Abstract
Plants regulate responses to salt stress using biological pathways, such as signal perception and transduction, photosynthesis, and energy metabolism. Little is known about the genetics of salt tolerance in Rosa chinensis. Tineke and Hiogi are salt-tolerant and salt-sensitive varieties of R. chinensis, respectively, and are good choices for studying salt-tolerance genes. We studied leaf and root tissues from 1-year-old Hiogi and Tineke plants simultaneously grown under the same conditions. A 0.4%-mmol/L salt ion mixture was added to the basic growth medium. Illumina sequencing was used to identify differentially expressed transcripts. GO and KEGG pathway enrichment analyses were performed to identify differentially expressed genes. We identified many differentially expressed genes associated with salt tolerance. The abscisic acid-dependent signaling pathway was the main pathway that mediated the salt stress response in R. chinensis. Two pathways (plant hormone signal transduction and glutathione metabolism) were also active in salt stress responses in R. chinensis. The difference in salt tolerance in the cultivars was due to different gene sensitivity to salt in these two pathways. Roots also play a role in salt stress response. The effects of salt stress in the roots are eventually manifested in the leaves, causing changes in processes such as photosynthesis, which eventually result in leaf wilting. In Tineke, Snrk2, ABF, HSP, GSTs, and GSH1 showed high activity during salt stress, indicating that these genes are markers of salt tolerance.
Introduction
Environmental stresses or abiotic factors, such as salinity, drought, heavy metals, and low temperature, often have a strong influence on plant survival and development. Plants have complex adaptive responses to abiotic stresses [1]. In their adaptation to salt stress, plants regulate many biological pathways, such as signal perception and transduction, photosynthesis, and energy metabolism [2]. Research on salt and drought has identified many stress-inducible regulators, including the upregulation of aquaporins, catalase, peroxidase and proline (PRO) accumulation [3]. In addition, abscisic acid (ABA)-dependent and -independent signaling pathways in plants are induced by stress [4]. Many transcription factors, such as ABA-binding factor (ABF) and drought-responsive elements are involved in the biological process of ABA [5]. Additionally, the accumulation of sodium and chloride in the cytoplasm results in cytotoxicity by reactive oxygen species (ROS) [6], which can lead to protein and lipid degradation and cell disruption [7]. Some salt-tolerant plant species have evolved mechanisms to cope with osmotic and ionic toxicity, including salt removal and extrusion of Na+ via an Na+/H+ antiporter in the vacuolar membrane [8]. Rosa chinensis (Chinese rose) is widely cultivated as an ornamental plant around the world. Although production of R. chinensis is increasing, environmental stress is an important factor limiting its growth. The successful cultivation of R. chinensis is influenced by salt stress [9], which is one of the most harmful factors affecting growth and productivity. Some studies have demonstrated the reduction of salt stress in R. chinensis, but more research is necessary. These studies have focused on petal development, flower bloom, and pests [10], but there is no information on the salt stress-related signaling pathways. Lack of molecular studies is an obstacle to understanding the molecular mechanisms underlying salt adaptation in R. chinensis. The development of next-generation sequencing technology has increased understanding of plant responses to drought and salt stress. Transcriptome sequencing techniques have been widely used in plant genomics research, including studies on the non-model plant tomato [11]. Therefore, we used RNA-seq technology to screen for salt-tolerance genes and to study the mechanisms of the salt stress response.
The Tineke rose is a salt-tolerant variety of R. chinensis, whereas the Hiogi rose is a salt-sensitive variety [12, 13]. The global transcriptome patterns in the roots and leaves of the two cultivars were compared to identify genes linked to salt tolerance. Sequence analysis revealed a large number of differentially expressed genes (DEGs) in R. chinensis under salt stress. Based on sequence similarities, we determined that genes related to salt responses and pathways are strongly represented within the DEGs.
Materials and methods
Plant material preparation and salt-stress treatment
Plant samples used were collected from the Tianjin Binhai New Area botanical garden with the approval of Tianjin Taida greening Group Co., Ltd. And no endangered or protected species involved in the experiment ground. One-year-old Hiogi and Tineke plants, planted at the same time and grown under the same natural environment conditions, were the study materials. Seedlings showing consistent growth were cultured in triangular cone flasks. Salt treatment was performed 10 d after normal growth conditions by adding a 1:1:3 ratio of 0.4% mmol/L NaCl : Na2SO4 : NaHCO3 to the base growth medium. Leaf and root changes were kept observing during salt stress. After 2 d the leaves began to wilt and the roots turned yellow and became soft. At 4 d, the leaves began to curl and the roots began to blacken and inactivate. At 7 d the leaves were yellow and the roots festering and at 9 d, the salt-sensitive Hiogi plants were dead, but the salt-tolerant Tineke plants remained alive. According to the phenotypic changes, the salt-tolerant and salt-sensitive properties can also be defined. Three young leaves and root tissues were taken for transcriptome sequencing at 2, 4, 7, and 9 d following salt treatment in Hiogi and Tineke, respectively (including 45 tissue samples, S1 Table). All fresh leaf and root tissues were frozen in liquid nitrogen and stored at −80°C for RNA extraction. For Hiogi, HL-0d, HL-2d, HL-4d, and HL-7d represent the control and salt treatment groups for each stage of the leaf tissue; HR-0d, HR-2d, HR-4d, and HR-7d represent the control and salt treatment groups for each stage of the root tissue. Thus, TL-0d and TL-2d, TL-4d, TL-7d and TL-9d represent the control and salt treatment groups for each stage in Tineke leaf tissue; TR-0d, TL-2d, TL-4d, TL-7d, and TL-9d represent the control and salt treatment groups for each stage of the root tissue.
Determination of physiological indexes, photosynthetic gas exchange and tissue NaCl content
Water content, superoxide dismutase (SOD), PRO, and malondialdehyde (MDA) levels were measured in leaf and root samples. Chlorophyll a and b levels were also measured in leaves. Tissue water content was calculated based on fresh and dry weight. Total levels of SOD (LP-P0296, Lanpaibio, China), Pro (LP-P0296, Lanpaibio, China), and MDA (LP-P0296, Lanpaibio, China) were determined by ELISA (enzyme-linked immunosorbent assay) according to the protocol. Standard curves were established in accordance with the reagent instructions (S2 Table). Chlorophyll a and chlorophyll b levels were measured by spectrophotometric methods that were used in a previous study [14]. Five samples were taken as biological repeats in each group. Because the roots were ulcerated after 7 d of salt treatment, only the control roots, 2 d, and 4 d samples were used for measurement. Hiogi died after 7 d of salt treatment therefore, no physiological indexes were measured after 4 d. Tineke died after 9 d of salt treatment therefore we measured the physiological indexes of Tineke at 9 d. Photosynthetic rate (A), stomatal conductance (Gs), intercellular CO2 concentration (Ci) and transpiration rate (E) were measured using a LCpro-SD portable photosynthesis system (ADC Bioscientific Ltd., UK) according to previous studies [15, 16]. In addition, the NaCl content in leaf was also measured with an atomic absorption spectrophotometer. Briefly, the dried Rosa chinensis leaf samples were pulverized and grinded, and 0.5g was placed in muffle furnace at 650 C ashing for 8h. The ash was dissolved in 5% nitric acid and was fixed to 100ml with deionized water. The content of Na+ was measured using an atomic absorption spectrophotometer (AAS) (TAS-990, Purkinje, China). Standard curve was established with Na+ standard liquid (S3 Table).
RNA extraction, library construction, and RNA sequencing
Total RNA extraction was performed using the TRIzol (Invitrogen, CA, USA) method, following the manufacturer instructions. RNA integrity was verified by 1% agarose gel electrophoresis and the concentration was measured using a Qubit Fluorometer (CA, USA). After RNA preparation, a total of at least 3 μg of RNA (treated with DNase) was used for the preparation of RNA sequencing libraries with a Lybay RNA Library Prep Kit (LB0014, Lybay, China). A total of 45 sequence libraries were constructed for each biological repeat sample. The Agilent Bioanalyzer 2100 system was used to assess library quality. Clustering of sequence libraries was performed on a cBot Cluster Generation System. After cluster generation, the enriched cDNA was sequenced using an Illumina Hiseq 4000 platform by Novogene Co., Ltd.
RNA sequence assembly and DEG analysis
Clean, high-quality reads from eight samples were merged together and assembled using the Trinity package to construct unique consensus sequences as RNA-seq reference sequences. The annotation of the assembled transcriptome was performed with NR, NT, Pfam, COG, KEGG, and GO. The specific software parameter settings were obtained from previous research [17]. For sequence data in each sample, the EdgeR software package was used to statistically analyze the read counts, adjusted through one scaling normalized factor. Gene expression levels for each sample were determined using RSEM [18]. To obtain global gene expression patterns in different conditions, we performed pairwise comparisons using the DEseq for screening DEGs [19].
Functional annotation and analysis of the transcripts
The annoated transcripts in NR database were further annotated with GO, and then classified into biological processes, molecular functions and cellular components using Blast2GO software [20, 21] according to previous studies [15, 22, 23]. After gene categories analysis, GO and KEGG enrichment were performed with GOseq software and KOBAS, respectively [24, 25]. GO terms (molecular function, cellular component, and biological process) and KEGG pathways with corrected p values (q value) of < 0.05 were considered significantly enriched. The KEGG Orthology database was used to statistically test the enrichment of DEGs in KEGG pathways.
Validation of gene expression using reverse transcriptase real-time PCR (qPCR)
The qPCR experiment was performed using a CFX96 real-time PCR detection system (Bio-Rad, Hercules, USA) with three biological repetitions and three technical repetitions for each sample. The PCR reaction mixture (20 μl) included 75 ng template RNA, 0.4 μM primers, and 10 μl RNA-direct SYBR® Green Realtime PCR Master Mix, according to the manufacturer’s instructions. The total RNA used in qPCR was treated with DNase. GAPDH was selected as reference gene according to previous studies [20, 21]. And the expression of GAPDH was verified to be stable with the forward primer ‘CTCAGACTCCTCCTTGATAGC’ and reverse primer ‘TTCTGCCTGCTCTCAATGG’ (S4 Table). Relative gene expression levels were calculated using the 2−ΔΔCT method, and regression analysis was performed between qPCR and RNA sequencing, including the salt tolerance candidate genes among Tineke and Hiogi.
Results
Effect of salt treatment on SOD, Pro, MDA, and other physiological indexes in R. chinensis
At the 2 d of salt treatment, the Tineke leaf showed slight curl and the root begins to turn yellow. But for Hiogi, the leaf was obviously curled and the root color turned yellow. At the 4 d of salt stress, the leaf in Tineke obviously curled with root color darkens. While the leaf in Hiogi start to turn yellow, and the root become black. At the 7 d, the Tineke leaf start to turn yellow, and the root died. For Hiogi, the leaf was withered with the root died. At the 9 d, only several leaves are alive in Tineke, and leaves on Hiogi were all died. The content of SOD, PRO, and MDA were measured base on standard curves. The result showed that SOD activity rapidly declined after 2nd day of salt treatment in Hiogi, but in Tineke, until 4 days salt treatment later there showed a precipitous decline in SOD (Table 1). The PRO and MDA concentrations of HL dropped firstly then raised after the 7th day. For TL, the PRO and MDA showed a significant decline (p<0.05) in 2nd day and then increased after 4th day and maintained a relatively stable content. For root tissue, both of PRO and MDA showed a continuous decreasing trend with the increase of salt treatment time. After salt treatment, water content significantly decreased (p<0.05) in all tissues, and in the later stage of salt treatment, the trend of decline tended to be gentle. The water content in Tineke root was higher than that in Hiogi in the 4d salt treatment. Chlorophyll a and b levels in Hiogi significantly decreased (p <0.05) after 2 d of salt stress, but the levels of Tineke significantly decreased (p<0.05) only after 4 d.
Table 1. Concentrations of physiological indexes.
| SOD Content (pg/g) |
PRO Content (ng/g) |
MDA Content (ng/g) |
Water content (%) | Chlorophyll a (mg/g) | Chlorophyll b (mg/g) | |
|---|---|---|---|---|---|---|
| HL-0d | 952.63 | 57.29 | 10.47 | 78.91 | 1695.99 | 629.94 |
| HL-2d | 72.56 | 32.03 | 3.19 | 73.02 | 1428.78 | 608.63 |
| HL-4d | 58.78 | 28.67 | 2.73 | 65.54 | 983.94 | 597.49 |
| HL-7d | 64.39 | 34.10 | 4.18 | 53.33 | 913.88 | 585.70 |
| HR-0d | 927.84 | 51.98 | 7.27 | 90.21 | — | — |
| HR-2d | 91.03 | 30.15 | 3.11 | 82.66 | — | — |
| HR-4d | 80.29 | 25.32 | 2.41 | 78.03 | — | — |
| TL-0d | 1032.35 | 63.56 | 9.08 | 82.10 | 1648.40 | 827.74 |
| TL-2d | 805.71 | 30.85 | 2.92 | 69.33 | 1632.52 | 729.67 |
| TL-4d | 59.32 | 31.02 | 5.67 | 60.17 | 1007.03 | 489.15 |
| TL-7d | 57.19 | 33.19 | 5.40 | 57.33 | 918.44 | 526.29 |
| TL-9d | 56.03 | 33.75 | 6.05 | 57.67 | 630.90 | 288.78 |
| TR-0d | 829.42 | 49.07 | 9.48 | 90.14 | — | — |
| TR-2d | 661.95 | 33.17 | 4.26 | 85.15 | — | — |
| TR-4d | 88.29 | 30.99 | 3.99 | 84.67 | — | — |
Effect of salt treatment on photosynthetic gas exchange and NaCl content in leaf
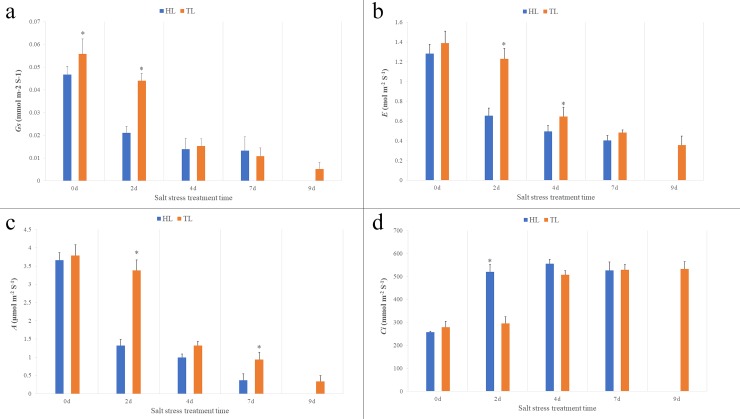
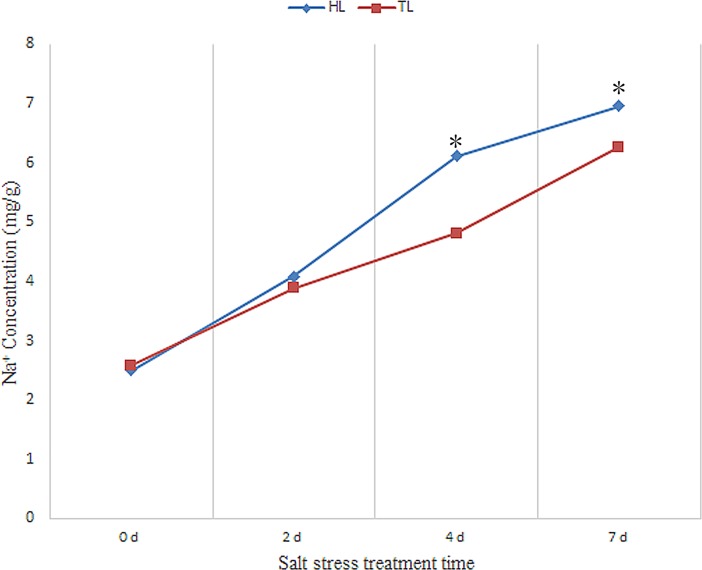
Photosynthetic gas exchange is an indication of plant health status, by which the salt stress damage to plants can be indirectly reflected. The photosynthesis rate (A), stomatal conductance (gs) and transpiration rate (E) reduced with the increasing salt treatment time (Fig 1A to 1C). The internal CO2 concentration (ci) increased with the increasing salt treatment time (Fig 1D). For Hiogi, the photosynthesis rate, stomatal conductance and transpiration rate sharply reduced in the 2nd day of salt treatment. While for Tineke, the sharply reduction occurred in the 4th day of salt stress. For the internal CO2 concentration, there were little changes after 2 days and 4 days salt treatment in Hiogi and Tineke, respectively. In addition, the NaCl content in leaf showed that the change trend of NaCl concentration, with the increase of salt stress time, were different between the two rose varieties (Fig 2). For Hiogi, the concentration of NaCl raised sharply after 2d. For Tineke, with the increase of salt treatment time, the concentration of NaCl gradually increased. At 4d and 7d, the NaCl content in Tineke was significantly lower than that in Hiogi.
Fig 1. Photosynthetic gas exchange in different salt stress treatment time.
a: Stomatal conductance (Gs), b: Transpiration rate (E), c: Photosynthetic rate (A), d: Internal CO2 concentration (Ci). The values are means +SD (Standard Deviation) (n = 5). The asterisk indicates a significant difference (p<0.05).
Fig 2. Na+ content in leaf tissue.
The values are means (n = 5). The asterisk indicates a significant difference (p<0.05).
RNA sequencing, assembly, and annotation
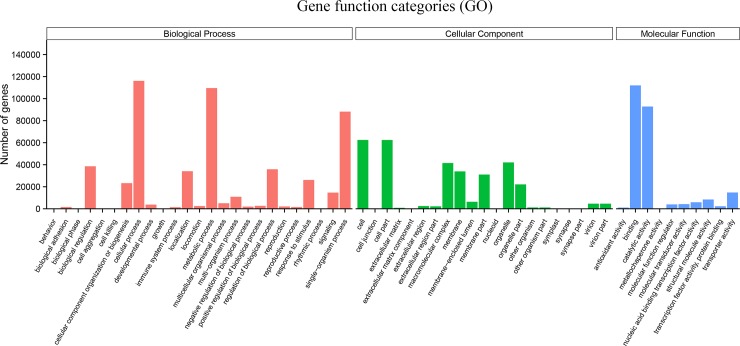
A total of 168.97 GB of raw data from the 45 libraries was generated with an average sequencing error rate of 0.02% and an average GC content of 45.98% (S5 Table). The data ratio of sequencing quality to Q20 and Q30 was 96.90% and 92.19%, respectively. All sequence data were uploaded to the SRA database with the SRA accession number SRP128235. As R. chinensis did not have an appropriate reference genome sequence, the Trinity method was used to assemble, de novo, all of the clean reads. In total, 1,126 million high-quality reads were assembled into 465,404 unigenes with an N50 length of 1,302 bp and an N90 length of 443 bp (Table 2). To obtain a general idea of the total sequenced mRNA, the transcripts were searched against the Nr, Nt, Pfam, KOG, Swiss-Prot, KEGG, and GO databases. Altogether, 319,798 unigenes were successfully matched with at least one database, with 239,038 (51.36%), 214,867 (46.16%), 102,631 (22.05%), 205,637 (44.18), 205,752 (44.2), 207,678 (44.62%), and 89,076 (19.13%) annotated transcripts (17.15%) showing a significant hit against the Nr, Nt, KEGG, Swiss-Prot, Pfam, GO, and KOG database, respectively. Functional annotation showed that biological processes, molecular functions, and cellular component categories occupied 47.98%, 29.42%, and 22.60% of the total sequence, respectively, including the annotated terms response to stress, response to abiotic stimulus, response to stimulus, and other terms. Based on a comparison with the KEGG database, 111,229 (34.78%) of the 319.798 annotated unigenes had significant matches and were thus assigned to 130 KEGG pathways, including “carbon metabolism” (5,455 unigenes), “ribosome” (6,549 unigenes), “purine metabolism” (2,460 unigenes), and “plant hormone signal transduction” (1,342 unigenes), which were all related to probable resistance responses. The GO classification results showed that 1,086,389 transcripts were divided into 3 categories: 521,204 (47.98%) in biological process, 319,655 (29.42%) in cellular components and 245,530 (22.60%) in molecular function (Fig 3). The GO classification analysis showed that these transcripts were mainly involved in the biological processes including signaling, negative regulation of biological process, response to stimulus, and cellular process; For cellular components, cell, cell part, and organelle were the top three GO terms; For molecular function categories, binding, catalytic activity, and transporter activity were mainly involved.
Table 2. Summary of transcriptome assembly.
| Category | Number | Total number | Mean length (bp) | N50 (bp) | N90 (bp) | |||
|---|---|---|---|---|---|---|---|---|
| 200–500 bp | 500bp–1kb | 1–2 kb | >2 kb | |||||
| transcripts | 845713 | 170289 | 91293 | 46459 | 1153754 | 539 | 771 | 234 |
| unigenes | 167193 | 160553 | 91199 | 46459 | 465404 | 941 | 1302 | 443 |
Fig 3. Gene function categories in GO.
DEG identification and GO and KEGG functional analysis in leaf tissue
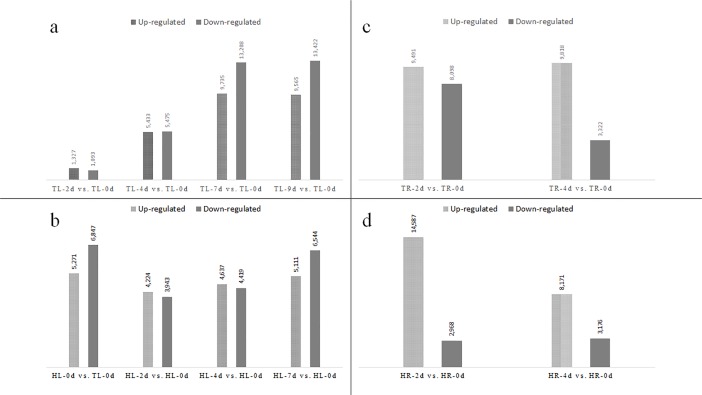
Comparison between Tineke and Hiogi was performed for the untreated and salt-treated groups. Among DEGs in HL-0d vs. TL-0d, HL-2d vs. TL-2d, HL-4d vs. TL-4d, and HL-7d vs. TL-7d, 1,734 DEGs overlapped (S1 Fig). There were 5,271 up regulated DEGs and 6,847 down regulated DEGs in HL-0d vs. TL-0d including PYR/PYL, PP2C, HEXA, crtB. In addition, ADP binding, protein binding, estrogen metabolic process, and steroid dehydrogenase activity were all enriched base on the down-regulated DEGs but no GO terms enriched base on up-regulated DEGs. This indicates that Tineke had higher activity in ADP binding, protein binding and other molecular function. The KEGG enrichment showed that Porphyrin and chlorophyll metabolism, Carotenoid biosynthesis, Plant hormone signal transduction, and RNA degradation were significantly enriched (p<0.05) base on the up regulated DEGs in HL-0d vs. TL-0d, which reveal that the expression of DEGs in these pathways of Hiogi were higher than that of Tineke. In the process of salt stress treatment, Hiogi was more affected than Tineke because the expression of unigenes in Hiogi, but not in Tineke, greatly changed in the first 2 d, and this continued until 7 d (Fig 4A). In contrast, the number of DEGs in Tineke slightly increased in the first 2 d and then increased with the continuation of salt treatment until 9 d (Fig 4B). This result indicates that the Tineke response to salt stress involved maintenance of a relatively stable internal state. There were more DEGs expressed in roots than in leaves (Fig 4C and 4D). In the comparison of HR-0d vs. TR-0d and HR-2d vs. TR-2d, 18,121 and 21,612 DEGs were found, respectively. DEGs in the comparison of HR-0d vs. TR-0d including MYC, ETR, LOX, and WRKY. The KEGG enrichment analysis showed that plant hormone signal transduction, ADP binding, protein binding, RNA degradation, and thiamine metabolism were the prominent pathways. Through the data, the expression pattern showed large difference in root tissue. Plant hormone signal transduction and ADP binding were the most important difference between the two Rosa chinensis varieties. For HR-4d vs. TR-4d, only 5,154 DEGs were found (S2 Fig). The DEG results showed that the numbers of upregulated and downregulated genes were similar in Tineke after 2 d of salt treatment and the greatest difference in the number of upregulated and downregulated genes occurred at 4 d. For Hiogi, the number of upregulated genes was much higher than the number of downregulated genes at 2 d of salt treatment (Fig 5). Among all the DEGs mentioned above, 2,132 DEGs overlapped including heat shock protein (HSP), CYP450, and ERFs, which were highly expressed in Tineke. The glutathione synthetase-related gene glutamate–cysteine ligase (GSH1) was highly expressed in Tineke, which enhanced the activity of the GSH metabolism pathway. Most DEGs were associated with secondary metabolites in the roots and some were annotated as transcription factors, protein modification and degradation factors, and phytohormone response factors.
Fig 4. Number of DEGs in Rosa chinensis.
Number of differential genes between different comparisons. a. Number of DEGs in Tineke leaf; b. Number of DEGs in Hiogi leaf; a. Number of DEGs in Tineke root; b. Number of DEGs in Hiogi root.
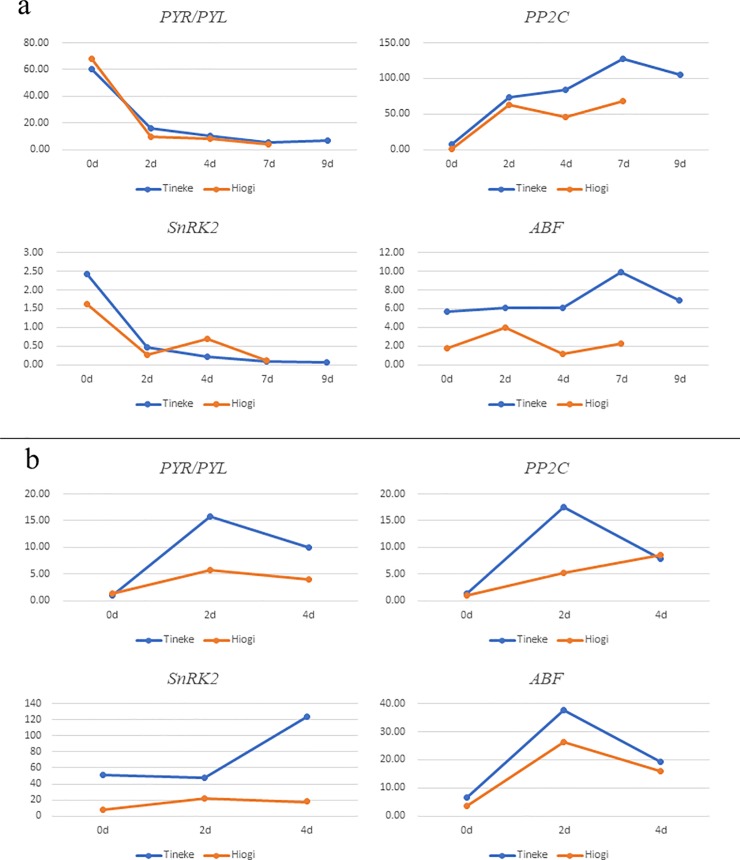
Fig 5. Expression of PYR/PYL, Snrk2, PP2C, and ABF.
a. PYR/PYL, Snrk2, PP2C, and ABF expression in leaf tissue. b. PYR/PYL, Snrk2, PP2C, and ABF expression in root tissue.
GO analysis showed that 25 and 7 GO terms were significantly enriched (p<0.05) in HL-0d vs. TL-0d and HL-2d vs. TL-2d, respectively, and no terms were enriched at 4 d and 7 d. The anaphase-promoting complex and the nuclear ubiquitin ligase complex were included. There were 502, 131, 143, and 113 enriched GOs in HL-2d, HL- 4d, and HL-7d when compared with HL-0d. The top 20 overlapped GOs included those related to catalytic activity, drug transporter activity, and oxidoreductase activity (S6 Table). In Tineke, 113, 210, 220, and 214 GOs were enriched in TL-2d, TL-t 4d, TL-7d, and TL-9d compared to the control. The most affected GO terms in Hiogi were related to catalytic activity, metabolic processes, oxidoreductase activity, and single-organism processes. The KEGG pathway enrichment result showed that 10, 12, 6, and 7 pathways were enriched in HL-0d, HL-2d, HL-4d, and HL-7d, respectively, compared with the corresponding time points in Tineke. Two pathways overlapped, including plant hormone signal transduction and carotenoid biosynthesis (S7 Table). Photosynthesis-related pathways were also significantly enriched (p≤0.05) in DEGs in Hiogi and Tineke, and the related genes were highly expressed in Hiogi. Moreover, 28, 22, and 29 significantly enriched (p<0.05) KEGG pathways were found in HL-2d, HL-4d, and HL-7d, respectively, when compared with TL-0d, including those related to plant hormone signal transduction and starch and sucrose metabolism.
Plant hormone signal transduction changes after salt treatment in R. chinensis
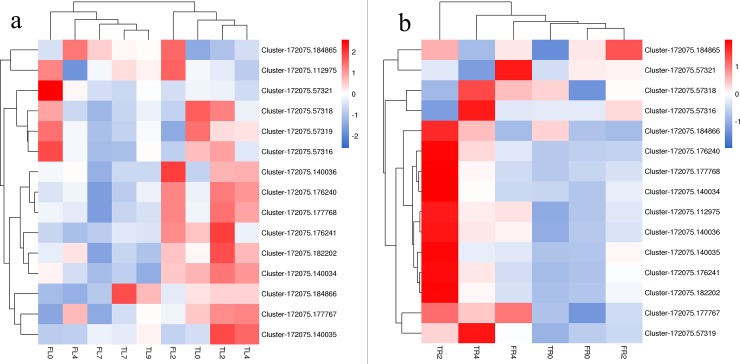
KEGG enrichment results showed that the plant hormone signal transduction pathway was significantly enriched (p<0.05) in the leaves and roots of the two R. chinensis varieties. The most obvious changes were to the ABA signal transduction process, and there were great differences in PYR/PYL, PP2C, Snrk2, and ABF between the different varieties. Fig 5 shows the expression pattern of these four genes in the leaves and roots of Hiogi and Tineke. PYR/PYL and Snrk2 showed a similar expression trend in all leaf tissues. PP2C expression level was higher in Tineke after 4 d of salinity stress, and the key transcription factor ABF expression level was also higher in the leaf tissue of Tineke compared to the leaf of Hiogi. For root tissue, the expression of PYR/PYL, Snrk2, PP2C, and ABF in the early salt treatment stage was significantly higher (p<0.05) in Tineke, and 4 d later, there was no significant difference in PP2C expression between Hiogi and Tineke. Fifteen transcripts of ABF were found and heatmaps were constructed to illustrate the expression pattern of ABF (Fig 6). Most of the ABF transcripts that were upregulated in Tineke leaves were downregulated (or not significantly different) in Hiogi. In roots, the expression level of these 15 transcripts in Tineke were significantly higher (p<0.05) than that in Hiogi and, at 2 d, the mRNA expression level was the highest in Tineke.
Fig 6. DEGs expression heatmap of transcripts of ABFs.
a. DEGs expression heatmap of transcripts of ABFs in leaf tissue. b. DEGs expression heatmap of transcripts of ABFs in root tissue.
Glutathione metabolic changes after salt treatment in R. chinensis
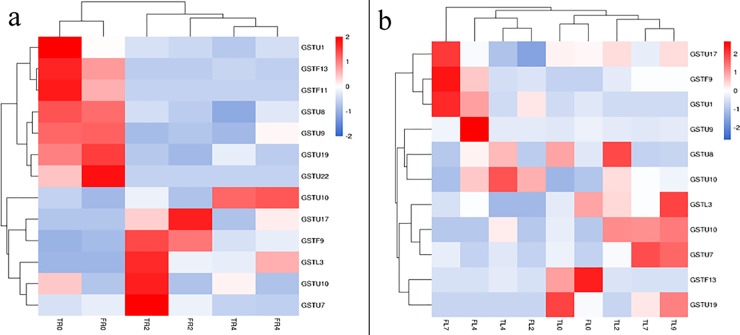
The glutathione metabolic pathway was significantly enriched (p<0.05) in both the leaves and roots of Tineke, but there was no significant enrichment in Hiogi (Table 3). In leaf tissue, the number of DEGs increased with increasing salt treatment time. The functional annotation of DEGs showed that most APX family members (APX6, APX3, APX2, APXT) were specifically upregulated in Tineke leaves and roots. In addition, 10 GST family members (GSTU1, GSTU8, GSTF11, GSTF13, GSTU9, GSTU19, GSTU22, GSTU17, GSTU10, GSTF9, GSTL3, GSTU7, and GSTU10) were differentially expressed after salt treatment. Some of these members were upregulated after salt treatment and some were downregulated (Fig 7). GSH1 plays an important role in glutathione synthesis. GSH1 expression was upregulated after salt treatment. For Hiogi, relatively few genes related to the glutathione metabolic pathway changed significantly (p<0.05) in expression. There were few differentially expressed GSTs in Hiogi after salt treatment (Table 3).
Table 3. DEGs in glutathione metabolic pathway.
| DEG number | Background number | P-value | Corrected P-value | Comparation |
|---|---|---|---|---|
| 33 | 1191 | 0.554203 | 1.00 | HL2 vs HL0 |
| 17 | 1191 | 0.117473 | 0.36 | HL4 vs HL0 |
| 54 | 1191 | 0.111765 | 0.34 | HL7 vs HL0 |
| 15 | 1191 | 0.047709 | 0.16 | TL2 vs TL0 |
| 65 | 1191 | 0.000286 | 0.00 | TL4 vs TL0 |
| 116 | 1191 | 0.001337 | 0.01 | TL7 vs TL0 |
| 107 | 1191 | 0.003009 | 0.01 | TL9 vs TL0 |
| 71 | 1191 | 0.994919 | 1.00 | HR2 vs HR0 |
| 63 | 1191 | 0.005869 | 0.06 | HR4 vs HR0 |
| 151 | 1191 | 2.61E-06 | 0.00 | TR2 vs TR0 |
| 68 | 1191 | 0.007885 | 0.01 | TR4 vs TR0 |
Fig 7. DEGs expression heatmap of members of GSTs falimy.
a. DEGs expression heatmap of members of GSTs falimy in leaf tissue. b. DEGs expression heatmap of members of GSTs falimy in root tissue.
Verification of transcript expression levels using qPCR
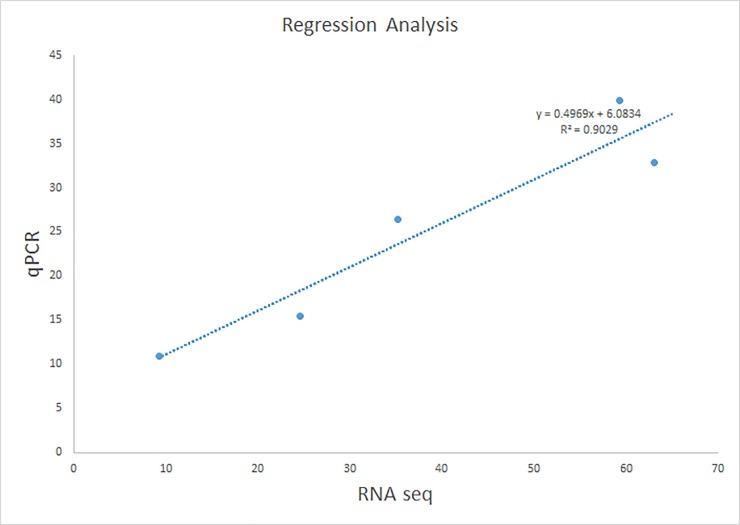
Five transcripts of PYR/PYL, PP2C, Snrk2, GSH1, and ABF were quantified by qPCR. All primers designed for qPCR are shown in Table 4. The qPCR results showed that gene expression patterns were consistent with transcriptome sequencing. PYR/PYL and Snrk2 showed similar expression in Tineke and Hiogi. With increased salt treatment time, the fold change of expression decreased. However, PP2C showed higher expression in Tineke than in Hiogi after 4 d of salt treatment. In addition, the fold changes of ABF expression in Tineke was also significantly higher (p<0.05) than that in Hiogi. This result shows that GSH1 was highly expressed in Tineke, whereas in Hiogi there were no obvious changes in GSH1 expression after salt treatment. The salt stress-induced changes in gene expression for the majority (~70%) of the tested genes were consistent between both approaches (RNA-seq and qPCR). The correlation coefficient (r2) between qPCR- and FPKM-derived expressions was 0.9029 (Fig 8), indicating high reliability of the RNA-seq data.
Table 4. Primers.
| Gene | Primer sequences (5ʹ-3ʹ) | Annealing temperature (°C) | Product length (bp) |
|---|---|---|---|
| PYR/PYL | F: TTGGCAAGAAGTCAACAAACAG | 57.5 | 146 |
| R: CACAGAAAAGATGGCTGGTAAC | 57.5 | ||
| PP2C | F: TAGTTGTCACCTATGGCTCTTG | 57.5 | 137 |
| R: CCTCTTTCCTCCGATCACAAG | 57.5 | ||
| SnRK2 | F: TTGGGAGTTGAAATGGATCG | 57.5 | 141 |
| R: GGTCTGCTTGTCCGTCATC | 57.5 | ||
| ABF | F: AGGTTATCACAGAAGCCATCC | 57.5 | 138 |
| R: CCAGTTTATTGCTGCCTCTTG | 57.5 | ||
| Gsh1 | F: ATTCCAGGCTCTTCACTTTCC | 57.5 | 141 |
| R: GTTTGGGTTCTTTGTTCAGG | 57.5 |
Fig 8. Correlation between qPCR and RNA sequencing for the 5 selected genes.
Each point represents a value of fold change of expression level.
Discussion
Plants typically accumulate small molecules, organic compounds, PRO, and protein protective agents in their cells to maintain osmotic homeostasis (PRO and glycine betaine) when subjected to salt stress [26, 27]. In this study, the contents of PRO and MDA in Tineke leaf rose earlier than Hiogi, but the accumulation of PRO and MDA did not reach the level before salt stress treatment. This was not in line with the laws of other plants [28–30]. This might be that the response to salt stress in Rosa chinensis did not depend on PRO and MDA. The rise of PRO and MDA content indicated that they also play a role in salt tolerance, but obviously not the most critical role. Also, the PRO and MDA result reflected the stronger tolerance of Tineke to salt stress. In addition, the NaCl result also proved that the NaCl contents regulation ability in Tineke was also better than that in Hiogi. Under salt stress, plant roots firstly receipt salt stress signal [31]. Root was affected by high permeability and caused the loss of water content [32]. The loss of water in roots further caused serious damage to the plants. The changes of water content in this study indicated that Tineke can control the loss of water better than Hiogi, which was is consistent with the salt tolerance characteristics of Tineke. Recent studies have demonstrated gene expression cascades in plants under abiotic stress. This is a defining feature of the ABA-dependent and ABA-independent signaling pathways and their association with stress responses [33]. After 2 d of salt treatment, plant hormone signal transduction was induced by the salt and played an exclusive role in Tineke and Hiogi responses to salt stress. PYR/PYL is a receptor of the ABA-signaling complex, and overexpression of PYR/PYL could suppress PP2Cs, which releases Snrk2s from the inhibition of PP2Cs, and subsequently activates the downstream target ABRE-binding factor [34]. ABF could activate the promoter of the DRE-binding protein 2A, which has important functions in the osmotic stress response [35]. In this study, there was no significant difference in ABF expression between Tineke, Hiogi, and the control. However, in root tissue, upregulation of PYR/PYL, Snrk2, and ABF transcripts was induced by salt treatment. The number of Snrk2 and ABF transcripts in Tineke was higher than in Hiogi, and this may have contributed to the greater salt tolerance of Tineke. Overexpression of ABF can enhance dehydration and stress tolerance via scavenging of ROS and modulating expression of stress-responsive genes [36]. In addition, HSPs play an important role in the ABF-induced ROS scavenging process [37], and the HSP expression observed in this study confirms this. GSH helps to alleviate salt stress in plants, so the damage from salt stress can be reduced with exogenous GSH supplementation [38]. The glutathione metabolism pathway was a critical pathway in Tineke after salt treatment. GSH1 is a key gene in the production of glutathione synthetase, but no studies have reported that GSH1 is related to salt tolerance in plants. However, we found 23 GSH1 transcripts that were highly expressed in Tineke roots, and qPCR showed that the expression level of GSH1 in the salt-tolerant varieties was significantly higher (p<0.05) than that of the salt-sensitive varieties. After salt treatment, the expression of GSH1 significantly increased (p<0.05). Because GSH1 is one of the key genes involved in the production of glutathione synthetase, high expression of GSH1 could increase glutathione content, which promotes the glutathione cycle. Gamma-L-glutamyl-L-2-aminobutyrylglycine (ophthalmate, OPH) is a potential indicator of salt-tolerant rose varieties. However, due to the difficulties of OPH detection, the OPH content in the salt-tolerant Tineke variety was not determined in this study. In addition, plant glutathione transferases play an important role in the detoxification of xenobiotics and toxic lipid peroxides via regulation of the glutathione-binding reaction [39]. GST also has important functions in biochemical reactions of primary metabolism and secondary products such as flavonoid derivatives [40]. Plant cells are continuously stressed by toxic ROS generated by oxygen consumption during metabolic respiration. When plants are exposed to salt or drought stress, large amounts of ROS are generated [41]. The upregulation of GST can counterbalance the production of ROS, which would reduce the harm caused by abiotic stresses [42]. We found that the activity of GSTs in Tineke was sensitive to salt stress (47 transcripts were differentially expressed), but in Hiogi, only 14 differentially expressed transcripts were found. In addition, we found great differences in the expression of GSH1 between the two R. chinensis cultivars when the plants were treated with salt, which has not been reported in previous salt stress studies. GSH1 had been reported to be related to environmental stress [43, 44]. A stress-related post-transcriptional regulation was reported to related to the expression of GSH1, which further affects glutathione metabolism [45]. The overexpression of GSH1 can promoted subsequent flowering and showed less responsive to SV treatment than in wild-type plant [46]. In this study, GSH1 act as a response factor for salt stress. The high expression of GSH1 has a certain linkage with salt tolerance. Where the GSH1 can be used as a bio-marker of salt tolerance in Rosa chinensis, more work is needed to do. But in this study, we did discover the relationship between GSH1 and salt tolerance. Chlorophyll content in plant leaf tissues typically decreases when subjected to salt stress [47]. In salt- and drought-resistant plants, the decline of chlorophyll content is not obvious, and in some species, chlorophyll levels may even increase [48]. The effects of salt stress on photosynthesis can be either direct or secondary, such as the oxidative stress arising from the superimposition of multiple stresses [49]. Chlorophyll levels in R. chinensis varieties suggested stronger salt tolerance in Tineke. An efficient response to the environment is particularly important for plants. In leaf tissue, when salt stimulation occurs, responses are triggered by primary osmotic stress signals or secondary signal metabolites (including hormones, ROS, and intracellular second messengers). These changes are consistent with our results. Salt and drought stress can induce stomatal closure and moderate increases in ROS levels, decreases in photosynthesis, and decreases in root growth rates [50]. The KEGG and GO enrichment results are again consistent with this. In Tineke, low concentrations of ROS maintained by plant hormone signal transduction and glutathione metabolism can reduce stomatal closure in leaves and maintain normal photosynthesis.
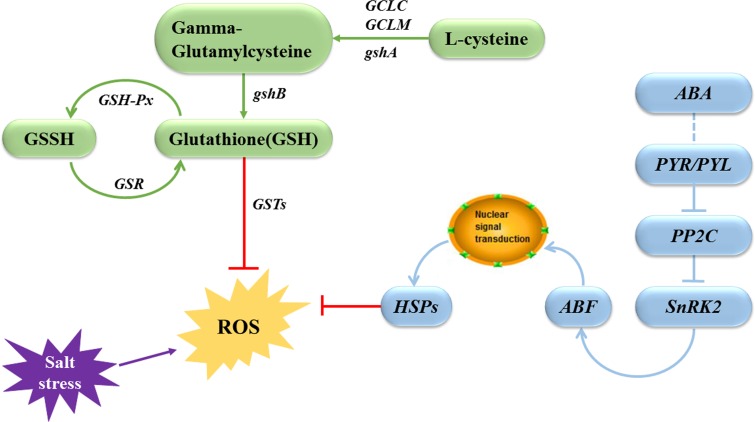
In conclusion, the ABA-dependent signaling pathway was the main pathway involved in the salt stress responses of R. chinensis. Responses to salt tolerance were mainly modulated via two pathways: plant hormone signal transduction and glutathione metabolism (Fig 9). The difference in salt tolerance between the cultivars Hiogi and Tineke was due to differences in gene sensitivity to salt in these two pathways. The effects of salt stress in the roots are eventually manifested in the leaves, causing a series of changes in the leaves, including a reduced photosynthesis rate, which eventually causes leaf wilting. In Tineke, Snrk2, ABF, HSP, GSTs, and GSH1 showed strong activity under salt stress, and these genes are markers of salt tolerance.
Fig 9. Schematic diagram of salt tolerance mechanism.
GSSH: oxidized form glutathione; GSR: glutathione reductase; GSH-Px: Glutathione peroxidase; GCLC: Glutamate-Cysteine Ligase Catalytic Subunit; GCLM: glutamate-cysteine ligase modifier subunit.
Supporting information
Different sets of colors represent different comparison.
(PDF)
Different sets of colors represent different comparison.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
This study was financially supported by the industrial platform construction of coastal saline ecological afforestation in Beijing-Tianjin-Hebei region (15YDLJSF00050).
Data Availability
All the sequence data had been uploaded to the SRA database (https://www.ncbi.nlm.nih.gov/sra) with the SRA accession SRP128235.
Funding Statement
This work was supported by industrial platform construction of coastal saline ecological afforestation in Beijing-Tianjin-Hebei region, 15YDLJSF00050 (http://www.tstc.gov.cn/zhengwugongkai/ztzl/kjgzhy/2016/dxfy/kjtd/201602/t20160205_87310.html) to XT. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zhu JK. Cell signaling under salt, water and cold stresses. Current opinion in plant biology. 2001;4(5):401–6. . [DOI] [PubMed] [Google Scholar]
- 2.Liska AJ, Shevchenko A, Pick U, Katz A. Enhanced photosynthesis and redox energy production contribute to salinity tolerance in Dunaliella as revealed by homology-based proteomics. Plant Physiol. 2004;136(1):2806–17. 10.1104/pp.104.039438 ; PubMed Central PMCID: PMC523343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jithesh MN, Prashanth SR, Sivaprakash KR, Parida A. Monitoring expression profiles of antioxidant genes to salinity, iron, oxidative, light and hyperosmotic stresses in the highly salt tolerant grey mangrove, Avicennia marina (Forsk.) Vierh. by mRNA analysis. Plant cell reports. 2006;25(8):865–76. 10.1007/s00299-006-0127-4 . [DOI] [PubMed] [Google Scholar]
- 4.Barrero JM, Rodriguez PL, Quesada V, Piqueras P, Ponce MR, Micol JL. Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant, cell & environment. 2006;29(10):2000–8. 10.1111/j.1365-3040.2006.01576.x . [DOI] [PubMed] [Google Scholar]
- 5.Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Current opinion in plant biology. 2003;6(5):410–7. . [DOI] [PubMed] [Google Scholar]
- 6.Ding W, Fang W, Shi S, Zhao Y, Li X, Xiao K. Wheat WRKY Type Transcription Factor Gene TaWRKY1 is Essential in Mediating Drought Tolerance Associated with an ABA-Dependent Pathway. Plant Molecular Biology Reporter. 2016;34(6):1–16. [Google Scholar]
- 7.Howell SH. Endoplasmic reticulum stress responses in plants. Annual review of plant biology. 2013;64:477–99. 10.1146/annurev-arplant-050312-120053 . [DOI] [PubMed] [Google Scholar]
- 8.Apse MP, Blumwald E. Na+ transport in plants. Febs Letters. 2007;581(12):2247–54. 10.1016/j.febslet.2007.04.014 [DOI] [PubMed] [Google Scholar]
- 9.Josine TL. Breeding and Expression of Arabidopsis thaliana AtDREB2A-CA in Rosa chinensis Jacq For Salinity Tolerance: Tianjin University; 2012. [Google Scholar]
- 10.Wu B, Hu Y, Huo P, Qian Z, Xin C, Zhang Z. Transcriptome analysis of hexaploid hulless oat in response to salinity stress. PloS one. 2017;12(2):e0171451 10.1371/journal.pone.0171451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Mao C, Shi Z, Kou X. The Amino Acid Metabolic and Carbohydrate Metabolic Pathway Play Important Roles during Salt-Stress Response in Tomato. Frontiers in plant science. 2017;8:1231 10.3389/fpls.2017.01231 ; PubMed Central PMCID: PMC5511834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zh Xiuzhen, Zh Zuoshuang. Rosa chinensis: China Forestry Publishing House; 2006. [Google Scholar]
- 13.Pan Xiaodong, Yang Hongxun. Production technology of Chinese rose: China Agricultural Publishing House; 2002. [Google Scholar]
- 14.Ergun E, Demirata B, Gumus G, Apak R. Simultaneous determination of chlorophyll a and chlorophyll b by derivative spectrophotometry. Anal Bioanal Chem. 2004;379(5–6):803–11. 10.1007/s00216-004-2637-7 . [DOI] [PubMed] [Google Scholar]
- 15.Al-Harrasi I, Al-Yahyai R, Yaish MW. Differential DNA methylation and transcription profiles in date palm roots exposed to salinity. PloS one. 2018;13(1):e0191492 10.1371/journal.pone.0191492 ; PubMed Central PMCID: PMC5774813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu J, Gates RN, Xu Y, Hu T. Diffusion limitations and metabolic factors associated with inhibition and recovery of photosynthesis following cold stress in Elymus nutans Griseb. J Photochem Photobiol B. 2016;163:30–9. 10.1016/j.jphotobiol.2016.08.008 [DOI] [PubMed] [Google Scholar]
- 17.Mattison CP, Rai R, Settlage R, Hinchliffe DJ, Madison C, Bland JM, et al. RNA-Seq Analysis of Developing Pecan (Carya illinoinensis) Embryos Reveals Parallel Expression Patterns Among Allergen and Lipid Metabolism Genes. Journal of Agricultural & Food Chemistry. 2017. [DOI] [PubMed] [Google Scholar]
- 18.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323 10.1186/1471-2105-12-323 ; PubMed Central PMCID: PMC3163565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–8. 10.1093/bioinformatics/btp612 . [DOI] [PubMed] [Google Scholar]
- 20.Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420–35. 10.1093/nar/gkn176 ; PubMed Central PMCID: PMC2425479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6. 10.1093/bioinformatics/bti610 . [DOI] [PubMed] [Google Scholar]
- 22.Yaish MW, Patankar HV, Assaha DVM, Zheng Y, Al-Yahyai R, Sunkar R. Genome-wide expression profiling in leaves and roots of date palm (Phoenix dactylifera L.) exposed to salinity. BMC Genomics. 2017;18(1):246 10.1186/s12864-017-3633-6 ; PubMed Central PMCID: PMC5423419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yaish MW, Al-Lawati A, Al-Harrasi I, Patankar HV. Genome-wide DNA Methylation analysis in response to salinity in the model plant caliph medic (Medicago truncatula). BMC Genomics. 2018;19(1):78 10.1186/s12864-018-4484-5 ; PubMed Central PMCID: PMC5781308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome biology. 2010;11(2):R14 10.1186/gb-2010-11-2-r14 ; PubMed Central PMCID: PMC2872874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Web Server issue):W316–22. 10.1093/nar/gkr483 ; PubMed Central PMCID: PMC3125809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoque MA, Banu MN, Okuma E, Amako K, Nakamura Y, Shimoishi Y, et al. Exogenous proline and glycinebetaine increase NaCl-induced ascorbate-glutathione cycle enzyme activities, and proline improves salt tolerance more than glycinebetaine in tobacco Bright Yellow-2 suspension-cultured cells. Journal of plant physiology. 2007;164(11):1457–68. 10.1016/j.jplph.2006.10.004 . [DOI] [PubMed] [Google Scholar]
- 27.Yaish MW. Proline accumulation is a general response to abiotic stress in the date palm tree (Phoenix dactylifera L.). Genetics & Molecular Research Gmr. 2015;14(3):9943–50. [DOI] [PubMed] [Google Scholar]
- 28.Chutipaijit S, Chaum S, Sompornpailin K. Proline accumulation and physiological responses of indica rice genotypes differing in tolerance to salt and drought stresses. Philippine Agriculturist. 2010;93(2):165–9. [Google Scholar]
- 29.Li W, Tang Z, Lv J, Ju B, Liu Z, Qin S. Protective effect of cerium against salinity-induced oxidative stress in Jerusalem artichoke (Helianthus tuberosus). International Journal of Agriculture & Biology. 2017;19(5):1201–6. [Google Scholar]
- 30.Wang XS, Han JG. Changes of Proline Content,Activity,and Active Isoforms of Antioxidative Enzymes in Two Alfalfa Cultivars Under Salt Stress. Agricultural Sciences in China. 2009;8(4):431–40. [Google Scholar]
- 31.Galvan-Ampudia CS, Testerink C. Salt stress signals shape the plant root. Curr Opin Plant Biol. 2011;14(3):296–302. 10.1016/j.pbi.2011.03.019 [DOI] [PubMed] [Google Scholar]
- 32.Skaggs TH, Genuchten MTV, Shouse PJ, Poss JA. Macroscopic approaches to root water uptake as a function of water and salinity stress. Agric Water Manage. 2006;86(1):140–9. [Google Scholar]
- 33.Fu J, Wu Y, Miao Y, Xu Y, Zhao E, Wang J, et al. Improved cold tolerance in Elymus nutans by exogenous application of melatonin may involve ABA-dependent and ABA-independent pathways. Scientific reports. 2017;7:39865 10.1038/srep39865 ; PubMed Central PMCID: PMC5206618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, et al. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Science signaling. 2013;6(270):rs8 10.1126/scisignal.2003509 . [DOI] [PubMed] [Google Scholar]
- 35.Kim JS, Mizoi J, Yoshida T, Fujita Y, Nakajima J, Ohori T, et al. An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant & cell physiology. 2011;52(12):2136–46. 10.1093/pcp/pcr143 . [DOI] [PubMed] [Google Scholar]
- 36.Huang XS, Liu JH, Chen XJ. Overexpression of PtrABF gene, a bZIP transcription factor isolated from Poncirus trifoliata, enhances dehydration and drought tolerance in tobacco via scavenging ROS and modulating expression of stress-responsive genes. BMC plant biology. 2010;10:230 10.1186/1471-2229-10-230 ; PubMed Central PMCID: PMC3017851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandey P, Ramegowda V, Senthil-Kumar M. Shared and unique responses of plants to multiple individual stresses and stress combinations: physiological and molecular mechanisms. Front Plant Sci. 2015;6:723 10.3389/fpls.2015.00723 ; PubMed Central PMCID: PMC4584981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teh CY, Mahmood M, Shaharuddin NA, Chai LH. In vitro rice shoot apices as simple model to study the effect of NaCl and the potential of exogenous proline and glutathione in mitigating salinity stress. Plant Growth Regulation. 2015;75(3):771–81. [Google Scholar]
- 39.Csiszar J, Horvath E, Vary Z, Galle A, Bela K, Brunner S, et al. Glutathione transferase supergene family in tomato: Salt stress-regulated expression of representative genes from distinct GST classes in plants primed with salicylic acid. Plant Physiol Biochem. 2014;78:15–26. 10.1016/j.plaphy.2014.02.010 . [DOI] [PubMed] [Google Scholar]
- 40.Nianiou-Obeidat I, Madesis P, Kissoudis C, Voulgari G, Chronopoulou E, Tsaftaris A, et al. Plant glutathione transferase-mediated stress tolerance: functions and biotechnological applications. Plant cell reports. 2017;36(6):791–805. 10.1007/s00299-017-2139-7 . [DOI] [PubMed] [Google Scholar]
- 41.Parida A, George S. Glutathione-s-transferase gene from prosopis juliflora confers abiotic stress tolerance in plants. EP; 2010. [Google Scholar]
- 42.Yang G, Xu Z, Peng S, Sun Y, Jia C, Zhai M. In planta characterization of a tau class glutathione S-transferase gene from Juglans regia (JrGSTTau1) involved in chilling tolerance. Plant cell reports. 2016;35(3):681–92. 10.1007/s00299-015-1912-8 . [DOI] [PubMed] [Google Scholar]
- 43.Bittsánszky A. Stress Capacity (Zn and Paraquat) of gsh1-Transgenic Poplar and Microsatellite Diversity of Androgenic Gametoclones.
- 44.Schellingen K, Straeten DVD, Remans T, Loix C, Vangronsveld J, Cuypers A. Ethylene biosynthesis is involved in the early oxidative challenge induced by moderate Cd exposure in Arabidopsis thaliana. Environmental & Experimental Botany. 2015;117:1–11. [Google Scholar]
- 45.Wachter A, Wolf S, Steininger H, Bogs J, Rausch T. Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant Journal. 2010;41(1):15–30. [DOI] [PubMed] [Google Scholar]
- 46.Hatanoiwasaki A, Ogawa KI. Overexpression of GSH1 gene mimics transcriptional response to low temperature during seed vernalization treatment of Arabidopsis. Plant Cell Physiol. 2012;53(7):1195 10.1093/pcp/pcs075 [DOI] [PubMed] [Google Scholar]
- 47.Santos CV. Regulation of chlorophyll biosynthesis and degradation by salt stress in sunflower leaves. Scientia Horticulturae. 2004;103(1):93–9. [Google Scholar]
- 48.Zhang ZH. Responses of different rice (Oryza sativa L.) genotypes to salt stress and relation to carbohydrate metabolism and chlorophyll content. African Journal of Agricultural Research. 2012;7(1). [Google Scholar]
- 49.Chaves MM, Flexas J, Pinheiro C. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot. 2009;103(4):551–60. 10.1093/aob/mcn125 ; PubMed Central PMCID: PMC2707345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bogeat-Triboulot MB, Brosche M, Renaut J, Jouve L, Le Thiec D, Fayyaz P, et al. Gradual soil water depletion results in reversible changes of gene expression, protein profiles, ecophysiology, and growth performance in Populus euphratica, a poplar growing in arid regions. Plant Physiol. 2007;143(2):876–92. 10.1104/pp.106.088708 ; PubMed Central PMCID: PMC1803728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Different sets of colors represent different comparison.
(PDF)
Different sets of colors represent different comparison.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All the sequence data had been uploaded to the SRA database (https://www.ncbi.nlm.nih.gov/sra) with the SRA accession SRP128235.