells plated on these surfaces showed that TCR-ligand complexes experienced tension across the cell junction and preferentially at the cell perimeter (Figure 3C and D). This spatial distribution of TCR tension is similar to that obtained using immobile tension probes.31 Line scan analysis across the cell-SLB contact revealed that Cy3B signal exceeded the A488 signal across the whole inter-membrane junction (Figure 3D). These results demonstrate that T-cells transmit pN forces to ligand-receptor complexes with highly hindered mobility.
Literature precedent revealed that pre-treatment of T-cells with blebbistatin not only retarded their ability to form the cSMAC,57,58 but also reduced IL-2 cytokine production.59 These observations identify myosin IIA as an essential component contributing to TCR transport and ultimately T-cell immune function. To investigate whether impairment of myosin IIA activity directly regulates TCR forces, we pretreated cells with 50 μM blebbistatin and plated these cells onto SLBs modified with tension probes for 30 min to allow for cell spreading. Under these conditions, T-cells formed limited clusters rather than the cSMAC (Figure 3E, A488 channel) and the tension density signal was dissipated (Figure 3E). This result shows that myosin IIA activity is required for mounting TCR tension during receptor clustering.
In summary, we report the general design of ratiometric tension probes for direct imaging of mechanical tension experienced by ligand-receptor complexes within intermembrane junctions. Our ratiometric tension probes showed pN tension experienced by TCRs undergoing clustering and translocation in a myosin IIA dependent fashion. We also revealed mechanical forces within the cSMAC, which is possibly associated with the endocytosis of TCRs for recycling. Our approach is broadly applicable for studying the interplay between force and receptor clustering for juxtacrine receptor signaling pathways such as those for B-cell receptors, Eph–ephrin, cadherins, and Notch–Delta.
Supplementary Material
ACKNOWLEDGMENTS
K.S. would like to thank for the financial support from the NIH (R01-GM097399), the Alfred P. Sloan Research Fellowship, the Camille-Dreyfus Teacher-Scholar Award, and the NSF CAREER Award (1350829). L.B. was supported by a postdoctoral fellowship from the National Multiple Sclerosis Society (F1963A1/1). B.E. was supported by NIH Grants R01-AI096879.
Footnotes
Supporting Information
Supporting Information contains DNA sequences used in this study, supporting schemes for fabrication of surfaces and supplementary data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- (1).Heldin C-H Cell 1995, 80, 213–223. [DOI] [PubMed] [Google Scholar]
- (2).Schlessinger J Cell 2002, 110, 669–672. [DOI] [PubMed] [Google Scholar]
- (3).Lemmon MA; Schlessinger J Cell 2010, 141, 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Grakoui A; Bromley SK; Sumen C; Davis MM; Shaw AS; Allen PM; Dustin ML Science 1999, 285, 221–227. [PubMed] [Google Scholar]
- (5).Bunnell SC; Hong DI; Kardon JR; Yamazaki T; McGlade CJ; Barr VA; Samelson LE J. Cell Biol 2002, 158, 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Yokosuka T; Sakata-Sogawa K; Kobayashi W; Hiroshima M; Hashimoto-Tane A; Tokunaga M; Dustin ML; Saito T Nat. Immunol 2005, 6, 1253–1262. [DOI] [PubMed] [Google Scholar]
- (7).Wu H Cell 2013, 153, 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Manz BN; Groves JT Nat. Rev. Mol. Cell Biol 2010, 11, 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wang N; Suo Z Biochem. Biophys. Res. Commun. 2005, 328, 1133–1138. [DOI] [PubMed] [Google Scholar]
- (10).van der Merwe PA; Dushek O Nat. Rev. Immunol 2011, 11, 47–55. [DOI] [PubMed] [Google Scholar]
- (11).Lanzavecchia A Nature 1985, 314, 537–539. [DOI] [PubMed] [Google Scholar]
- (12).Friedl P; Bröcker E-B Immunol. Rev. 2002, 186, 83–89. [DOI] [PubMed] [Google Scholar]
- (13).Varma R; Campi G; Yokosuka T; Saito T; Dustin ML Immunity 2006, 25, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Yu Y; Smoligovets AA; Groves JT J. Cell Sci 2013, 126, 1049–1058. [DOI] [PubMed] [Google Scholar]
- (15).Ritter AT; Angus KL; Griffiths GM Immunol. Rev 2013, 256, 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ma Z; Finkel TH Trends Immunol. 2010, 31, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Chen W; Zhu C Immunol. Rev 2013, 256, 160–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kim ST; Takeuchi K; Sun Z-YJ; Touma M; Castro CE; Fahmy A; Lang MJ; Wagner G; Reinherz EL J. Biol. Chem 2009, 284, 31028–31037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Li Y-C; Chen B-M; Wu P-C; Cheng T-L; Kao L-S; Tao M-H; Lieber A; Roffler SR J. Immunol 2010, 184, 5959–5963. [DOI] [PubMed] [Google Scholar]
- (20).Liu Z; Liu Y; Chang Y; Seyf HR; Henry A; Mattheyses AL; Yehl K; Zhang Y; Huang Z; Salaita K Nat. Meth 2015, 13, 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Das DK; Feng Y; Mallis RJ; Li X; Keskin DB; Hussey RE; Brady SK; Wang J-H; Wagner G; Reinherz EL; Lang MJ Proc. Natl. Acad. Sci. USA 2015, 112, 1517–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Liu B; Chen W; Evavold, Brian D.; Zhu, C. Cell 2014, 157, 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Bashour KT; Gondarenko A; Chen H; Shen K; Liu X; Huse M; Hone JC; Kam LC Proc. Natl. Acad. Sci. USA 2014, 111, 2241–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Mossman KD; Campi G; Groves JT; Dustin ML Science 2005, 310, 1191–1193. [DOI] [PubMed] [Google Scholar]
- (25).Stabley DR; Jurchenko C; Marshall SS; Salaita KS Nat. Meth 2012, 9, 64–67. [DOI] [PubMed] [Google Scholar]
- (26).Jurchenko C; Salaita KS Mol. Cell. Biol 2015, 35, 2570–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Galior K; Liu Y; Yehl K; Vivek S; Salaita K Nano Lett. 2016, 16, 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Zhang Y; Ge C; Zhu C; Salaita K Nat. Commun 2014, 5, 5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Blakely BL; Dumelin CE; Trappmann B; McGregor LM; Choi CK; Anthony PC; Duesterberg VK; Baker BM; Block SM; Liu DR; Chen CS Nat. Meth 2014, 11, 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Liu Y; Yehl K; Narui Y; Salaita KJ Am. Chem. Soc 2013, 135, 5320–5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Morimatsu M; Mekhdjian AH; Adhikari AS; Dunn AR Nano Lett. 2013, 13, 3985–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Liu Y; Medda R; Liu Z; Galior K; Yehl K; Spatz JP; Cavalcanti-Adam EA; Salaita K Nano Lett. 2014, 14, 5539–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ma VP-Y; Liu Y; Yehl K; Galior K; Zhang Y; Salaita K Angew. Chem. Int. Ed 2016, 55, 5488–5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Liu Y; Blanchfield L; Ma VP-Y; Andargachew R; Galior K; Liu Z; Evavold BD; Salaita K Proc. Natl. Acad. Sci. USA 2016, 113, 5610–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Katsumi A; Orr AW; Tzima E; Schwartz MA J. Biol. Chem 2004, 279, 12001–12004. [DOI] [PubMed] [Google Scholar]
- (36).Cremer PS; Boxer SG J. Phys. Chem. B 1999, 103, 2554–2559. [Google Scholar]
- (37).Castellana ET; Cremer PS Surf. Sci. Rep 2006, 61, 429–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Kaizuka Y; Douglass AD; Varma R; Dustin ML; Vale RD Proc. Natl. Acad. Sci. USA 2007, 104, 20296–20301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Hartman NC; Nye JA; Groves JT Proc. Natl. Acad. Sci. USA 2009, 106, 12729–12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Manz BN; Jackson BL; Petit RS; Dustin ML; Groves J Proc. Natl. Acad. Sci. USA 2011, 108, 9089–9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).O’Donoghue GP; Pielak RM; Smoligovets AA; Lin JJ; Groves JT eLife 2013, 2, e00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Caculitan NG; Kai H; Liu EY; Fay N; Yu Y; Lohmüller T; O’Donoghue GP; Groves JT Nano Lett. 2014, 14, 2293–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Biswas KH; Hartman KL; Yu C.-h.; Harrison OJ; Song H; Smith AW; Huang WYC; Lin W-C; Guo Z; Padmanabhan A; Troyanovsky SM; Dustin ML; Shapiro L; Honig B; Zaidel-Bar R; Groves JT Proc. Natl. Acad. Sci. USA 2015, 112, 10932–10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Salaita K; Nair PM; Petit RS; Neve RM; Das D; Gray JW; Groves JT Science 2010, 327, 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Greene Adrienne C.; Lord Samuel J.; Tian A; Rhodes C; Kai H; Groves Jay T. Biophys. J 2014, 106, 2196–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Narui Y; Salaita K Biophys. J 2013, 105, 2655–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Yu C.-h.; Law JBK; Suryana M; Low HY; Sheetz MP Proc. Natl. Acad. Sci. USA 2011, 108, 20585–20590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Yu C.-h.; Rafiq Nisha Bte M.; Krishnasamy A; Hartman Kevin L.; Jones Gareth E.; Bershadsky Alexander D.; Sheetz Michael P. Cell Reports 2013, 5, 1456–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Yu C.-h.; Rafiq NBM; Cao F; Zhou Y; Krishnasamy A; Biswas KH; Ravasio A; Chen Z; Wang Y-H; Kawauchi K; Jones GE; Sheetz MP Nat. Commun 2015, 6, 8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Woodside MT; Behnke-Parks WM; Larizadeh K; Travers K; Herschlag D; Block SM Proc. Natl. Acad. Sci. USA 2006, 103, 6190–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Ha T; Enderle T; Ogletree DF; Chemla DS; Selvin PR; Weiss S Proc. Natl. Acad. Sci. USA 1996, 93, 6264–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Yun CS; Javier A; Jennings T; Fisher M; Hira S; Peterson S; Hopkins B; Reich NO; Strouse GF J. Am. Chem. Soc 2005, 127, 3115–3119. [DOI] [PubMed] [Google Scholar]
- (53).Bendix PM; Pedersen MS; Stamou D Proc. Natl. Acad. Sci. USA 2009, 106, 12341–12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Vardhana S; Choudhuri K; Varma R; Dustin ML Immunity 2010, 32, 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Martínez-Martín N; Fernández-Arenas E; Cemerski S; Delgado P; Turner M; Heuser J; Irvine Darrell J.; Huang B; Bustelo Xosé R.; Shaw A; Alarcón B Immunity 2011, 35, 208–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Dustin ML; Cooper JA Nat. Immunol 2000, 1, 23–29. [DOI] [PubMed] [Google Scholar]
- (57).Ilani T; Vasiliver-Shamis G; Vardhana S; Bretscher A; Dustin ML Nat. Immunol 2009, 10, 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Yu Y; Fay NC; Smoligovets AA; Wu H-J; Groves JT PLoS ONE 2012, 7, e30704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Judokusumo E; Tabdanov E; Kumari S; Dustin Michael L.; Kam Lance C. Biophys. J 2012, 102, L5–L7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.