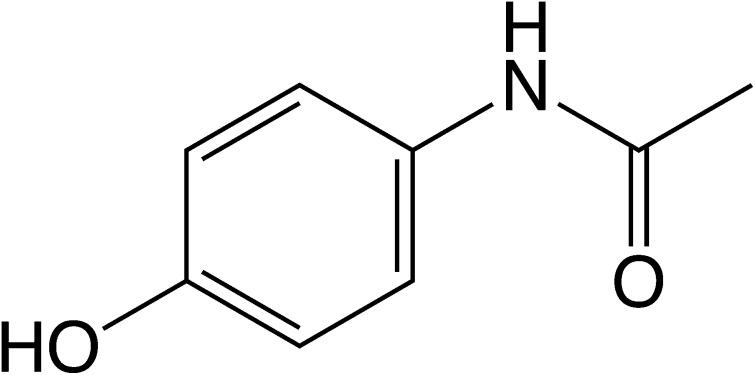
 After over 60 years of therapeutic use in the UK, paracetamol (acetaminophen, N-acetyl-p-aminophenol, APAP) remains the subject of considerable research into both its mode of action and toxicity.
After over 60 years of therapeutic use in the UK, paracetamol (acetaminophen, N-acetyl-p-aminophenol, APAP) remains the subject of considerable research into both its mode of action and toxicity.
Abstract
After over 60 years of therapeutic use in the UK, paracetamol (acetaminophen, N-acetyl-p-aminophenol, APAP) remains the subject of considerable research into both its mode of action and toxicity. The pharmacological properties of APAP are the focus of some activity, with the role of the metabolite N-arachidonoylaminophenol (AM404) still a topic of debate. However, that the hepatotoxicity of APAP results from the production of the reactive metabolite N-acetyl-p-benzoquinoneimine (NAPQI/NABQI) that can deplete glutathione, react with cellular macromolecules, and initiate cell death, is now beyond dispute. The disruption of cellular pathways that results from the production of NAPQI provides a source of potential biomarkers of the severity of the damage. Research in this area has provided new diagnostic markers such as the microRNA miR-122 as well as mechanistic biomarkers associated with apoptosis, mitochondrial dysfunction, inflammation and tissue regeneration. Additionally, biomarkers of, and systems biology models for, glutathione depletion have been developed. Furthermore, there have been significant advances in determining the role of both the innate immune system and genetic factors that might predispose individuals to APAP-mediated toxicity. This perspective highlights some of the progress in current APAP-related research.
1. Introduction
First described in 1878 the analgesic and antipyretic drug paracetamol (acetaminophen, N-acetyl-p-aminophenol, APAP) was little used clinically until the withdrawal of phenacetin from the market on account of observed renal toxicity. At the time of writing, APAP is probably the most widely available and commonly used drug worldwide and represents a very important analgesic;1 indeed it is included in the 20th World Health Organization Model List of Essential Medicines as updated in March 2017.2 Some 60 years after its introduction as a therapeutic agent in the UK, APAP remains a compound of great interest, not only because of its therapeutic properties but also for its ability to cause hepatotoxicity in overdose; it was early recognised that APAP was hepatotoxic at high doses3 but that there were marked species differences in sensitivity.4 Further, these early studies established that a metabolite of APAP, rather than the drug itself, was the cause of its hepatotoxicity. Mitchell and colleagues showed that, whilst the major routes of biotransformation for APAP are indeed detoxication reactions, that form the phenolic sulfate and glucuronide conjugates, a minor route was the oxidative metabolism of the drug via the P450 system to an electrophilic intermediate, most likely N-acetyl-p-benzoquinoneimine (NAPQI, also commonly termed NABQI), which rapidly depleted hepatic glutathione (GSH).5 Subsequently, it was shown that acetaminophen can be activated by non-P450-dependent metabolism, for example by cyclooxygenases6 and myeloperoxidases,7 although the contribution of this to the toxicity of APAP is unclear. When intracellular GSH concentrations reached a low level, toxicity ensued.8 These early studies found a strong relationship between covalent binding of APAP to tissue proteins and cytotoxicity, leading to the proposal that these were causally linked.9 Early studies also demonstrated that freshly isolated hepatocytes were a good model for the metabolism and toxicity of APAP, including large species differences in sensitivity.10 It has now been very well established that the toxicity of APAP results from metabolic activation and there is also evidence that some of the analgesic properties of the drug may derive from in vivo biotransformation to an arachidonic acid conjugate of p-aminophenol (N-arachidonoylphenolamine, or AM404) via fatty acid amide hydrolase.11–13
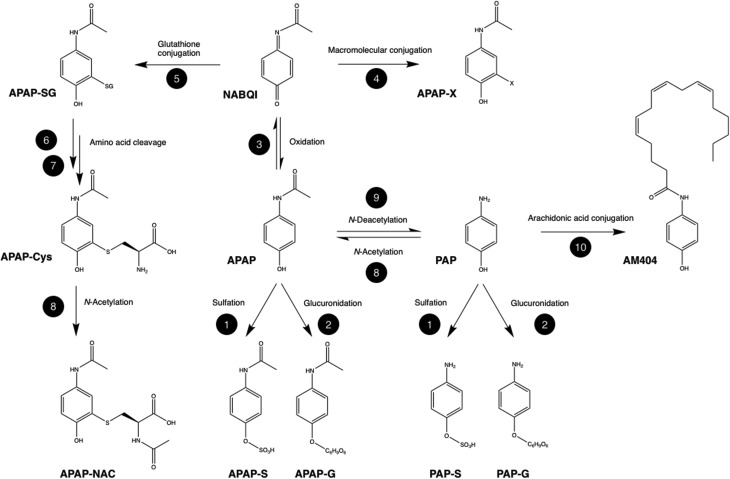
The metabolism of APAP is illustrated in Fig. 1 for the biotransformation reactions that may be significant for its efficacy or toxicity. However, it should be noted the metabolic fate of APAP is surprisingly complex with a large number of minor metabolites in addition to those shown in the figure.
Fig. 1. Major metabolic biotransformations of paracetamol related to therapeutic efficacy and observed hepatotoxicity. Metabolites: APAP: acetyl-p-aminophenol, acetaminophen, paracetamol; PAP: p-aminophenol; AM404: n-arachidonoylaminophenol; NABQI, NAPQI: n-acetyl-p-benzoquinoneimine; APAP-X: macromolecular conjugate of acetaminophenol; APAP-SG: glutathione conjugate of acetaminophen; APAP-Cys: cysteinyl acetaminophen; APAP-NAC: mercapturate of acetaminophen; APAP-S: acetaminophen sulfate; APAP-G: acetaminophen sulfate; PAP-S: p-aminophenol sulfate; PAP-G: p-aminophenol sulfate. Biotransformation enzymes and cofactors: 1: sulfotransferase, 3′-phosphoadenosine-5′-phosphosulfate (PAPS); 2: uridine 5′-diphospho-glucuronosyltransferase (UGT), uridine 5′-diphospho-glucuronic acid (UDPGA); 3: cytochrome P450, O2, NADPH; 4: non-enzymatic electrophilic addition; 5: glutathione-s-transferase (GST), gluthathione (GSH); 6: gamma-glutamyl transpeptidase; 7: cysteinyl glycinase; 8: n-acetyltransferase (NAT); 9: n-deacetylase; 10: fatty acid amide hydrolase (FAAH).
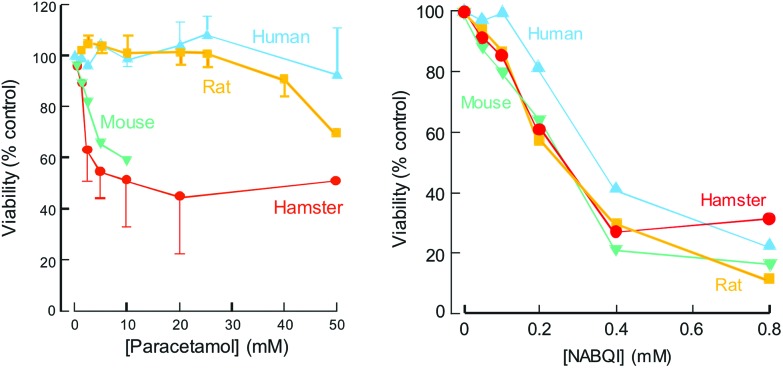
If APAP were being developed today, faced with such observations on its toxicity, a key question would be the relative sensitivity of humans, as compared to the laboratory species studied. An obvious approach would be to compare the effects of APAP incubation with freshly isolated hepatocytes from a number of species including humans. When such studies were undertaken10 they revealed that the relative sensitivity of freshly isolated hepatocytes from mouse, rat and hamster reflected that in vivo and that human hepatocytes were relatively resistant to the cytotoxicity of APAP (Fig. 2). The availability of chemically defined NAPQI enabled these studies to establish that differences in species sensitivity were due to differences in the rate of metabolic conversion of APAP to NAPQI.14
Fig. 2. Toxicity of paracetamol and NABQI to freshly isolated hepatocytes from mouse, hamster, rat and human (means ± SEM, n ≥ 4). Produced from data published in Tee et al. (1987).10.
The aforementioned in vitro studies were performed using hepatocytes from a relatively small number of donors (n = 4–11). Hence, an important issue was the extent to which APAP toxicity might vary within the population. If APAP were being developed today, an early indication of this variability could be obtained from a knowledge of the cytochrome P450 (CYP) enzymes responsible for its metabolic activation and their population variability. Studies with human CYP enzymes determined that several can metabolise APAP to NAPQI, the most important of which are CYP1A2, CYP2E1 and CYP3A4.15,16 These studies were later extended17 to demonstrate that, at high concentrations of APAP, metabolism via CYP2E1 predominates. It is perhaps interesting to note, given the association of paracetamol induced drug-induced liver injury (DILI) hepatotoxicity and mitochondrial toxicity18,19 that CYP2E1 is located not only in the endoplasmic reticulum but also within mitochondria. A study where this was investigated in hepatocytes expressing CYP2E1 in both locations or exclusively in mitochondria showed a range of adverse effects in both types of cell (despite lower cellular CYP2E1 in the latter). These results led the authors to suggest that mitochondrial CYP2E1 was able to cause the oxidative stress and cytotoxicity resulting from APAP exposure.20
Immunoblotting of liver samples from 30 different human donors using a panel of form-specific anti-CYP antibodies revealed substantial inter-individual variability in the amounts of these CYPs expressed in liver.21 Hence, it would be predicted that humans would show considerable inter-individual variability in their susceptibility to the hepatotoxicity of the drug. Parallel studies in a large number of healthy volunteers22 using a therapeutic dose of APAP confirmed the considerable variation in CYP-dependent metabolism of APAP to NAPQI, with greater than ten-fold variation amongst ∼200 individuals,22 confirming previous findings.23 These results are consistent with the observation that, whilst an overdose of APAP can cause hepatotoxicity in humans, there is a wide range in sensitivity amongst individuals and many subjects are at relatively low risk.24
When NAPQI was first synthesised and its properties studied it was found to be not only an electrophile but also a strong thiol oxidant.14 This observation gave rise to the question of the relative role played by covalent binding and thiol oxidation in the toxicity of APAP. In order to investigate this, a two-phase model of APAP toxicity in freshly isolated hamster hepatocytes was developed;25 in phase 1, metabolic activation of APAP occurred with depletion of GSH, but no loss of cell viability; in phase 2, in fresh medium with no APAP present, there was progressive morphological damage, leading ultimately to cell death. The addition of the thiol reductant, dithiothreitol, at the start of phase 2, prevented and reversed the toxicological damage that would otherwise occur, in the absence of any reduction in covalent binding or resynthesis of GSH. It was concluded that the toxicity of APAP to hepatocytes is largely through reversible oxidation of thiol groups in key enzymes. Studies with the antidotes N-acetylcysteine (NAC) and methionine, revealed that whereas NAC was able to prevent and reverse the toxicity of APAP in phase 2, methionine was essentially without effect, despite the fact that both were effective in phase 1. It was notable that when NAC was added in phase 2, there was restoration of GSH concentrations whereas this did not occur with methionine. Hence, it is likely that NAPQI leads to oxidation of one or more enzymes involved in the conversion of methionine to cysteine, preventing GSH resynthesis from this precursor in phase 2, whereas with NAC such inhibition does not prevent GSH resynthesis.26 Recent studies have provided evidence that cystathionine beta-synthase and cystathionine gamma-lyase, necessary for the conversion of methionine to cysteine, are targets for thiol oxidation by NAPQI.27 Following treatment with NAC, the resulting GSH can act both to detoxify NAPQI by conjugation and to reverse toxicity by thiol reduction. In support of this there is some (slight) evidence that administration of NAC to APAP overdose patients is effective at later times than methionine.28
2. Biomarkers: monitoring, understanding and predicting APAP hepatotoxicity in humans
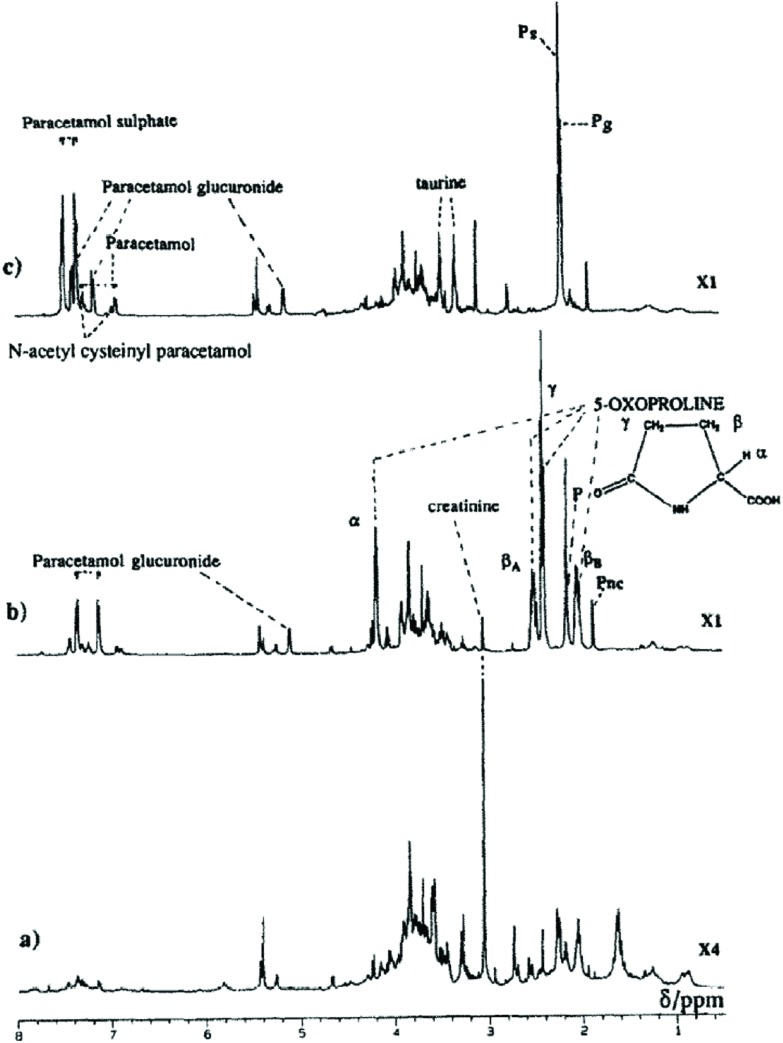
As well as the formation of GSH conjugates, covalent binding to macromolecules and GSH depletion the effects of APAP administration have been seen in changes in a variety of endogenous metabolites; both 5-oxoproline (pyroglutamate, 5OXP)29 and ophthalmic acid (OPA), a tripeptide analog of GSH in which the cysteine group is replaced by l-2-aminobutyrate, have been found to reflect the effects of reactive metabolite production on the GSH depletion seen with APAP toxicity.30 For example, the excretion in the urine of large quantities (up to concentrations of 1 M) of 5OXP was observed after three weeks of feeding APAP in the diet (1%) to young rats.29 These results are illustrated in Fig. 3 below.
Fig. 3. Single pulse 1H NMR spectra of pooled urine from (a) control rats, (b) rats fed APAP in the diet (1%, 10 weeks) and (c) diet containing both APAP and methionine (both 1%). Key P, PS and PG = n-acetyl signals for APAP, APAP sulphate and APAP glucuronide respectively. PNC is the resolved n-acetyl on the side chain of the mercapturate. Reproduced from Ghauri et al. (1993).29.
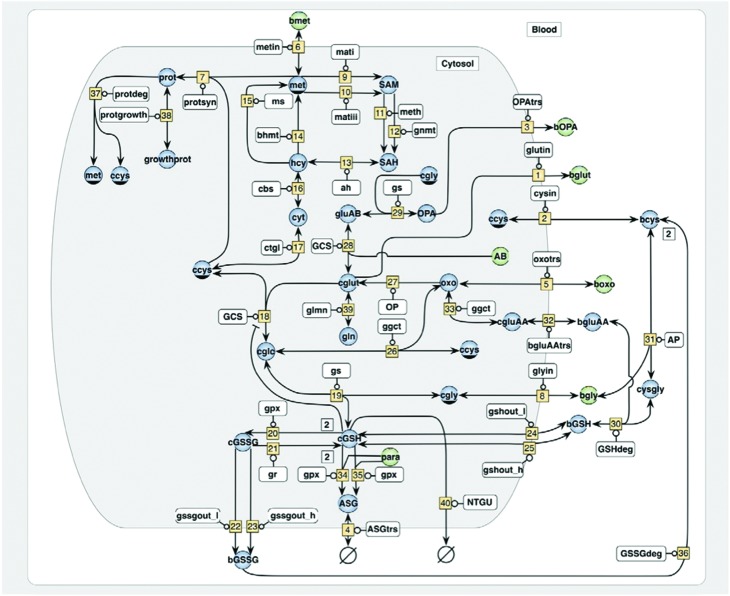
This dosing regimen also resulted in effects on growth, with the animals failing to gain weight.31 Both of these effects were prevented by the administration of methionine in the diet leading to the suggestion that both the 5-oxoprolinuria and growth inhibition resulted from a lack of sulfur-containing amino acids due to GSH consumption to detoxify NAPQI, with severe disruption of the gamma-glutamyl cycle. Similarly, studies in the mouse showed that OPA was also produced in response to APAP. Both 5OXP and OPA are likely general biomarkers for oxidative stress and GSH deficiency rather than APAP specific biomarkers, but elevated urinary 5OXP is seen in humans suffering from APAP overdose and is often clinically reported as anion gap metabolic acidosis.32–34 A systems biology approach, developed using in vitro studies in THLE-2E1 cells (CYP2E1-transfected SV40 large T-antigen immortalized human liver epithelial cells) resulted in a computational model that used the relative concentrations of OPA and 5OXP to predict intracellular concentrations of GSH (Fig. 4); increased 5OXP was associated with increased biosynthesis whilst OPA production indicated the depletion of the sulfur-containing amino acids essential for GSH biosynthesis.35
Fig. 4. Schematic of computational model of glutathione homeostasis. The network embraces pathways of methionine catabolism, glutathione metabolism, 5-oxoproline and ophthalmic acid synthesis and glutathione mediated detoxification. Variable metabolites are indicated by blue circles, fixed metabolites are indicated by green circles, enzymes are shown in white rectangles and rate numbers are shown in yellow squares. The shaded area refers to the intracellular space, while the prefixes ‘c’ and ‘b’ denote substances in the cell and the blood, respectively. The enzymes which catalyze the numbered reactions are: v[9]-mati-methionine adenosyl transferase i-2516; v[10]-matiii-methionine adenosyl transferase iii-2516; v[11]-meth-glycine n-methyltransferase-21120; v[12]-gnmt-dnamethyltransferase-21172; v[13]-ah-s-adenosyl-homocysteine hydrolase-3311; v[14]-bhmt-betaine-homocysteine methyltransferase-2115, v[15]-ms-methionine synthase-21113; v[16]-cbs-cystathionine gamma-synthase-42122; v[17]-ctgl-cystathionase-4411; v[18]-gcs-glutamylcysteine synthetase-6322; both v[19] and v[27]-gs-glutathione synthetase-6323; v[20]-gpx-glutathione peroxidase-11119; v[21]-gr-glutathione reductase-1817; v[26]-ggct-gamma-glutamylcyclotransferase-2324; v[25]-op-5-oxoprolinase-3529, v[27]-gcsglutamylcysteine synthetase-6322; v[31]-ap-aminopeptidase-34112; v[33]-ggct-gamma-glutamylcyclotransferase-2324; v[34]-gpx-glutathione s-transferase-25118; v[35]-gpx-glutathione s-transferase-25118. Metabolites assumed to be present at variable concentrations are: met—methionine; SAM—s-adenosyl-methionine; SAH—s-adenosylhomocysteine; hcy—homocysteine; cyt—cystathionine, ccys—cytosolic cysteine; bcys—blood cysteine; glc—glutamyl-cysteine; cGSH—cytosolic glutathione; bGSH—blood glutathione; cGSSG—cytosolic glutathione disulfide; bgssg—blood glutathione disulfide; cgly—cytosolic glycine; cglut—cytosolic glutamate; opa—ophthalmic acid; n—[n-(γ-glutamyl)-α-aminobutyryl]glycine; oxo—oxoproline = pyroglutamic acid; asg—acetaminophen glutathione adduct; gluab—glutamyl aminobutyrate; bgluaa—blood glutamyl amino acid; gln—glutamine; prot—protein. Metabolites assumed to be present at fixed concentrations are: ab—2-aminobutyrate; bet—betaine; bgly—blood glycine; bglut—blood glutamate; bmet—blood methionine; CNADPH—nicotinamide adenine dinucleotide phosphate; cser —cytosolic serine; H2O2—cellular hydrogen peroxide; bopa—blood ophthalmic acid; n-[n-(γ-glutamyl)-α-aminobutyryl]glycine; boxo—blood oxoproline, pyroglutamic acid; basg—blood acetaminophen glutathione adduct; para—paracetamol (acetaminophen). Reproduced from Geenen et al. (2013).35.
Whilst poor nutritional status may predispose some subjects to toxicity via a reduced ability to produce GSH for NAPQI-detoxication other metabolic factors may also come into play. For example, Clostridium spp. that are present in the gut, convert tyrosine and phenylalanine to p-cresol for which the metabolic detoxication in humans is, as for APAP, predominantly via sulfation. However, the capacity of sulfation is limited and competition between APAP and p-cresol will limit the detoxication of both by this route, increasing the likely formation of NAPQI as metabolism is pushed towards CYP2E1. In addition, p-cresol is also likely to deplete GSH as, like APAP, it is a substrate for oxidative metabolism, forming reactive metabolites (RMs) via CYP P450-mediated metabolism (both a quinone methide via CYP2D6, 2C19, 1A2, 1A1, and 2E1 and a 4-methyl-o-hydroquinone that is further oxidized to 4-methyl-1,2-benzoquinone, mainly via CYP2E1 but also CYP1A1, 1A2, and 2D6)36 potentially reducing the ability of the host to simultaneously detoxify APAP-derived RMs. In both rats and humans administered APAP, the endogenous quantities of p-cresol sulfate in the urine were correlated with liver effects (rat)37 and the APAP-sulfate/APAP-glucuronide ratio (humans),38 directly leading to the concept of pharmacometabonomics.37,38 Similarly, urinary concentrations of p-cresol and p-cresol sulfate have been found to be elevated in children with autism39 and when the urinary APAP-sulfate/APAP-glucuronide ratio was determined for 20 “low functioning” autistic children receiving the drug it was found to be significantly lower than for the controls.40
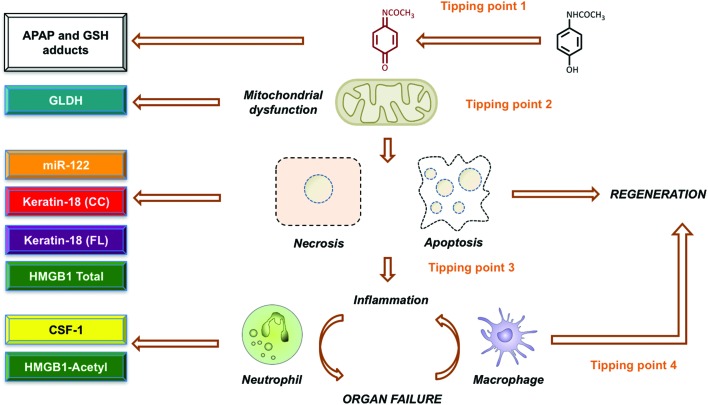
The prediction of acute APAP-induced liver injury in humans, and consequent treatment needs or patient outcomes during hepatotoxicity remains difficult. A lack of sensitivity and specificity of currently used biomarkers of organ injury coupled with a poor understanding of the mechanistic basis of hepatotoxicity remain contributory factors.41 One approach to dealing with such issues adopts a ‘tissue to periphery’ strategy in which novel and targeted bioanalytical technologies are integrated with traditional methods of pathological assessment to identify improved biomarkers that can be quantified in blood/urine that reflect changes in less accessible tissues.42 Results of both clinical and pre-clinical studies of APAP overdose have demonstrated the identification and development of circulating oligonucleotide and protein biomarkers that provide enhanced hepatic specificity (miR-122) and/or can inform on mechanistic events such as necrosis (keratin-18, HMGB1), apoptosis (caspase-cleaved keratin-18), mitochondrial dysfunction (glutamate dehydrogenase), inflammation (acetyl-HMGB1) and tissue regeneration (CSF-1).41–47 Recently it was demonstrated in large (n > 1500) prospective studies of APAP overdose patients, encompassing the entire spectrum of the disease, that markers such as miR-122 provide a sensitive and early identification of liver injury at hospital presentation that permits stratified treatment (Fig. 5). Furthermore, in studies of APAP hepatotoxicity, in which patients have been transferred to a tertiary liver unit, the prognostic ability of elevated plasma acetyl-HMGB1 and KIM-1 (kidney injury) to predict the need for liver transplant, and of elevated CSF-1 to predict spontaneous survival, has been demonstrated.48 Key outcomes from these studies and from collaborations with the IMI funded SAFE-T consortium have resulted in the Letter of Support status for the further qualification of these biomarkers across the spectrum of drug-induced liver injury from the FDA (July 2016)49 and EMA (September 2016).50 By using hepatocyte specific conditional knock-out mice it has also been shown that the inflammatory mediator and biomarker, HMGB1, plays an integral role in the mechanism of toxicity by linking cell death to inflammation.51 In addition, the therapeutic potential of chimeric anti-HMGB1 to block post-injury inflammation and reduce APAP hepatotoxicity, at a time when NAC is ineffective, has recently been demonstrated.52
Fig. 5. Paracetamol mechanistic complexity offers new opportunities for biomarker and therapeutic development. Based on decades of preclinical and clinical evidence, the mechanism of paracetamol induced hepatotoxicity has been largely defined. Key tipping points have been identified that modulate the overall response and may serve as crucial targets for therapeutic intervention. This include bioactivation of paracetamol to naqpi (1), mitochondrial dysfunction (2), cell death mode dynamics (3) and the balance between inflammation resulting in either enhanced injury or hepatic regeneration (4). Recently, it has been shown that these key events can also be reported by circulating biomarkers that offer improved diagnosis, prediction and prognostication during paracetamol toxicity. For example, monitoring APAP and GSH adducts may serve to report bioactivation, GLDH can be considered a marker of mitochondrial injury, cell death mode dynamics can be reported with kertin-18/caspase cleaved keratin-18 release and the balance between pro-inflammatory and pro-regenerative events can be assessed by HMGB1-acetyl and CSF-1 respectively.
3. The innate immune response to APAP-induced liver injury
The clinical syndrome of APAP induced acute liver failure is one of innate immune dysfunction. Massive hepatocyte death over the course of hours provokes rapid and comprehensive innate immune activation. Kupffer cells, the tissue resident macrophages of the liver, are activated through toll-like receptors by damage associated molecular patterns (DAMPs) released from necrotic hepatocytes. The Kupffer cells respond by secreting pro-inflammatory cytokines and chemokines that drive a systemic inflammatory response and the recruitment of myeloid effector cells to the liver.53 Neutrophils have been reported to be among the earliest cells to arrive and may contribute to local tissue damage through the production of reactive oxygen species and the release of proteases.54 Other studies have failed to replicate this finding, and the role of neutrophils in early tissue injury remains controversial.55 Infiltrating monocytes, initially proinflammatory in function, mature within the liver to monocyte derived macrophages which have important tissue repair functions.56 In the systemic compartment, patients exhibit a numerical and functional deficit in circulating monocytes that renders them susceptible to infections.57 Understanding the innate immune response to liver injury – the balance between pro-inflammatory and tissue restorative functions and between local hepatic and systemic effects – will be vital to improving treatments and outcomes for patients with acute liver failure.58
In vivo imaging, while not a new technique, has been revolutionised over the past decades by improvements in fluorescence microscopy and surgical preparation. Laser scanning confocal microscopy allows high-quality live imaging of fluorescently labelled cells and tissues using a pinhole to exclude light out-with the focal plane. Optical sectioning through the tissue permits the generation of 3D images and videos. A number of recent studies have utilised such in vivo imaging to study the immunopathology of APAP-induced acute liver failure (reviewed by Marques et al. (2015)).59 Researchers have been able to demonstrate sinusoidal DNA deposition, neutrophil accumulation and CCR2+ monocyte recruitment to be key factors in the pathogenesis of propagating liver injury following APAP hepatotoxicity.60–62
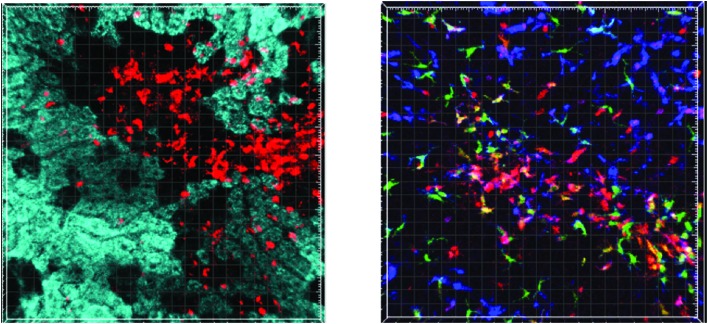
Through the application of laser scanning confocal microscopy a number of the features of innate immune activation in APAP-induced acute liver injury have been demonstrated in a mouse model. Hepatocyte necrosis within the centrilobular region can be visualised by an absence of rhodamine 123 staining and acquisition of nuclear propidium iodide staining. Compromise of the sinusoidal endothelium is evident as intravascular contrast (labelled albumin) extravasates into the peri-venular space. Within 6–8 hours of injury anti-Ly6G-labelled neutrophils are seen to migrate towards and accumulate within the centrilobular area. Using a Cx3cr1gfp/+ Ccr2rfp/+ transgenic reporter mouse, the temporo-spatial characteristics of monocyte migration into the injured liver and in situ maturation can be followed (see Fig. 6).
Fig. 6. Left: Still from a video showing neutrophil migration into the centrilobular region 8 hours after APAP dosing in a wild type mouse. Rhodamine 123 (cyan) is sequestered in mitochondria with an intact membrane potential and thus, stains viable hepatocytes. Black areas show an absence of rhodamine 123 staining in non-viable hepatocytes. Anti-Ly6G antibody (red) shows migration of neutrophils towards and their accumulation within the area of necrosis. Right: Still from a video showing mature inflammatory infiltrate around the centrilobular area 72 hours after APAP in a transgenic Cx3cr1gfp/+ Ccr2rfp/+ reporter mouse. Kupffer cells (blue, anti-f4/80) line the sinusoids throughout the lobule. Large monocyte derived macrophages with mixed CCR2 (red) Cx3cr1 (green) f4/80 (blue) signal are seen in the centrilobular region. CCR2 positive monocytes scan the sinusoids.
Highly mobile CCR2+CX3CR1– monocytes scan the sinusoids throughout the whole hepatic lobule, showing significantly greater displacement in their migration paths than CCR2+CX3CR1+ cells, which are largely restricted to small movements within the centrilobular region. These techniques, such as intravital microscopy offer a powerful tool to explore immune cell function in minimally perturbed in vivo models of disease. Its application to the study of APAP-induced acute liver injury has already yielded novel insights into the immunopathology of this condition. Ongoing work aims to identify key features of innate immune cell migration and interactions within the injured liver, to identify potential targets for immunotherapy.
4. Genetic factors in APAP-induced DILI
As discussed above, the hepatotoxicity of APAP has long been recognized. It is now well established as an important cause of acute liver failure (ALF), which has an incidence of approx. 10 cases per million people annually in developed countries. Indeed, 50% of ALF cases in the USA and Europe are due to drug exposure with 80% of these cases relating to APAP overdose and the remaining 20% to injury from drugs other than APAP where the drug was taken at the prescribed dose.63 DILI due to APAP is an inevitable consequence if sufficient drug is consumed and there is no medical intervention but it remains possible that some individuals are more susceptible to this toxicity than others, especially at lower levels of overdose. There is a rapid effect (several days) involving predominantly hepatocyte necrosis. Unlike many forms of DILI which are idiosyncratic, the underlying mechanism for APAP toxicity is unlikely to involve T cell response but, as discussed above, there is a role for the innate immune system.64
There has been good progress to date on identifying genetic risk factors for idiosyncratic DILI and, in particular, various human leukocyte antigen (HLA) alleles have been demonstrated to be strong risk factors.65 However, for the concentration-dependent intrinsic DILI seen in APAP overdose, progress on identifying genetic risk factors is more limited. The underlying mechanism for the cellular necrosis seen in APAP overdose may involve reactive metabolite formation followed by protein interaction and hepatocyte injury. Hepatocyte injury releases compounds such as NAD and ATP. This triggers inflammasome activation and an innate immune response.64 It is therefore possible that genetic susceptibility to APAP overdose could be determined by the ability of an individual to form the toxic imine metabolite determined by genotype for certain drug metabolism genes but also genotype for genes affecting the ability of cells to respond to oxidative stress, the innate immune system generally, and damage repair. Such factors might be particularly relevant to APAP-induced DILI where the extent of overdose is limited (e.g. consumption of approx. 10 g) with a relatively low plasma drug concentration yet the patient still goes on to develop serious liver toxicity. Genetic studies on APAP-induced DILI in humans to date have involved three different approaches: (i) studies in healthy volunteers given APAP at normal doses but showing transient transaminase elevation; (ii) studies on patients who have suffered liver failure following APAP overdose; (iii) administration of the imine metabolite NAPQI to human lymphoblastoid cell lines combined with a genome-wide association study (GWAS).
Approx. 19% of volunteers taking 4 g APAP per day showed transient alanine aminotransferase (ALT) > 5 times upper limit of normal (ULN) after 7 to 10 days.66 These individuals may serve as a surrogate for those more sensitive to toxicity in overdose. Using data from mouse models where evidence for genetic variation in susceptibility to APAP DILI has also been obtained, polymorphisms in a set of candidate genes were genotyped in approximately 120 individuals for whom data on circulating ALT following APAP administration at 4 g per day for 7 days was available.67 Polymorphisms in CD44 (rs1467558), which codes for a protein involved in lymphocyte adhesion and activation, and in CAPN10 (rs3749166) which encodes a protease released following hepatocyte damage, were predictive for the extent of ALT elevation with borderline significant effects seen for those carrying rs1467558 or rs3749166.
An entirely separate candidate gene study involving 275 patients who had suffered acute liver failure after either deliberate or accidental APAP overdose also reported some borderline significant associations.68 For the rs1467558 CD44 variant discussed above, there was a trend towards significance in those who had overdosed unintentionally. A more significant finding emerged from the intentional overdose group where rs776746, a polymorphism that predicts CYP3A5 expression and is likely to be relevant to imine production, was more common. Other polymorphisms in drug metabolising genes relevant to APAP toxicity, including CYP2E1 and various UGT and SULT genes, showed no significant difference in cases compared with controls. A study using 176 human lymphoblastoid cell lines exposed to the imine NAPQI used a novel approach to identify risk factors for APAP DILI.69 The IC50 for APAP toxicity in each cell line was examined in relation to the genome-wide genotype for the cell line. This GWAS approach resulted in one genome-wide significant signal with rs2880961 on chromosome 3 found to be the strongest genome-wide significant marker. However, this signal is in a “gene desert” with no nearby genes and is currently difficult to interpret.
In summary, genetic risk factors for development of serious DILI at relatively low levels of APAP overdose seem likely to exist but the limited studies performed to date need to be followed up by genome-wide approaches using larger numbers of confirmed DILI cases or possibly further studies on volunteers showing transient ALT elevation. At present, the most plausible findings relate to CD44 and CYP3A5.
5. Perspective
As indicated in the introduction, APAP has been in clinical use in the UK for over 60 years, over which time much has been learnt about its toxicity. If the “thought experiment” is performed whereby APAP is considered as a recent compound that was being progressed through drug development today, it would be clear that it is reproducibly hepatotoxic at high doses in laboratory species, but that there are marked species differences in sensitivity (mouse ≫ rat). Hepatotoxicity would be shown to be a consequence of CYP-mediated metabolism to NAPQI and differences in this metabolic pathway are a key determinant of relative sensitivity to its hepatotoxicity. Studies with isolated hepatocytes would establish that human cells are less sensitive than those from mice, and that this difference is due to the relative rates of NAPQI formation. APAP activation would be shown to be catalysed mainly by CYP2E1, CYP1A2 and CYP3A4, with CYP2E1 predominating at high concentrations. The expression of these enzymes is known to vary appreciably amongst individuals and hence inter-individual differences in susceptibility would be anticipated.
Depending on the importance of APAP to such a hypothetical development portfolio, in-depth mechanistic studies might be undertaken at this point. These would establish the mode of action, involving reversible thiol-group oxidation (and/or covalent binding) as a key event, leading to mitochondrial perturbation and ultimately to necrotic cell death. Based on this knowledge, one would envisage the development of antidotes such as NAC, which would be effective even for some time after the metabolic phase of toxicity was complete. However, it would be apparent from such studies that the relationships between dose, GSH concentrations and toxicity are complex. Hence, even now, and despite the knowledge that APAP can cause hepatotoxicity in overdose, and the extensive research that has been conducted on its pharmacokinetics and mechanisms of toxicity over the years, there is still no quantitative model for predicting de novo the dose level at which hepatotoxicity will occur in humans and how this will vary with metabolic activity.70 Ultimately the decision to progress the development of APAP would then come down to potential risk versus benefit. It is not at all clear with the current regulatory concerns regarding DILI that a drug with the properties of APAP would be advanced for approval and, if approved, that it would ever be considered acceptable for sale over the counter. And yet, as noted in the opening paragraph of this perspective, despite all of this, APAP remains in the 20th World Health Organization Model List of Essential Medicines.2
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c7tx00340d
‡This article summarises the main contributions to the recent Drug Metabolism Group (DMG) meeting held in 2017 “60 years of paracetamol: what do we really know?”. For more information about the DMG and the meeting see ESI.
References
- Sharma C. V., Mehta V. Critical Care Pain. 2013:1–6. doi: 10.1093/bjaceaccp/mkt049. [DOI] [Google Scholar]
- http://www.who.int/medicines/publications/essentialmedicines/en/ .
- Mitchell J. R., Jollow D. J., Potter W. Z., Davis D. C., Gillette J. R., Brodie B. B. J. Pharmacol. Exp. Ther. 1973;187:185–194. [PubMed] [Google Scholar]
- Davis D. C., Potter W. Z., Jollow D. J., Mitchell J. R. Life Sci. 1974;14:2099–2109. doi: 10.1016/0024-3205(74)90092-7. [DOI] [PubMed] [Google Scholar]
- Mitchell J. R., Jollow D. J., Potter W. Z., Gillette J. R., Brodie B. B. J. Pharmacol. Exp. Ther. 1973;187:211–217. [PubMed] [Google Scholar]
- Potter D. W., Hinson J. A. J. Biol. Chem. 1987;262:974–980. [PubMed] [Google Scholar]
- van Zyl J. M., Basson K., van der Walt B. J. Biochem. Pharmacol. 1989;38:161–165. doi: 10.1016/0006-2952(89)90163-9. [DOI] [PubMed] [Google Scholar]
- Potter W. Z., Thorgeirsson S. S., Jollow D. J., Mitchell J. R. Pharmacology. 1974;12:129–143. doi: 10.1159/000136531. [DOI] [PubMed] [Google Scholar]
- Moldéus P. Biochem. Pharmacol. 1978;27:2859–2863. doi: 10.1016/0006-2952(78)90201-0. [DOI] [PubMed] [Google Scholar]
- Tee L. B., Davies D. S., Seddon C. E., Boobis A. R. Biochem. Pharmacol. 1987;36:1041–1045. doi: 10.1016/0006-2952(87)90412-6. [DOI] [PubMed] [Google Scholar]
- Hogestatt E. D., Jonsson B. A., Ermund A., Andersson D. A., Bjork H., Alexander J. P., Cravatt B. F., Basbaum A. I., Zygmunt P. M. J. Biol. Chem. 2005;280:31405–31412. doi: 10.1074/jbc.M501489200. [DOI] [PubMed] [Google Scholar]
- Anderson B. J. Paediatr. Anaesth. 2008;18:915–921. doi: 10.1111/j.1460-9592.2008.02764.x. [DOI] [PubMed] [Google Scholar]
- Graham G. G., Davies M. J., Day R. O., Mohamudally A., Scott K. F. Inflammopharmacology. 2012;21:201–232. doi: 10.1007/s10787-013-0172-x. [DOI] [PubMed] [Google Scholar]
- Blair I. A., Boobis A. R., Davies D. S., Cresp T. M. Tetrahedron Lett. 1980;21:4947–4950. [Google Scholar]
- Raucy J. L., Lasker J. M., Lieber C. S., Black M. Arch. Biochem. Biophys. 1989;271:270–283. doi: 10.1016/0003-9861(89)90278-6. [DOI] [PubMed] [Google Scholar]
- Patten C. J., Thomas P. E., Guy R. L., Lee M., Gonzalez F. J., Guengerich F. P., Yang C. S. Chem. Res. Toxicol. 1993;6:511–518. doi: 10.1021/tx00034a019. [DOI] [PubMed] [Google Scholar]
- Laine J. E., Auriola S., Pasanen M., Juvonen R. O. Xenobiotica. 2009;39:1–21. doi: 10.1080/00498250802512830. [DOI] [PubMed] [Google Scholar]
- Moore M., Thor H., Moore G., Nelson S., Moldéus P., Orrenius S. J. Biol. Chem. 1985;260:13035–13040. [PubMed] [Google Scholar]
- Xie Y., McGill M. R., Du K., Dorko K., Kumer S. C., Schmitt T. M., Ding W. X., Jaeschke H. Toxicol. Appl. Pharmacol. 2015;289:213–222. doi: 10.1016/j.taap.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knockaert L., Descatoire V., Vadrot N., Fromenty B., Robin M. A. Toxicol. in Vitro. 2011;25:475–484. doi: 10.1016/j.tiv.2010.11.019. [DOI] [PubMed] [Google Scholar]
- Edwards R. J., Adams D. A., Watts P. S., Davies D. S., Boobis A. R. Biochem. Pharmacol. 1998;56:377–387. doi: 10.1016/s0006-2952(98)00033-1. [DOI] [PubMed] [Google Scholar]
- Boobis A. R., Fawthrop D. J., Seddon C. E., Speirs C. J. and Davies D. S., Variability in the pharmacokinetics and metabolism of acetaminophen, in Pharmacogenetics of Drug Metabolism, ed. W. Kalow, Pergamon Press, New York, 1992, pp. 791–812. [Google Scholar]
- Critchley J. A., Nimmo G. R., Gregson C. A., Woolhouse N. M., Prescott L. F. Br. J. Clin. Pharmacol. 1986;22:649–657. doi: 10.1111/j.1365-2125.1986.tb02953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott L. F. Am. J. Ther. 2000;7:99–114. doi: 10.1097/00045391-200007020-00007. [DOI] [PubMed] [Google Scholar]
- Tee L. B., Boobis A. R., Huggett A. C., Davies D. S. Toxicol. Appl. Pharmacol. 1986;83:294–314. doi: 10.1016/0041-008x(86)90307-8. [DOI] [PubMed] [Google Scholar]
- Boobis A. R., Tee L. B., Hampden C. E., Davies D. S. Food Chem. Toxicol. 1986;24:731–736. doi: 10.1016/0278-6915(86)90172-9. [DOI] [PubMed] [Google Scholar]
- McGarry D. J., Chakravarty P., Wolf C. R., Henderson C. J. J. Pharmacol. Exp. Ther. 2015;355:137–144. doi: 10.1124/jpet.115.227389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brok J., Buckley N., Gluud C. Cochrane Database Syst. Rev. 2009;2:CD003328. doi: 10.1002/14651858.CD003328.pub2. [DOI] [PubMed] [Google Scholar]
- Ghauri F. Y., McLean A. E. M., Beales D., Wilson I. D., Nicholson J. K. Biochem. Pharmacol. 1993;46:953–957. doi: 10.1016/0006-2952(93)90506-r. [DOI] [PubMed] [Google Scholar]
- Soga T., Baran R., Suematsu M., Ueno Y., Ikeda S., Sakurakawa T., Kakazu Y., Ishikawa T., Robert M., Nishioka T., Tomita M. J. Biol. Chem. 2006;281:16768–16776. doi: 10.1074/jbc.M601876200. [DOI] [PubMed] [Google Scholar]
- McLean A. E. M., Armstrong G. R., Beales D. Biochem. Pharmacol. 1989;38:347–352. doi: 10.1016/0006-2952(89)90048-8. [DOI] [PubMed] [Google Scholar]
- McGregor A., Laight N., Nolan S. J. Intensive Care Soc. 2012;13:54–56. [Google Scholar]
- Armenian P., Gerona R., Blanc P. D., Wu A. H., Mookherjee S. J. Emerg. Med. 2012;43:54–57. doi: 10.1016/j.jemermed.2011.06.017. [DOI] [PubMed] [Google Scholar]
- Fenves A. Z., Kirkpatrick III H. M., Patel V. V., Sweetman L., Emmett M. Clin. J. Am. Soc. Nephrol. 2006;1:441–447. doi: 10.2215/CJN.01411005. [DOI] [PubMed] [Google Scholar]
- Geenen S., du Preez F. B., Snoep J. L., Foster A. J., Sarda S., Kenna J. G., Wilson I. D., Westerhoff H. V. Biochim. Biophys. Acta. 2013;1830:4943–4959. doi: 10.1016/j.bbagen.2013.04.014. [DOI] [PubMed] [Google Scholar]
- Yan Z., Zhong H. M., Maher N., Torres R., Leo G. C., Caldwell G. W., Huebert N. Drug Metab. Dispos. 2005;33:1867–1876. doi: 10.1124/dmd.105.006387. [DOI] [PubMed] [Google Scholar]
- Clayton T. A., Lindon J. C., Cloarec O., Antti H., Charuel C., Hanton G., Provost J.-P., Le Net J.-L., Baker D., Walley R. J., Everett J. R., Nicholson J. K. Nature. 2006;440:1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- Clayton T. A., Baker D., Lindon J. C., Everett J. R., Nicholson J. K. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persico A. M., Napolioni V. Neurotoxicol. Teratol. 2012;36:82–90. doi: 10.1016/j.ntt.2012.09.002. [DOI] [PubMed] [Google Scholar]
- Alberti A., Pirrone P., Elia M., Waring R. H., Romano C. Biol. Psychiatry. 1999;46:420–424. doi: 10.1016/s0006-3223(98)00337-0. [DOI] [PubMed] [Google Scholar]
- Antoine D. J., Dear D. J. Expert Rev. Clin. Pharmacol. 2016;9:633–635. doi: 10.1586/17512433.2016.1154786. [DOI] [PubMed] [Google Scholar]
- Clarke J. I., Dear J. W., Antoine D. J. Expert Opin. Drug Saf. 2016;15:625–634. doi: 10.1517/14740338.2016.1160057. [DOI] [PubMed] [Google Scholar]
- Antoine D. J., Williams D. P., Kipar A., Jenkins R. E., Regan S. L., Sathish J. G., Kitteringham N. R., Park B. K. Toxicol. Sci. 2009;112:521–531. doi: 10.1093/toxsci/kfp235. [DOI] [PubMed] [Google Scholar]
- Antoine D. J., Jenkins R. E., Dear J. W., Williams D. P., McGill M. R., Sharpe M. R., Craig D. G., Simpson K. J., Jaeschke H., Park B. K. J. Hepatol. 2012;56:1070–1079. doi: 10.1016/j.jhep.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Stutchfield B. M., Antoine D. J., Mackinnon A. C., Gow D. J., Bain C. C., Hawley C. A., Hughes M. J., Francis B., Wojtacha D., Man T. Y., Dear J. W., Devey L. R., Mowat A. M., Pollard J. W., Park B. K., Jenkins S. J., Simpson K., Hume D. A., Wigmore S. J., Forbes S. J. Gastroenterology. 2015;149:1896–1909. doi: 10.1053/j.gastro.2015.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey Lewis P. J., Dear J., Platt V., Simpson K. J., Craig D. G., Antoine D. J., French N. S., Dhaun N., Webb D. J., Costello E. M., Neoptolemos J. P., Moggs J., Goldring C. E., Park B. K. Hepatology. 2011;54:1767–1776. doi: 10.1002/hep.24538. [DOI] [PubMed] [Google Scholar]
- Antoine D. J., Dear J. W., Lewis P. S., Platt V., Coyle J., Masson M., Thanacoody R. H., Gray A. J., Webb D. J., Moggs J. G., Bateman D. N., Goldring C. E., Park B. K. Hepatology. 2013;58:777–787. doi: 10.1002/hep.26294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine D. J., Sabbisetti V. S., Francis B., Jorgensen A. L., Craig D. G., Simpson K. J., Bonventre J. V., Park d B. K., Dear J. W. Hepatology. 2015;62:591–599. doi: 10.1002/hep.27857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA, http://www.fda.gov/Drugs/DevelopmentApprovalProcess/ucm434382.htm.
- EMA, http://www.ema.Europa.eu/docs/en_GB/document_library/Other/2016/09/WC500213479.pdf.
- Huebener P., Pradere J. P., Hernandez C., Gwak G. Y., Caviglia J. M., Mu X., Loike J. D., Jenkins R. E., Antoine D. J., Schwabe R. F. J. Clin. Invest. 2015;125:539–550. doi: 10.1172/JCI76887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundback P., Lea J. D., Sowinska A., Ottosson L., Furst C. M., Steen J., Aulin C., Clarke J. I., Kipar A., Klevenvall L., Yang H., Palmblad K., Park B. K., Tracey K. J., Blom A. M., Andersson U., Antoine D. J., Erlandsson Harris H. Hepatology. 2016;64:1699–1710. doi: 10.1002/hep.28736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenkel O., Mossanen J. C., Tacke F. C. Hepatobiliary Surg. Nutr. 2014;3:331–343. doi: 10.3978/j.issn.2304-3881.2014.11.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques P. E., Amaral S. S., Pires D. A., Nogueira L. L., Soriani F. M., Lima B. H., Lopes G. A., Russo R. C., Avila T. V., Melgaço J. G., Oliveira A. G., Pinto M. A., Lima C. X., De Paula A. M., Cara D. C., Leite M. F., Teixeira M. M., Menezes G. B. Hepatology. 2012;56:1971–1982. doi: 10.1002/hep.25801. [DOI] [PubMed] [Google Scholar]
- Williams C. D., Bajt M. L., Sharpe M. R., McGill M. R., Farhood A., Jaeschke H. Toxicol. Appl. Pharmacol. 2014;275:122–133. doi: 10.1016/j.taap.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond E., Samia-Grinberg S., Pasmanik-Chor M., Brazowski E., Shibolet O., Halpern Z., Varol C. J. Immunol. 2014;193:344–353. doi: 10.4049/jimmunol.1400574. [DOI] [PubMed] [Google Scholar]
- Antoniades C. G., Berry P. A., Davies E. T., Hussain M., Bernal W., Vergani D., Wendon J. Hepatology. 2006;44:34–43. doi: 10.1002/hep.21240. [DOI] [PubMed] [Google Scholar]
- Possamai L. A., Thursz M. R., Wendon J. A., Antoniades C. G. J. Hepatol. 2014;61:439–445. doi: 10.1016/j.jhep.2014.03.031. [DOI] [PubMed] [Google Scholar]
- Marques P. E., Oliveira A. G., Chang L., Paula-Neto H. A., Menezes G. B. J. Hepatol. 2015;63:733–742. doi: 10.1016/j.jhep.2015.05.027. [DOI] [PubMed] [Google Scholar]
- Marques P. E., Oliveira A. G., Pereira R. V., David B. A., Gomides L. F., Saraiva A. M., Pires D. A., Novaes J. T., Patricio D. O., Cisalpino D., Menezes-Garcia Z., Leevy W. M., Chapman S. E., Mahecha G., Marques R. E., Guabiraba R., Martins V. P., Souza D. G., Mansur D. S., Teixeira M. M., Leite M. F., Menezes G. B. Hepatology. 2015;61:348–360. doi: 10.1002/hep.27216. [DOI] [PubMed] [Google Scholar]
- Marques P. E., Amaral S. S., Pires D. A., Nogueira L. L., Soriani F. M., Lima B. H., Lopes G. A., Russo R. C., Avila T. V., Melgaço J. G., Oliveira A. G., Pinto M. A., Lima C. X., De Paula A. M., Cara D. C., Leite M. F., Teixeira M. M., Menezes G. B. Hepatology. 2012;56:1971–1982. doi: 10.1002/hep.25801. [DOI] [PubMed] [Google Scholar]
- Mossanen J. C., Krenkel O., Ergen C., Govaere O., Liepelt A., Puengel T., Heymann F., Kalthoff S., Lefebvre E., Eulberg D., Luedde T., Marx G., Strassburg C. P., Roskams T., Trautwein C., Tacke F. Hepatology. 2016;64:1667–1682. doi: 10.1002/hep.28682. [DOI] [PubMed] [Google Scholar]
- Bernal W., Wendon J. N. Engl. J. Med. 2013;369:2525–2534. doi: 10.1056/NEJMra1208937. [DOI] [PubMed] [Google Scholar]
- Hoque R., Sohail M. A., Salhanick S., Malik A. F., Ghani A., Robson S. C., Hoque R., Sohail M. A., Salhanick S., Malik A. F., Ghani A., Robson S. C., Mehal W. Z. Am. J. Physiol.: Gastrointest. Liver Physiol. 2012;302:G1171–G1179. doi: 10.1152/ajpgi.00352.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban T. J., Daly A. K., Aithal G. R. Semin. Liver Dis. 2014;34:123–133. doi: 10.1055/s-0034-1375954. [DOI] [PubMed] [Google Scholar]
- Watkins P. B., Kaplowitz N., Slattery J. T., Colonese C. R., Colucci S. V., Stewart P. W., Harris S. C. J. Am. Med. Assoc. 2006;296:87–93. doi: 10.1001/jama.296.1.87. [DOI] [PubMed] [Google Scholar]
- Harrill A. H., Watkins P. B., Su S., Ross P. K., Harbourt D. E., Stylianou I. M., Boorman G. A., Russo M. W., Sackler R. S., Harris S. C., Smith P. C., Tennant R., Bogue M., Paigen K., Harris C., Contractor T., Wiltshire T., Rusyn I., Threadgill D. W. Genome Res. 2009;19:1507–1515. doi: 10.1101/gr.090241.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court M. H., Peter I., Hazarika S., Vasiadi M., Greenblatt D. J., Lee W. M. Drug Metab. Dispos. 2014;42:28–32. doi: 10.1124/dmd.113.053546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer A. M., Fridley B. L., Jenkins G. D., Batzler A. J., Pelleymounter L. L., Kalari K. R., Ji Y., Chai Y., Nordgren K. K., Weinshilboum R. M. Toxicol. Sci. 2011;120:33–41. doi: 10.1093/toxsci/kfq375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurlinden T. J., Reisfeld B. Eur. J. Drug Metab. Pharmacokinet. 2016;41:267–280. doi: 10.1007/s13318-015-0253-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.