 Ambient fine particulate matter (PM2.5) is a complex mixture associated with lung cancer risk.
Ambient fine particulate matter (PM2.5) is a complex mixture associated with lung cancer risk.
Abstract
Ambient fine particulate matter (PM2.5) is a complex mixture associated with lung cancer risk. PM2.5-bound nitro-polycyclic aromatic hydrocarbons (NPAHs) have been demonstrated to possess mutagenicity and carcinogenicity. Previous studies showed that PM2.5 induced DNA damage, whereas there is little knowledge of whether 9-nitroanthracene (9-NA), a typical compound of NPAHs in PM2.5, causes DNA damage. Also, the regulating mechanisms of PM2.5 and 9-NA in DNA damage and repair are not yet fully established. Here we sought to investigate the molecular mechanisms of DNA damage and repair in the lungs of male Wistar rats exposed to PM2.5 (1.5 mg per kg body weight) or three different dosages of 9-NA. And then DNA strand breaks, 8-OH-dG formation, DNA–protein crosslink and DNA repair gene expressions in rat lungs were analyzed. In addition, alteration in oxidative stress factors and metabolic enzymes were detected. The results showed that (1) PM2.5 and higher dosage 9-NA (4.0 × 10–5 and 1.2 × 10–4 mg per kg body weight) significantly caused lung DNA damage, accompanied by increasing OGG1 expression while inhibiting MTH1 and XRCC1 expression, elevating the levels of GADD153, hemeoxygenase-1 and malondialdehyde, and promoting the activities of CYP450 isozymes and glutathione S-transferase. (2) 1.3 × 10–5 mg kg–1 9-NA exposure couldn't cause DNA damage and oxidative stress. (3) At the approximately equivalent dose level, PM2.5-induced DNA damage effects were more obvious than 9-NA with positive correlation. It suggests that DNA damage caused by PM2.5 and 9-NA may be mediated partially through influencing the DNA repair capacity and enhancing oxidative stress and biotransformation, and this negative effect of 9-NA might be related to the PM2.5-induced lung genotoxicity.
Introduction
Ambient fine particulate matter (PM2.5) is an important air pollutant worldwide. It has been featured with multiple components [such as metals, water-soluble ions, polycyclic aromatic hydrocarbons (PAHs), organic carbons, etc.] and diversified sources (coal combustion, vehicle exhaust, biomass burning, ground dust, etc.). Different exposure levels of PM2.5 at various times and spaces have different degrees of toxicities. Recent studies have revealed that PM2.5 can aggravate the morbidity/mortality of pulmonary diseases and the risk of lung cancer in urban areas.1,2 Due to the complexity of PM2.5, scientists focused on not only the toxicity of PM2.5 itself, but also the toxicity contributions from the PM2.5 associated chemical components, especially when evaluating the respiratory health effects induced by ambient PM2.5.
PAHs and their nitro-derivatives, which are considered as classic persistent organic pollutants, are noticeable components of urban air PM2.5. PAHs such as benzo(a)pyrene and benz(a)anthracene are mutagenic and carcinogenic environmental pollutants,3 and they could increase the lung cancer risk in the Chinese population.4 PM2.5-bound nitro-polycyclic aromatic hydrocarbons (NPAHs) are mainly released from the incomplete combustion of fossil fuels and the photochemical reactions between PAHs and nitrogen dioxide. Some NPAHs like 1-nitropyrene and 1,6-dinitropyrene are particularly of great concern because they possess much higher direct-acting mutagenicity and carcinogenicity than the parent PAHs,5,6 although the exposure concentrations of NPAHs in the air environment are far lower than those of PAHs.
9-Nitroanthracene (9-NA) in the particulate phase is widespread in coal combustion and biomass burning processes.7,8 It was reported that the mass concentrations of 9-NA in all the PM2.5-bound NPAHs were the highest in East Asia including China, Russia, Korea and Japan.8–10 Although 9-NA is currently classified by the International Association for Research and Cancer (IARC) as “not classifiable as to its carcinogenicity to humans” (“Group 3”),3 it had been found that 9-NA was mutagenic in tester strains TA98 and TA100 minus S9, while it exhibited potent mutagenic activity in the L5178Y mammalian cell mutagenicity assay in the presence of S9.11 Also, PM2.5 and PAHs positively correlated with the increased incidence of lung cancer on the basis of epidemiological data.2,4 However, the cancer risks to humans and toxicological effects because of their exposure to 9-NA have not been fully investigated and assessed so far.
DNA damage is considered to be a key triggering mechanism of lung cancer.12 It mainly includes DNA base damage, DNA strand breaks, DNA oxidative damage, DNA-adducts, DNA–protein crosslinking (DPC), etc. Many human, animal and cell studies showed that PM2.5 induced DNA strand breaks13–15 and that PM2.5 exposure elevated the levels of 8-hydroxy-2′-deoxyguanosine (8-OH-dG), a biomarker of oxidative DNA damage, in the urine of workers in Boston of USA.16 Additionally, PM2.5 extracts including multiple NPAHs could cause DNA strand breaks in human A549 cells5 and NPAHs induced urinary oxidative DNA damage.17 As far as we know, whether 9-NA causes DNA damage remains unclear. Accordingly, DNA strand breaks, 8-OH-dG levels, and DPC coefficients were detected in the lungs of rats exposed to PM2.5 and 9-NA.
From another perspective, it is a remarkable fact that multiple DNA damage repair genes such as nucleotide excision repair (NER) gene, 8-oxoguanine DNA glycosylase (OGG1), MutT Homolog 1 (MTH1), and X-ray repair cross-complementing group 1 (XRCC1) are involved in mammalian nucleotide excision repair and play important roles in DNA repair processes.18–20 It was reported that PM2.5 greatly inhibited NER, in turn suppressing DNA repair and enhancing DNA replication errors,21 but the regulating mechanisms of PM2.5 and 9-NA in DNA damage and repair are not yet fully established. Therefore, in this study, we focused on the DNA damage effects and DNA repair gene expression in rat lungs induced by PM2.5 and 9-NA.
Previous research studies have reported that growth arrest- and DNA damage-inducible gene 153 (GADD153) can be highly promoted when environmental chemicals initiate DNA damage or oxidative stress, while heme oxygenase 1 (HO-1), superoxide dismutase (SOD) and malonaldehyde (MDA) may be clearly induced when oxidative stress (including lipid peroxidation) occurs in the cells under the oxide stimulus.22–24 In the present study, GADD153, HO-1, SOD and MDA were used as the inducible factors of oxidative stress to explore the oxidative stress in rat lungs induced by PM2.5 and 9-NA. Besides, the biotransformation process and the metabolic enzymes (phase I and phase II enzymes) play key roles in the NPAH metabolic process.25,26 Thus, the changes of cytochrome P450 (CYP450) isoforms CYP1A1 and 1A2 as well as glutathione S-transferase (GST) in rat lungs were investigated to indicate the lung biotransformation characteristic of PM2.5 and 9-NA.
Exogenous stimuli may increase reactive oxygen species (ROS) levels or weaken the anti-oxidative system, resulting in oxidative stress, which would induce oxidative damage to DNA, lipids and proteins, further destroying the normal metabolism and physiological functions.27 A previous study indicated that the genotoxicity of environmental pollutants was linked to oxidative stress and DNA damage caused by them.28 What we focus on is that oxidative stress may induce DNA damage,29 and DNA repair genes play important roles in DNA damage repair processes. If normal DNA repair processes fail, DNA damage may occur.30 Also, the genotoxicity is related to the upregulation of metabolic enzymes and detoxifying enzymes,31 in which some metabolically active intermediates may mediate oxidative damage and reactions with DNA to a great extent.32 The lung is the major target organ of environmental pollutants, and the lung damage and genotoxicity induced by chemicals are comprehensive and complex effects, in which oxidative stress, metabolic disturbance and DNA damage are interrelated. The logical relationship among these indicators may be speculated and described as follows: oxidative stress-induced DNA damage in lungs appears to be an important mechanism of the genotoxicity induced by PM2.5, while metabolic enzymes may affect oxidative stress and be associated with genotoxicity. Our data will clarify the toxicological roles in DNA damage and repair, oxidative stress and metabolic activation induced by PM2.5 and 9-NA in depth and will provide new insight into the evaluation of the genotoxicity of PM2.5-bound 9-NA exposure.
Experimental
PM2.5 sample preparation
PM2.5 samples were collected on March 2014 in Taiyuan, China. The protocols of the PM2.5 sample pre-process and the preparation of PM2.5 normal saline suspensions have been described in our previous reports.10,33 Briefly, PM2.5 samples were collected on quartz fiber filters (QFFs). PM2.5 samples and blank QFFs were cut into small pieces and submerged in Milli-Q water with sonication. The suspensions of PM2.5 and blank filter were dried under freeze vacuum and made into powder, and then they were instilled with physiological saline under sonication prior to the treatment. In the preliminary experiment, the blank filter suspension did not induce pathological alterations nor affect the levels of 8-OH-dG and DPC in the lungs of the rats, and no statistical difference was observed between the normal control and blank filter groups. Accordingly, the physiological saline group was used as the normal control group in this study.
During sampling, the 24-h mean mass concentration of PM2.5 was 80.5 μg m–3. The concentrations ranged from 39 to 121 μg m–3, in which the concentrations of 66.7% samples were higher than the China National Ambient Quality Standard for PM2.5 (75 μg m–3). The mass concentrations of Σ-nitropyrene, Σ-nitrofluorene, Σ-nitrochrysene and Σ-nitroanthracene on the collected PM2.5 samples were detected at levels of 0.38 to 3.04 ng m–3, 0.21 to 0.43 ng m–3, 0.19 to 2.38 ng m–3, and 9.55 to 16.52 ng m–3, respectively.10 Also, the exposure levels of heavy metals and water-soluble constituents during sampling were detected as follows: the contents of As, Cd, Cu, Ni, Pb and Zn were 11.7, 2.2, 23.4, 10.9, 98.9 and 321.4 ng m–3, while the contents of F–, Cl–, NO3–, SO4–, Na+, NH4+, K+, Mg2+, and Ca2+ were 0.13, 0.25, 13.55, 15.56, 0.62, 8.76, 0.89, 0.16 and 2.40 μg m–3, respectively.
Animal and treatment protocols
We selected male Wistar rats (body weight: 180–200 g) as the experimental animals, which were purchased from the Animal Center of Hebei Medical University (Shijiazhuang, China) and bred in the Animal House in the Institute of Environmental Science of Shanxi University (Taiyuan, China) under the standard conditions (24 °C ± 2 °C and 50% ± 5% humidity). The rats were randomly organized into five parallel groups with five animals for each group: (1) the dimethylsulfoxide (DMSO) control (5% DMSO), (2) 1.5 mg per kg body weight (b.w.) PM2.5 group, and (3)–(5) 1.3 × 10–5, 4.0 × 10–5, and 1.2 × 10–4 mg per kg b.w. 9-NA in DMSO. The rats were administered using 5% DMSO, PM2.5 suspensions and 9-NA solutions by intratracheal instillation respectively for one/two days enduring for 10 days. The animals were maintained in accordance with the guidelines of the Ministry of Health People's Republic of China, Beijing, China, and approved by the institutional ethical committee (IEC) of Shanxi University with permission no. IEC 201510010.
In this study, the mass concentrations of Σ-nitroanthracene on the collected PM2.5 samples were detected at levels of 9.55 to 16.52 ng m–3.10 Based on these data, the concentration of 16.52 ng m–3 was used to estimate the 9-NA instillation dosage for each rat every 2 days as 4.0 × 10–5 mg per kg b.w. by taking the respiratory volume limit of an adult rat (200 mL min–1) into account. Moreover, according to the orange alert criterion of haze PM2.5 in China (500 μg m–3), the PM2.5 instillation dosage of 1.5 mg per kg b.w. was calculated in this study, which was in agreement with the PM2.5 dosage in the previous study in our lab.33 Meanwhile, given that the approximate proportion of the mass concentration of NPAHs in wintertime PM2.5 ranged from 9 × 10–6 to 1 × 10–4,34 1.3 × 10–5, 4.0 × 10–5 and 1.2 × 10–4 mg per kg b.w. of 9-NA dosages were comparable to the dosage of 1.5 mg per kg b.w. PM2.5 when the proportions of 9 × 10–6, 2.7 × 10–5 and 8.1 × 10–5 9-NA in PM2.5 were chosen in this study for the requirement of both dose–response relationship experiment design and 9-NA toxicity evaluation.
After the last treatment, the rats in different groups were euthanized and sacrificed. Then a piece of fresh lung tissue per rat was minced and ground for the comet assay, and another piece was fixed in 4% paraformaldehyde in PBS for hematoxylin and eosin (HE) staining analysis. Besides, partial lung tissue was obtained and homogenized for ELISA and biochemical analysis, and the rest was quickly frozen in liquid nitrogen and then stored at –80 °C for mRNA and protein measurement.
Comet assay
The alkaline comet assay was performed as follows. (1) Single cell suspensions were prepared in ice-cold phosphate-buffered saline (PBS) from a piece of lung (see Animal and treatment protocols), (2) “Sandwich gel” was prepared. The first layer was 1% normal melting-point agarose (NMA) in a slide; the second layer was a mixture of cell/0.65% molten low melting-point agarose (LMA), and the third layer was 0.65% LMA. (3) The slides containing the “sandwich gel” were transferred to cold lysis solution (2.5 mM NaCl, 100 mM EDTA, 1% sodium sarcosinate and 10 mM Tris, pH 10.0, to which 1% Triton X-100 and 10% DMSO were freshly added) for 60 min at 4 °C to cause denaturation. (4) The slides were then subjected to electrophoresis with cold electrophoresis buffer (300 mM NaOH, 1 mM EDTA, pH 13.0) at 25 V for 30 min at 4 °C, and immersed in Tris buffer (0.4 M Tris, pH 7.5) to neutralize the excess alkali. Subsequently the slides were air dried. (5) DNA was stained with 100 μL 4S Red Plus (Shengon, Shanghai, China, 1 : 10 000) for 20 min and immediately rinsed with Milli-Q water and air dried. (6) The slides were examined at 400× magnifications using an Olympus BX51 fluorescence microscope (Olympus, Tokyo, Japan). 20–30 randomly acquired images of microscopic fields relative to each sample were recorded to enable the analysis of 100–150 cells. (7) DNA damage indexes including comet tail DNA %, tail length, and olive tail moment (OTM) were assessed by a Comet Assay Software Project (CASP, CASP, version 1.2.3 beta1).
Real time quantitative RT-PCR
Lung tissues (see Animal and treatment protocols) were used for mRNA extraction and quantitative RT-PCR analysis according to our previous methods.33 The iCycler iQ Real Time PCR Detection System (Bio-Rad, Richmond, CA, USA) with the Quantitect SYBRGreen I PCR kit was employed for conducting real-time PCR. All the GenBank accession numbers, the sequences and the annealing temperatures of the primers are listed in Table 1. The relative quantification of the expression of the target genes was performed using the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA as an internal control. Mean expression in each treated group is shown as changed fold compared to the mean expression in the control group which has been assigned an arbitrary value of 1.
Table 1. Primer information used in real-time RT-PCR.
| Genes | Accession no. | Sequence |
|
| MTH1 | NM_057120 | F | 5′-AGTGAAGAAATGCGCCCTCA-3′ |
| Products | 148 bp, 60 °C | R | 5′-TGAGGATGGTGTCCTGACCA-3′ |
| GADD153 | NM_001109986 | F | 5′-GTCACAAGCACCTCCCAAAG-3′ |
| Products | 110 bp, 60 °C | R | 5′-CCACTCTGTTTCCGTTTCCT-3′ |
| XRCC1 | NM_053435 | F | 5′-GATGGGGAACAGTCAGAAGGAC-3′ |
| Products | 195 bp, 60 °C | R | 5′-AATTGGCAGGTCAGCCTCTG-3′ |
| OGG1 | NM_030870 | F | 5′-CAACATTGCTCGCATCACTGG-3′ |
| Products | 195 bp, 60 °C | R | 5′-ATGGCTTTAGCACTGGCACATACA-3′ |
| HO-1 | BC091164 | F | 5′-GTCAAGCACAGGGTGACAGA-3′ |
| Products | 77 bp, 58 °C | R | 5′-ATCACCTGCAGCTCCTCAAA-3′ |
| CYP1A1 | NM_012540 | F | 5′-TAACTCTTCCCTGGATGCCTTCAA-3′ |
| Products | 109 bp, 56 °C | R | 5′-GTCCCGGATGTGGCCCTTCTCAAA-3′ |
| CYP1A2 | NM_012541 | F | 5′-ACCCTGAGTGAGAAGGTGAT-3′ |
| Products | 99 bp, 56 °C | R | 5′-GAGGATGGCTAAGAAGAGGA-3′ |
| GAPDH | NM_017008 | F | 5′-ATGTATCCGTTGTGGATCTGAC-3′ |
| Products | 78 bp, 56 °C | R | 5′-CCTGCTTCACCACCTTCTTG-3′ |
Western blotting
Total proteins of lung tissues from different groups were extracted by a protein extraction kit (Beyotime, Shanghai, China) according to the manufacturer's instructions. Protein samples were mixed with loading buffer and denatured for 5 min at 95 °C. Western blot analysis of MTH1, GADD153, XRCC1 and actin was performed as described previously.33 The rabbit polyclonal primary antibodies were used as the second anti-body for the detection of MTH1 (Sc-67291, dilution ratio: 1 : 100), GADD153 (Sc-575, dilution ratio: 1 : 100), XRCC1 (Sc-11429, dilution ratio: 1 : 100) (Santa Cruz, CA, USA) and actin (AB10024, dilution ratio: 1 : 3000; Sangon, Shanghai, China) at 4 °C overnight, whereas the infrared-labeled goat anti-rabbit secondary antibody (AlexaFlor 680 goat anti-rabbit IgG (H+L), USA) was adopted with a concentration of 1 : 20 000 at room temperature for 1.5 h. The western blot results were quantified and recorded by using an Odyssey Infrared Imaging System (Li-COR Biosciences, USA).
ELISA assay
The measured lung tissues were homogenized in ice-cold 0.9% physiological saline and then centrifuged for 10 min at 3000 rpm (4 °C). The supernatants were carefully collected for the late analysis. The levels of CYP1A1, CYP1A2, OGG1, HO-1 and 8-OH-dG in the lung homogenates were measured using a rat ELISA kit (R&D Company Ltd, USA), and the level of CYP450s was detected using the rat ELISA kit from the Beijing Fangcheng Biochemistry, China, according to the manufacturer's instructions.
Measurement of SOD, MDA, GST and DPC
The biological activities of SOD, MDA and GST in the lung supernatants were measured using the corresponding kits (Jiancheng Biochemistry, Nanjing, China) according to the manufacturer's protocols. Lung supernatant's DPC levels were detected totally as described previously.35
Statistical analysis
Data were analysed with one-way ANOVA using the SPSS19.0. The statistical analysis of difference between the groups was determined by post hoc tests and Fisher's least significant difference (LSD) test. A level of P < 0.05 was accepted as statistically significant. The correlations of lung damage effects between the PM2.5 group and the 9-NA group were evaluated by using correlation analysis. A positive correlation is indicated by a correlation coefficient (r) >0.8.
Results and discussion
Histopathology and DNA damage induced by PM2.5 and 9-NA in rat lungs
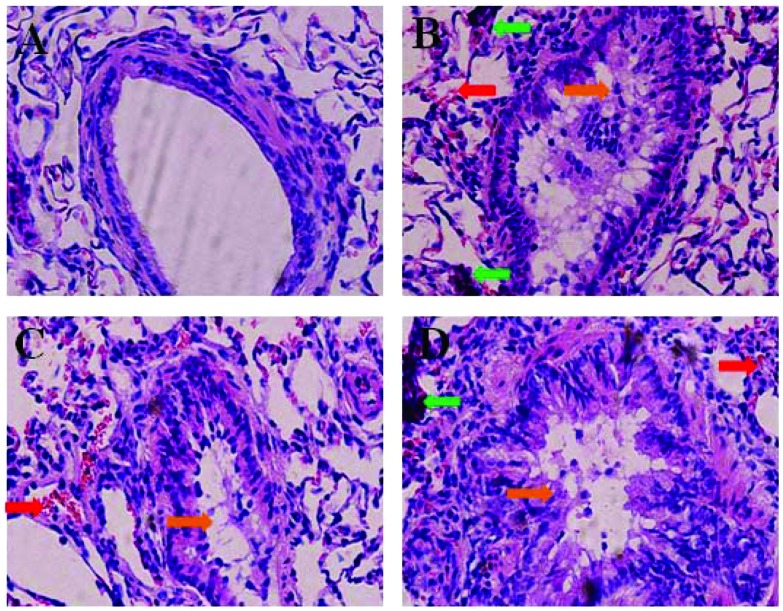
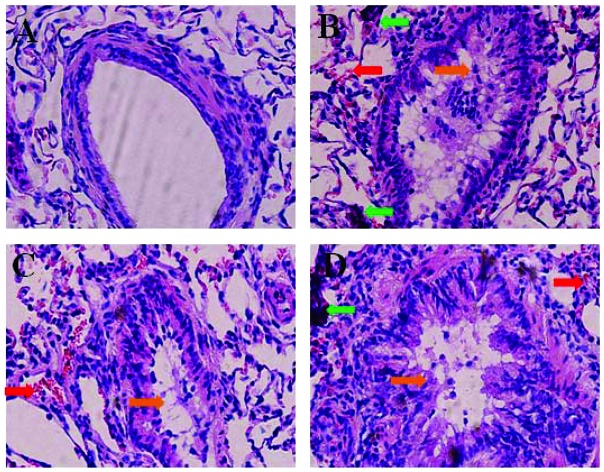
The representative HE staining images are shown in Fig. 1. No histopathological abnormalities were observed in the control (Fig. 1A) and 1.3 × 10–5 mg kg–1 9-NA group animals (data not shown). In the 1.5 mg per kg b.w. PM2.5 group, hyperemia, inflammatory cells and bronchial epithelial hyperplasia were observed in the lungs (Fig. 1B). In the 4.0 × 10–5 and 1.2 × 10–4 mg per kg b.w. 9-NA groups, different degrees of hyperemia, inflammatory cell infiltration, diminished alveolar spaces, and bronchial epithelial hyperplasia existed in the lungs (Fig. 1C and D), in which pathological changes were more serious in the 1.2 × 10–4 mg per kg b.w. 9-NA group.
Fig. 1. HE staining results in the lungs of rats from a physiological saline control (A), 1.5 mg per kg b.w. PM2.5 (B), 4.0 × 10–5 mg per kg b.w. 9-NA (C) and 1.2 × 10–4 mg per kg b.w. 9-NA (D) groups, ×400 magnification. The red, green and orange arrows indicate the sites of hyperemia, inflammatory cell infiltration and bronchial epithelial hyperplasia, respectively.
PM2.5 contains many inorganic and organic components, many of which can adversely influence human respiratory health. PM2.5 and PM2.5-bound PAHs/NPAHs are of great concern because of their toxicities (carcinogenicity or mutagenicity) to humans.1–4,36 9-NA, as a typical PM2.5-bound NPAH, was proven to have some mutagenicity, but few epidemiological or toxicological studies are available to evaluate the carcinogenicity of 9-NA to humans or experimental animals until now.
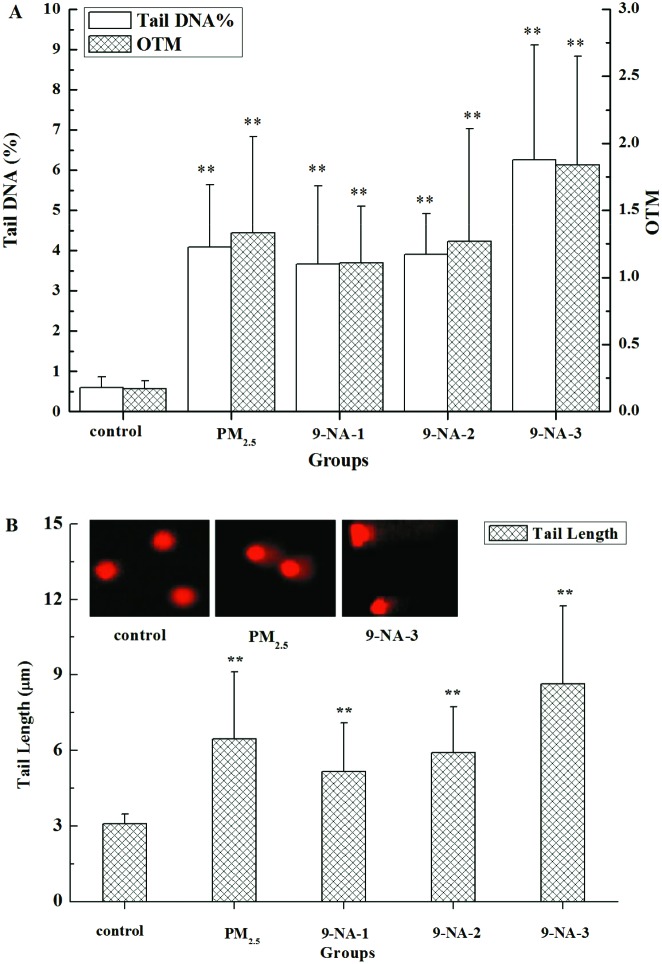
In Fig. 2A and B, PM2.5 and 9-NA at all doses tested significantly increased the values of three DNA damage markers (tail DNA %, OTM and tail length values) in lung cells compared with the control (P < 0.01). 9-NA at all doses tested caused significant increases in tail DNA %, OTM and tail length values of lung cells in a dose-dependent manner (r > 0.99).
Fig. 2. DNA damage from the comet assay results obtained from the lungs of rats of different groups; changes of tail DNA %, OTM (A) and tail length (B). The values are mean ± SD from three individual samples. Using one-way ANOVA, comparing with the control group, a significant difference is indicated by **P < 0.01. Control: 5% DMSO; PM2.5: 1.5 mg per kg b.w. PM2.5 suspension; 9-NA-1: 1.3 × 10–5 mg per kg b.w. 9-NA solution; 9-NA-2: 4.0 × 10–5 mg per kg b.w. 9-NA solution; and 9-NA-3: 1.2 × 10–4 mg per kg b.w. 9-NA solution, the same as the following figures and tables.
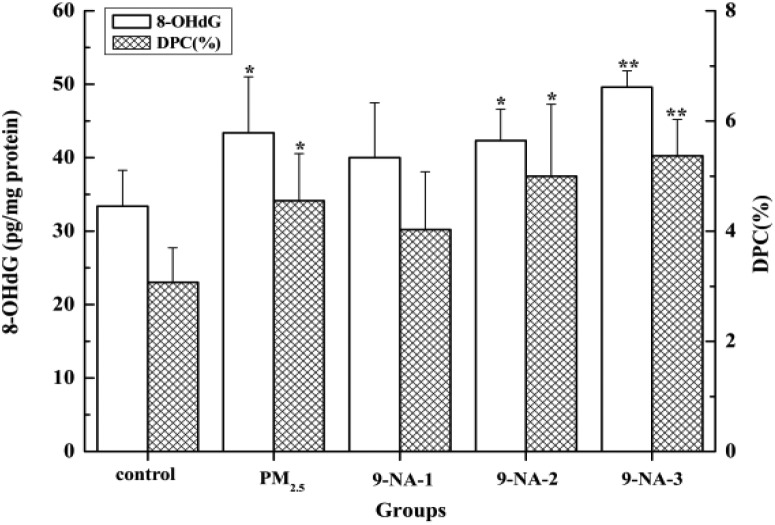
Fig. 3 displays that PM2.5 and 9-NA at higher doses (4.0 × 10–5 and 1.2 × 10–4 mg per kg b.w., respectively) significantly raised the 8-OH-dG levels in the lungs compared with the control (P < 0.05 or P < 0.01), and such increases induced by 9-NA had a obvious positive concentration–effect relationship (r = 0.94). Also, the influences of PM2.5 and 9-NA on the DPC formation in the lungs are shown in Fig. 3. The DPC background level was 3.07% in the control group, and PM2.5 significantly caused an increase in DPC formation with 4.55% (P < 0.05). For 9-NA, no significant difference was observed in the presence of 9-NA at the dosage of 1.3 × 10–5 mg kg–1. After 4.0 × 10–5 and 1.2 × 10–4 mg per kg b.w. of 9-NA exposure, DPC average levels were statistically increased to 5.00% (P < 0.05) and 5.37% (P < 0.01), respectively. 9-NA caused an increase in DPC formation with a concentration-dependent property (r = 0.85).
Fig. 3. Levels of 8-OH-dG and DPC in the lungs of rats of different groups. The values are mean ± SD from five individual samples. The values are mean ± SD from five individual samples. Using one-way ANOVA, comparing with the control group, a significant difference is indicated by*P < 0.05 and **P < 0.01.
Fig. 2 and 3 also display the differences in some markers of DNA damage induced by PM2.5 (1.5 mg per kg b.w.) and 9-NA (4.0 × 10–5 mg per kg b.w.). On the basis of the results, 4.0 × 10–5 mg kg–1 9-NA induced adverse effects were relatively less than PM2.5. The great increases of tail DNA%, tail length, OTM, 8-OHdG and DPC levels between PM2.5 and 9-NA exposure have positive correlations, and the values of r were 0.80, 0.81, 0.82, 0.96 and 0.97, respectively. We hypothesize that 9-NA in PM2.5 might be related to PM2.5-induced lung DNA damage.
A report about DNA breakage induced by crude extract and NPAH fractionated extracts of PM2.5 in BEAS-2B cells demonstrated that OTM and micronucleus (MN) formation were significantly induced by the crude extract and NPAH fractionated extracts at the same dosage (50 μg cm–2) compared to the control,37 indicating that the NPAH fractionated extracts were the biologically active fractions of PM2.5 responsible for the genotoxic effects. Besides, the effects of OTM and MN of the NPAH fractionated extracts in BEAS-2B cells were lower than those of the crude extract of PM2.5. In our study, also, we found that the DNA damage responses (DNA tail length, OTM and DPC) in lungs induced by PM2.5 were higher than those of 9-NA, which may indicate that the toxicity of PM2.5 might be more than that of 9-NA under the present experimental conditions. This result was in agreement with the above viewpoint.37 It may be because PM2.5 probably contains many of the complicated components compared to single 9-NA. Further studies on the precise mechanism(s) of DNA damage by the lower dose and longer exposure of PM2.5 or 9-NA and the contribution of 9-NA to PM2.5 toxicity would help us understand the underlying biological mechanisms of PM2.5-induced lung diseases.
DNA repair gene expressions induced by PM2.5 and 9-NA in rat lungs
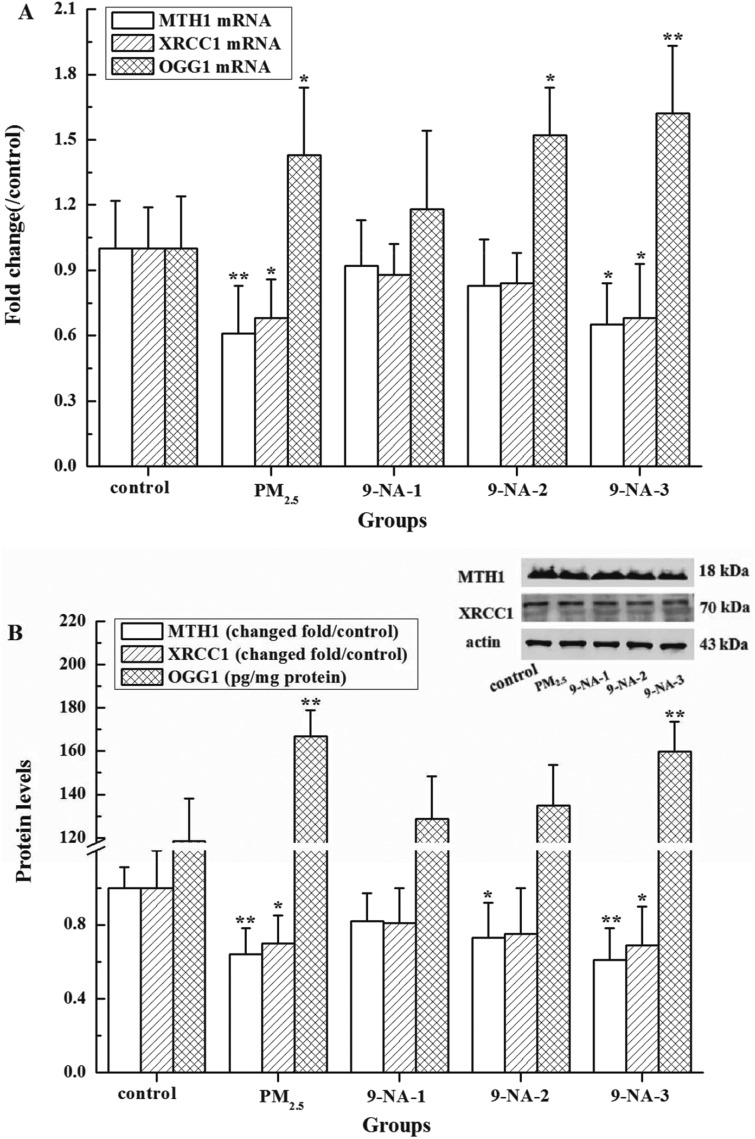
As observed in Fig. 4A and B, the OGG1 mRNA and protein levels in the lungs of rats exposed to PM2.5 at the dosage of 1.5 mg per kg b.w. were significantly increased whereas the MTH1 and XRCC1 expressions were markedly suppressed compared with the control (P < 0.05 or P < 0.01). As for 9-NA, 4.0 × 10–5 and 1.2 × 10–4 mg kg–1 of 9-NA decreased obviously MTH1 protein levels in the lungs of rats compared to the control (P < 0.05 or P < 0.01). 1.2 × 10–4 mg per kg b.w. 9-NA increased the OGG1 expression while inhibiting the XRCC1 expression compared with the control (P < 0.05 or P < 0.01). No significant changes of the MTH1 and XRCC1 were observed in the rats exposed to 9-NA at the concentrations of 1.3 × 10–5 and 4.0 × 10–5 mg per kg b.w. compared to the control (P > 0.05). 9-NA caused the elevations in OGG1 mRNA and protein expressions in lung cells in a dose-dependent manner (r = 0.87–0.99), while it gave rise to the decreases in MTH1 and XRCC1 expressions with a concentration-dependent property (r = 0.80–0.98).
Fig. 4. Expression of mRNA (A) and protein (B) of MTH1, XRCC1 and OGG1 in rat lungs treated with PM2.5 or 9-NA. The values are mean ± SD from five individual samples. Using one-way ANOVA, comparing with the control group, a significant difference is indicated by*P < 0.05 and **P < 0.01.
Our present results revealed, under the experimental conditions, that PM2.5 and higher dosage 9-NA could induce DNA damage in the lung of rats, whose underlying mechanisms may be linked to the regulation of DNA repair genes. If DNA damage occurs and it is not repaired before replication, the accumulation of DNA damage along with the unrepaired or mispairing bases can cause mutations, possibly leading to disease.38 DNA repair genes are involved in the early biological effects of DNA-damaging agents and are able to repair massive amounts of damaged DNA. OGG1, as a DNA glycosylase, may recognize and remove the altered base like 8-OH-dG in the base excision repair (BER) pathway.39 MTH1 protein may effectively catalyze the hydrolysis of 8-oxo-dGTP to 8-oxodGMP, thereby preventing the 8-oxo-dGTP misincorporation into DNA.40 As a scaffolding protein, XRCC1 plays a major role in BER and single strand break repair (SSBR) pathways via its ability to interact with multiple enzymatic components of repair reactions such as DNA polymerase beta, DNA ligase III and poly(ADP-ribose) polymerases (PARP),41 which have a positive regulation role in the DNA repair process.42 Based on our results, PM2.5 and higher dosage 9-NA caused OGG1 over-expression that could enhance the capability of removing 8-OH-dG from the lungs of rats. On the other hand, the decreasing expression of MTH1 and XRCC1 suppressed the roles of catalyzing the hydrolysis from 8-oxo-dGTP to 8-oxodGMP and repairing BER and SSBR, along with DNA strand breaks and 8-OH-dG formation. It is speculated that the inhibition roles of MTH1 and XRCC1 caused by PM2.5 or 9-NA were greater than the enhancement roles of OGG1 did by PM2.5 or 9-NA on the basis of the DNA damage results (see Fig. 2 and 3). It was found that PM2.5 caused OGG1 over-expression that had a significant repair effect on DNA strand breaks in BEAS-2B cells caused by PM2.5.43 PM2.5 at the dosage of 1 µg cm–2 increased MTH1 mRNA expression in the cultured epithelial cells of rat lungs.44 Ambient PM2.5 weakly induced the XRCC1 mRNA expression whereas its nanoparticles down-regulated XRCC1 in human lung cells.45,46 Considering the complicated roles of DNA repair genes in the regulation of DNA damage responses, further in-depth work is needed to investigate the underlying molecular mechanisms of PM2.5 and NPAHs on DNA damage and repair.
Effects of PM2.5 on markers of oxidative stress in rat lungs
In Table 2, the GADD153, HO-1 and MDA levels in the lungs of rats exposed to PM2.5 at the dosage of 1.5 mg per kg b.w. were significantly increased whereas SOD enzyme activities were obviously decreased compared with the control (P < 0.05 or P < 0.01). As for 9-NA, the mRNA and protein levels of GADD153 showed an obvious increase in response to the higher dose exposure to 9-NA (4.0 × 10–5 and/or 1.2 × 10–4 mg per kg b.w.) compared to the control (P < 0.05 or P < 0.01), whereas the GADD153 gene expression was not significantly changed in the presence of 9-NA at the dose of 1.3 × 10–5 mg per kg b.w. (P > 0.05). Also, the mRNA and protein levels of HO-1 showed an obvious increase in response to the higher dose exposure to 9-NA (4.0 × 10–5 and/or 1.2 × 10–4 mg per kg b.w.) compared to the control (P < 0.05 or P < 0.01), which shows similar trends to the GADD153 expression induced by 9-NA. Besides, as shown in Table 2, relative to the control, the highest dose of 9-NA (1.2 × 10–4 mg per kg b.w.) markedly enhanced the MDA contents and decreased the SOD activities (P < 0.05 or P < 0.01) in rat lungs. The level changes of GADD153, HO-1, MDA and SOD in the presence of the lowest dose 9-NA (1.3 × 10–5 mg per kg b.w.) were not statistically significant compared with the control group (P > 0.05).
Table 2. GADD153, HO-1 mRNA and protein levels, SOD activities and MDA contents in rat lungs treated with PM2.5 or 9-NA.
| Groups | Control | PM2.5 | 9-NA-1 | 9-NA-2 | 9-NA-3 |
| HO-1 mRNA (changed fold) | 1.00 ± 0.19 | 1.33 ± 0.22* | 1.23 ± 0.25 | 1.41 ± 0.23* | 1.55 ± 0.24** |
| HO-1 protein (ng mg–1 protein) | 3.69 ± 1.05 | 6.23 ± 1.02* | 4.89 ± 1.45 | 5.39 ± 2.88 | 6.76 ± 1.29* |
| GADD153 mRNA (changed fold) | 1.00 ± 0.19 | 1.34 ± 0.27* | 1.22 ± 0.19 | 1.40 ± 2.25* | 1.47 ± 0.24** |
| GADD153 protein (changed fold) | 1.00 ± 0.20 | 1.36 ± 0.21* | 1.11 ± 0.14 | 1.22 ± 0.33 | 1.33 ± 0.27* |
| SOD (U mg–1 protein) | 28.2 ± 1.81 | 24.6 ± 1.66* | 27.2 ± 0.39 | 26.2 ± 2.18 | 23.7 ± 3.25** |
| MDA (nmol mg–1 protein) | 1.08 ± 0.15 | 1.39 ± 0.21* | 1.24 ± 0.23 | 1.31 ± 0.19 | 1.49 ± 0.27** |
Previous studies revealed that the induction of GADD153 is highly responsive to endoplasmic reticulum stress, oxidative stress and DNA damage.47–49 GADD153 also may be activated through an activator protein-1 element in its promoter in the presence of ultraviolet irradiation or hydrogen peroxide.50 Specially, the GADD153 expression may be a valuable prognostic factor of early-stage non-small cell lung cancer in patients and has potential clinical significance.51 Recent studies showed that GADD153 levels were increased in the PM2.5-induced apoptosis of the lung cells of rats via endoplasmic reticulum stress, and GADD153 mRNA expressions were up-regulated in the gene expression profiles from BEAS-2B cells exposed to PM10 for 1 day.52,53 Laing et al. (2010) reported that ambient PM2.5 exposure activates GADD153 in the lungs of mice.54 In this study, accompanied by DNA damage, the GADD153 expression in rat lungs was highly induced by PM2.5 and high concentration 9-NA compared with the control. This experimental result was consistent with the previous reports. It suggests that PM2.5 and high dosage 9-NA can mount an active response to DNA damage through an increase in the GADD153 expression, which can be used as a marker for the evaluation of the DNA damage induced by PM2.5 and 9-NA.
HO-1, the rate-limiting enzyme in charge of heme degradation, is an inducible antioxidant enzyme that exerts cytoprotective effects in various cells,23 because an excess of heme can catalyze ROS formation. The high-expression of HO-1 means, to some extent, that ROS accumulation and stress response occur. As we know, the anti-oxidative enzyme SOD can scavenge superoxide radicals and GST participates in the reduction of organic hydroperoxides and ROS quenching.25 As observed in Table 2, under PM2.5 or 9-NA exposure conditions, the inhibition of SOD, the activation of HO-1 and the elevation of MDA (a typical lipid peroxidation product) occur. The increase of GST may be a kind of stress response to ROS and may exert an important metabolic function. The occurrence of oxidative stress and excessive ROS production would induce DNA strand breaks, DNA oxidative damage and formation of DPC,24 increasing the risk of inducing genotoxicity and involving in the aetiology of cancer.55,56 Actually, it had been reported that PM2.5 could produce semiquinone radicals, superoxide radicals and hydroxyl radicals,57–59 and that NPAHs including 9-NA were proven to generate reactive intermediates including ROS (singlet oxygen and superoxide) and free radicals during ultraviolet irradiation, regulating the occurrence of lipid peroxidation.4 Thus, the elevation of HO-1 and MDA levels induced by PM2.5 or 9-NA is the fundamental mechanism of PM2.5/9-NA-mediated lung oxidative stress and DNA damage.
Effects of PM2.5 and 9-NA on metabolic enzymes in lungs of rats
As shown in Table 3, PM2.5 at the doses of 1.5 mg per kg b.w. significantly increased the levels of CYP450, CYP1A1, CYP1A2 and GST in the lungs of rats compared with the control group (P < 0.05 or P < 0.01). As for 9-NA, CYP450 activity and CYP1A2 expression in the presence of 9-NA at 1.2 × 10–4 mg per kg b.w. concentration were statistically high versus the control, whereas no statistic changes of them in the lungs at 1.3 × 10–5 and 4.0 × 10–5 mg per kg b.w. 9-NA were observed compared with the control. The mRNA and protein levels of CYP1A1 showed an obvious increase in response to the higher dose exposure to 9-NA (4.0 × 10–5 and/or 1.2 × 10–4 mg per kg b.w.) compared to the control (P < 0.05 or P < 0.01). In addition, the GST activity was 32.4 U mg–1 protein in the presence of 4.0 × 10–5 mg per kg b.w. 9-NA and 36.5 U mg–1 protein in the presence of 1.2 × 10–4 mg per kg b.w. 9-NA, which were higher than that of the control (P < 0.01). The level changes of metabolic enzyme activities in the presence of the lowest dose 9-NA (1.3 × 10–5 mg per kg b.w.) were not statistically significant compared with the control group (P > 0.05). 9-NA increased phase I enzyme CYP450 and phase II enzyme GST activities in a dose-dependent manner respectively (r = 0.99 and 0.95).
Table 3. mRNA and protein levels of CYP1A1 and 1A2 and activities of CYP450s and GSTs in rat lungs treated with PM2.5 or 9-NA.
| Groups | CYP1A1 mRNA (changed fold) | CYP1A1 protein (ng mg–1 protein) | CYP1A2 mRNA (changed fold) | CYP1A1 protein (ng mg–1 protein) | GST (U mg–1 protein) | CYP450 (U mg–1 protein) |
| Control | 1.00 ± 0.26 | 22.7 ± 3.9 | 1.00 ± 0.32 | 2 ± 0.14 | 26.1 ± 2.5 | 0.071 ± 0.008 |
| PM2.5 | 1.44 ± 0.25* | 29.7 ± 4* | 1.31 ± 0.29 | 2.61 ± 0.3** | 34.8 ± 4.3** | 0.103 ± 0.016* |
| 9-NA-1 | 1.25 ± 0.27 | 25.5 ± 4.4 | 1.16 ± 0.31 | 2.17 ± 0.2 | 29 ± 3 | 0.079 ± 0.013 |
| 9-NA-2 | 1.47 ± 0.37* | 27.4 ± 4.2 | 1.38 ± 0.34 | 2.23 ± 0.28 | 32.4 ± 3** | 0.086 ± 0.01 |
| 9-NA-3 | 1.59 ± 0.46* | 31.1 ± 4.9* | 1.44 ± 0.32* | 2.68 ± 0.21** | 36.5 ± 3.3** | 0.12 ± 0.037** |
The CYP1A1 and 1A2 are CYP450 isoforms and important phase I enzymes involved in the metabolism of several pharmacological compounds, environmental pollutants, toxins, etc.26 They can mediate the PAH oxidation to epoxide and diol-epoxide intermediates, and convert pro-carcinogens like PAHs into full carcinogens, whose gene polymorphisms are associated with the risk of cancers.60 Previous studies revealed that PM2.5 induced ROS production and DNA damage in A549 cells,57,61,62 which was likely related to the activation of CYP enzymes (CYP1A1, CYP2E1 and CYP1B1) in response to PAHs adsorbed on the particle surface.61,62 CYP450 enzymes may convert PAH epoxide intermediates to the ultimate carcinogenic metabolites, such as diol-epoxides.63 Phase II enzyme GSTs may catalyze the conjugation of glutathione together with the intermediates of xenobiotics from the phase I reaction, which can promote its elimination from the organism and exert a detoxification role.64 The high expression levels of GSTs partly mean that the excess of the harmful metabolites has been eliminated. A research study indicated that the metabolic activation of PM2.5 with high-expression of CYP1A1, CYP2E1 and GST-pi1 is one of the underlying mechanisms of action closely involved in its cytotoxicity in human alveolar macrophages.65 NPAHs, such as 1-nitropyrene, 2-nitrofluoranthene, 3-nitrofluoranthene and 6-nitrochrysene, could induce CYP1A1 and CYP1A2 expression in various human tissue-derived cell lines.66 To our knowledge, little information about the effects of 9-NA on the metabolic enzymes in the lungs has been reported. In the present study, PM2.5 or higher doses of 9-NA markedly increased the activities of CYP450 isozymes and GST in rat lungs (see Table 3). Combining this result with the responses of oxidative damage and DNA damage mediated by metabolic enzymes31,32 implied that PM2.5 or 9-NA may activate phase I and phase II enzymes and disturb the lung metabolic process, further promoting the lung oxidative DNA damage.
Notably, our research clearly confirmed that PM2.5 and 9-NA induced a significant increase of lung DNA damage accompanied by decreasing DNA repair capacity and that DNA damage, oxidative stress and metabolic enzyme activation may be associated with the genotoxic effects, which may have the potency to induce the pathogenesis of lung diseases.
Conclusions
In conclusion, this study suggests that PM2.5 and high dosage 9-NA cause DNA damage, inhibit DNA repair gene expression, and enhance the levels of oxidative stress factors and metabolic enzymes in rat lungs. We propose that these effects are derived from three mechanisms: (1) the inhibition effects of the MTH1 and XRCC1 expression induced by PM2.5 and 9-NA exceed the scavenging role of OGG1 to damaged DNA, (2) PM2.5 and 9-NA significantly induce the levels of oxidative stress factors like GADD153, HO-1 and MDA, along with SOD suppression, leading to oxidative stress, and (3) PM2.5 and 9-NA markedly activate GST and CYP450 as well as CYP1A1 and CYP1A2, disturbing the biotransformation. These data indicate that the PM2.5-induced DNA damage in the lungs of rats is mediated partly by both the DNA repair dysfunction and the activation of oxidative stress and metabolic enzymes, and the results also suggest that the negative effect of 9-NA might be related to the PM2.5-induced lung genotoxicity.
Conflict of interest
There are no conflicts of interest to declare.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (no. 91543202 and 21575084), the HKBU Strategic Development Fund (15-1012-P04), the Nature Science Foundation of Shanxi Province in China (no. 2014011036-2), and the Foundation of Educational Committee of Shanxi Province in China (no. 2014110).
References
- Li P., Xin J., Wang Y., Wang S., Li G., Pan X., Liu Z., Wang L. Environ. Sci. Pollut. Res. Int. 2013;20:6433–6444. doi: 10.1007/s11356-013-1688-8. [DOI] [PubMed] [Google Scholar]
- Vinikoor-Imler L. C., Davis J. A., Luben T. J. Int. J. Environ. Res. Public Health. 2011;8:1865–1871. doi: 10.3390/ijerph8061865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC (International Agency for Research on Cancer), IARC monographs on the evaluation of carcinogenic risk to humans, 2015, vol. 112, p. 27. [Google Scholar]
- Zhang Y. X., Tao S., Shen H. Z., Ma J. Proc. Natl. Acad. Sci. U. S. A. 2009;106:21063–21067. doi: 10.1073/pnas.0905756106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Jariyasopit N., Schrlau J., Jia Y., Tao S., Yu T. W., Dashwood R. H., Zhang W., Wang X., Simonich S. L. Environ. Sci. Technol. 2011;45:6887–6895. doi: 10.1021/es201443z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Igarashi K., Tang N., Lin J. M., Wang W., Kameda T., Toriba A., Hayakawa K. Mutat. Res. 2010;695:29–34. doi: 10.1016/j.mrgentox.2009.10.010. [DOI] [PubMed] [Google Scholar]
- Albinet A., Leoz-Garziandia E., Budzinski H., Viilenave E. Sci. Total Environ. 2007;384:280–292. doi: 10.1016/j.scitotenv.2007.04.028. [DOI] [PubMed] [Google Scholar]
- Bandowe B. A., Meusel H., Huang R. J., Ho K., Cao J., Hoffmann T., Wilcke W. Sci. Total Environ. 2014;473–474:77–87. doi: 10.1016/j.scitotenv.2013.11.108. [DOI] [PubMed] [Google Scholar]
- Hayakawa K. Chem. Pharm. Bull. 2016;64:83–94. doi: 10.1248/cpb.c15-00801. [DOI] [PubMed] [Google Scholar]
- Ma N., Bian W., Li R., Geng H., Zhang J., Dong C., Shuang S., Cai Z. Anal. Methods. 2015;7:3967–3971. [Google Scholar]
- Fu P. P., Von Tungeln L. S., Chou M. W. Carcinogenesis. 1985;6:753–757. doi: 10.1093/carcin/6.5.753. [DOI] [PubMed] [Google Scholar]
- Kastan M. B. Mol. Cancer Res. 2008;6:517–524. doi: 10.1158/1541-7786.MCR-08-0020. [DOI] [PubMed] [Google Scholar]
- Meng Z., Zhang Q. Food Chem. Toxicol. 2007;45:1368–1374. doi: 10.1016/j.fct.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Chu M., Sun C., Chen W., Jin G., Gong J., Zhu M., Yuan J., Dai J., Wang M., Pan Y., Song Y., Ding X., Guo X., Du M., Xia Y., Kan H., Zhang Z., Hu Z., Wu T., Shen H. Toxicol. Lett. 2015;235:172–178. doi: 10.1016/j.toxlet.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Gao Z. X., Song X. L., Li S. S., Lai X. R., Yang Y. L., Yang G., Li Z. J., Cui Y. H., Pan H. W. Ophthalmol. Visual Sci. 2016;57:3093–3102. doi: 10.1167/iovs.15-18839. [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Mukherjee S., Ngo L. C., Christiani D. C. Environ. Health Perspect. 2004;112:666–671. doi: 10.1289/ehp.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neophytou A. M., Hart J. E., Chang Y., Zhang J. J., Smith T. J., Garshick E., Laden F. Toxics. 2014;2:377–390. doi: 10.3390/toxics2030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabeppu Y. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:75–94. doi: 10.1016/s0079-6603(01)68091-7. [DOI] [PubMed] [Google Scholar]
- Sugasawa K. DNA Repair. 2016;S1568–S7864:30096–30099. [Google Scholar]
- Thacker J., Zdzienicka M. Z. DNA Repair. 2003;2:655–672. doi: 10.1016/s1568-7864(03)00062-4. [DOI] [PubMed] [Google Scholar]
- Mehta M., Chen L. C., Gordon T., Rom W., Tang M. S. Mutat. Res. 2008;657:116–121. doi: 10.1016/j.mrgentox.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luethy J. D., Holbrook N. J. Cancer Res. 1994;54:1902s–1906s. [PubMed] [Google Scholar]
- Takahashi T., Morita K., Akagi R., Sassa S. Curr. Med. Chem. 2004;11:1545–1561. doi: 10.2174/0929867043365080. [DOI] [PubMed] [Google Scholar]
- Evans M. D., Dizdaroglu M., Cooke M. S. Mutat. Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Sharma R., Yang Y., Sharma A., Awasthi S., Awasthi Y. C. Antioxid. Redox Signaling. 2004;6:289–300. doi: 10.1089/152308604322899350. [DOI] [PubMed] [Google Scholar]
- Guengerich F. P. Chem. Res. Toxicol. 2007;21:70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- Marnett L. J. Toxicology. 2002;181–182:219–222. doi: 10.1016/s0300-483x(02)00448-1. [DOI] [PubMed] [Google Scholar]
- Hanot-Roy M., Tubeuf E., Guilbert A., Bado-Nilles A., Vigneron P., Trouiller B., Braun A., Lacroix G. Toxicol. In Vitro. 2016;33:125–135. doi: 10.1016/j.tiv.2016.01.013. [DOI] [PubMed] [Google Scholar]
- Risom L., Møller P., Loft S. Mutat. Res. 2005;592:119–137. doi: 10.1016/j.mrfmmm.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Sugasawa K. DNA Repair. 2016;44:110–117. doi: 10.1016/j.dnarep.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Shah U. K., Seager A. L., Fowler P., Doak S. H., Johnson G. E., Scott S. J., Scott A. D., Jenkins G. J. Mutat. Res. 2016;808:8–19. doi: 10.1016/j.mrgentox.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Huang S., Hu J. Chin. J. Ind. Hyg. Occup. Dis. 2014;32:549–554. [PubMed] [Google Scholar]
- Li R., Kou X., Geng H., Xie J., Yang Z., Zhang Y., Cai Z., Dong C. Chem. Res. Toxicol. 2015;28:408–418. doi: 10.1021/tx5003723. [DOI] [PubMed] [Google Scholar]
- Lin Y., Ma Y., Qiu X., Li R., Fang Y., Wang Y., Zhu Y., Hu D. J. Geophys. Res.: Atmos. 2015;120:7219–7228. [Google Scholar]
- Xie J., Fan R., Meng Z. Inhalation Toxicol. 2007;19:759–765. doi: 10.1080/08958370701399885. [DOI] [PubMed] [Google Scholar]
- Tuominen J., Salomaa S., Pyysalo H., Skytta E., Tikkanen L., Nurmela T., Sorsa M., Pohjola V., Sauri M., Himberg K. Environ. Sci. Technol. 1998;22:1228–1234. doi: 10.1021/es00175a017. [DOI] [PubMed] [Google Scholar]
- Oh S. M., Kim H. R., Park Y. J., Lee S. Y., Chung K. H. Mutat Res. 2011;723:142–151. doi: 10.1016/j.mrgentox.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Clancy S. Nature Educ. 2008;1:103. [Google Scholar]
- Boiteux S., Radicella J. P. Arch. Biochem. Biophys. 2000;377:1–8. doi: 10.1006/abbi.2000.1773. [DOI] [PubMed] [Google Scholar]
- Svensson L. M., Jemth A. S., Desroses M., Loseva O., Helleday T., Högbom M., Stenmark P. FEBS Lett. 2011;585:2617–2621. doi: 10.1016/j.febslet.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Hanssen-Bauer A., Solvang-Garten K., Akbari M., Otterlei M. Int. J. Mol. Sci. 2012;13:17210–17229. doi: 10.3390/ijms131217210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott K. W., Aoufouchi S., Johnson P., Shall S. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wang Y., Lin Z., Zhou X., Chen T., He H., Huang H., Yang T., Jiang Y., Xu W., Yao W., Liu T., Liu G. Exp. Mol. Pathol. 2015;99:365–373. doi: 10.1016/j.yexmp.2015.08.005. [DOI] [PubMed] [Google Scholar]
- Choi J. H., Kim J. S., Kim Y. C., Kim Y. S., Chung N. H., Cho M. H. J. Vet. Sci. 2004;5:11–18. [PubMed] [Google Scholar]
- Traversi D., Cervella P., Gilli G. Environ. Sci. Pollut. Res. Int. 2015;22:1279–1289. doi: 10.1007/s11356-014-3435-1. [DOI] [PubMed] [Google Scholar]
- Asharani P., Sethu S., Lim H. K., Balaji G., Valiyaveettil S., Hande M. P. Genome Integr. 2012;3:2. doi: 10.1186/2041-9414-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S., Mori M. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- Fontanier-Razzaq N., McEvoy T. G., Robinson J. J., Rees W. D. Biol. Reprod. 2001;64:1386–1391. doi: 10.1095/biolreprod64.5.1386. [DOI] [PubMed] [Google Scholar]
- Tang J. R., Nakamura M., Okura T., Takata Y., Watanabe S., Yang Z. H., Liu J., Kitami Y., Hiwada K. Biochem. Biophys. Res. Commun. 2002;290:1255–1259. doi: 10.1006/bbrc.2002.6336. [DOI] [PubMed] [Google Scholar]
- Guyton K. Z., Xu Q., Holbrook N. J. Biochem. J. 1996;314:547–554. doi: 10.1042/bj3140547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Lee M. G., Choi K. C., Kang H. M., Chang Y. S. Oncol. Lett. 2012;4:408–412. doi: 10.3892/ol.2012.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Shamy M., Kluz T., Muñoz A. B., Zhong M., Laulicht F., Alghamdi M. A., Khoder M. I., Chen L. C., Costa M. Toxicol. Appl. Pharmacol. 2012;265:147–157. doi: 10.1016/j.taap.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D., Liu X. J., Du Y. F., Wang T. T., Zhu J. N., Liu L. F. Int. J. Resp. 2016;36:907–914. [Google Scholar]
- Laing S., Wang G., Briazova T., Zhang C., Wang A., Zheng Z., Gow A., Chen A. F., Rajagopalan S., Chen L. C., Sun Q., Zhang K. Am. J. Physiol.: Cell Physiol. 2010;299:C736–C749. doi: 10.1152/ajpcell.00529.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkena K. V., Young C. Y., Tindall D. J. Obstet. Gynecol. Int. 2010;2010:302051. doi: 10.1155/2010/302051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poljšak B., Fink R. Oxid. Med. Cell. Longevity. 2014;2014:671539. doi: 10.1155/2014/671539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagai M., Saito H., Ichinose T., Kodama M., Mori Y. Free Radical Biol. Med. 1993;14:37–47. doi: 10.1016/0891-5849(93)90507-q. [DOI] [PubMed] [Google Scholar]
- Dellinger B., Pryor W. A., Cueto R., Squadrito G. L., Hegde V., Deutsch W. A. Chem. Res. Toxicol. 2001;14:1371–1377. doi: 10.1021/tx010050x. [DOI] [PubMed] [Google Scholar]
- Gehling W. M., Khachatryan L., Dellinger B. Environ. Sci. Technol. 2014;48:4266–4272. doi: 10.1021/es401770y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Feng S. Curr. Drug Metab. 2015;16:850–863. doi: 10.2174/138920021610151210164501. [DOI] [PubMed] [Google Scholar]
- Billet S., Garçon G., Dagher Z., Verdin A., Ledoux F., Cazier F., Courcot D., Aboukais A., Shirali P. Environ. Res. 2007;105:212–223. doi: 10.1016/j.envres.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Gualtieri M., Longhin E., Mattioli M., Mantecca P., Tinaglia V., Mangano E., Proverbio M. C., Bestetti G., Camatini M., Battaglia C. Toxicol. Lett. 2012;209:136–145. doi: 10.1016/j.toxlet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- Shimada T., Fujii-Kuriyama Y. Cancer Sci. 2004;95:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Miyamoto M., Murakami A., Ohigashi H., Osawa T., Uchida K. A. Biochem. Biophys. Res. Commun. 2003;302:593–600. doi: 10.1016/s0006-291x(03)00219-5. [DOI] [PubMed] [Google Scholar]
- Saint-Georges F., Abbas I., Billet S., Verdin A., Gosset P., Mulliez P., Shirali P., Garçon G. Toxicology. 2008;244:220–230. doi: 10.1016/j.tox.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Iwanari M., Nakajima M., Kizu R., Hayakawa K., Yokoi T. Arch. Toxicol. 2002;76:287–298. doi: 10.1007/s00204-002-0340-z. [DOI] [PubMed] [Google Scholar]