 The proteasome inhibitor bortezomib is associated with the development of peripheral neuropathy in patients, but the mechanism is not fully understood.
The proteasome inhibitor bortezomib is associated with the development of peripheral neuropathy in patients, but the mechanism is not fully understood.
Abstract
The proteasome inhibitor bortezomib is associated with the development of peripheral neuropathy in patients, but the mechanism by which bortezomib can induce peripheral neuropathy is not fully understood. One study suggested that off-target inhibition of proteases other than the proteasome, particularly HtraA2/Omi, may be the underlying mechanism of the neuropathy. The same study also concluded that carfilzomib, a second proteasome inhibitor that is associated with less peripheral neuropathy in patients than bortezomib, showed no inhibition of HtrA2/Omi. The goal of the work described here was to determine whether either proteasome inhibitors truly affected HtrA2/Omi activity. A variety of methods were used to test the effects of both bortezomib and carfilzomib on HtrA2/Omi activity that included in vitro recombinant enzyme assays, and studies with the human neuroblastoma SH-SY5Y cell line and HtrA2/Omi-knockout mouse embryonic fibroblasts. The compound ucf-101 was used to assess the effects of specific HtrA2/Omi inhibition. In contrast to previously published data, our results clearly demonstrated that neither bortezomib nor carfilzomib inhibited HtrA2/Omi activity in recombinant enzyme assays at concentrations up to 100 μM, while the specific inhibitor ucf-101 did inhibit the enzyme. The proteasome inhibitors did not inhibit HtrA2/Omi activity in either SH-SY5Y cells or mouse embryonic fibroblasts, as determined by expression of the HtrA2/Omi substrates eIF4G1 and UCH-L1. Based on our biochemical and cell-based assays, we conclude that neither bortezomib nor carfilzomib inhibited HtrA2/Omi activity. Therefore, it is unlikely that bortezomib associated peripheral neuropathy is a direct result of off-target inhibition of HtrA2/Omi.
Introduction
The proteasome inhibitor (PI) bortezomib has become the cornerstone of multiple myeloma treatment as a result of the drug's unprecedented efficacy.1 Nevertheless, bortezomib causes peripheral neuropathy in a subset of patients,2 and this side effect can limit the drug's potential benefit for those patients. Although the mechanism of bortezomib-associated peripheral neuropathy is unknown, we have previously demonstrated that it is related to the drug's mechanism of action.3 Peptide boronic acids are known to inhibit proteases other than the proteasome, such as leukocyte elastase, pancreatic elastase, cathepsin G, and chymotrypsin, but the role of these proteases in the homeostasis of sensory neurons is not known.4
A recent study reported that multiple proteases (cathepsin A, cathepsin G, chymase, dipeptidyl peptidase II, and HtrA2/Omi) were inhibited by bortezomib (a peptide boronate PI) and not by carfilzomib (a peptide epoxyketone PI).5 Of note, with the exception of HtrA2/Omi, bortezomib was much less potent against these purified enzymes than against the LMP7 subunit of the proteasome.5 However, a second study that used a similar purified protein assay showed that bortezomib up to 100 μM did not affect HtrA2/Omi activity.6 This second study also used high-throughput screening of both of bortezomib and carfilzomib to show no inhibition of HtrA2/Omi by either PI at concentrations up to 33 μM.6 A third study presented suggestive results that bortezomib partially inhibited recombinant human HtrA2/Omi in vitro, but the peptide boronate PI ixazomib citrate (NINLARO®) did not.7 Ixazomib citrate is the first marketed oral PI for the treatment of patients with multiple myeloma who have received at least one prior therapy. Ixazomib citrate is a modified peptide boronic acid and is the citrate ester of ixazomib, the biologically active moiety. Ixazomib citrate rapidly hydrolyzes to ixazomib upon contact with aqueous solution or plasma.8
Emerging clinical data9,10 have demonstrated that among these proteases, only cathepsin A and HtrA2/Omi are involved in neurodegenerative diseases. Specifically, cathepsin A is involved in galactosialidosis9 and HtrA2/Omi in Parkinson's disease,10 although neither were implicated in the etiology of peripheral neuropathy. HtrA2/Omi is a serine protease that is involved in mitochondrial homeostasis and induction of apoptosis,11 and has a limited number of known physiological substrates.12,13 Since HtrA2/Omi has been implicated in neuronal survival,14 it was suggested that off-target inhibition of HtrA2/Omi by bortezomib and not by carfilzomib could be the mechanism underlying bortezomib-associated peripheral neuropathy.5
The objective of the present study was to investigate the effects of bortezomib and carfilzomib on HtrA2/Omi activity. No inhibition of HtrA2/Omi activity was noted with bortezomib and carfilzomib across a variety of in vitro and cell-based assays. These results suggest that the cause of PI-associated peripheral neuropathy may not be due to off-target inhibition of HtrA2/Omi activity. The unambiguous identification of proteins that are targeted by PI in the peripheral nervous system is absolutely necessary in order to understand the mechanism of PI-induced peripheral neuropathy. Our results represent significant contributions to this goal.
Materials and methods
Test and control articles
Stock solutions of proteasome inhibitors bortezomib (VELCADE®, Millennium Pharmaceuticals, Inc. (a wholly owned subsidiary of Takeda Pharmaceutical Company Limited) Cambridge, MA) and carfilzomib (KYPROLIS®, Onyx Pharmaceuticals, San Francisco, CA), and ixazomib (NINLARO®, Millennium Pharmaceuticals, Inc) were dissolved in dimethyl sulfoxide (DMSO). The HtrA2/Omi inhibitor ucf-101 (Calbiochem, Billerica, MA) was dissolved in DMSO immediately before use.
Recombinant enzyme assay
Recombinant human HtrA2/Omi (R&D Systems, Minneapolis, MN) activity assay using the model substrate β-Casein (Sigma-Aldrich, St Louis, MO) was performed according to manufacturer's instructions. The cleavage assay in 50 μl total volume was incubated at 45 °C for 1 hour and stopped by the addition of 50 μl of 2× SDS (sodium dodecyl sulphate) loading buffer. For the enzyme inhibition assay, recombinant HtrA2/Omi was pre-incubated with bortezomib, carfilzomib, or ucf-101 for 45 minutes at room temperature and then transferred to reaction tubes and incubated at 45 °C for 1 hour. Samples were boiled for 5 minutes, and then 20 μL was resolved on a 4% to 20% denaturing SDS gel (Invitrogen, Carlsbad, CA). The gel was then stained with SimplyBlue Safe Stain (Invitrogen) for 2 hours and destained with water overnight.
HtrA2/Omi fluorescence-based biochemical assays
The fluorescence-based assay for HtrA2/Omi activity was adapted from Martins et al.15 A solution with final concentrations of 31 pM HtrA2/Omi and 10 μM of the fluorescent substrate Mca-IRRVSYSF(K-Dnp)K (Innovagen, Lund, Sweden) were incubated in reaction buffer (50 mM Tris pH 8, 0.5 mM EDTA, 1 mM DTT (dithyothreitol), 0.01% BSA). Test compounds dispensed as 300 nL DMSO solutions into solid black 384 well plates (Corning Inc., Corning, NY). Ucf-101 was used as control. The plates were read on a BMG PolarStar Galaxy at 30 °C for 1 hour. The excitation wavelength was 320 nm and the emission wavelength was 405 nm and gain 30. Because the reaction proceeded in a linear fashion for 1 hour, rate data from 0 to 45 minutes were used for the inhibition calculations. Results from this assay are presented as the mean of four independent experiments.
ClpXP and LonP1 fluorescence-based biochemical assays
The fluorescence-based assay for ClpXP activity was adapted from Akopian et al.16 For ClpP, C-terminal-His6-ClpP_P57-T277, lacking the N-terminal putative mitochondrial signal sequence, was expressed in E. coli and purified by Ni-NTA affinity chromatography (Qiagen, Valencia, CA) followed by size-exclusion chromatography. For ClpX, C-terminal-StrepII-tagged ClpX_A65-S633, lacking the N-terminal putative mitochondrial signal sequence, was expressed in E. coli and purified by Strep-Tactin affinity chromatography (Qiagen), followed by anion-exchange and size-exclusion chromatography. A solution with final concentrations of 10 nM ClpXP complex (6 ClpX·14 ClpP), 50 μM Ac-Trp-Leu-Ala-AMC and 2 mM ATP-Mg were incubated in reaction buffer 50 mM Tris-HCl (pH 8.0), 100 mM KCl, 5 mM MgCl2, 0.01% BSA, 0.005% Triton-X 100 and 1 mM DTT.
The fluorescence-based assay for LonP1 activity was adapted from Fishovitz et al.17 C-terminal-His6-Lonp1_F68-R959, lacking the N-terminal putative mitochondrial signal sequence, was expressed in E. coli and purified by Ni-NTA affinity chromatography (Qiagen) followed by size-exclusion chromatography. A solution with final concentrations of 24 nM LonP1 monomer, 100 μM of the fluorescent substrate FRET 89–98 Abu [(3nitro) Tyr-Arg-Gly-Ile-Thr-Abu-Ser-Gly-Arg-Gln-Lys (Abz)], and 1 mM ATP-Mg were incubated in reaction buffer 50 mM Tris-HCl (pH 8.0), 100 mM KCl, 5 mM MgCl2, 0.01% BSA, 0.005% Triton-X 100 and 1 mM DTT.
For ClpXP and LonP1 assays, fluorescence was monitored continuously in Pherastar or Clariostar plate readers at either 30 °C (LonP1) or 37 °C (ClpXP) for 30 minutes after stabilization of the reaction slopes. The excitation wavelength was 350 nm and the emission wavelength was 450 nm. Raw slopes and r2 were calculated by linear fit using MARS software (BMG Labtech, Cary, NC). The concentration producing 50% inhibition (IC50) were calculated using the Hill equation relative to 0 and 100 percent inhibition controls using Pipeline Pilot (Accelrys, San Diego, CA) and Condoseo (Genedata, Lexington, MA) software. The geometric average was calculated from three independent experiments.
Cell culture and treatments
SH-SY5Y cells and cell culture medium and reagents were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and were cultured in 1 : 1 mixture of DMEM and F12 ATCC-formulated Eagle's Minimal Essential Medium as recommended by the supplier at 37 °C in 5% CO2. Wild type and HtrA2/Omi-knockout mouse embryonic fibroblasts (MEFs) were a gift from Julian Downward (Signal Transduction Laboratory, Cancer Research Institute, London, UK). MEFs were cultured in Dulbecco's modified Eagle's medium (DMEM) with 4.5 g L–1d-glucose supplemented with 10% (8.5% in staurosporine-related experiments),18 heat-inactivated fetal bovine serum (FBS), 25 mM HEPES (Invitrogen), 50 μg mL–1 penicillin, and 50 μg mL–1 streptomycin. MEFs were incubated at 37 °C in 5% CO2. SH-SH5Y cells and MEFs were plated into 14 cm petri dishes at a density of 5 × 106 cells per 40 mL tissue culture medium and kept in the incubator for 2 days before treatment with the investigated compounds. Unless indicated otherwise, cells were treated for 12 hours with the indicated concentrations of compounds. In SH-SY5Y cells, the concentration producing 90% inhibition (IC90) of the chymotryptic activity of the 20S proteasome was determined to be 10 nM for bortezomib and 34 nM for carfilzomib (data not shown).
Preparation of protein extracts
For SDS whole cell extracts, cells were collected by scraping, washed with 50 mL of 1× PBS three times, and then collected in 1.5 mL Eppendorf tubes. Cells were lysed at 2 × 108 cells per mL in SDS lysis buffer (125 mM Tris-HCl pH 6.8, 4% SDS, and Protease Inhibitor Cocktail (Thermo Fisher Scientific, Waltham, MA) at room temperature. Samples were sonicated for 30 seconds with a Virsonic Ultrasonic Cell Disruptor 1000 (VirTis, Gardiner, NY), and then centrifuged at room temperature (10 000g, 10 min). Protein concentration in the supernatant was determined using the DC protein assay kit (Bio-Rad, Hercules, CA). After quantification, an equal volume of dilution buffer (20% glycerol, 0.01% bromophenol blue in water) was added to each sample.
For cell extract with RIPA buffer, cells were collected from 4 × 15 cm petri dishes by scraping, washed with 50 mL of 1× PBS three times, and then collected in 1.5 mL snap-top tubes. Cells were lysed on ice for 30 minutes in 1.25 × cell pellet volume of RIPA buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% sodium deoxycholate, 0.1% lithium dodecyl sulfate [LDS], 1% Triton X-100) containing protease inhibitor cocktail and EDTA. The supernatant was collected after centrifugation at 20 000g at 4 °C for 20 minutes, to which was added 0.1× volume of 10% LDS. Samples were sonicated for 30 seconds and then centrifuged once more at 12 000 rpm at room temperature for 10 minutes. Protein concentration in the supernatant was determined using the DC protein assay kit (Bio-Rad).
Western blot analysis and antibodies
Protein samples were reduced with 100 mM DTT (final concentration) and heated at 95 °C for 5 minutes. For the analysis of large proteins (eIF4G1 and α-Fodrin), protein extracts were resolved on a NuPAGE® Novex 3%–8% Tris-Acetate gel (Invitrogen). For the analysis of other proteins, protein extracts were resolved on a Novex® 4%–20% Tris-Glycine Mini Gel (Invitrogen). After electrophoresis, proteins were transferred to 0.2 μm nitrocellulose membranes (Invitrogen) and incubated with the appropriate primary antibody (ESI Table 1†). Membranes were blocked with BLOTTO [1× PBS containing 0.05% Tween-20, 5% nonfat dry milk from Bio-Rad (Blotting-Grade Blocker)] solution for 1 h and incubated with the primary antibodies in BLOTTO overnight at 4 °C. Membranes were washed three times (15 min, 10 min, 5 min) with 1× PBS containing 0.05% Tween-20 and incubated with a HRP-conjugated secondary antibody for 1 hour at room temperature. Membranes were washed three times (15 min, 10 min, 5 min) with 1× PBS containing 0.05% Tween-20, and bands were visualized with Western Blotting Luminol Reagent (Santa Cruz Biotechnology, Santa Cruz, CA). Blots were exposed to BioMax MR double emulsion films (Eastman Kodak, Rochester, NY).
Results
The inhibitor ucf-101, but not proteasome inhibitors bortezomib and carfilzomib, inhibited HtrA2/Omi activity
A recent report proposed that both bortezomib and carfilzomib inhibit several serine proteases.5 This study concluded that bortezomib specifically inhibited HtrA2/Omi, a serine protease, at a pharmacologically relevant concentration while carfilzomib did not. For bortezomib, the IC50 for HtrA2/Omi activity was 3 nM. However, another study found that bortezomib did not inhibit HtrA2/Omi activity at concentrations up to 100 μM.6 The goal of the investigation presented here was to provide clarification on whether or not bortezomib was a bona fide inhibitor of HtrA2/Omi activity.
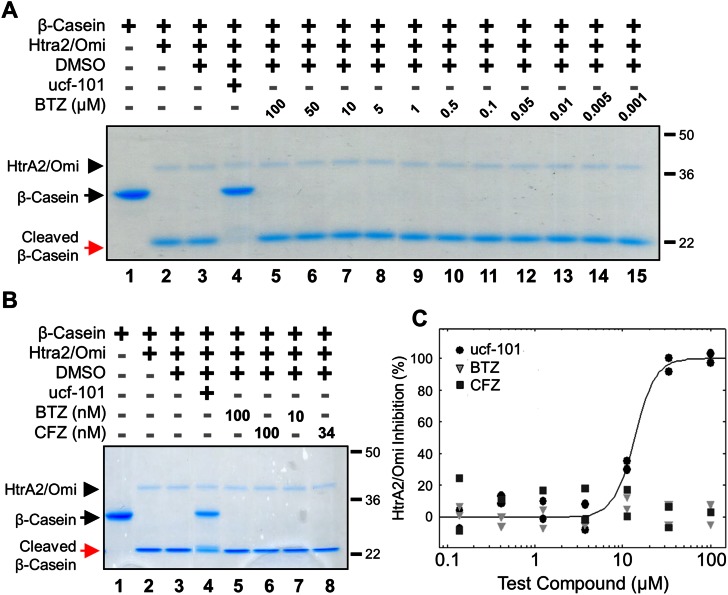
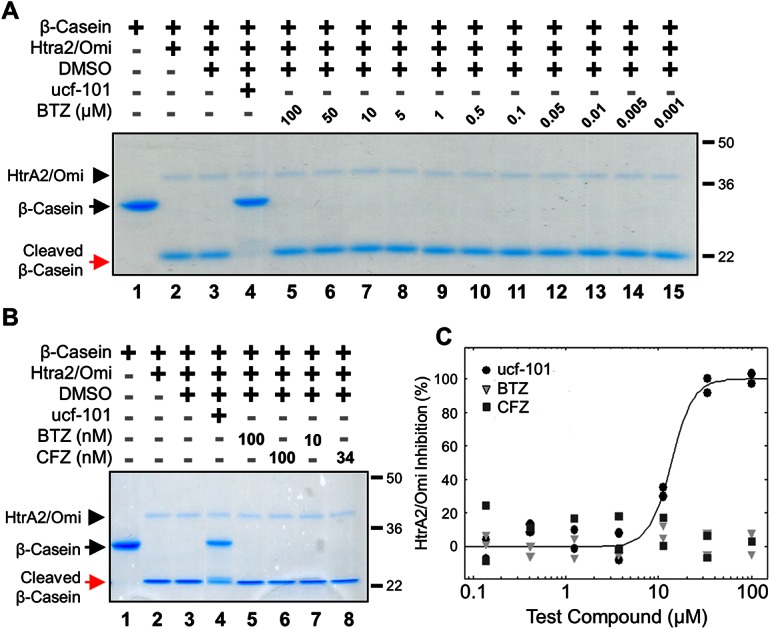
The effects of bortezomib and carfilzomib on recombinant human HtrA2/Omi activity were assessed with a gel-based assay using the substrate β-casein. In this simple model, active HtrA2/Omi cleaved the 29 kiloDalton (kDa) β-casein protein into a to a 21 kDa fragment; inhibition of HtrA2/Omi prevented this cleavage. As shown with a Commassie Brilliant Blue stain, a broad range of bortezomib (1 nM to 100 μM) had no effect on β-casein cleavage (Fig. 1A; red arrow). In contrast, ucf-101, a specific HtrA2/Omi inhibitor,19 completely prevented β-casein cleavage (black arrow). HtrA2/Omi levels were comparable between all treatments (black arrowhead). A direct comparison was then made between bortezomib and carfilzomib at both high (100 nM) and biologically-relevant (10 and 34 nM, respectively) concentrations. Neither bortezomib nor carfilzomib inhibited HtrA2/Omi (Fig. 1B, red versus black arrows). A biochemical assay with the fluorescent substrate peptide (Mca-IRRVSYSF (Dnp)K)15 was also used. This assay demonstrated that only ucf-101 inhibited HtrA2/Omi (IC50 = 11 μM) while bortezomib and carfilzomib had no effects at concentrations ranging from 0.1 to 100 μM (Fig. 1C). Taken together, these results strongly indicate that neither bortezomib nor carfilzomib are HtrA2/Omi inhibitors.
Fig. 1. Neither bortezomib nor carfilzomib inhibits recombinant HtrA2/Omi in vitro (A) gel-based enzyme assay of recombinant human HtrA2/Omi protease using β-casein as a model substrate. Recombinant human HtrA2/Omi was pre-incubated with 100 μM ucf-101 (Lane 4), or bortezomib (BTZ) up to 100 μM (Lanes 5–15), each dissolved in DMSO, for 45 minutes at room temperature before 1 μg mL–1 β-casein was added. After incubation at 45 °C for 1 hour, the reaction components were separated on a denaturing SDS gel and the separated proteins were visualized by Coomassie Brilliant Blue staining. (B) Recombinant human HtrA2/Omi was pre-incubated with the BTZ, carfilzomib (CFZ), or ucf-101, and analyzed in a gel-based enzyme assay using β-casein as a model substrate. (C) Peptide cleavage was assayed by using the fluorescent optimal substrate Mca-IRRVSYSF (K-Dnp) K. In the assays 10 μM fluorescent substrate was incubated with 30 pM human recombinant HtrA2/Omi for 60 minutes in the presence of BTZ, CFZ, or ucf-101. The solid line indicates a dose response curve for ucf-101 plotted against the % inhibition of HtrA2/Omi; dose response curves for either BTZ or CFZ could not be determined.
Proteasome inhibitors do not prevent the cleavage of HtrA2/Omi substrates in SH-SY5Y cells
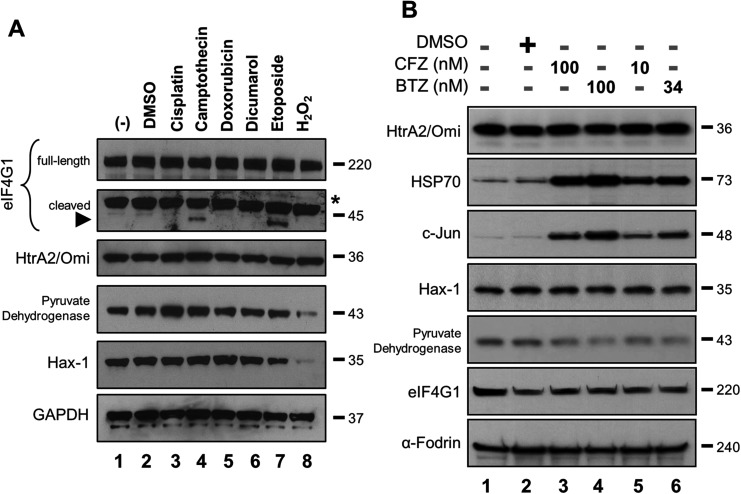
It was also reported that bortezomib but not carfilzomib prevented the cleavage of the eIF4G1 in human neuroblastoma SH-SY5Y cells.5 The full-length eIF4G1 protein is a putative HtrA2/Omi substrate that has not been fully validated by in vitro experiments.13 Detection of HtrA2/Omi-specific cleavage products is complicated since eIF4G1 is also cleaved by proteases other than HtrA2/Omi.20 A western blot was performed to detect the native eIF4G1 protein (220 kDa) and one of its HtrA2/Omi-associated cleavage products. SH-SY5Y cells were treated with compounds known to activate HtrA2/Omi [cisplatin (Lane 3), camptothecin (Lane 4), doxorubicin (Lane 5) and etoposide (Lane 7)] or regulate the expression level of eIF4G1 [dicumarol (Lane 6) and H2O2 (Lane 8)].21 Lysates were then probed for various proteins, including eIF4G1. While there was no appreciable change in the levels of the native eIF4G1 protein, a 45 kDa cleavage product was detected after camptothecin or etoposide treatment (Fig. 2A; black arrowhead). However, the reported putative HtrA2/Omi-associated cleavage products12 with molecular weight of 166, 63, and 10 kDa were not detected, which at some level may be cell line-specific (ESI Fig. 1†). Levels of two other HtrA2/Omi substrates, Hax-1 and pyruvate dehydrogenase,13,22 did not change after camptothecin or etoposide treatment. The H2O2 effects on these markers may be directly related to cell stress or death.
Fig. 2. Identification of eIF4G1 and its HtrA2/Omi linked cleavage products by western blots in SH-SY5Y cells. (A) Cells were treated with DMSO (0.25% for 12 h), cisplatin (30 mg ml–1 for 16 h), camptothecin (86 nM for 16 h), doxorubicin (5 μM for 8 h), dicumarol (300 μM for 10 h), etoposide (20 μM for 8 h), or H2O2 (100 μM for 6 h). Lysates were analyzed by western blot for the endogenous levels of native eIF4G1 protein (220 kDa) and the 45 kDa HtrA2/Omi linked cleavage product with a monoclonal eIF4G1 antibody (Abcam), or for the indicated proteins. An asterisk indicates non-specific binding (B) Protease inhibitor treatment does not change the expression level of HtrA2/Omi substrate proteins. SH-SY5Y cells were exposed to 0.25% DMSO, BTZ or CFZ for 12 hours. Whole cell extracts were analyzed by western blots for the endogenous levels of the indicated proteins. Results are representatives of two independent experiments.
If either bortezomib or carfilzomib were bona fide inhibitors of HtrA2/Omi activity, then the expression levels of eIF4G1, Hax-1 and pyruvate dehydrogenase should increase. To test this hypothesis, SH-SY5Y cells were treated with pharmaceutically relevant concentrations of both bortezomib and carfilzomib. Levels of HtrA2/Omi were unchanged, while both c-Jun and HSP70 accumulated, consistent with their known role in the ubiquitin-proteasome pathway (Fig. 2B).23 Neither compound affected levels of either eIF4G1 or Hax-1 (Lanes 3–6 versus Lane 2). Of note, α-Fodrin was used as a large molecular-weight control for eIF4G1. Interestingly, levels of pyruvate dehydrogenase decreased with PI treatment, but this may be a cell line-specific finding that has not been reported elsewhere. Co-treatment with ucf-101 was not successful in this cell line probably due to compound instability in the media (ESI Fig. 2†). Taken together, these results indicate that the two PIs do not affect HtrA2/Omi activity because expression levels of these known substrates do not increase.
Staurosporine-induced HtrA2/Omi-dependent cleavage of eIF4G1 and UCH-L1 was not abrogated by bortezomib or carfilzomib in mouse embryonic fibroblasts
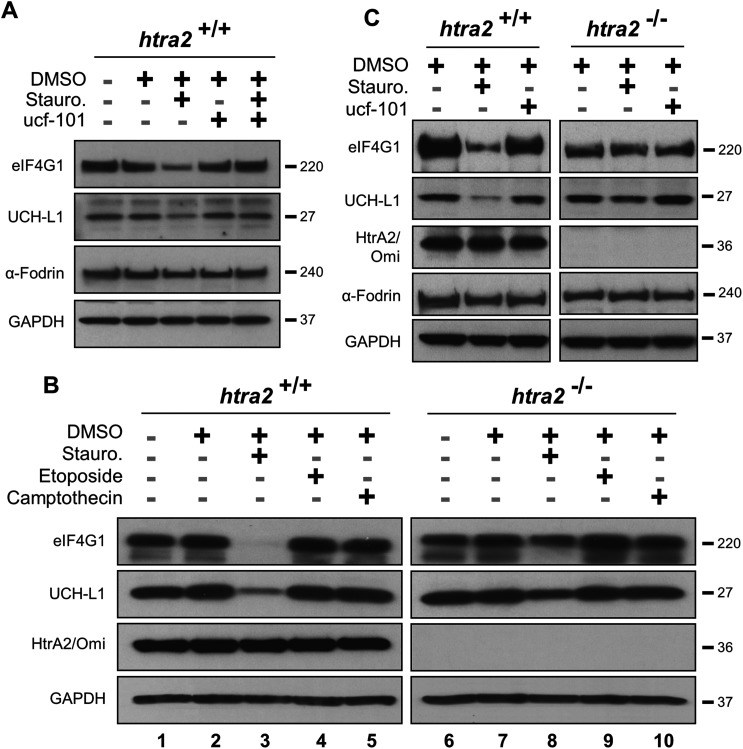
Since eIF4G1 can be cleaved by multiple proteases in cells,21,24,25 its HtrA2/Omi-dependent proteolysis was confirmed by using MEFs. UCH-L1 is a second validated HtrA2/Omi substrate18 and was utilized as a positive control. Cells were pre-incubated with ucf-101 and then treated with staurosporine, a compound known to activate HtrA2/Omi,26 to prevent the degradation of both eIF4G1 and UCH-L1 (Fig. 3A). Exposure to staurosporine or ucf-101 did not affect α-Fodrin levels. MEFs were then isolated from HtrA2/Omi-knockout mice.27 Cells were treated with one of three compounds known to activate HtrA2/Omi: staurosporine, etoposide, or camptothecin.26 Western blots of lysates from these cells (Fig. 3B) indicated that only staurosporine induced the degradation of either eIF4G1 or UCH-L1 when HtrA2/Omi was expressed (Lane 3 versus 8). To confirm the observations in wild-type and HtrA2/Omi-knockout MEFs that eIF4G1 and UCH-L1 are substrates of HtrA2/Omi, both MEFs were treated with ucf-101 (Fig. 3C). Exposure to ucf-101 had no effect on the protein levels of UCH-L1, eIF4G1, or α-Fodrin in wild type or knockout cells at baseline (e.g., without staurosporin), but the protein levels of UCH-L1 and eIF4G1 were decreased in an HtrA2/Omi-dependent manner in staurosporine-treated wild type cells.
Fig. 3. Cleavage of eIF4G1 and UCH-L1 in mouse embryonic fibroblasts is HtrA2/Omi dependent. (A) Wild type MEF cells were treated with 500 nM staurosporine (Stauro.) for 8 h and/or ucf-101 for 6 h. (B) Wild type (htra2 +/+) and HtrA2/Omi-deficient (htra2 –/–) mouse embryonic fibroblast (MEF) cells were treated with staurosporine, 86 nM camptothecin, or 20 μM etoposide. Endogenous level of full length eIF4G1, UCH-L1, HtrA2/Omi, and GAPDH was assayed by western blots in cell extracts. (C) Wild type and HtrA2/Omi-deficient MEFs were treated with 500 nM staurosporine or ucf-101 for 8 hours.
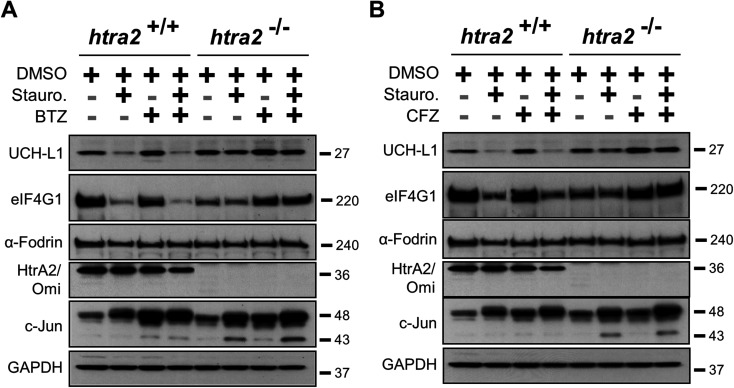
The ability of PIs to block the ability of HtrA2/Omi-mediated cleavage of eIF4G1 and UCH-L1 in MEFs was next investigated. Staurosporine-treated wild type and HtrA2/Omi-knockout MEFs were exposed to bortezomib and carfilzomib. The pharmacological effect of the PIs was demonstrated by evaluating c-Jun induction.23 In contrast to ucf-101 treatment, when MEFs were pre-incubated for one hour with either bortezomib (Fig. 4A) or carfilzomib (Fig. 4B) followed by staurosporine stimulation, the degradation of UCH-L1 and eIF4G1was not prevented. As a control, staurosporine had no effect on either UCH-L1 or eIF4G1in the HtrA2/Omi-knockout MEFs. Taken together, these findings further demonstrate that neither bortezomib nor carfilzomib was an inhibitor of HtrA2/Omi in cells.
Fig. 4. Staurosporine induced HtrA2/Omi dependent cleavage of eIF4G1 and UCH-L1 proteins was not abrogated by bortezomib or carfilzomib. Wild type and HtrA2/Omi-deficient MEFs were pre-incubated with 100 nM BTZ (A) or 100 nM CFZ (B) for 1 hour and then treated with 500 nM staurosporine for 10 hours. Endogenous level of full length eIF4G1, UCH-L1, α Fodrin, c-Jun, HtrA2/Omi, and GAPDH was assayed in cell extracts by western blots. Results are representatives of two independent experiments.
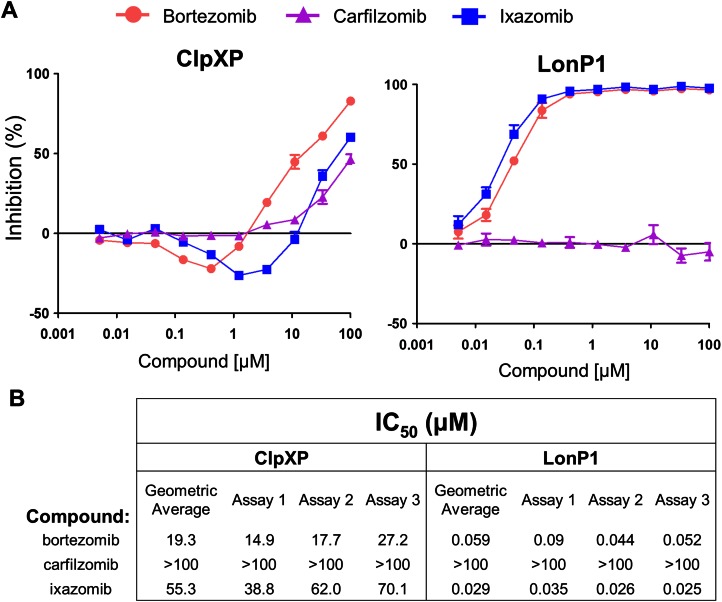
As part of a screening strategy we also tested whether bortezomib, carfilzomib or ixazomib could inhibit the activity of ClpXP and LonP1, two other serine proteases located in the mitochondria.26 All three PIs were relatively weak versus ClpXP, showing incomplete inhibition at 100 μM (Fig. 5A, left). Notably, there was significant activation by the boronic acids, bortezomib and ixazomib, prior to the onset of inhibition. This may be due to partial occupancy of the 14 ClpP sites that might have served to organize and activate the others. These two PIs, but not carfilzomib, also inhibited LonP1 (Fig. 5A, right). Additionally, none of the three PIs inhibited HtrA2/Omi activity (ESI Fig. 3†). Based upon a geometric average across three independent experiments, both ixazomib and bortezomib appeared to be inhibitors of LonP1 in vitro while carfilzomib was not (Fig. 5B). The biological relevance of this finding is unclear at this point, and as evidenced by the HtrA2/Omi controversy, it is imperative to validate these findings with other types of assays.
Fig. 5. Proteasome inhibitors have off-target effects on the serine proteases ClpXP and LonP1. Peptide cleavage assays were performed for ClpXP and LonP1 with their fluorogenic substrates in the presence of a proteasome inhibitor (bortezomib, carfilzomib, or ixazomib). (A) Graphs showing the percent inhibition of ClpXP (left) and LonP1 (right) across various proteasome inhibitor concentrations. Representative data are for a single experiment. Error bars indicate standard error. (B) A geometric average was taken from the individual IC50's (in μM) from three separate experiments. For ClpXP, activation interfered with the IC50 fit, and those points were deleted (generally the two showing most activation) to aid with calculations.
Discussion
The cause and progression of bortezomib-related peripheral neuropathy is a very complicated issue. Interestingly, changing the route of administration from intravenous to subcutaneous has decreased peripheral neuropathy as a side effect of bortezomib.28 One controversial proposal was that bortezomib caused peripheral neuropathy, in part, due to the inhibition of HtrA2/Omi.5 The work presented here thoroughly investigated this hypothesis and instead found no bortezomib off-target HtrA2/Omi inhibition. The absence of a bortezomib-HtrA2/Omi link was also supported by another independent investigation that used similar methodologies.6
The effects of bortezomib and carfilzomib on the enzymatic activity of the protease HtrA2/Omi were assessed through established gel-based and fluorogenic assays. The HtrA2/Omi inhibitor ucf-101 inhibited the activity of the protein in both types of assays (Fig. 1). However, neither bortezomib nor carfilzomib affected HtrA2/Omi activity, even at concentrations up to 100 μM (Fig. 1C).
The effects of PIs on HtrA2/Omi were tested in the SH-SY5Y neuroblastoma cell line. These cells were used in a previous report that found that bortezomib but not carfilzomib decreased the expression of the putative HtrA2/Omi substrates eIF4G1 and pyruvate dehydrogenase.5 Western blots for the HtrA2/Omi substrate eIF4G1 did not produce any of the expected cleavage products (166, 63 and 10, kDa) identified in the literature,12 although camptothecin or etoposide treatment produced a 45 kDa fragment (Fig. 2A). The human eIF4G1 protein is often phosphorylated and has at least five different isoforms,29 and its cleavage can produce many products that lead to complex western blot results.30 Thus, in the absence of validated, reliable eIF4G1 cleavage products in SH-SY5Y cells, the study endpoints were instead the native full length eIF4G1 protein. Neither bortezomib nor carfilzomib affected the native full length eIF4G1 protein levels at concentrations up to 100 nM (Fig. 2B). PI treatment is also known to result in a decrease of pyruvate dehydrogenase.22 Here, expression of pyruvate dehydrogenase (Fig. 2B) decreased after exposure to either bortezomib or carfilzomib. Taken together, these results starkly contrast previous data that showed that bortezomib, but not carfilzomib, decreased the expression levels of these proteins within SH-SY5Y cells.5
HtrA2/Omi-deficient MEF cells were used to characterize the relationships between HtrA2/Omi, eIF4G1 protein, and its cleavage products. Wild type (htra2 +/+) and HtrA2/Omi-knockout MEFs (htra2 –/–) were used to determine if drug-stimulated cleavage of eIF4G1 is HtrA2/Omi-dependent. There was an HtrA2/Omi component to the staurosporine-induced cleavage of eIF4G1, as indicated by the loss of full length protein in MEFs, since degradation was significantly less in knockout cells (Fig. 3B). This observation provided the opportunity to study the changes in the expression levels of the native eIF4G1 protein in the wild type MEFs without the need for investigating the expression levels of its elusive degradation products. When MEFs were treated with ucf-101, staurosporine-induced degradation of the native eIF4G1 was prevented, supporting the notion that HtrA2/Omi has a role in the regulation of eIF4G1 expression levels (Fig. 3C). In contrast to ucf-101, bortezomib and carfilzomib did not prevent staurosporine-induced HtrA2/Omi-dependent degradation of eIF4G1 (Fig. 4A and B). The ucf-101 inhibitor also prevented the staurosporine-induced degradation of the verified HtrA2/Omi substrate UCH-L1 (Fig. 3B and C). Neither bortezomib nor carfilzomib were able to prevent this degradation (Fig. 4), which further suggests that the two PIs do not affect HtrA2/Omi activity.
Recently, other proteases such as LonP1 were reported to be inhibited by PIs including bortezomib, MG132, and lactacystin (cLβL), but not by epoxomicin.31–33 In addition, two recent peptidomic analysis reports on the effects of bortezomib and epoxomicin on HEK293T and SH-SY5Y cells suggested the potential presence of off-target proteases not only for bortezomib but for epoxomicin, from which carfilzomib is derived.34,35 These enzymes remain to be identified and investigated for their differential inhibition by various PIs and their role in the genesis of PI-associated peripheral neuropathy. This study is the first to report that the boronic acids, bortezomib and ixazomib, inhibited the activity of the serine protease LonP1 in vitro and only weakly inhibits ClpXP in vitro (Fig. 5). To date there are no reports linking LonP1 inhibition to peripheral neuropathy. In the clinic ixazomib was associated with low rates of peripheral neuropathy.36 Thus, the biological relevance of this finding remains unclear.
In summary, both the recombinant enzyme assays and cell-based assays consistently demonstrated that neither bortezomib nor carfilzomib caused inhibition of HtrA2/Omi. These data indicate that it is highly unlikely that inhibition of this enzyme plays a causative role in PI-induced peripheral neuropathy. These results were in contrast to results published previously by others5,7 that indicated bortezomib but not carfilzomib or ixazomib was an inhibitor of HtrA2/Omi. The reason for these disparate results is not known, but in regards to the eIF4G1 cleavage shown elsewhere,5 it is concerning that the size of the protein, as well as the various cleavage products were not presented. In conclusion, it is clear from our analysis that neither bortezomib nor carfilzomib inhibits the enzymatic activity of HtrA2/Omi.
Author's contributions
Experimental design and execution: Vilmos Csizmadia, Paul Hales, Christopher Tsu, Larry Dick, and Francis Wolenski; reagent development: Jingya Ma and Jiejin Chen; data analysis: Pooja Shah, Paul Fleming, Joseph Senn, and Vivek Kadambi; manuscript preparation: Vilmos Csizmadia and Francis Wolenski.
Conflicts of interest
All authors were compensated employees of Takeda Pharmaceutical Company Limited when this research was conducted.
Supplementary Material
Acknowledgments
We thank Dr Allison Berger, Cheryl Riel and Benjamin Snow for their suggestions and critical reading of the manuscript.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c6tx00220j
References
- Esseltine D. L., Mulligan G. Semin. Hematol. 2012;49:196–206. doi: 10.1053/j.seminhematol.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Argyriou A. A., Iconomou G., Kalofonos H. P. Blood. 2008;112:1593–1599. doi: 10.1182/blood-2008-04-149385. [DOI] [PubMed] [Google Scholar]
- Csizmadia V., Csizmadia E., Silverman L., Simpson C., Raczynski A., O'Brien L., Gallacher M., Cardoza K., Kadambi V. J., Fedyk E. R., Alden C. L. Vet. Pathol. 2010;47:358–367. doi: 10.1177/0300985809358423. [DOI] [PubMed] [Google Scholar]
- Kettner C. A., Shenvi A. B. J. Biol. Chem. 1984;259:15106–15114. [PubMed] [Google Scholar]
- Arastu-Kapur S., Anderl J. L., Kraus M., Parlati F., Shenk K. D., Lee S. J., Muchamuel T., Bennett M. K., Driessen C., Ball A. J., Kirk C. J. Clin. Cancer Res. 2011;17:2734–2743. doi: 10.1158/1078-0432.CCR-10-1950. [DOI] [PubMed] [Google Scholar]
- Bachovchin D. A., Koblan L. W., Wu W., Liu Y., Li Y., Zhao P., Woznica I., Shu Y., Lai J. H., Poplawski S. E., Kiritsy C. P., Healey S. E., DiMare M., Sanford D. G., Munford R. S., Bachovchin W. W., Golub T. R. Nat. Chem. Biol. 2014;10:656–663. doi: 10.1038/nchembio.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan D., Tian Z., Zhou B., Kuhn D., Orlowski R., Raje N., Richardson P., Anderson K. C. Clin. Cancer Res. 2011;17:5311–5321. doi: 10.1158/1078-0432.CCR-11-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupperman E., Lee E. C., Cao Y., Bannerman B., Fitzgerald M., Berger A., Yu J., Yang Y., Hales P., Bruzzese F., Liu J., Blank J., Garcia K., Tsu C., Dick L., Fleming P., Yu L., Manfredi M., Rolfe M., Bolen J. Cancer Res. 2010;70:1970–1980. doi: 10.1158/0008-5472.CAN-09-2766. [DOI] [PubMed] [Google Scholar]
- d'Azzo A., Bonten E. Biochem. Soc. Trans. 2010;38:1453–1457. doi: 10.1042/BST0381453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss K. M., Martins L. M., Plun-Favreau H., Marx F. P., Kautzmann S., Berg D., Gasser T., Wszolek Z., Muller T., Bornemann A., Wolburg H., Downward J., Riess O., Schulz J. B., Kruger R. Hum. Mol. Genet. 2005;14:2099–2111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- Gray C. W., Ward R. V., Karran E., Turconi S., Rowles A., Viglienghi D., Southan C., Barton A., Fantom K. G., West A., Savopoulos J., Hassan N. J., Clinkenbeard H., Hanning C., Amegadzie B., Davis J. B., Dingwall C., Livi G. P., Creasy C. L. Eur. J. Biochem. 2000;267:5699–5710. doi: 10.1046/j.1432-1327.2000.01589.x. [DOI] [PubMed] [Google Scholar]
- Vande Walle L., Van Damme P., Lamkanfi M., Saelens X., Vandekerckhove J., Gevaert K., Vandenabeele P. J. Proteome Res. 2007;6:1006–1015. doi: 10.1021/pr060510d. [DOI] [PubMed] [Google Scholar]
- Zurawa-Janicka D., Skorko-Glonek J., Lipinska B. Expert Opin. Ther. Targets. 2010;14:665–679. doi: 10.1517/14728222.2010.487867. [DOI] [PubMed] [Google Scholar]
- Jones J. M., Datta P., Srinivasula S. M., Ji W., Gupta S., Zhang Z., Davies E., Hajnoczky G., Saunders T. L., Van Keuren M. L., Fernandes-Alnemri T., Meisler M. H., Alnemri E. S. Nature. 2003;425:721–727. doi: 10.1038/nature02052. [DOI] [PubMed] [Google Scholar]
- Martins L. M., Turk B. E., Cowling V., Borg A., Jarrell E. T., Cantley L. C., Downward J. J. Biol. Chem. 2003;278:49417–49427. doi: 10.1074/jbc.M308659200. [DOI] [PubMed] [Google Scholar]
- Akopian T., Kandror O., Tsu C., Lai J. H., Wu W., Liu Y., Zhao P., Park A., Wolf L., Dick L. R., Rubin E. J., Bachovchin W., Goldberg A. L. J. Biol. Chem. 2015;290:11008–11020. doi: 10.1074/jbc.M114.625640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishovitz J., Li M., Frase H., Hudak J., Craig S., Ko K., Berdis A. J., Suzuki C. K., Lee I. ACS Chem. Biol. 2011;6:781–788. doi: 10.1021/cb100408w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D. W., Nam M. K., Rhim H. Biochem. Biophys. Res. Commun. 2011;415:24–29. doi: 10.1016/j.bbrc.2011.09.148. [DOI] [PubMed] [Google Scholar]
- Cilenti L., Lee Y., Hess S., Srinivasula S., Park K. M., Junqueira D., Davis H., Bonventre J. V., Alnemri E. S., Zervos A. S. J. Biol. Chem. 2003;278:11489–11494. doi: 10.1074/jbc.M212819200. [DOI] [PubMed] [Google Scholar]
- Morley S. J., Coldwell M. J., Clemens M. J. Cell Death Differ. 2005;12:571–584. doi: 10.1038/sj.cdd.4401591. [DOI] [PubMed] [Google Scholar]
- Alard A., Fabre B., Anesia R., Marboeuf C., Pierre P., Susini C., Bousquet C., Pyronnet S. Mol. Cell. Biol. 2010;30:1097–1105. doi: 10.1128/MCB.00868-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F., Kaplitt M. G. PLoS One. 2009;4:e7100. doi: 10.1371/journal.pone.0007100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meriin A. B., Gabai V. L., Yaglom J., Shifrin V. I., Sherman M. Y. J. Biol. Chem. 1998;273:6373–6379. doi: 10.1074/jbc.273.11.6373. [DOI] [PubMed] [Google Scholar]
- Vosler P. S., Gao Y., Brennan C. S., Yanagiya A., Gan Y., Cao G., Zhang F., Morley S. J., Sonenberg N., Bennett M. V., Chen J. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18102–18107. doi: 10.1073/pnas.1112635108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marissen W. E., Lloyd R. E. Mol. Cell. Biol. 1998;18:7565–7574. doi: 10.1128/mcb.18.12.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Walle L., Lamkanfi M., Vandenabeele P. Cell Death Differ. 2008;15:453–460. doi: 10.1038/sj.cdd.4402291. [DOI] [PubMed] [Google Scholar]
- Martins L. M., Morrison A., Klupsch K., Fedele V., Moisoi N., Teismann P., Abuin A., Grau E., Geppert M., Livi G. P., Creasy C. L., Martin A., Hargreaves I., Heales S. J., Okada H., Brandner S., Schulz J. B., Mak T., Downward J. Mol. Cell. Biol. 2004;24:9848–9862. doi: 10.1128/MCB.24.22.9848-9862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride A., Ryan P. Y. Expert Rev. Anticancer Ther. 2013;13:339–358. doi: 10.1586/era.13.9. [DOI] [PubMed] [Google Scholar]
- Bradley C. A., Padovan J. C., Thompson T. L., Benoit C. A., Chait B. T., Rhoads R. E. J. Biol. Chem. 2002;277:12559–12571. doi: 10.1074/jbc.M111134200. [DOI] [PubMed] [Google Scholar]
- Bushell M., McKendrick L., Janicke R. U., Clemens M. J., Morley S. J. FEBS Lett. 1999;451:332–336. doi: 10.1016/s0014-5793(99)00614-6. [DOI] [PubMed] [Google Scholar]
- Bayot A., Basse N., Lee I., Gareil M., Pirotte B., Bulteau A. L., Friguet B., Reboud-Ravaux M. Biochimie. 2008;90:260–269. doi: 10.1016/j.biochi.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Granot Z., Kobiler O., Melamed-Book N., Eimerl S., Bahat A., Lu B., Braun S., Maurizi M. R., Suzuki C. K., Oppenheim A. B., Orly J. Mol. Endocrinol. 2007;21:2164–2177. doi: 10.1210/me.2005-0458. [DOI] [PubMed] [Google Scholar]
- Lu B., Lee J., Nie X., Li M., Morozov Y. I., Venkatesh S., Bogenhagen D. F., Temiakov D., Suzuki C. K. Mol. Cell. 2013;49:121–132. doi: 10.1016/j.molcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker L. D., Gelman J. S., Castro L. M., Gozzo F. C., Ferro E. S. J. Proteome Res. 2012;11:1981–1990. doi: 10.1021/pr2012076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman J. S., Dasgupta S., Berezniuk I., Fricker L. D. Biochim. Biophys. Acta. 2013;1834:2408–2417. doi: 10.1016/j.bbapap.2013.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. K., Berdeja J. G., Niesvizky R., Lonial S., Laubach J. P., Hamadani M., Stewart A. K., Hari P., Roy V., Vescio R., Kaufman J. L., Berg D., Liao E., Di Bacco A., Estevam J., Gupta N., Hui A. M., Rajkumar V., Richardson P. G. Lancet Oncol. 2014;15:1503–1512. doi: 10.1016/S1470-2045(14)71125-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.