 Occupational and environmental exposure to uranium has been confirmed to cause tissue injury and carcinogenesis.
Occupational and environmental exposure to uranium has been confirmed to cause tissue injury and carcinogenesis.
Abstract
Occupational and environmental exposure to uranium has been confirmed to cause tissue injury and carcinogenesis. As a heavy metal from actinide series, the chemical and radiological toxicities of uranium jointly induce the detrimental effects. However, the mutual action and mechanism of both forms of toxicities still need to be further elucidated. DNA double-strand break (DSB) is a fundamental cause of cell death or genomic instability induced by ionizing radiation. Herein, we investigate the effect of uranyl nitrate on the cellular function of DNA damage response and intrinsic DSB repair on the aspect of chemical toxicity. The results indicated that uranyl ion increased the accumulation of nuclear DNA DSBs in a dose-dependent manner. Both homologous recombination (HR) and non-homologous end joining (NHEJ) pathways of DSB repair were affected by the uranyl ion. The inhibition of DSB repair efficiency is attributed to the depression of a set of critical repair proteins, particularly those for the HR pathway such as ATM, BRCA1, RPA80 and EXO1. The available data enable us to imagine that the chemical toxicity of uranium leads to inhibition of cellular DNA repair capability, which can further aggravate its radiological toxicity.
1. Introduction
With the development of nuclear energy and nuclear power plant, the demand for uranium is increasing. During processes of nuclear fuel cycle such as mining of uranium, production of nuclear weapons, spent fuel reprocessing, nuclear waste treatment, disposal, etc., radionuclides are likely to contaminate the environment through migration, diffusion, transfer and transformation, subsequently posing a threat to the organisms directly or indirectly. The presence of uranium (DU) contaminants in air, water and land has also been reported to be the result of the use of DU ammunition during the military actions, such as the First Gulf War in Balkans.1–3 Uranium is the heaviest naturally occurring element and has both radiotoxic and chemotoxic properties. Uranium includes three radioactive isotopes: 238U (99.27%), 235U (0.72%) and 234U (0.0055%).4 The major pathways of uranium exposure include inhalation, ingestion and penetration through wounds. Through inhalation, uranium particles can get deposited in the respiratory tract and lung tissues. Once uranium enters the human blood, primarily as uranyl (UO22+) salts or in complex with proteins5, it quickly gets distributed to other tissues and organs, like the kidneys, bones, livers and spleen, through blood circulation.
The uranyl ion is a highly toxic and carcinogenic metal ion,6 but the detailed molecular mechanisms are poorly understood. The first epidemiological study of the lung carcinoma incidence in uranium miners was carried out in Schneeberg, Saxonia, and Jáchymov, West Bohemia, in the 1920s and 1930s.7 Similar studies were conducted in other parts of the world later on.4,8,9 The studies suggested that radon released during the decay of 226Ra and 238U is the major risk factor for lung cancer.6 The radon daughters generated from the decay of radon emit α particles that induce DNA damage including double-strand breaks (DSBs). In recent years, a series of studies have reported that increased production of reactive oxygen species (ROS) by heavy metals, which include the radioactive nucleotide uranium, caused oxidative stress as a main factor to induce DNA damage, apoptosis, autophagy and genotoxicity.10–16 In addition, there are some indirect evidences that uranium may disrupt the DNA repair system. Several studies found that uranium exposure was associated with the induction of DNA damage or disturbance of DNA repair.17–20 However, the mechanism is not well established. Therefore, our study originates from the perspective of DNA repair to address the carcinogenic mechanism of uranyl ions.
A series of proteins in mammalian cells have been identified to be dedicated for the maintenance of genomic stability through multiple DNA repair pathways. DSB is the most detrimental form of DNA damage if not properly repaired, which can trigger cell death, acute tissue damage and malignant transformation.21–23 Homologous recombination (HR) and non-homologous end joining (NHEJ) are two major repair pathways of DSBs.24 DNA-dependent protein kinase (DNA-PK) complex, composed of Ku70, Ku80 and the catalytic subunit DNA-PKcs, is the critical component initiating the NHEJ pathway of DSB repair. In the HR pathway, DSBs can be recognized by the MRE11–RAD50–NBS1 complex, which promotes the activation of ataxia-telangiectasia mutated (ATM) protein and the end processing of DNA damage for HR. DNA end resection, which is essential for HR, is regulated by a number of proteins including ATM and BRCA1.25 DSB resection is primarily triggered in the S and G2 phases of the cell cycle, when sister chromatids can be used for HR. BRCA1 promotes homologous recombination in S and G2. BRCA1 negatively regulates 53BP1, an inhibitory component on DSB resection of the HR pathway. In S phase, BRCA1 directs the recruitment of the E3 ubiquitin ligase UHRF1 to DSBs, where UHRF1 mediates K63-linked polyubiquitination of RIF1. Consequently, RIF1 dissociates from 53BP1, facilitating HR initiation of DSBs.26 RAP80, an 80 kDa nuclear protein, is responsible for recruitment of the BRCA1 A complex (BRCA1, BARD1, BRCC36, Abraxas, and RAP80) to the sites of DNA damage.27 EXO1 nuclease induces formation of 3′ ssDNA ends compatible for RPA accumulation in the processing of DSB resection.28 Furthermore, BRCA2 mediates the displacement of RPA from the 3′ ssDNA ends and assembly of RAD51 filaments, leading to strand invasion into homologous DNA sequences. D loop structure is formed after strand invasion. Finally, HR repair is completed. Based on information given above, we investigated the effects of uranyl nitrate (UN) on DNA damage repair capability and the related DNA repair protein changes.
2. Materials and methods
2.1. Chemicals, materials, cell lines and plasmids
Uranyl nitrate (UO2(NO3)2·6H2O) (China National Nuclear Corporation, Beijing, China) was dissolved in double-distilled water (ddH2O) to prepare 40 mM stock solution and stored at room temperature. LHC-8 culture medium was purchased from Gibco (California, USA). Heat-inactivated fetal bovine serum (FBS) and trypsin–EDTA solution were purchased from Sigma (Darmstadt, Germany). Human bronchial epithelial cell line (BEP2D) was originally obtained from Dr CC Harris (Laboratory of Human Carcinogenesis Division of Basic Science, National Cancer Institute, NIH, USA). Homologous recombination and non-homologous end joining reporter plasmids were gifted by Dr Zhenkun Lou (Division of Oncology Research, Mayo Clinic Rochester, USA).
2.2. Cell culture, uranyl nitrate (UN) treatment and irradiation
BEP2D cells were cultured in a serum-free LHC-8 medium in an incubator at 37 °C in an atmosphere of 5% CO2. The prepared 40 mM uranyl nitrate solution (UO2(NO3)2·6H2O), which is equivalent to 40 mM UO22+, was used as the stock solution. Working solution was diluted to 10 mM before use. In the UN exposure group, for every 10 ml of culture medium, 1, 5, 10, 20 and 40 μl UN working solution was added. Then, 40 μl ddH2O was added to the culture medium in the control group. For the UN exposure group, the cells were treated with 1, 5, 10, 20 and 40 μM UN for 24 h. Cells were irradiated with 60Co γ-rays at a dose rate of 1.98 Gy min–1 at room temperature.
2.3. Cytotoxicity assay
When the cultured cells reached a confluence of approximately 80%–90%, the cells were seeded into 96-well plates at a density of 1 × 104 cells per well and treated with different concentrations of uranyl nitrate (1, 5, 10, 20, 40, 50, 75, 100 μM) for 24 h, or ddH2O as the solvent control. The number of viable cells was determined using the Cell Counting Kit-8 (CCK-8) Proliferation Cytotoxicity Assay Kit (Dojindo, Japan), according to the manufacturer's instruction.
2.4. Colony-forming ability assay
Cells were seeded into 6 cm culture dishes at a density of 8 × 102 cells per dish. After attachment, cells were incubated with different concentrations of UN for 24 h or irradiated with 2, 4 Gy of γ-rays. The medium was replaced with a fresh medium and cells were cultured for another 14 days until visible colonies formed. The colonies were fixed with 70% ethanol for 30 min, stained with Giemsa's solution for 20 min and counted. The average number was used to determine the formation ability.
2.5. Apoptosis analysis
Apoptosis was determined using Annexin V-FITC/PI Apoptosis Kit according to the manufacturer's instruction (Beyotime, Shanghai, China). After treating with different concentrations of UN for 24 h, BEP2D cells were collected, washed twice with cold PBS, and incubated with 5 μl FITC-Annexin V and 1 μl PI working solution (100 μg ml–1) for 30 min in the dark at room temperature. Finally, the apoptotic cells were measured by flow cytometry analysis.
2.6. Reactive oxygen species (ROS) detection
ROS was determined using Reactive Oxygen Species Assay Kit according to the manufacturer's instruction (Beyotime, Shanghai, China). After the BEP2D cells were treated with different concentrations of UN for 24 h, the medium was removed and washed with PBS. Cells were collected and incubated with 10 μM DCFH-DA in PBS at 37 °C for 15 min. Cellular fluorescence was measured by flow cytometry analysis.
2.7. Western blot analysis
The western blot was carried out according to the standard procedure. Briefly, after sample preparation, cells were lysed in protein extraction reagent (Thermo, Massachusetts, USA) containing protease inhibitor (Roche, Basel, Switzerland) and phosphatase inhibitor (Roche, Basel, Switzerland). Protein concentrations were measured using BCA Protein Assay Kit (Thermo USA). Then, 25 μg proteins were loaded onto SDS-polyacrylamide gel, subjected to electrophoresis, and blotted onto NC membrane (Pall Corporation, New York, USA). After blotting, the following antibodies were used. The primary antibodies were anti-P53, anti-γH2AX, anti-ATM, anti-Ku80 (Santacruz, Texas, USA), anti-BRCA1, anti-Ku70, anti-RAP80, anti-DNA-PKcs, anti-EXO1, anti-53BP1, and anti-RAD51 (Abcam, Cambridge, UK). Immunohybridization bands were visualized with a chemiluminescent substrate (Thermo, Massachusetts, USA). Images were captured using the LAS-5000 luminescent image analyzer (GE, Connecticut, USA).
2.8. Neutral comet assay
The neutral comet assay was performed to detect DNA DSBs according to the manual of the Comet Assay Kit (Trevigen, Maryland, USA). After propidium iodide (Sigma, Darmstadt, Germany) staining, the images of the comet slides were obtained by fluorescence microscopy. The comet parameter tail moment was gauged for at least 50 cells in each experiment using the Comet Score software.
2.9. In vivo HR assay
BEP2D cells were seeded into 12-well plates. After attachment, cells were transfected with 1 μg DR-GFP, 1 μg pCBA-SceI and 1 μg RFP expressing plasmids and mixed with 7.5 μl Lipofectamine 2000 (Invitrogen, California, USA) following the manual. After 5 h, the medium was changed and different concentrations of uranyl nitrate were added for 67 h. Cells were harvested by trypsinization and resuspended in phosphate buffer saline (PBS). Cellular fluorescence was measured by flow cytometry analysis.
2.10. In vivo NHEJ assay
Before transfection, NHEJ-GFP plasmid was digested with HindIII enzyme overnight and recovered by gel extraction kit. BEP2D cells were seeded in 12-well plates. After attachment, cells were transfected with 1 μg pCherry and 1 μg digested NHEJ-GFP plasmid and mixed with 5 μl Lipofectamine 2000 (Invitrogen, California, USA) following the manual. After 4 h, the medium was changed and different concentrations of uranyl nitrate were added for 20 h. Cells were harvested by trypsinization and resuspended in phosphate buffer saline (PBS). Cellular fluorescence was measured by flow cytometry analysis.
2.11. Statistical analysis
Data are expressed as mean ± SD from at least three independent experiments. Statistical comparisons between different groups were done by one-way ANOVA, followed by the Student–Newman–Keuls test.
3. Results
3.1. Cytotoxicity of uranyl nitrate to BEP2D cells
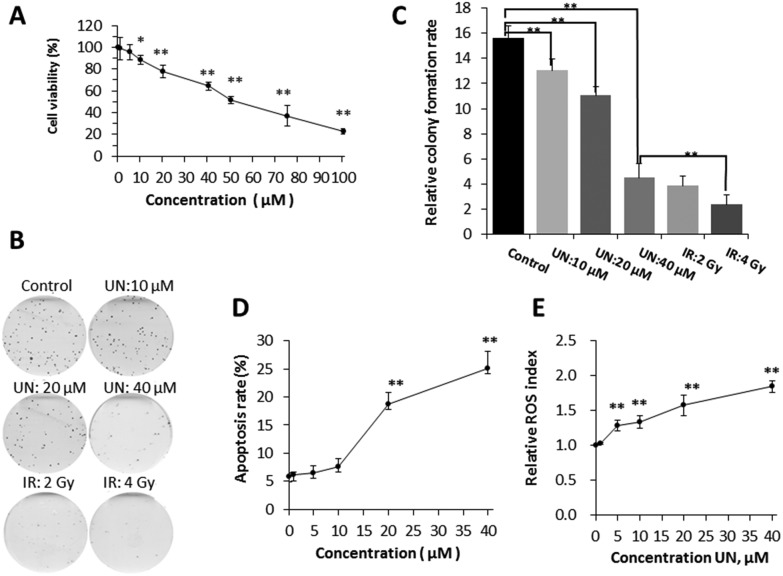
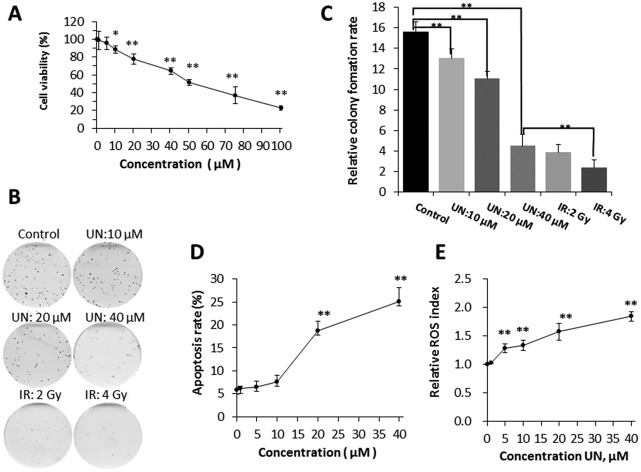
When BEP2D cells were acutely exposed to different concentrations (0, 1, 5, 10, 20, 40, 50, 75, 100 μM) of uranyl nitrate for 24 h, a dose-dependent inhibition of cell proliferation was demonstrated, and the half maximal inhibitory concentration (IC50) was approximately 50 μM based on CCK8 detection. The cell viability at concentrations of 1 and 5 μM were over 90% and no significant difference (p > 0.05) was observed compared with control (0 μM), suggesting that 1 and 5 μM of uranyl nitrate has minimum toxicity on BEP2D cells. The decrease in cell viability at a concentration of 10 μM and higher was statistically significant compared with control (p < 0.05) (Fig. 1A). Moreover, the colony formation assay showed that the decrease in colony-forming ability was also significant compared with the control (p < 0.05) (Fig. 1B and C). It was observed that 10 μM of uranyl nitrate started to produce significant toxicity on BEP2D cells, and the survival rate of colony forming assay was 83.4%. This concentration was considered as the low toxicity dose group for the next research. The cell viability of 20 μM concentration was further decreased to approximately 78% and was significantly different (p < 0.01) from control (Fig. 1A). Moreover, the relative colony formation rate was also significantly different (p < 0.01) from control (Fig. 1B and C). This result suggested that 20 μM of UN has moderate toxicity on BEP2D cells as a medium-dose group for the further research. The cell viability of 40 μM treatment group for 24 h was approximately 65% and was significantly different (p < 0.01) from control (Fig. 1A). The colony forming assay showed that the survival rate was significantly decreased (28.7%) and was close to the survival level as that of 2-Gy-irradiated cell group (40 μM vs. 2 Gy group, p > 0.05) (Fig. 1B and C).
Fig. 1. Effects of uranyl nitrate on cytotoxicity, apoptosis, and ROS. (A) Cell viability assay. BEP2D cells were treated with different concentrations of uranyl nitrate for 24 h. The CCK8 working solution was added directly to the cultures for proliferation assay. (B) Cell-colony-forming ability assay. After treatment with 0, 10, 20 and 40 μM of uranyl nitrate for 24 h or exposure to 2 and 4 Gy of γ-rays, BEP2D cells were seeded into 60 mm dishes and cultured in a normal growth medium for 14 days, and then fixed with ethanol and stained with Giemsa's solution. (C) Cell survival rates were presented for the treated cells. (D) Apoptosis induction. BEP2D cells were treated with 0, 1, 5, 10, 20 and 40 μM uranyl nitrate for 24 h. Apoptosis was measured by flow cytometry analysis. (E) ROS detection. BEP2D cells were treated with 0, 1, 5, 10, 20 and 40 μM uranyl nitrate for 24 h. ROS was measured as described in the Materials and methods section. *P < 0.05, **p < 0.01.
3.2. Uranyl nitrate-induced apoptosis and ROS production
BEP2D cells were exposed to different concentrations (0, 1, 5, 10, 20, 40 μM) of uranyl nitrate for 24 h, and we found that a sharply increased apoptosis was induced at the concentration of 20 μM. At the concentration of 40 μM, the apoptosis rate was 25% (Fig. 1D). ROS analysis showed that uranyl nitrate increased the production of ROS in BEP2D cells from the concentration of 5 μM in a dose-dependent manner (Fig. 1E).
3.3. Uranyl nitrate increased the yield of DNA double-strand breaks
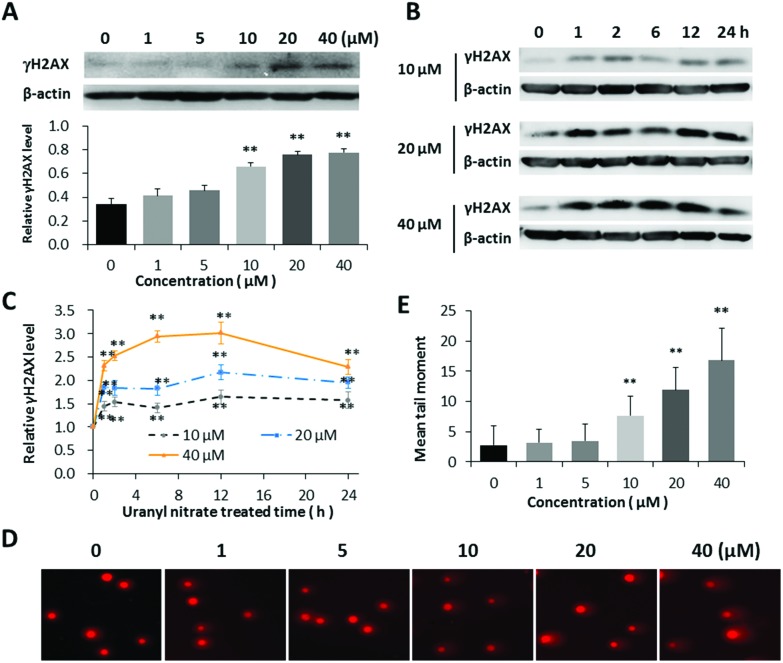
The histone H2AX is rapidly phosphorylated at S139 (γH2AX) once the double-strand break is produced in nuclear genomic DNA, which has been widely accepted as a biomarker of DSBs.29 The induction of DNA damage by uranyl nitrate was first assessed indirectly via western blot analysis with anti-γH2AX antibody. A dose-dependent increased expression of γH2AX was observed in BEP2D cells after treatment with 0–40 μM of uranyl nitrate for 24 h (Fig. 2A). The γH2AX level increased as early as 1 h after treatment with uranyl nitrate and reached the peak at 12 h (Fig. 2B and C). We further evaluated and confirmed the DNA DSBs induced by uranyl nitrate using the neutral comet assay. As shown in Fig. 2D and E, the tail moment of comet cells were markedly increased at the concentration of 10 μM, and the yield of DSBs increased with the increasing uranyl nitrate concentration.
Fig. 2. Increased yield of DNA double-strand breaks by uranyl nitrate. (A) Effect of different concentrations of uranyl nitrate on the expression of γH2AX protein. The γH2AX protein level was detected by the western blot in BEP2D after treatment with 0, 1, 5, 10, 20 and 40 μM of uranyl nitrate for 24 h. The expression intensities were quantified using the Quantity One software. β-Actin was used as a loading control. (B) Effect of different treatment times of uranyl nitrate on γH2AX protein expression. (C) The intensities of γH2AX expression western blot bands were quantified using the Quantity One software. β-Actin was used as a loading control, and the data of ratio were standardized by control. (D) Comet images of DSBs detected by neutral cell gel electrophoresis. (E) The statistical histogram of comet tail moment. *P < 0.05, **p < 0.01.
3.4. Effects of uranyl nitrate on DNA double-strand break repair
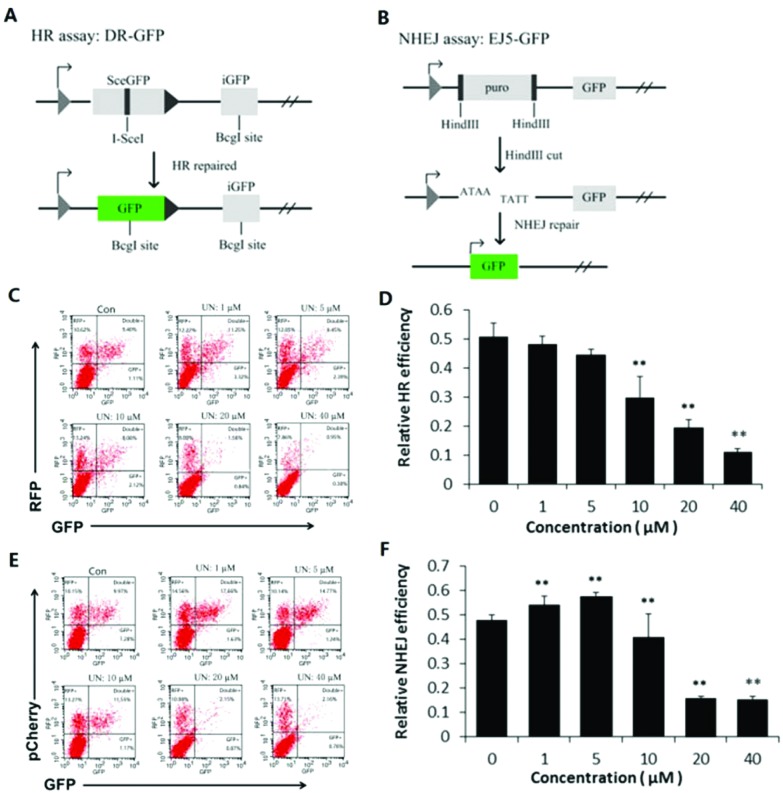
The oxidative stress could be one of the reasons leading to the increased level of DNA damage in the uranyl nitrate-treated cells. To ascertain whether the intrinsic DNA repair efficiency was also affected, we utilized DR-GFP and EJ5-GFP assay systems to determine the influence of uranyl nitrate exposure on the DSB repair efficiency in BEB-2D cells. The plasmids and principles for the assay of the HR and NHEJ pathways of DNA DSB repair are shown in Fig. 3A and B, respectively. The representatives of the flow cytometry measurements for HR and NHEJ pathway activities are shown in Fig. 3C (HR) and Fig. 3E (NHEJ). In this assay, both the pRFP and pCherry plasmids expressed the red fluorescence protein (RFP), and the transfection efficiency for them was about 20%, whereas the transfection efficiency for DR-GFP and EJ5-GFP was 10%. As shown in Fig. 3D, the relative efficiency of DSB in the HR pathway, as represented by the percentage of GFP+ cells among RFP+ cells, declined in a dose–effect relationship due to uranyl nitrate. The inhibition effect was statistically significant at a dose of 10 μM and at higher doses. A dose-dependent inhibitory effect on NHEJ efficiency was also found in BEP2D cells as a result of uranyl nitrate at a concentration of 10 μm and above (Fig. 3F). However, the result showed that an increased NHEJ efficiency was induced as a result of low doses (1, 5 μM) of uranyl nitrate, suggesting a potential hormetic effect of low-dose uranyl nitrate.
Fig. 3. Effect of uranyl nitrate on DNA double-strand break repair. (A) Diagram of the homologous recombination (HR) reporter assay. The DR-GFP plasmid carries a non-functional mutant GFP (SceGFP) and an internal truncate of GFP (iGFP). When transfected with an I-SceI along with an RFP-expressing plasmid to control transfection efficiencies, I-SceI excised SceGFP to induce DSBs.30 Only by HR repair with iGFP as the homologous template could restore the damaged SceGFP to a functional GFP. GFP+ cells were HR-repair cells. (B) Diagram of the NHEJ reporter assay. EJ5-GFP contains a promoter that is separated from a GFP-coding cassette by a puro gene that is flanked by two HindIII sites. Before transfection, the EJ5-GFP plasmid was linearized by HindIII enzyme digestion. Only by NHEJ repair could join the promoter and the GFP sequence, and GFP+ cells were NHEJ-repaired cells.31 (C) A representative of the flow cytometry measurements for HR pathway activity. (D) Quantification of the HR activity. Relative HR efficiency was measured by the percentage of GFP+ cells among RFP+ cells. (E) A representative of the flow cytometry measurements for NHEJ pathway activity. The pCherry plasmids also expressed the red fluorescence protein (RFP). (F) Quantification of the NHEJ assay. Relative NHEJ efficiency was measured by the percentage of GFP+ cells among pCherry+ cells.*P < 0.05, **p < 0.01.
3.5. Effects of uranyl nitrate on expression of DNA repair proteins
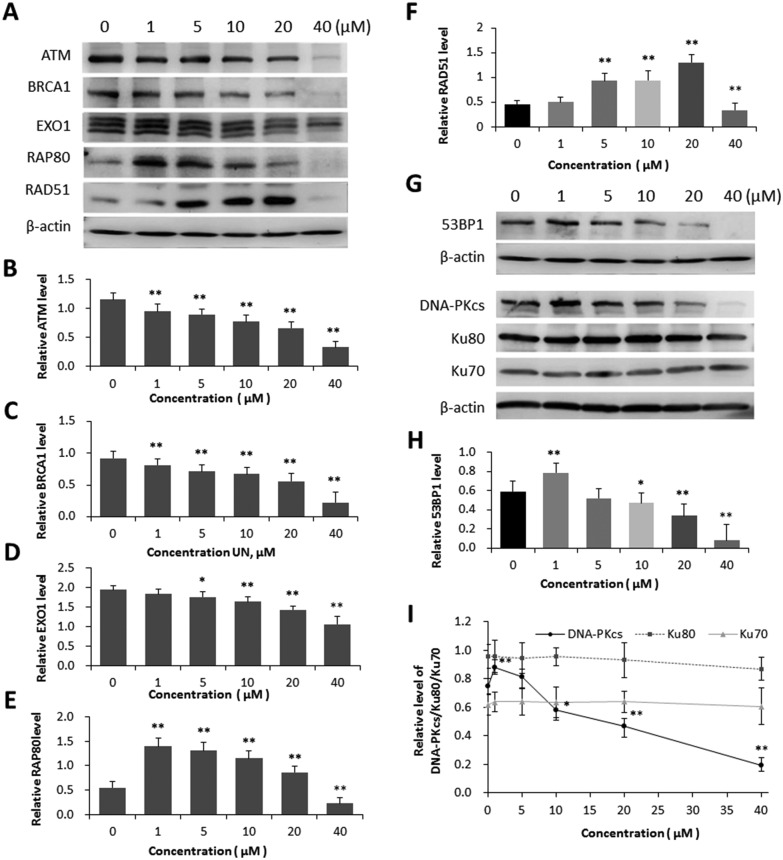
Inhibition of DNA DSB repair efficiency is certainly a critical reason for the increased level of DNA DSBs in the cells treated with uranyl nitrate. In order to determine the mechanism by which uranyl nitrate affected the cellular DNA repair system, we used the western blot analysis to detect the expression changes in a series of DNA repair proteins in both HR and NHEJ pathways. The results indicated that the expression levels of HR pathway proteins ATM, BRCA1 and EXO1 were decreased in BEP2D cells due to uranyl nitrate with increasing concentrations (Fig. 4A, B, C and D). Although the expression of HR pathway proteins RAP80 and RAD51 was decreased by uranyl nitrate at a dose of 40 μM or a higher dose, an increased expression was observed for both proteins at doses lower than 20 μM (Fig. 4A, E and F). The detection of NHEJ pathway proteins showed that the expressions of 53BP1 (Fig. 4G and H) and DNA-PKcs (Fig. 4G and I) were significantly increased due to uranyl nitrate at a dose of 5 μM and lower doses. The expression of DNA-PKcs and 53BP1 proteins declined when the treatment doses were increased to 10 μM and higher. There was no significant change in the expression of Ku80 and Ku70, another two components of DNA-PK complex, in BEP2D cells treated with uranyl nitrate at the tested concentrations of 1–40 μM.
Fig. 4. Effect of uranyl nitrate on the expression of DNA repair proteins. (A) Effect of different concentrations of uranyl nitrate on HR pathway proteins in BEP2D cells, detected by western blot analysis. (B)–(F) The densitometric quantitation of the expression of proteins by western blot analyses for ATM (B), BRCA1 (C), EXO1 (D), RAP80 (E) and RAD51 (F). (G) Effect of different concentrations of uranyl nitrate on NHEJ pathway proteins in BEP2D cells, detected by western blot analysis. (H) The densitometric quantitation of the expression of proteins by western blot analyses for 53BP1. (I) The densitometric quantitation of the expression of proteins by western blot analyses for DNA-PK complex. The intensities of western blot hybridization bands were quantified using the Quantity One software. β-Actin was used as the sample loading control. *P < 0.05, **p < 0.01.
4. Discussion
Uranyl nitrate was considered as a weakly radioactive compound, which consists of depleted form of 238U with a specific activity of approximately 0.3 μCi g–1. However, the radiation level from its radioactive decay is very low, and the radiological toxicity was not taken into account in this study. Therefore, the cell killing is attributed to its chemical toxicity rather than the radioactive toxicity. The assay of clonogenic cell death demonstrated that 40 μM uranyl nitrate resulted in approximately 60% cell reproductive death (Fig. 1C), while the apoptotic cell death was about 25% (Fig. 1D), implying that multiple types of cell death could be induced by uranyl nitrate, e.g. mitotic cell death/catastrophe and pyroptosis. Cell killing due to several toxic environmental chemicals is associated with oxidative stress. Although uranyl nitrate could increase the cellular ROS, the augmented level was very limited. A high level of DNA damage has been detected in uranyl nitrate-treated cells (Fig. 2D and E). The increased expression of γH2AX also demonstrated that uranyl nitrate triggered the DNA damage response in BEP2D cells (Fig. 2A–C). We considered that the increased level of DNA damage was due to its chemical toxicity through inhibiting cellular DNA repair activity, which consequently led to the accumulation of spontaneous DNA damage.
Importantly, inactivation of intrinsic DNA repair pathways could sharply enhance the yield of endogenous or autonomous DNA damage as well as the residual DNA damage level after exposure to exogenous genotoxic agents such as ionizing radiation, ultraviolet light and various chemicals. As mentioned earlier, western blot analysis of the DNA damage biomarker γH2AX and the neutral comet assay demonstrated that uranyl nitrate increased the level of DNA DSBs in BEP2D cells in a dose-dependent manner. The increased ROS level might be one of the causes for the cytotoxicity and genotoxicity of uranyl nitrate, but not the main cause as the increased level of ROS is limited.
Some epidemiological studies have also found that elevated blood uranium levels were associated with increased DNA damage32,33 and decreased DNA repair response in human populations.17–19 However, the mechanism of uranium interfering intrinsic DNA repair activity is still not clear. The higher level of DNA damage in the uranium-exposed cells could be due to either increased induction as a consequence of oxidative stress or the inhibition of repair capability. Our results indicated that uranyl nitrate inhibited both HR and NHEJ repair pathways of DNA double-strand breaks. Although a potential hormetic effect on NHEJ activity was displayed by a low dose (<5 μM) of uranyl nitrate, its mechanism and biological significance remained yet to be explored. HR is a precise repair pathway, which can restore the genomic integrity and fidelity of the broken DNA by utilizing sister chromatids as a template for repair. NHEJ is an inaccurate repair pathway through direct religation of DSBs without the presence of homology DNA. Therefore, deregulated overexpression of NHEJ activity could also lead to genomic instability and be dangerous for the cells. We utilized HR and NHEJ reporter plasmids to analyse the effect of uranyl nitrate on DNA repair and found that a low dose of uranyl nitrate inhibited HR but promoted NHEJ activity, whereas a high dose of uranyl nitrate inhibited both HR and NHEJ. This effect of uranyl nitrate on DNA repair is similar to what has been reported for other carcinogenic and co-carcinogenic metals such as arsenic.34,35 Importantly, DNA double-strand break is a critical type of DNA damage induced by ionizing radiation. The effect of uranyl nitrate on DSB repair pathways can further increase the radiological toxicity of the radioactive uranium.
Previous reports suggested that some zinc finger-containing DNA repair proteins such as C2H2 zinc finger proteins Sp1, Aart and XPA could be inhibited by uranium.36,37 Furthermore, the uranyl ion inhibited the activity of DNA repair protein PARP-1 and resulted in zinc loss from the protein.37 This inhibition could be attributed to the direct binding action of uranyl ion to the zinc finger motif, which disturbs the activity of the protein or enzyme. However, no evidence was found to support that uranium might also inhibit non-zinc finger DNA-binding proteins.36 Our results indicated that uranyl nitrate caused the inhibition of expression of a set of DNA repair proteins of the HR pathway, including ATM, BRCA1, RAP80 and EXO1. ATM is a key kinase mediating its downstream target recruitment to DNA damage sites and contributes to the activation of DNA damage response signal.24 EXO1 is involved in the progressive stage of DNA end resection.24 RAP80 forms a complex with BRCA1 and targets BRCA1 to DNA damage sites. BRCA1 promotes HR by activating end resection. In contrast, 53BP1 forms a barrier that inhibits DNA end resection.24 Cells depleted of ATM, BRCA1, RPA80 and EXO1 exhibited the phenotype of genomic and chromosomal instability.38–40 We also found that RAP80, another regulator of the HR pathway, was inhibited by a high dose of uranyl nitrate. Rad51 is also a key component of HR and it forms Rad51 recombinase filaments around the broken single-stranded DNA to promote HR. Unexpectedly, our result indicated that 5, 10 and 20 μM of uranyl nitrate increased the expression of RAD51 although HR activity was inhibited at the doses levels. It is worth noting that a previous study suggested that RAD51 did not affect uranium-induced cytotoxicity or genotoxicity.41 Moreover, a high level of RAD51 was involved in tumour progression by destabilizing the genome.42
In the NHEJ pathway, DNA-PKcs plays a critical role in stabilizing DSB ends and prevents end resection. 53BP1 mentioned earlier has been suggested to promote NHEJ by increasing the stability and mobility of DSBs to find each other for productive ligation.24 DNA-PKcs and 53BP1 were found to be induced by a low dose of uranyl nitrate but inhibited by high doses, suggesting that the abnormal NHEJ activity induced by uranyl nitrate could be associated with the alterations of DNA-PKcs and 53BP1.
Uranyl ion is a confirmed highly toxic carcinogenic metal ion. Numerous epidemiological studies found a strong association between high levels of uranium exposure and increased risk for lung cancer.8,9 This study confirmed that uranyl nitrate increased the cellular DNA damage level as a consequence of inhibition of DNA DSB repair efficiency, particularly the HR pathway. Our data have provided further mechanistic explanation for the increased risks of uranium on genomic instability and carcinogenesis.
5. Conclusion
This study provided evidence that uranyl nitrate inhibits both HR and NHEJ pathways of DNA DSB repair, which leads to increased DNA damage and genotoxicity as a result of the accumulation of spontaneous DNA damages. The expression of multiple HR pathway proteins including ATM, BRCA1, RPA80 and EXO1 was depressed by uranyl nitrate. The expression of NHEJ pathway proteins DNA-PKcs and 53BP1 was also depressed. Further investigation is required to determine how uranyl nitrate affects the expression of the abovementioned proteins. The available data enable us to imagine that the chemical toxicity of uranium leads to inhibition of cellular DNA repair capacity, which can aggravate its radiological toxicity.
Conflicts of interest
There is no conflict to be declared.
Acknowledgments
This study was supported by the grants from National Key Basic Research Program (973 Program) of MOST, China (Grant No. 2015CB910601), the National Natural Science Foundation of China (Grant No. 31370843, 31500681) to P. K. Zhou and the National Natural Science Foundation, China (31570853 and 81602799) to T. Ma.
References
- Sansone U., Stellato L., Jia G., Rosamilia S., Gaudino S., Barbizzi S., Belli M. Radiat. Prot. Dosim. 2001;97:317–320. doi: 10.1093/oxfordjournals.rpd.a006679. [DOI] [PubMed] [Google Scholar]
- Jia G., Belli M., Sansone U., Rosamilia S., Gaudino S. Appl. Radiat. Isot. 2005;63:381–399. doi: 10.1016/j.apradiso.2005.05.042. [DOI] [PubMed] [Google Scholar]
- Besic L., Muhovic I., Asic A., Kurtovic-Kozaric A. J. Environ. Radioact. 2017;172:207–217. doi: 10.1016/j.jenvrad.2017.03.013. [DOI] [PubMed] [Google Scholar]
- Song Y., Salbu B., Heier L. S., Teien H. C., Lind O. C., Oughton D., Petersen K. Aquat. Toxicol. 2012;112–113:62–71. doi: 10.1016/j.aquatox.2012.01.019. [DOI] [PubMed] [Google Scholar]
- Eidson A. F. Health Phys. 1994;67:1–14. doi: 10.1097/00004032-199407000-00001. [DOI] [PubMed] [Google Scholar]
- Miller A. C., Stewart M., Rivas R. Biochimie. 2009;91:1328. doi: 10.1016/j.biochi.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Tesinska E. Prague Med. Rep. 2009;110:165–172. [PubMed] [Google Scholar]
- Gilliland F. D., Hunt W. C., Archer V. E., Saccomanno G. Health Phys. 2000;79:365. doi: 10.1097/00004032-200010000-00004. [DOI] [PubMed] [Google Scholar]
- Archer V. E., Coons T., Saccomanno G., Hong D. Y. Health Phys. 2004;87:480–489. doi: 10.1097/01.hp.0000133216.72557.ab. [DOI] [PubMed] [Google Scholar]
- Yazzie M., Gamble S. L., And E. R. C., Stearns D. M. Chem. Res. Toxicol. 2003;16:524–530. doi: 10.1021/tx025685q. [DOI] [PubMed] [Google Scholar]
- Periyakaruppan A., Kumar F., Sarkar S., Sharma C. S., Ramesh G. T. Arch. Toxicol. 2007;81:389–395. doi: 10.1007/s00204-006-0167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periyakaruppan A., Sarkar S., Ravichandran P., Sadanandan B., Sharma C. S., Ramesh V., Hall J. C., Thomas R., Wilson B. L., Ramesh G. T. Arch. Toxicol. 2009;83:595–600. doi: 10.1007/s00204-008-0396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmash S. A., Smirnova V. S., Karp O. E., Usacheva A. M., Berezhnov A. V., Ivanov V. E., Chernikov A. V., Bruskov V. I., Gudkov S. V. J. Environ. Radioact. 2014;127:163. doi: 10.1016/j.jenvrad.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Zhang S. M., Shang Z. F., Zhou P. K. Toxicol. Res. 2015;4:613–622. [Google Scholar]
- Xu Y. M., Zhou Y., Chen D. J., Huang D. Y., Chiu J. F., Lau A. T. Toxicol. Res. 2013;2:280–287. [Google Scholar]
- Huang J., Wu G., Zeng R., Wang J., Cai R., Ho J. C., Zhang J., Zheng Y. Toxicol. Res. 2017;6:324–332. doi: 10.1039/c6tx00372a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legator M. S. Environ. Health Perspect. 1995;103:466–470. doi: 10.1289/ehp.95103466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au W. W., McConnell M. A., Wilkinson G. S., Ramanujam V. M. S., Alcock N. Mutat. Res., Fundam. Mol. Mech. Mutagen. 1998;405:237–245. doi: 10.1016/s0027-5107(98)00141-9. [DOI] [PubMed] [Google Scholar]
- Au W. W., Giri A. K., Ruchirawat M. Int. J. Hyg. Environ. Health. 2009;213:32–39. doi: 10.1016/j.ijheh.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Wilson J., Zuniga M. C., Yazzie F., Stearns D. M. J. Appl. Toxicol. 2015;35:338–349. doi: 10.1002/jat.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode E. L., Ulrich C. M., Potter J. D. Cancer Epidemiol., Biomarkers Prev. 2002;11:1513. [PubMed] [Google Scholar]
- Smith T. R., Levine E. A., Freimanis R. I., Akman S. A., Allen G. O., Hoang K. N., Liumares W., Hu J. J. Carcinogenesis. 2008;29:2132. doi: 10.1093/carcin/bgn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavragani I. V., Laskaratou D. A., Frey B., Candéias S. M., Gaipl U. S., Lumniczky K., Georgakilas A. G. Toxicol. Res. 2016;5:12–33. doi: 10.1039/c5tx00222b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., Elledge S. J. Mol. Cell. 2010;40:179. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono M., Niimi A., Oike T., Hagiwara Y., Sato H., Sekine R., Yoshida Y., Isobe S. Y., Obuse C., Nishi R., Petricci E., Nakada S., Nakano T., Shibata A. Cell Rep. 2017;18:520–532. doi: 10.1016/j.celrep.2016.12.042. [DOI] [PubMed] [Google Scholar]
- Kumar R., Cheok C. F. DNA Repair. 2014;15:54–59. doi: 10.1016/j.dnarep.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Kim H., Chen J., Yu X. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- Tomimatsu N., Mukherjee B., Catherine Hardebeck M., Ilcheva M., Vanessa Camacho C., Louise Harris J., Porteus M., Llorente B., Khanna K. K., Burma S. Nat. Commun. 2014;5:3561. doi: 10.1038/ncomms4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J., Huang Y. C., Xu Q. Z., Zhou L. J., Shang Z. F., Huang B., Wang Y., Liu X. D., Wu D. C., Zhou P. K. BMC Mol. Biol. 2010;11:18. doi: 10.1186/1471-2199-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend L. E. M., Jasin M., Krawczyk P. M. Methods Enzymol. 2013;546:175. doi: 10.1016/B978-0-12-801185-0.00009-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo N., Cheng A., Huang N., Stark J. M. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp W., Plappert U., Muller W. U., Rehn B., Schneider J., Braun A., Bauer P. C., Vahrenholz C., Presek P., Brauksiepe A. Radiat. Environ. Biophys. 2000;39:275–282. doi: 10.1007/s004110000072. [DOI] [PubMed] [Google Scholar]
- Lourenço J., Pereira R., Pinto F., Caetano T., Silva A., Carvalheiro T., Guimares A., Gonalves F., Paiva A., Mendo S. Toxicology. 2013;305:89–98. doi: 10.1016/j.tox.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Beyersmann D., Hartwig A. Arch. Toxicol. 2008;82:493–512. doi: 10.1007/s00204-008-0313-y. [DOI] [PubMed] [Google Scholar]
- Morales M. E., Derbes R. S., Ade C. M., Ortego J. C., Stark J., Deininger P. L., Roy-Engel A. M. PLoS One. 2016;11:e0151367. doi: 10.1371/journal.pone.0151367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsock W. J., Cohen J. D., Segal D. J. Chem. Res. Toxicol. 2007;20:784–789. doi: 10.1021/tx600347k. [DOI] [PubMed] [Google Scholar]
- Cooper K. L., Dashner E. J., Tsosie R., Cho Y. M., Lewis J., Hudson L. G. Toxicol. Appl. Pharmacol. 2016;291:13. doi: 10.1016/j.taap.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derheimer F. A., Kastan M. B. FEBS Lett. 2010;584:3675. doi: 10.1016/j.febslet.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Petit S. A., Ficarro S. B., Toomire K. J., Xie A., Lim E., Cao S. A., Park E., Eck M. J., Scully R. Cancer Discovery. 2014;4:1430–1447. doi: 10.1158/2159-8290.CD-13-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono M., Niimi A., Oike T., Hagiwara Y., Sato H., Sekine R., Yoshida Y., Isobe S. Y., Obuse C., Nishi R. Cell Rep. 2014;18:520. doi: 10.1016/j.celrep.2016.12.042. [DOI] [PubMed] [Google Scholar]
- Holmes A. L., Joyce K., Xie H., Falank C., Hinz J. M., Sr J. P. W. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2014;762:1–9. doi: 10.1016/j.mrfmmm.2014.02.001. [DOI] [PubMed] [Google Scholar]
- Richardson C., Stark J. M., Jasin M. Oncogene. 2004;23:546–553. doi: 10.1038/sj.onc.1207098. [DOI] [PubMed] [Google Scholar]