 Our data represent the first demonstration that oxy-PAHs can be potent inhibitors of CYP1 expression and function.
Our data represent the first demonstration that oxy-PAHs can be potent inhibitors of CYP1 expression and function.
Abstract
Oxygenated polycyclic aromatic hydrocarbons (oxy-PAHs) are found in the environment together with PAHs. However, less is known concerning their biological activity including their impact on aryl hydrocarbon receptor (AHR) signalling and the subsequent modulation of the cytochrome P450 monooxygenases (CYP). In this study, the effects of 15 environmentally relevant oxy-PAHs on the induction and activity of the CYP1 enzymes were determined in vitro by measuring gene expression levels and enzyme activity. We found that nine of the tested oxy-PAHs significantly induced CYP1A1 and CYP1B1 gene expression in human keratinocytes (HaCaT cells) while only five of these also were potent inducers of CYP1-dependent ethoxyresorufin-O-deethylase (EROD) activity suggesting that some of the oxy-PAHs are both activators of AHR signalling and inhibitors of CYP1 function. Using a recombinant human CYP1A1 enzyme we showed that eleven of the oxy-PAHs potently inhibited enzyme activity with benz[a]anthracene-7,12-quinone (7,12-BAQ) and benzo[a]fluorenone (BFLO) being the most potent inhibitors (IC50 = 0.037 and 0.061 μM, respectively). We further exposed HaCaT cells to binary mixtures of oxy-PAHs and the model AHR agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) to investigate potential interaction effects. The results showed that oxy-PAHs can interfere with the TCDD-mediated effects leading to reduced CYP1A1 and 1B1 expression and EROD activity. These data represent the first demonstration that oxy-PAHs can be potent inhibitors of CYP1 expression and function and make important contributions towards understanding the mechanisms through which oxy-PAHs can contribute to the overall risk of polycyclic aromatic compounds.
Introduction
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous environmental pollutants of which many are classified as human carcinogens.1 A related group of compounds of toxicological concern is the oxygenated PAHs (oxy-PAHs), which are emitted from the same primary sources as PAHs but can also be formed through secondary oxidation of PAHs in the environment.2,3 As a result they are found in the environment together with the PAHs and frequently at similar or even higher levels.4,5 The oxy-PAHs include polycyclic aromatic quinones and ketones and, in contrast to the well-studied PAHs, the biological effects of oxy-PAHs are not as well characterized. However, recent studies have reported cytotoxicity, genotoxicity, mutagenicity and teratogenicity of some oxy-PAHs in different in vitro and in vivo systems.6–9 Since the oxy-PAHs are present in the environment and pose a possible risk for humans and wild-life, there is a need to characterize the mode of action of oxy-PAH toxicity.
Several PAHs are activators of the aryl hydrocarbon receptor (AHR) and the subsequent induction and action of cytochrome P450 monooxygenases (CYP) 1A1 and 1B1 are important in mediating many of the biological effects of PAHs, including carcinogenicity and developmental defects.10 In addition, several PAHs are potent inhibitors of the CYP1 family enzymes11,12 which in combination with AHR activation can lead to synergistic biological effects. This has been observed in fish embryos as severe developmental toxicity13–17 and in mammalian in vivo and in vitro systems as increased levels of DNA adducts18–20 in response to mixtures of AHR activators such as benzo[a]pyrene (B[a]P) or β-naphthoflavone (βNF), and CYP1 inhibitors such as fluoranthene (FL) or α-naphthoflavone (αNF).
In contrast to PAHs, the potency of oxy-PAHs to activate the AHR and its role in mediating the toxicity has not been studied in detail. Data from cellular reporter assays show however that oxy-PAHs in general are weaker AHR activators than their parent compounds and, as observed with PAHs, that larger (four-ring) oxy-PAHs elicit the strongest activation.21–24 We and others have also shown that the developmental toxicity of oxy-PAHs in zebrafish embryos is at least partially AHR-dependent.7,25,26 However, to the best of our knowledge, there is no published data on the ability of oxy-PAHs to inhibit CYP1 enzymes and the potential to thereby contribute to mixture effects of PAHs and other environmental pollutants.
To determine the impact of oxy-PAHs on CYP1 function, we examined the induction and inhibition of human CYP1 by 15 environmentally relevant oxy-PAHs by measuring the gene expression levels of CYP1A1 and 1B1 and enzyme activity. The systems used were human HaCaT keratinocytes and microsomes containing human recombinant CYP1A1. HaCaT cells were used as a model system since dermal exposure to PAHs is a cause of major concern for occupational settings and contaminated soil due to their association with increased cancer risk.27,28 These cells were further chosen since they express significant levels of the AHR and have previously been used by us and others to study AHR activation and signalling.29–31 We first determined the ability of oxy-PAHs, as single compounds, to influence the CYP1 function and gene expression in a dose- and time-dependent manner. We further exposed HaCaT cells to binary mixtures of oxy-PAHs and the model AHR agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) to investigate potential interaction effects.
Materials and methods
Caution: PAHs are carcinogens, and thus, experimental handling must be carried out under special safety conditions, for example, those outlined in the NCI guidelines.
Chemicals
1-Indanone (1-INO), 1H-phenalen-1-one (1H-PHO), acenaphthylene-1,2-quinone (1,2-ACNQ), chrysene-1,4-quinone (1,4-CHRQ), 2-methylanthracene-9,10-quinone (2-MAQ), 2,3-dimethylanthracene-9,10-quinone (2,3-DMAQ), naphthacene-5,12-quinone (5,12-NQ), 7H-benz[de]anthracene-7-one (7H-BAO), 9-fluorenone (9-FLO), phenanthrene-9,10-quinone (9,10-PQ), anthracene-9,10-quinone (9,10-AQ) and benz[a]anthracene-7,12-quinone (7,12-BAQ) all with purity ≥95% and from Sigma Aldrich (Stockholm, Sweden) were kindly provided by Dr Staffan Lundstedt at Umeå University. 4H-Cyclopenta[def]phenanthrene-4-one (4H-CPO), benzo[a]fluorenone (BFLO) and 6H-benzo[cd]pyren-6-one (6H-BPO), all with purity ≥99% were obtained from the Institute for Reference Materials and Measurements (EC-JRC-IRMM, Geel, Belgium). 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD; CASRN 1746-01-6, 98%) was obtained from Cambridge Isotope Laboratory (Andover, MA). Benzo[a]pyrene (B[a]P; CASRN 50-32-8, ≥96%), 7-ethoxyresorufin, fluorescamine, dihydronicotinamide-adenine dinucleotide phosphate (NADPH) and microsomes containing recombinant human CYP1A1 and NADPH-P450 reductase were obtained from Sigma-Aldrich (Stockholm, Sweden). All cell culture reagents were purchased from Gibco (Life Technologies, Stockholm, Sweden).
Cell culture and exposure
Human immortalized keratinocytes (HaCaT) were kindly provided by N. E. Fusenig (DKFZ, Heidelberg, Germany). HaCaT cells were cultured in DMEM supplemented with 10% fetal bovine serum, penicillin (100 units per mL) and streptomycin (0.1 mg mL–1) and maintained at 37 °C in 5% CO2. Prior to exposure cells were seeded at 1 × 104 cells per well in 96-well plates and cultured for 3 d to reach high confluency. The cells were exposed, in fresh DMEM medium, to solvent control (0.1–0.18% DMSO), oxy-PAHs (0.1–10 μM), B[a]P (0.1–10 μM) or TCDD (5 nM) alone or to binary mixtures of oxy-PAH and TCDD respectively for up to 48 h.
Cell viability assay
The cell viability was assessed using the Alamar Blue assay which is based on the reduction potential of metabolically active cells. Following exposure, a 10% solution of AlamarBlue® reagent (Invitrogen) in DMEM medium was added to each well and incubated for 1 h at 37 °C. The fluorescence was measured at excitation/emission wavelengths 560/590 nm using a Victor 3 Wallac plate reader (Perkin Elmer, MA, US). Three separate experiments were analysed. The results were expressed as percentage cell viability versus DMSO control.
EROD assays
CYP1-dependent ethoxyresorufin-O-deethylase (EROD) activity in cells was measured as previously described with some modifications.32 Exposures were terminated at the indicated time points by removing the medium and rinsing the cells with PBS. The EROD reaction was initiated by addition of 100 μL of 2 μM 7-ethoxyresorufin into sodium phosphate buffer (50 mM, pH 8.0) followed by 20 min of incubation at 37 °C. The reaction was terminated by addition of 75 μL of cold fluorescamine solution (0.15 mg mL–1 in acetonitrile), and the formation of resorufin was measured in triplicate using a GENios Pro plate reader (Tecan, Männedorf, Switzerland) at excitation/emission wavelengths 535/590 nm. Enzymatic activity was normalized for the cellular protein content determined by fluorescamine fluorescence at excitation/emission wavelengths 390/485 nm.
The EROD activity of microsomes containing human recombinant CYP1A1 (2.5 nM) was assayed as described earlier with minor changes.30 After pre-incubation with DMSO (final concentration <0.5%) or oxy-PAH (0.00064–20 μM) for 5 min, 7-ethoxyresorufin (0.5 μM) and NADPH (0.4 mM) were added. Formation of resorufin was measured in triplicate over time as described above with repeated measurements every 2 seconds. Kinetic parameters for inhibition of EROD activities were estimated by nonlinear regression analysis using GraphPad Prism 6.05 (GraphPad, San Diego, CA) with the EROD activity of DMSO-control set as 100% and the assay without addition of NADPH as 0%.
Real-time qRT-PCR
Gene expression was assessed using the Ambion® Cells-to-CT™ 1-Step Power SYBR® Green kit (Life Technologies, Stockholm, Sweden) according to the protocol. In brief, 2 μL cell lysate and 200 nM primers were used per 20 μL reaction mixture. Three separate experiments were analysed in duplicate using an Applied Biosystems 7500 Real-Time PCR System (Life Technologies, Stockholm, Sweden). Cycling conditions were 48 °C for 30 min, 95 °C for 10 min and then 40 cycles of 95 °C for 15 s and 60 °C for 1 min followed by melt curve analysis. Relative gene expression quantification was based on the comparative threshold cycle method (2–ΔΔCt) using beta-2-microglobulin (B2M) as the housekeeping gene.30 Primer sequences and efficiencies (%) were as follows: CYP1A1 forward 5′-CACCATCCCCCACAGCAC-3′ and reverse 5′-ACAAAGACACAACGCCCCTT-3′ (102%), CYP1B1 forward 5′-AGTGCAGGCAGAATTGGATCA-3′ and reverse 5′-AGGACATAGGGCAGGTTGGG-3′ (111%), B2M forward 5′-GATGAGTATGCCTGCCGTGTG-3′ and reverse 5′-CAATCCAAATGCGGCATCT-3′ (108%).
Statistical analysis
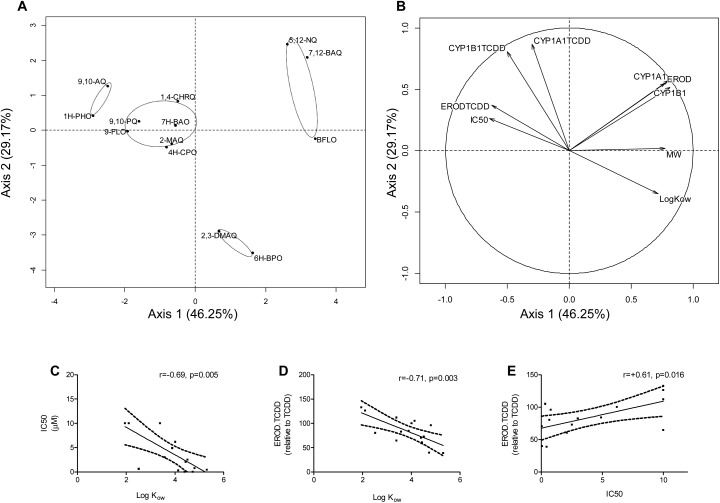
All experiments were performed in triplicate and data presented are means ± SE. Statistical analyses were performed with GraphPad Prism 6.05 unless otherwise stated. P value <0.05 was considered significant. Viability, EROD activities and CYP1 gene expression in response to oxy-PAHs and/or TCDD were analysed using two-way ANOVA followed by Bonferroni's post hoc test. Principal component analysis (PCA, two axes) of the different measured endpoints was performed with the R software (FactoMineR package, ; http://cran.r-project.org/). This analysis included the 13 oxy-PAHs with data for all endpoints at 1 μM and 6 h post exposure in addition to IC50 (inhibition of CYP1A1-mediated EROD activity), log Kow and MW values. Pearson correlation (two-tailed) was also calculated to further confirm the relationship between the EROD, qRT-PCR, IC50, log Kow or MW data.
Results
Impact on cellular EROD activity and gene expression of CYP1A1 and 1B1 by oxy-PAHs
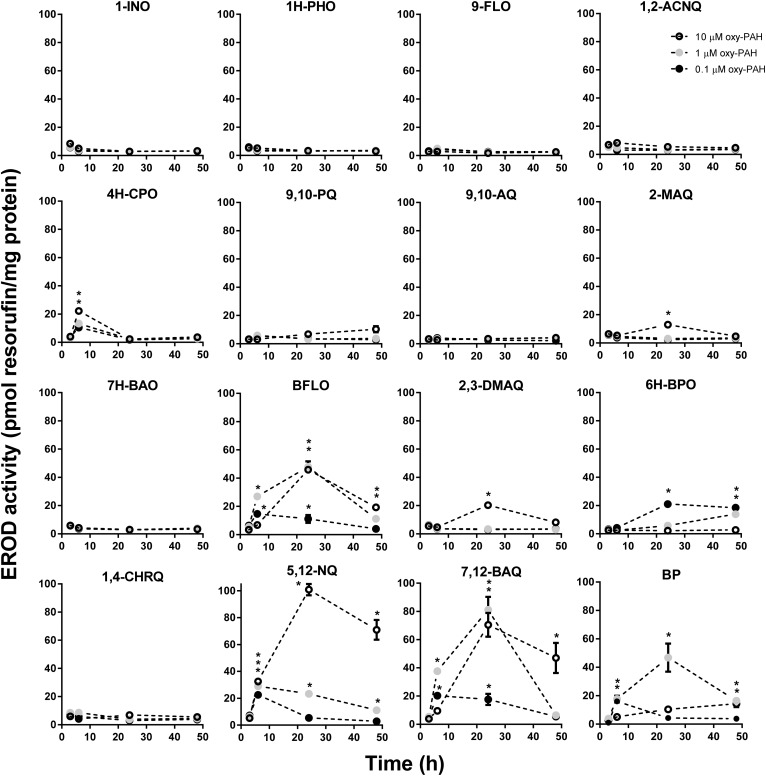
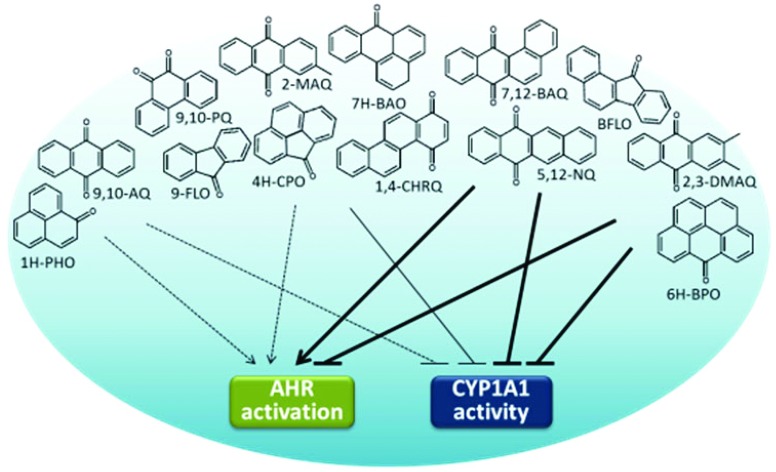
We first investigated the ability of oxy-PAHs to induce CYP1 enzyme activity and gene expression in HaCaT cells by determining the levels of EROD activity and CYP1A1 and 1B1 mRNA. As shown in Fig. 1, the oxy-PAHs showed clear differences in the potency of inducing EROD activity. In general, the oxy-PAHs displayed time- and dose-dependent effects with transient increase of enzyme activity peaking at 24 h. The most potent inducers were BFLO, 5,12-NQ, and 7,12-BAQ, which displayed similar or higher levels compared to equimolar levels of B[a]P. The four oxy-PAHs 4H-CPO, 2-MAQ, 2,3-DMAQ, and 6H-BPO displayed significantly increased levels of enzyme activity compared to the DMSO control but lower than B[a]P. Notably, 6H-BPO displayed the highest activity in response to the lowest dose of 0.1 μM and much lower activity in response to higher doses, suggesting that 6H-BPO is an inhibitor of CYP1 activity or transcription. Similar trends were observed also for BFLO and 7,12-BAQ of which the middle dose (1 μM) showed higher induction at 6 h compared to 10 μM. The remaining 8 oxy-PAHs had no significant effect on cellular EROD activity.
Fig. 1. Oxy-PAHs induce a time- and dose-dependent increase of cellular EROD activity. HaCaT cells were exposed to 0.1, 1.0 or 10 μM oxy-PAH or B[a]P and CYP1-dependent EROD activity was measured at 3, 6, 24 and 48 h after exposure. Data points represent means ± SE, n = 3. *p < 0.05 as compared with DMSO control by two-way ANOVA.
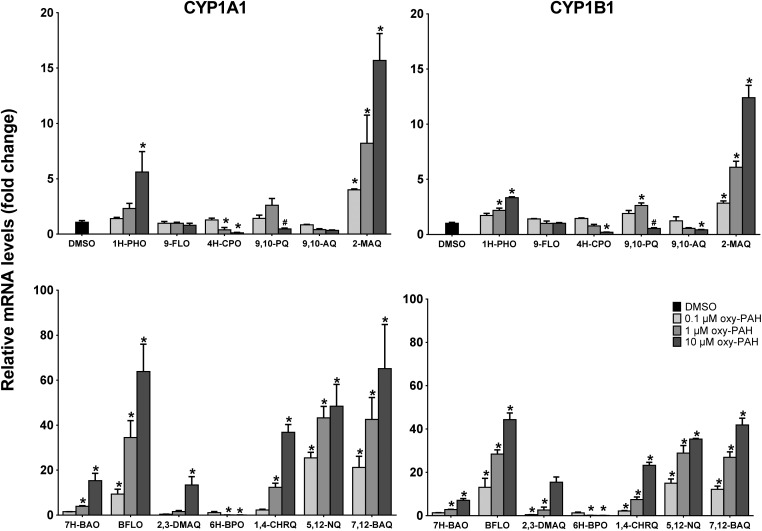
Assessing the change in the mRNA levels of CYP1A1 and 1B1 at 6 h in response to the different oxy-PAHs showed a differential induction which in general correlated with the EROD activity data at 24 h (Fig. 2). The strongest effect was observed in response to BFLO, 1,4-CHRQ, 5,12-NQ, and 7,12-BAQ causing up to about 60- and 40-fold increase of the CYP1A1 and CYP1B1 mRNA levels respectively compared to the DMSO control. In general, the oxy-PAHs induced slightly higher expression levels of CYP1A1 compared to 1B1, although with similar dose–response trends. Even though 1,4-CHRQ did not induce EROD activity, the gene expression levels were significantly increased suggesting that 1,4-CHRQ inhibits the CYP1 function. Notably, the mRNA level data for 4H-CPO, 9,10-AQ and 6H-BPO revealed significant dose-dependent reduction, up to 15-fold, of the CYP1A1 and CYP1B1 mRNA levels compared to the DMSO control suggesting that these oxy-PAHs may act as inhibitors of AHR activation. The effect of 10 μM 9,10-PQ was deemed to most likely be due to the observed increased cytotoxicity (ESI Fig. S1†) and in agreement with previous observations.33
Fig. 2. Oxy-PAHs induce CYP1 gene expression. HaCaT cells were exposed to 0.1, 1.0 or 10 μM oxy-PAH and effects on gene expression of CYP1A1 (left panels) and CYP1B1 (right panels) were determined by qRT-PCR at 6 h after exposure. Data points represent means ± SE, n = 3. #Increased cytotoxicity. *p < 0.05 and >2-fold change as compared with DMSO control by two-way ANOVA.
Inhibition of CYP1A1 activity by oxy-PAHs
Next, we examined the inhibition of CYP1A1 enzyme activity by oxy-PAHs using microsomes expressing human CYP1A1. The different oxy-PAHs showed clear differences in the potency of inhibiting CYP1A1 enzyme activity. Examples of inhibition curves obtained from BFLO, 6H-BPO and 1,4-CHRQ are shown in Fig. 3 and all inhibition curves are shown in ESI Fig. S2.† Of the 15 oxy-PAHs tested, BFLO and 7,12-BAQ were the most potent inhibitors with IC50 values of 0.061 and 0.037 μM, respectively. 4H-CPO, 9,10-PQ, 7H-BAO and 6H-BPO were found to inhibit CYP1A1 with IC50 values ranging between 0.32 and 0.77 μM. 9-FLO, 2-MAQ, 2,3-DMAQ, 1,4-CHRQ, and 5,12-NQ inhibited CYP1A1 with IC50 values ranging between 2.1 and 6.2 μM while 1-INO, 1H-PHO, 1,2-ACNQ, and 9,10-AQ all displayed IC50 values >10 μM. Determined IC50 values are shown in Table 1.
Fig. 3. Oxy-PAHs inhibit CYP1A1-mediated EROD activity. Plots are shown for inhibition by BFLO ([black circle]), 6H-BPO (◆), and 1,4-CHRQ (■). Data points represent means ± SE, n = 3. See Table 1 for IC50 values.
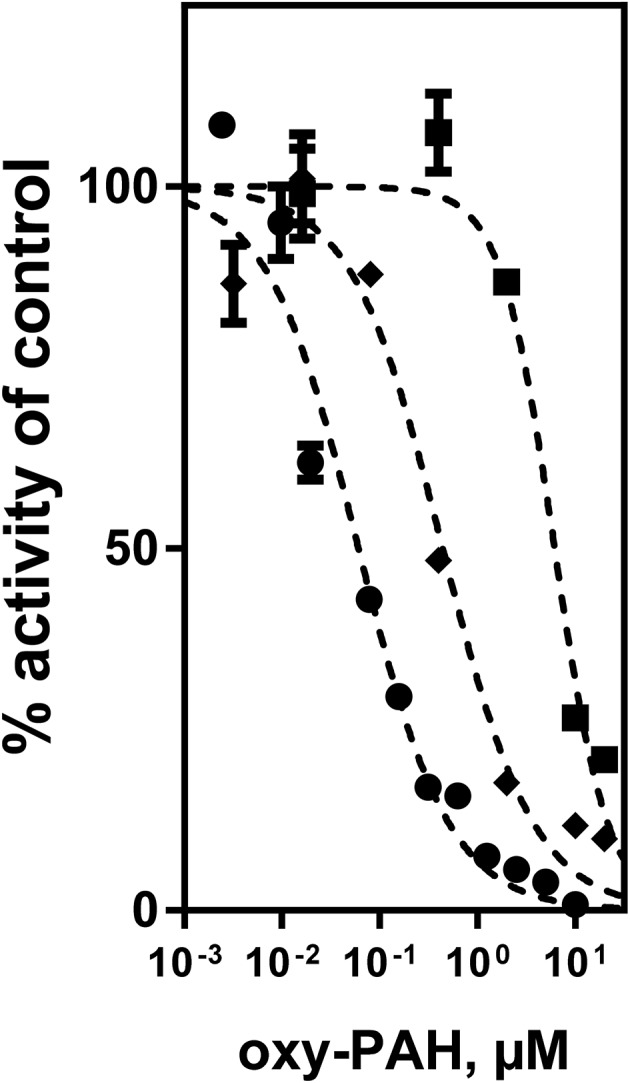
Table 1. Oxy-PAHs used in this study with abbreviations, CAS numbers, molecular weight (MW), logarithm of octanol-water partition coefficient (log Kow) and inhibition of CYP1A1-mediated EROD activity (IC50 with 95% CI).
| Oxy-PAHs | Abbr. | CAS no. | MW (g mol–1) | log Kow a | IC50 (μM) | 95% CI b |
| 1-Indanone | 1-INO | 83-33-0 | 132.2 | 2.11 | >10 | — |
| 1H-Phenalen-1-one | 1H-PHO | 548-39-0 | 180.2 | 3.39 | >10 | — |
| 9-Fluorenone | 9-FLO | 486-25-9 | 180.2 | 3.58 | 3.0 | 2.3–4.0 |
| Acenaphthylene-1,2-quinone | 1,2-ACNQ | 82-86-0 | 182.2 | 1.95 | >10 | — |
| 4H-Cyclopenta[def]phenanthrene-4-one | 4H-CPO | 5737-13-3 | 204.2 | 4.14 | 0.32 | 0.27–0.39 |
| Phenanthrene-9,10-quinone | 9,10-PQ | 84-11-7 | 208.2 | 2.52 | 0.65 | 0.50–0.86 |
| Anthracene-9,10-quinone | 9,10-AQ | 84-65-1 | 208.2 | 3.39 | >10 | — |
| 2-Methylanthracene-9,10-quinone | 2-MAQ | 84-54-8 | 222.2 | 3.89 | 4.9 | 3.5–6.9 |
| 7H-Benz[de]anthracene-7-one | 7H-BAO | 82-05-3 | 230.3 | 4.81 | 0.77 | 0.62–0.96 |
| Benzo[a]fluorenone | BFLO | 479-79-8 | 230.3 | 4.73 | 0.061 | 0.049–0.075 |
| 2,3-Dimethylanthracene-9,10-quinone | 2,3-DMAQ | 20054-39-1 | 236.3 | 4.44 | 2.1 | 1.3–3.2 |
| 6H-Benzo[cd]pyren-6-one | 6H-BPO | 3074-00-8 | 254.3 | 5.31 | 0.44 | 0.32–0.62 |
| Chrysene-1,4-quinone | 1,4-CHRQ | 100900-16-1 | 258.3 | 4.01 | 6.2 | 4.8–8.0 |
| Naphthacene-5,12-quinone | 5,12-NQ | 1090-13-7 | 258.3 | 4.52 | 2.5 | 1.5–4.1 |
| Benz[a]anthracene-7,12-quinone | 7,12-BAQ | 2498-66-0 | 258.3 | 4.40 | 0.037 | 0.021–0.065 |
aEstimated using KOWWIN in EPI suite v 4.11, US Environmental Protection Agency.
b95% confidence interval of IC50 values.
Effects of oxy-PAHs on the induction and activity of CYP1 by TCDD
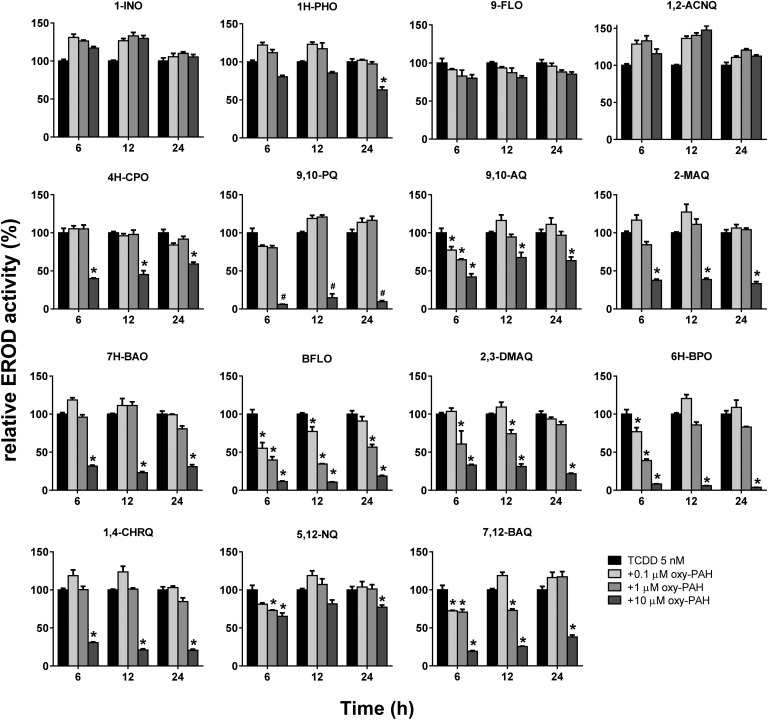
Having established the potency of oxy-PAHs to inhibit CYP1 enzyme activity we wanted to investigate the ability of oxy-PAHs to affect the expression and function of CYP1 induced by another environmentally relevant pollutant and AHR agonist, TCDD. This was performed by assaying the impact on cellular EROD activity and the CYP1A1 and 1B1 mRNA levels in HaCaT cells co-exposed to TCDD (5 nM) and each of the 15 oxy-PAHs. The EROD activity in response to TCDD alone was linear between 6 and 24 h after exposure (data not shown) and was for comparative purposes set as 100% activity. The results showed that most oxy-PAHs either displayed no inhibition or significant inhibition of EROD activity only in response to the highest dose of 10 μM (Fig. 4). In agreement with the microsome results, BFLO and 7,12-BAQ were the most potent inhibitors of TCDD-mediated EROD activity, with BFLO causing significant inhibition in response to all doses and time points except at 24 h and 0.1 μM. Further, significant inhibition, and in general transient, in response to the lower doses was observed for 9,10-AQ, 2,3-DMAQ, 6H-BPO and 5,12-NQ. 6H-BPO decreased the EROD activity the most, with more than 90% inhibition for up to 24 h in response to the highest dose. This effect was not associated with increased cytotoxicity (ESI Fig. S1†).
Fig. 4. Oxy-PAHs inhibit TCDD induced cellular EROD activity. HaCaT cells were exposed to TCDD alone (5 nM) or in combination with 0.1, 1.0 or 10 μM oxy-PAH and effects on CYP1-dependent EROD activity was measured at 6, 12 and 24 h after exposure. 100% is the EROD activity induced by TCDD alone. Data points represent means ± SE, n = 3. #Increased cytotoxicity. *p < 0.05 as compared with TCDD alone by two-way ANOVA.
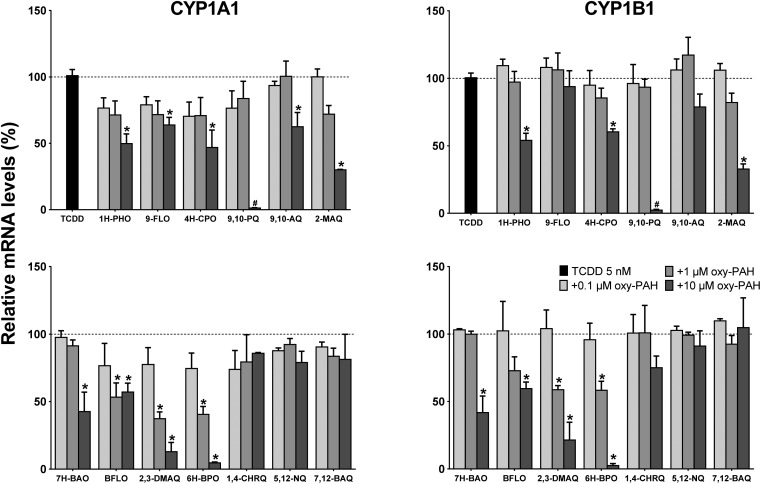
The gene expression results showed the differing ability of the oxy-PAHs to inhibit TCDD mediated induction of the CYP1A1 and 1B1 mRNA levels (Fig. 5). As with the EROD data, the expression levels in response to TCDD alone was set as 100% (corresponding to 138- and 36-fold increase of the CYP1A1 and 1B1 mRNA levels respectively compared to the DMSO control). In general, the CYP1A1 and CYP1B1 levels were inhibited to a similar extent by the different oxy-PAHs. The most efficient inhibitors were BFLO, 2,3-DMAQ and 6H-BPO which significantly reduced the mRNA levels by around 50% for both CYP1A1 and 1B1 in response to 1 μM. The high inhibiting potency of 2,3-DMAQ and 6H-BPO is in agreement with the observed strong inhibition of CYP1 gene expression in response to the oxy-PAH alone (cf.Fig. 2). The three four-ring oxy-PAHs, 1,4-CHRQ, 5,12-NQ and 7,12-BAQ had no effect at all on the expression levels while the remaining oxy-PAHs in general only showed significant effects in response to 10 μM. As mentioned above, the effect of 10 μM 9,10-PQ on both endpoints was deemed to be most likely due to the observed increased cytotoxicity.
Fig. 5. Oxy-PAHs inhibit TCDD induced CYP1 gene expression. HaCaT cells were exposed to TCDD alone (5 nM) or in combination with 0.1, 1.0 or 10 μM oxy-PAH and effects on gene expression of CYP1A1 (left panels) and CYP1B1 (right panels) were determined by qRT-PCR at 6 h after exposure. 100% is the mRNA level induced by TCDD alone. Data points represent means ± SE, n = 3. #Increased cytotoxicity. *p < 0.05 as compared with TCDD alone by two-way ANOVA.
Analysis of differences in CYP1 modulation between oxy-PAHs
In Table 2 the impact of the 15 oxy-PAHs on the different endpoints is summarized. To further distinguish the extent of differentiation or grouping of the different oxy-PAHs with regard to potency and mode of CYP1 modulation, a Principal Component Analysis (PCA) was conducted on the data. The first and second principal components explained 75.42% of the variance in the data. The individual plot highlighted four different groups of compounds (Fig. 6A). The first group included 1H-PHO and 9,10-AQ which, compared to the other oxy-PAHs, were determined to be weak AHR activators and CYP1A1 inhibitors and moderate inhibitors of TCDD-mediated effects (cf.Table 2). The second group consisted of 9-FLO, 4H-CPO 9,10-PQ, 2-MAQ, 7H-BAO and 1,4-CHRQ, all determined to be weak AHR activators and moderate CYP1A1 and TCDD inhibitors. The third group included the strong AHR activators and strong CYP1A1 inhibitors BFLO, 5,12-NQ and 7,12-BAQ. The last group included 2,3-DMAQ and 6H-BPO which were inhibitors of CYP1A1 and strong inhibitors of TCDD-mediated effects. Group 1 gathered compounds with low molecular weight (MW) and lipophilicity (log Kow), group 2 moderate MW and log Kow, while both the third and fourth group consisted of compounds presenting higher MW and log Kow (cf.Table 1). The variables plot (Fig. 6B) showed strong correlation between CYP1A1 and 1B1 gene expression (CYP1A1, CYP1B1) and cellular EROD activity (EROD) as well as between log Kow and CYP1A1 inhibition (IC50) or inhibition of TCDD (ERODTCDD). Further a correlation was observed between CYP1A1 inhibition (IC50) and inhibition of TCDD (ERODTCDD). Indeed, Pearson correlation analysis confirmed that these variables were all significantly correlated (Fig. 6C–E, correlation data not shown for CYP1A1, CYP1B1, EROD or MW).
Table 2. Overview of the obtained results for the 15 oxy-PAHs and their effects on the five assayed endpoints.
| AHR activator (EROD) a | AHR activator (CYP1 mRNA) b , c | CYP1A1 inhibitor (IC50) d | TCDD interaction (EROD) e | TCDD interaction (CYP1 mRNA) e , c | |
| 1-INO | = | ND | = | = | ND |
| 1H-PHO | = | ↑ | = | ↓ | ↓ |
| 9-FLO | = | = | ↓ | = | ↓ |
| 1,2-ACNQ | = | ND | = | = | ND |
| 4H-CPO | ↑ | ↓ | ↓↓ | ↓ | ↓ |
| 9,10-PQ | = | ↑ | ↓↓ | = | = |
| 9,10-AQ | = | ↓ | = | ↓↓ | ↓ |
| 2-MAQ | ↑ | ↑ | ↓ | ↓ | ↓ |
| 7H-BAO | = | ↑ | ↓↓ | ↓ | ↓ |
| BFLO | ↑↑ | ↑↑ | ↓↓ | ↓↓ | ↓↓ |
| 2,3-DMAQ | ↑ | ↑ | ↓ | ↓↓ | ↓↓ |
| 6H-BPO | ↑ | ↓ | ↓↓ | ↓↓ | ↓↓ |
| 1,4-CHRQ | = | ↑↑ | ↓ | ↓ | = |
| 5,12-NQ | ↑↑ | ↑↑ | ↓ | ↓↓ | = |
| 7,12-BAQ | ↑↑ | ↑↑ | ↓↓ | ↓↓ | = |
a↑↑Strong inducer (≥BP and p < 0.05), ↑inducer (p < 0.05), = no significant effect.
b↑↑Strong inducer (≥20-fold and p < 0.05), ↑inducer (<20-fold and p < 0.05), = no significant effect, ↓inhibitor (p < 0.05), ND not determined.
cSince the effects on the CYP1A1 and 1B1 mRNA levels were very similar they are described together as CYP1 mRNA.
d↓↓Strong inhibitor (IC50 < 1 μM), ↓inhibitor (1 μM < IC50 < 10 μM), = no effect (IC50 >10 μM).
e↓↓Strong inhibitor (p < 0.05 at ≤1 μM oxy-PAH), ↓inhibitor (p < 0.05 at 10 μM oxy-PAH), = no significant effect, ND not determined.
Fig. 6. Inhibition and induction of CYP1 by oxy-PAHs are driven by lipophilicity. Principal component analysis representing normalized coefficients for the 13 oxy-PAHs with data at 1 μM exposure and 6 h post exposure on the first two axes (axis 1 + axis 2: 75.42%). The individual factor map is shown in (A) and the variables factor map in (B). EROD refers to cellular EROD activity of single exposure (Fig. 1), CYP1A1 and CYP1B1 refer to gene expression of single oxy-PAH exposure (Fig. 2), ERODTCDD to cellular EROD activity of oxy-PAHs in combination with TCDD (Fig. 4), CYP1A1TCDD and CYP1B1TCDD refer to gene expression in combination with TCDD (Fig. 5), MW to molecular weight and, IC50 to CYP1A1 IC50 values (both in Table 1). Pearson correlation analysis between log Kow, IC50 and/or ERODTCDD data are shown in panels (C)–(E), respectively. Plots show linear regression with 95% confidence bands, Pearson's r and p-value. For more details see Materials and methods.
Discussion
CYP1 enzymes have an established role in bioactivation of procarcinogens such as PAHs and inhibition of these enzymes has therefore been considered as an effective approach for chemoprevention. However, recent studies show that the induced CYP1 enzyme is protective. It is needed for rapid clearance of carcinogens like B[a]P34 and CYP1A inhibition has been shown to amplify the carcinogenic and teratogenic effects of PAHs.35,36 This emphasizes the complex relationship between AHR activation, CYP1 induction and toxicity and suggests that the combined effects on AHR activation and CYP1 function may contribute to the increased toxicity of chemical mixtures.
To further investigate the impact of oxy-PAHs on AHR signalling and CYP1 function, and to discriminate between the effects on enzyme activity and CYP1 transcription, we performed a series of experiments using recombinant enzymes and human cells. AHR activation is most often assessed by measuring induction of CYP1 gene expression or enzyme activity, or by using luciferase reporter assays. However, since CYP1 enzymes have been shown to be inhibited by a large number of compounds,37 measuring enzyme activity alone may give false negative results. Accordingly, in this study we found that nine of the tested oxy-PAHs significantly induced CYP1 expression while only three of these, BFLO, 5,12-NQ and 7,12-BAQ, also were strong inducers of cellular EROD activity suggesting that some of the oxy-PAHs are both activators of AHR signalling and inhibitors of CYP1 catalytic function. Notably, some of the EROD-inducing oxy-PAHs showed reverse dose–responses with higher induction at lower concentrations, especially at early time points, further supporting CYP1-inhibiting properties. This was corroborated by the enzyme kinetic parameters determined using an recombinant CYP1A1 enzyme where eleven of the oxy-PAHs tested showed clear inhibition of enzyme activity with IC50 values ranging from 6.2 to 0.037 μM. Among these 7,12-BAQ and BFLO presented the lowest IC50 values (0.037 and 0.061 μM respectively), placing them among the most potent CYP1 inhibiting PAHs such as dibenz[a,j]acridine, pyrene and benz[a]anthracene and the flavone αNF, which all have reported IC50 values in the sub 100 nM range.11,12 When calculating IC50 values a wide concentration range of the potential enzyme inhibitor is tested in combination with a fixed concentration of the substrate and the enzyme. One should therefore be cautious when comparing IC50 values with other published values since other experimental conditions may have been used in these studies. Nonetheless, our results clearly show that oxy-PAHs can be potent CYP1A1 inhibitors and thereby impact on their own metabolism or the metabolism of other compounds such as PAHs and endogenous CYP1 substrates.
Previous studies have shown that various naturally occurring naphtho- and anthraquinones can be inhibitors of CYP enzymes37 but these data represent the first demonstration that oxy-PAHs found as environmental pollutants can be potent inhibitors of CYP1 enzymes. Furthermore our results showed significant levels of induction of CYP1B1 gene expression by nine of the fifteen oxy-PAHs, to a similar extent as CYP1A1, which demonstrates that CYP1B1 probably also makes an important contribution to the cellular response to this group of compounds. This is moreover in accord with the ability of non-polar PAHs to induce CYP1B1 and further adds to the important role of CYP1B1 in mediating the effects of polycyclic aromatic compounds (PACs).38,39 Three of the oxy-PAHs, 4H-CPO, 9,10-AQ and 6H-BPO, caused dose-dependent suppression of CYP1A1 and/or 1B1 gene expression, indicating AHR antagonistic properties that may at least partly explain the observed EROD inhibition by these compounds. In the co-exposure to TCDD, nine and seven of the thirteen oxy-PAHs tested caused suppressed expression of CYP1A1 and CYP1B1 respectively, further indicating the AHR antagonistic effects of these compounds, while none of the compounds displayed the additive effects on gene expression.
Notably, several of the oxy-PAHs shown to inhibit the recombinant CYP1A1, including the most potent inhibitors 7,12-BAQ and BFLO, caused induced EROD activity in the cell-based studies. The lack of EROD-inhibition with these compounds may be explained by the more complex situation in the cell-based assays compared to using the recombinant enzymes and low uptake into the cells. Another explanation could however be the indirect AHR-activation previously described by us where several other CYP1-inhibitors were shown to activate AHR signalling by blocking the metabolic clearance of the high-affinity AHR ligand 6-formylindolo[3,2-b]carbazole (FICZ) present in the cell culture medium.30
The oxy-PAHs, together with other polar PACs, are an emerging group of compounds with high significance in the environment and most likely also of high significance in relationship to effects on human health.40–42 The 15 oxy-PAHs included in this study have all been detected at significant amounts, often at similar or even higher levels than PAHs, in several environmental matrices including air particulate matter, contaminated soil, water, sediment and diesel particulate.3–5,43,44 Human exposure to oxy-PAHs is not well investigated but available studies report significant occupational exposure and a correlation between exposure to oxy-PAHs and health effects.45,46 The presence of carbonylic oxygen in the oxy-PAHs results in higher water solubility and thereby higher bioavailability compared to the PAHs, which may impact on their biological activity.47,48 Exposure to oxy-PAHs occurs in complex mixtures with numerous other PACs and we recently showed that interactions between different PACs in soil extracts are likely to be important for the observed toxicity.25 Here we demonstrated that binary exposures to oxy-PAHs with TCDD, a prototypical halogenated aromatic hydrocarbon (HAH) and the AHR activator resulted in a compound-, time- and dose-dependent inhibition of TCDD-mediated EROD activity and CYP1 expression. In contrast, there are several reports showing synergistic developmental toxicity in fish embryos after combined exposure to an AHR activator and a CYP1-inhibitor. These effects were shown to be AHR-dependent13,49 and to involve antioxidant responses.50,51 This discrepancy appears to relate to the type of AHR activator since it was previously shown that the combination of a CYP1 inhibitor with a HAH-type activator (e.g. PCB126) caused reduced toxicity while a PAH-type activator (e.g. B[a]P) caused synergistic toxicity.16 In a similar manner the carcinogenic potency of PAH mixtures depend on interactions with the AHR/CYP signalling pathways by the compounds present in the mixture and may be an important mechanism by which non-carcinogenic PAHs can contribute to cancer development.20,39,52,53 Considering the growing list of physiological functions associated with AHR signalling these results suggest that exposure to chemical mixtures including CYP1-inhibitors and AHR activators may contribute to immune related ailments, endocrine disruption and cancer development.
In the present study no clear relationship was highlighted between the position of the oxygen atom(s) on the benzene ring and the observed biological effects. The ketones 1H-PHO, 7H-BAO, 4H-CPO, BFLO and 6H-BPO; and the paraquinones with central diones 9,10-AQ, 2-MAQ, 2,3-DMAQ, 5,12-NQ and 7,12-BAQ presented large differences in potency to activate AHR or inhibit CYP1A1. These data are in agreement with a recent study by Knecht et al. (2013)7 where 38 oxy-PAHs were screened for their developmental toxicity in zebrafish. The authors concluded that the oxy-PAHs containing adjacent diones on 6-carbon moieties or terminal paraquinones on multi-ring structures were the most toxic while the toxicity of paraquinones with central diones varied considerably.7 Based on the 15 oxy-PAHs screened in the present study we can conclude that the activation of the AHR, the inhibition of CYP1A1, as well as the interaction with TCDD was strongly correlated with lipophilicity and molecular weight. This correlation could be of concern for human health since larger and more lipophilic oxy-PAHs are found in significant levels in soil, air and water.3–5,43,44 The fact that lipophilicity and molecular weight of both PAHs and oxy-PAHs are important determinants of the ability to activate the AHR have been shown previously.21,54 However, PACs which are larger than the AHR ligand binding domain will likely have a lower binding affinity and activation ability, as has been shown for the six-ring dibenzopyrenes.55 The same relationship has also been observed with PAHs and their ability to inhibit various CYP1 enzymes which suggests a similar mechanism of inhibition for both PAHs and oxy-PAHs.11,12
In conclusion, we show for the first time that oxy-PAHs, in addition to activation of the AHR, can be potent inhibitors of CYP1, both on the level of gene expression and enzyme activity. Further we show that oxy-PAHs can interact with other environmental pollutants leading to non-additive effects on AHR activation. Our results make an important contribution to the knowledge base about polar PACs and their mode of action. Further in vivo studies would complement these findings and investigate their implications on oxy-PAH toxicity at the organism level.
Abbreviations
- AHR
Aryl hydrocarbon receptor
- B[a]P
Benzo[a]pyrene
- CYP
Cytochrome P450 monooxygenase
- EROD
Ethoxyresorufin O-deethylase
- FL
Fluoranthene
- HAH
Halogenated aromatic hydrocarbon
- log Kow
Octanol-water partition coefficient
- MW
Molecular weight
- αNF
α-Naphthoflavone
- βNF
β-Naphthoflavone
- PAC
Polycyclic aromatic compound
- oxy-PAH
Oxygenated PAH
- PAH
Polycyclic aromatic hydrocarbon
- PCA
Principal component analysis
- TCDD
2,3,7,8-Tetrachlorodibenzo-p-dioxin
Supplementary Material
Acknowledgments
This work was supported by the Swedish Research Council Formas (EW), the Cancer and Allergy foundation and the ÅForsk foundation (FLB, KD).
Footnotes
†Electronic supplementary information (ESI) available: Fig. S1 (Alamar Blue viability assay) and Fig. S2 (CYP1A1 oxy-PAH inhibition curves). See DOI: 10.1039/c6tx00004e
References
- IARC, Some non-heterocyclic polycyclic aromatic hydrocarbons and some related occupational exposures, IARC Press, Lyon, France, 2010, distributed by World Health Organization. [Google Scholar]
- Lundstedt S., White P. A., Lemieux C. L., Lynes K. D., Lambert I. B., Öberg L., Haglund P., Tysklind M. Ambio. 2007;36:475. doi: 10.1579/0044-7447(2007)36[475:sfatho]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Layshock J. A., Wilson G., Anderson K. A. Environ. Toxicol. Chem. 2010;29:2450. doi: 10.1002/etc.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandowe B. A., Wilcke W. J. Environ. Qual. 2010;39:1349. doi: 10.2134/jeq2009.0298. [DOI] [PubMed] [Google Scholar]
- Walgraeve C., Demeestere K., Dewulf J., Zimmermann R., Van Langenhove H. Atmos. Environ. 2010;44:1831. [Google Scholar]
- Dasgupta S., Cao A., Mauer B., Yan B., Uno S., McElroy A. Environ. Sci. Pollut. Res. Int. 2014;21:13867. doi: 10.1007/s11356-014-2586-4. [DOI] [PubMed] [Google Scholar]
- Knecht A. L., Goodale B. C., Truong L., Simonich M. T., Swanson A. J., Matzke M. M., Anderson K. A., Waters K. M., Tanguay R. L. Toxicol. Appl. Pharmacol. 2013;271:266. doi: 10.1016/j.taap.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Zhang L., Jiang Y., Li Y., Lu P. Chemosphere. 2014;100:42. doi: 10.1016/j.chemosphere.2013.12.079. [DOI] [PubMed] [Google Scholar]
- Sen S., Bhojnagarwala P., Francey L., Lu D., Penning T. M., Field J. Chem. Res. Toxicol. 2012;25:2117. doi: 10.1021/tx300201p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert D. W., Dalton T. P., Okey A. B., Gonzalez F. J. J. Biol. Chem. 2004;279:23847. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- Shimada T., Guengerich F. P. Chem. Res. Toxicol. 2006;19:288. doi: 10.1021/tx050291v. [DOI] [PubMed] [Google Scholar]
- Shimada T., Yamazaki H., Foroozesh M., Hopkins N. E., Alworth W. L., Guengerich F. P. Chem. Res. Toxicol. 1998;11:1048. doi: 10.1021/tx980090+. [DOI] [PubMed] [Google Scholar]
- Billiard S. M., Timme-Laragy A. R., Wassenberg D. M., Cockman C., Di Giulio R. T. Toxicol. Sci. 2006;92:526. doi: 10.1093/toxsci/kfl011. [DOI] [PubMed] [Google Scholar]
- Timme-Laragy A. R., Cockman C. J., Matson C. W., Di Giulio R. T. Aquat. Toxicol. 2007;85:241. doi: 10.1016/j.aquatox.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenberg D. M., Nerlinger A. L., Battle L. P., Di Giulio R. T. Environ. Toxicol. Chem. 2005;24:2526. doi: 10.1897/04-440r1.1. [DOI] [PubMed] [Google Scholar]
- Wassenberg D. M., Di Giulio R. T. Environ. Health Perspect. 2004;112:1658. doi: 10.1289/ehp.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M. L., Getzinger G. J., Cooper E. M., Clark B. W., Garner L. V. T., Di Giulio R. T., Ferguson P. L., Stapleton H. M. Environ. Toxicol. Chem. 2014;33:2767. doi: 10.1002/etc.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaga T. J., Jecker L., Bracken W. M., Weeks C. E. Cancer Lett. 1979;7:51. doi: 10.1016/s0304-3835(79)80076-2. [DOI] [PubMed] [Google Scholar]
- Rice J. E., Defloria M. C., Sensenhauser C., Lavoie E. J. Chem. – Biol. Interact. 1988;68:127. doi: 10.1016/0009-2797(88)90011-7. [DOI] [PubMed] [Google Scholar]
- Staal Y. C., Hebels D. G., van Herwijnen M. H., Gottschalk R. W., van Schooten F. J., van Delft J. H. Carcinogenesis. 2007;28:2632. doi: 10.1093/carcin/bgm182. [DOI] [PubMed] [Google Scholar]
- Larsson M., Hagberg J., Giesy J. P., Engwall M. Environ. Toxicol. Chem. 2014;33:943. doi: 10.1002/etc.2517. [DOI] [PubMed] [Google Scholar]
- Machala M., Ciganek M., Blaha L., Minksova K., Vondracek J. Environ. Toxicol. Chem. 2001;20:2736. [PubMed] [Google Scholar]
- Misaki K., Kawami H., Tanaka T., Handa H., Nakamura M., Matsui S., Matsuda T. Environ. Toxicol. Chem. 2007;26:1370. doi: 10.1897/06-465r.1. [DOI] [PubMed] [Google Scholar]
- Bekki K., Takigami H., Suzuki G., Tang N., Hayakawa K. J. Health Sci. 2009;55:601. [Google Scholar]
- Wincent E., Jönsson M. E., Bottai M., Lundstedt S., Dreij K. Environ. Sci. Technol. 2015;49:3869. doi: 10.1021/es505588s. [DOI] [PubMed] [Google Scholar]
- Goodale B. C., La Du J., Tilton S. C., Sullivan C. M., Bisson W. H., Waters K. M., Tanguay R. L. Toxicol. Sci. 2015;147:397. doi: 10.1093/toxsci/kfv139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knafla A., Phillipps K. A., Brecher R. W., Petrovic S., Richardson M. Regul. Toxicol. Pharmacol. 2006;45:159. doi: 10.1016/j.yrtph.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Knafla A., Petrovic S., Richardson M., Campbell J., Rowat C. Regul. Toxicol. Pharmacol. 2011;59:101. doi: 10.1016/j.yrtph.2010.09.011. [DOI] [PubMed] [Google Scholar]
- Ono Y., Torii K., Fritsche E., Shintani Y., Nishida E., Nakamura M., Shirakata Y., Haarmann-Stemmann T., Abel J., Krutmann J., Morita A. Exp. Dermatol. 2013;22:349. doi: 10.1111/exd.12148. [DOI] [PubMed] [Google Scholar]
- Wincent E., Bengtsson J., Mohammadi Bardbori A., Alsberg T., Luecke S., Rannug U., Rannug A. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4479. doi: 10.1073/pnas.1118467109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolpmann K., Brinkmann J., Salzmann S., Genkinger D., Fritsche E., Hutzler C., Wajant H., Luch A., Henkler F. Cell Death Dis. 2012;3:e388. doi: 10.1038/cddis.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hestermann E. V., Stegeman J. J., Hahn M. E. Toxicol. Sci. 2000;53:316. doi: 10.1093/toxsci/53.2.316. [DOI] [PubMed] [Google Scholar]
- Motoyama Y., Bekki K., Chung S. W., Tang N., Kameda T., Toriba A., Taguchi K., Hayakawa K. J. Health Sci. 2009;55:845. [Google Scholar]
- Nebert D. W., Shi Z., Galvez-Peralta M., Uno S., Dragin N. Mol. Pharmacol. 2013;84:304. doi: 10.1124/mol.113.086637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis I. W. H., Dreij K., Mattsson Å., Jernström B., Stenius U. Toxicology. 2014;321:27. doi: 10.1016/j.tox.2014.03.012. [DOI] [PubMed] [Google Scholar]
- Billiard S. M., Meyer J. N., Wassenberg D. M., Hodson P. V., Di Giulio R. T. Toxicol. Sci. 2008;105:5. doi: 10.1093/toxsci/kfm303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Sridhar J., Foroozesh M. Molecules. 2013;18:14470. doi: 10.3390/molecules181214470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddens L. K., Bunde K. L., Harper Jr. T. A., McQuistan T. J., Lohr C. V., Bramer L. M., Waters K. M., Tilton S. C., Krueger S. K., Williams D. E., Baird W. M. Toxicol. Appl. Pharmacol. 2015;287:149. doi: 10.1016/j.taap.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan B., Marston C. P., Luch A., Dashwood W. M., Brooks E., Pereira C., Doehmer J., Baird W. M. Int. J. Cancer. 2007;120:1161. doi: 10.1002/ijc.22466. [DOI] [PubMed] [Google Scholar]
- Lemieux C. L., Long A. S., Lambert I. B., Lundstedt S., Tysklind M., White P. A. Environ. Sci. Technol. 2015;49:1787. doi: 10.1021/es504465f. [DOI] [PubMed] [Google Scholar]
- Mutlu E., Warren S. H., Matthews P. P., King C., Linak W. P., Kooter I. M., Schmid J. E., Ross J. A., Gilmour M. I., Demarini D. M. Environ. Mol. Mutagen. 2013;54:719. doi: 10.1002/em.21812. [DOI] [PubMed] [Google Scholar]
- Umbuzeiro G. A., Franco A., Martins M. H., Kummrow F., Carvalho L., Schmeiser H. H., Leykauf J., Stiborova M., Claxton L. D. Mutat. Res. 2008;652:72. doi: 10.1016/j.mrgentox.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Lundstedt S., Bandowe B. A. M., Wilcke W., Boll E., Christensen J. H., Vila J., Grifoll M., Faure P., Biache C., Lorgeoux C., Larsson M., Frech Irgum K., Ivarsson P., Ricci M. TrAC, Trends Anal. Chem. 2014;57:83. [Google Scholar]
- Tidwell L. G., Allan S. E., O'Connell S. G., Hobbie K. A., Smith B. W., Anderson K. A. Environ. Sci. Technol. 2015;49:141. doi: 10.1021/es503827y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- O'Connell S. G., Kincl L. D., Anderson K. A. Environ. Sci. Technol. 2014;48:3327. doi: 10.1021/es405022f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Han I. K., Hu M., Shao M., Zhang J. J., Tang X. Chemosphere. 2010;81:1280. doi: 10.1016/j.chemosphere.2010.08.055. [DOI] [PubMed] [Google Scholar]
- Arp H. P., Lundstedt S., Josefsson S., Cornelissen G., Enell A., Allard A. S., Kleja D. B. Environ. Sci. Technol. 2014;48:11187. doi: 10.1021/es5034469. [DOI] [PubMed] [Google Scholar]
- Josefsson S., Arp H. P., Kleja D. B., Enell A., Lundstedt S. Chemosphere. 2015;119:1268. doi: 10.1016/j.chemosphere.2014.09.102. [DOI] [PubMed] [Google Scholar]
- Van Tiem L. A., Di Giulio R. T. Toxicol. Appl. Pharmacol. 2011;254:280. doi: 10.1016/j.taap.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner L. V., Di Giulio R. T. Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol. 2012;155:573. doi: 10.1016/j.cbpc.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme-Laragy A. R., Van Tiem L. A., Linney E. A., Di Giulio R. T. Toxicol. Sci. 2009;109:217. doi: 10.1093/toxsci/kfp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courter L. A., Musafia-Jeknic T., Fischer K., Bildfell R., Giovanini J., Pereira C., Baird W. M. Toxicol. Sci. 2007;95:63. doi: 10.1093/toxsci/kfl137. [DOI] [PubMed] [Google Scholar]
- Tarantini A., Maitre A., Lefebvre E., Marques M., Rajhi A., Douki T. Toxicology. 2011;279:36. doi: 10.1016/j.tox.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Piskorska-Pliszczynska J., Keys B., Safe S., Newman M. S. Toxicol. Lett. 1986;34:67. doi: 10.1016/0378-4274(86)90146-3. [DOI] [PubMed] [Google Scholar]
- Machala M., Vondracek J., Blaha L., Ciganek M., Neca J. V. Mutat. Res. 2001;497:49. doi: 10.1016/s1383-5718(01)00240-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.