 We employed an in vivo assay system of Caenorhabditis elegans to determine if and which microRNAs (miRNAs) were dysregulated upon exposure to coal combustion related fine particulate matter (PM2.5) by profiling the miRNAs using SOLiD sequencing.
We employed an in vivo assay system of Caenorhabditis elegans to determine if and which microRNAs (miRNAs) were dysregulated upon exposure to coal combustion related fine particulate matter (PM2.5) by profiling the miRNAs using SOLiD sequencing.
Abstract
We employed an in vivo assay system of Caenorhabditis elegans to determine if and which microRNAs (miRNAs) were dysregulated upon exposure to coal combustion related fine particulate matter (PM2.5) by profiling the miRNAs using SOLiD sequencing. From this, expression of 25 miRNAs was discovered to become dysregulated by exposure to PM2.5. Using the corresponding C. elegans deletion mutants, 5 miRNAs (mir-231, mir-232, mir-230, mir-251 and mir-35) were found to be involved in the control of PM2.5 toxicity. Furthermore, mutation of mir-231 or mir-232 induced a resistance to PM2.5 toxicity, whereas mutation of mir-230, mir-251, or mir-35 induced a susceptibility to PM2.5 toxicity. SMK-1, an ortholog of the mammalian SMEK protein, was identified as a molecular target for mir-231 in the regulation of PM2.5 toxicity. In addition, the genes of sod-3, sod-4 and ctl-3, which are necessary for protection against oxidative stress, were determined to be important downstream targets of smk-1 in the regulation of PM2.5 toxicity. The triggering of this mir-231-SMK-1-SOD-3/SOD-4/CTL-3 signaling pathway may be a critical molecular basis for the role of oxidative stress in the induction of coal combustion related PM2.5 toxicity.
Introduction
Epidemiological studies have suggested the statistical associations between fine particulate matter (PM) emission and human health.1,2 For example, an elevation in PM air pollution was associated with the increased risk of all-cause, cardiopulmonary, and lung cancer mortality.1 PM less than 2.5 μm in aerodynamic size (PM2.5) is responsible for air pollution caused by coal combustion to a great degree. Coal combustion related PM2.5 has been suggested to be closely associated with lung dysfunction, cancer, and induction of oxidative stress.3–5 The cytotoxicity assay further indicated that exposure to coal combustion related PM2.5 could result in a decrease in cell viability, increase in global DNA methylation, and oxidative DNA damage in vascular endothelial cells.6
Nematode Canorhabditis elegans, an important model animal, can be maintained in the laboratory, where it is grown on agar plates or liquid cultures with Escherichia coli OP50 as the food source.7 Numerous basic physiological processes, stress responses, and molecular regulation mechanisms have been proven to be conserved between C. elegans and human beings or mammals.8C. elegans has been employed as an important alternative model animal for toxicological study, and it can provide an assay system for asking in vivo questions.9–11 Recently, C. elegans has been successfully used for the toxicity assessment and the toxicological study of PM2.5, including the coal combustion related PM2.5.12–16 Exposure to coal combustion related PM2.5 can damage the functions of both primary targeted organs, such as intestine, and secondary targeted organs, such as neurons, and reproductive organs in nematodes.13,15
MicroRNAs (miRNAs) are a large class of short noncoding RNAs found in various organisms, which usually act to post-transcriptionally inhibit the gene expression via sequence-specific base pairing with target mRNAs.17,18 Previous studies have suggested that the miRNAs could participate in the regulation of toxicity formation of certain environmental toxicants, such as engineered nanomaterials of graphene oxide and multi-walled carbon nanotubes.19–23 However, the role of miRNAs in the toxicity formation of coal combustion related PM2.5 is still largely unclear in nematodes. In this study, we employed the in vivo assay system of C. elegans to identify the dysregulated miRNA profiling induced by coal combustion related PM2.5 using the SOLiD sequencing technique. We further investigated the role of miRNAs in the regulation of the toxicity of coal combustion related PM2.5 in nematodes. Moreover, we focused on one of the miRNAs, mir-231, to examine the underlying molecular mechanism for its role in regulating the toxicity of coal combustion related PM2.5. Our results will highlight the crucial role of miRNAs in the induction of coal combustion related PM2.5 toxicity in organisms.
Materials and methods
Sample collection
Coal combustion related PM2.5 was acquired from a site of coal production in the Datong, Shanxi province, China. The PM2.5 samples were first collected on glass fiber membranes, and then organic compounds within the samples were removed. Our previously published report contains the detailed protocols by which the PM2.5 was prepared for experimental analysis, as well as an analysis of its chemical components.13 Briefly, these chemical components included at least 2 mg m–3 of Fe, Pb and Zn, and less than 1 mg m–3 of As, Cd, Cr, Cu and Ni.13 In addition, there were also 14 types of polycyclic aromatic hydrocarbons (PAHs) identified, where most were present in the PM2.5 at concentrations of at least 1 ng m–3.13
C. elegans maintenance
Several of the C. elegans strains used in this study were obtained from Caenorhabditis Genetics Center. The nematodes were maintained at 20 °C in plates containing nematode growth medium (NGM) and Escherichia coli OP50 as previously described.7 The L1-larvae were kept synchronized by lysing gravid nematodes with a bleaching mixture of 0.45 mol L–1 NaOH and 2% HOCl.24
Exposure
Previously published research has shown that long term exposure of nematodes to more than 1 mg L–1 of PM2.5 results in significant decreases in locomotion and brood size, and an increase in reactive oxygen species (ROS) production in the intestines.13 In this study, 1 mg L–1 of PM2.5 was dissolved in K-medium (32 mmol L–1 KCl, 5 mmol L–1 NaCl) and used as the working concentration in experiments. Prolonged exposure to PM2.5 from L1-larvae to young adults (approximately 2.5 days) was performed in the liquid K medium in sterile tissue culture plate wells in the presence of OP50 food at 20 °C. The control nematodes were grown in the liquid K medium in sterile tissue culture plate wells in the presence of OP50 food at 20 °C without the addition of PM2.5. Toxicity was assessed based on the endpoints of locomotion and intestinal ROS production.
Intestinal ROS production
The endpoint of intestinal ROS production was used to reflect the functional state of a primary targeted organ, the intestine, in nematodes.25 Intestinal ROS production was analyzed as described previously.26,27 Following the incubation in PM2.5, the exposed nematodes were transferred into sterile tissue culture plate wells and incubated in 1 μmol L–1 CM-H2DCFDA in the dark for 3 h. The nematodes were then evaluated by microscopy after mounting them on 2% agar pads and then examining the relative fluorescence intensity of the intestine under a laser scanning confocal microscope. Relative fluorescence intensity of the intestine was semi-quantified using relative fluorescence units (RFU). For each treatment, 30 nematodes were evaluated in 3 replicates.
Locomotion behavior
Nematode locomotion is an indicative parameter of the functional state of the motor neurons.28 In this study, locomotion was assessed based on examination of head thrash and body bend under a dissecting microscope by visual inspection as previously published.29,30 The head thrash and body bend were defined as directional changes in bending at the mid-body and pharynx posterior bulb, respectively, along the y axis when the nematode is moving along the x axis. For each treatment, 20 nematodes were assessed in 6 replicates.
Small RNA extraction, SOLiD sequencing, and bioinformatics analysis
Extraction of small RNAs and SOLiD sequencing were executed as previously described.31 Briefly, the control and PM2.5 (1 mg L–1) treated nematodes were lysed and small RNAs were extracted using a mirVana™ miRNA isolation kit (Ambion) based on the manufacturer's protocol. These small RNAs were then reverse transcribed into a double-stranded cDNA library, on which adaptor ligation was then performed. Importantly, this library was created to be compatible with the high-throughput sequencing system used, the Applied Biosystems SOLiD™ system. SOLiD sequencing results were collected as nucleotide sequences and their coverage. The resulting sequences were compared against the Genbank (; http://www.ncbi.nlm.nih.gov/Genbank) and miRNAbase (; http://www.mirbase.org) databases, and the registered miRNAs were analyzed using all the collected sequences.
A total of three independent biological replicates were performed. The average expression of each of the miRNAs was compared between nematodes exposed to PM2.5 and the control. Dysregulation of miRNA expression was detected in PM2.5-exposed nematodes by DESeq, which is an R package capable of estimating variance and testing for differential expression. The expression of each miRNA was denoted as up- or down-regulated, where a 2-fold, statistically significant (P < 0.05) change was the cutoff; the results were then normalized and displayed as a scatter diagram. The predicted target genes of dysregulated miRNAs that were the most notably affected by exposure to PM2.5 were then classified based on their gene ontology and the KEGG pathways using the bioinformatics tools at ; http://www.geneontology.org and ; http://www.pantherdb.org, respectively.
Reverse-transcription and quantitative real-time polymerase chain reaction (PCR)
Total RNA was isolated from the nematodes using Trizol (Invitrogen, UK) according to the manufacturer's protocol. The extracted RNA was used as a template for the synthesis of cDNA. The relative expression levels of the RNA were measured by real-time PCR (RT-PCR) in an ABI 7500 with Evagreen (Biotium, USA). Three biological replicates were performed. The expression of each miRNA is presented as the relative expression ratio between each miRNA and F35C11.9, which encodes a small nuclear RNA U6. Quantification of each gene target was expressed as the relative expression ratio between the gene of interest and the reference gene tba-1, which encodes a tubulin protein. The sequences of the primers used for the RT-PCR of the miRNAs are presented in Tables S1 and S2,† while those used to amplify genes are in Table S3.†
RNA interference (RNAi)
To generate nematodes with specific knock-down, RNAi was used through the E. coli strain HT115, carrying double-stranded RNA corresponding to each target, and was fed to the nematodes. Primer sequences for RNAi carrying the E. coli strain construction are presented in Table S4.† First, RNAi carrying E. coli strains were incubated overnight at 37 °C and then were transferred to NGM plates. L1 larvae were then added to these wells and incubated for 2 days at 20 °C until the nematodes became gravid. The resulting eggs were incubated at 20 °C until the offspring became young adults and were then used for the subsequent assays.
Statistical analysis
Results in this article are all presented as mean ± standard deviation (SD). Cohorts were analyzed for statistically significant differences by ANOVA using SPSS 12.0 (SPSS Inc., Chicago, USA), where P < 0.05 and P < 0.01 were considered significant.
Results
SOLiD sequencing
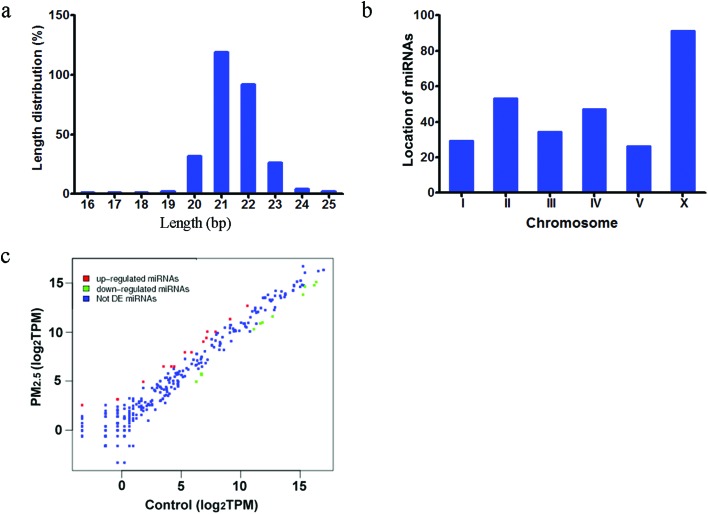
To generate miRNA expression profiles so that comparisons could be made between control and PM2.5 (1 mg L–1) exposed nematodes, SOLiD sequencing was performed. It was found that the miRNA sequences ranged in size from 19–25 nucleotides, and the most frequent lengths were 20 and 23 nucleotides, which are the lengths of mature miRNAs according to the miRNA database (Fig. 1a). In addition, all of the detected miRNAs mapped across all chromosomes, including both the autosomal and the X chromosomes (Fig. 1b). The coverage of the miRNAs was visualized by a scatter diagram, and both up- and down-regulated miRNAs were found following exposure to PM2.5 (Fig. 1c). Overall, this data suggest that RNAomics sequencing is a feasible method to identify the miRNAs with potential roles in regulating the in vivo response to coal combustion related PM2.5 toxicity.
Fig. 1. Basic SOLiD sequencing results. (a) Length distribution of miRNAs. (b) Chromosome distribution of miRNAs. (c) Scatter diagram of miRNAs. DE miRNAs: differentially expressed miRNAs. TMP: transcripts per million. The concentration of coal combustion related PM2.5 was 1 mg L–1. Prolonged exposure was performed from L1-larvae to young adults at 20 °C in the presence of food.
Identification of miRNAs dysregulated by exposure to coal combustion related PM2.5
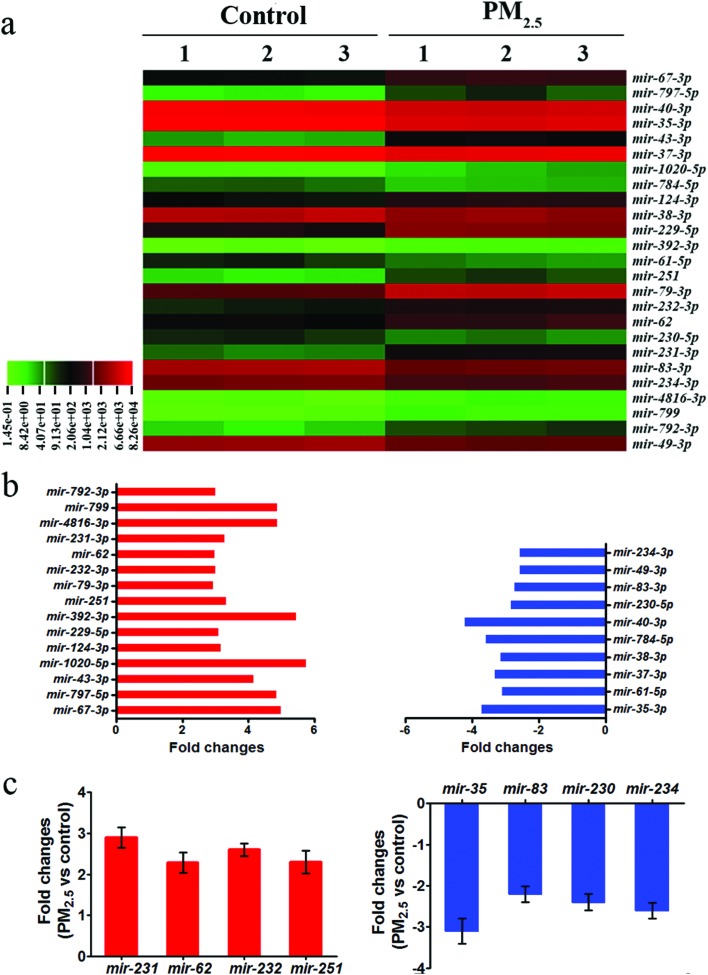
To identify miRNAs with changes in expression triggered by exposure to PM2.5, fold changes between control and PM2.5 exposed miRNAs were calculated, where a 2.0-fold change cutoff was considered statistically significant. To assign annotations to the resulting genes of interest, the miRNA sequences identified were compared against the Genbank and miRbase databases. Twenty-five miRNAs were found to be differentially expressed in nematodes exposed to PM2.5 compared to the control (Fig. 2 and Table S5†). The molecular signature of the 25 miRNAs that were found to be dysregulated following exposure to PM2.5 was displayed through hierarchical clustering (Fig. 2a). Of these 25 miRNAs, 15 were up-regulated, i.e. mir-43, mir-62, mir-67, mir-79, mir-124, mir-229, mir-231, mir-232, mir-251, mir-392, mir-792, mir-797, mir-799, mir-1020 and mir-481 (Fig. 2b and Table S5†). Moreover, the remaining 10 were downregulated, consisting of mir-35, mir-37, mir-38, mir-40, mir-49, mir-61, mir-83, mir-230, mir-234 and mir-784 (Fig. 2b and Table S5†).
Fig. 2. Identification of miRNAs involved in the control of coal combustion related PM2.5 toxicity. (a) Hierarchical clustering assay of miRNAs expression. (b) Fold changes of miRNAs in coal combustion related PM2.5 exposed nematodes. (c) Expression pattern of mature miRNAs detected by real-time PCR after coal combustion related PM2.5 exposure. The concentration of coal combustion related PM2.5 was 1 mg L–1. Prolonged exposure was performed from L1-larvae to young adults at 20 °C in the presence of food. Bars represent mean ± SD.
To confirm the SOLiD sequencing results, changes in the expression levels of several miRNAs that were dysregulated upon prolonged exposure to PM2.5 (1 mg L–1) were measured by qRT-PCR. Compared to the control, the miRNAs mir-62, mir-231, mir-232, and mir-251 were found to be significantly up-regulated, while mir-35, mir-83, mir-230, and mir-234 were significantly down-regulated (Fig. 2c). Therefore, the alterations in expression for these candidate miRNAs were similar by both SOLiD sequencing and qRT-PCR.
Gene ontology and signaling pathways of the predicted gene targets of the miRNAs dysregulated in response to coal combustion related PM2.5
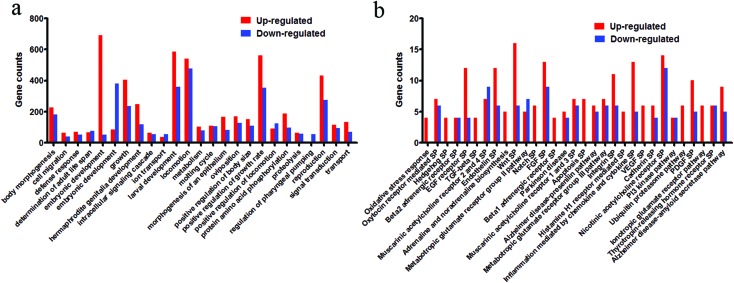
To characterize the functions of the miRNAs that were dysregulated in response to PM2.5 in the nematodes, the genes targeted by these miRNAs were predicted using the TargetScan database. Overall, 1456 and 959 genes were found to be the putative targets of the down- and up-regulated miRNAs, respectively. Analysis of the ontology of these target genes was performed to generate putative biological processes for these genes,8 and thus, a possible role for these miRNAs (Tables S6 and S7†). The functions of the target genes were found to fall into several categories including defense, cell migration, development, growth, locomotion, reproduction, metabolism, longevity, transport, and signal transduction (Fig. 3a).
Fig. 3. Gene ontology terms and signaling pathways based on predicted targets of dysregulated miRNA induced by coal combustion related PM2.5. (a) Gene ontology terms. Gene counts >40. (b) KEGG signaling pathways. Gene counts >4. SP: signaling pathway.
Furthermore, signaling pathways mediated by the genes that were predicted to be targeted by the dysregulated miRNAs were identified using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database. In nematodes, the up-regulated miRNAs were found to influence 105 signaling pathways, while the down-regulated miRNAs influenced 95 signaling pathways (signaling pathways with gene counts more than 4 have been shown in Fig. 3b, Tables S8 and S9†). These signaling pathways were predominantly related to oxidative stress response, Wnt, TGF-beta, PI3 kinase pathway, VEGF, Parkinson disease, Alzheimer disease-amyloid secretase pathway, p53, apoptosis, Notch signaling pathways, and so on (Fig. 3b). Identifying these important signaling pathways increases our understanding of the molecular mechanisms of action during miRNA regulation of coal combustion derived PM2.5 toxicity.
Loss-of-function mutations of candidate miRNAs altered responses to coal combustion related PM2.5 in nematodes
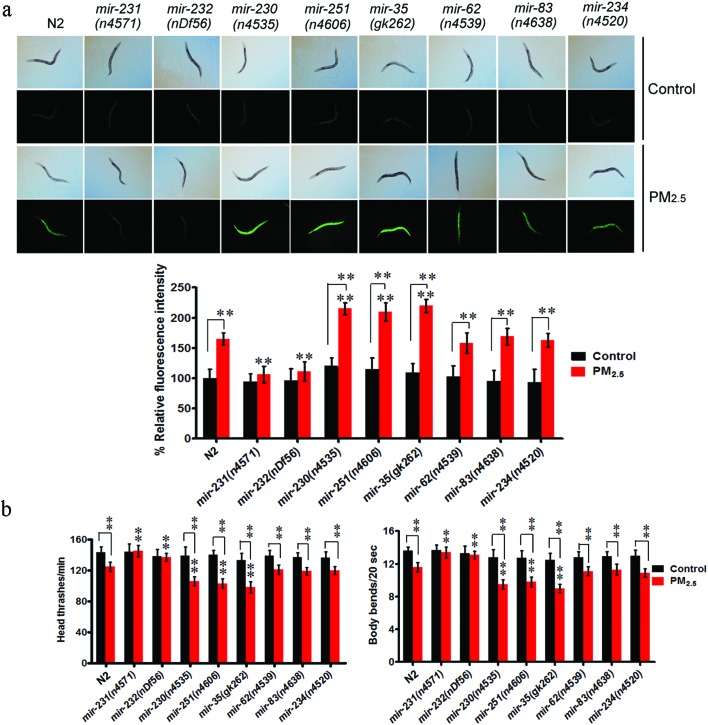
Loss-of-function miRNA mutants are powerful tools to study the function of miRNAs in the in vivo C. elegans model.32 Due to signaling pathways mediated by the dysregulated miRNAs including the oxidative stress response and neurodegeneration, such as Parkinson's disease and Alzheimer disease in Fig. 3b, intestinal ROS production and locomotion in nematodes with loss-of-function mutations in candidate miRNAs were assessed compared to wild-type N2 nematodes after exposure to PM2.5. For intestinal ROS production, it was found that nematodes with loss of function of mir-230, mir-251, or mir-35 had higher levels of ROS when exposed to PM2.5 compared to wild-type N2, while the reverse occurred in mir-231 and mir-232 mutants (Fig. 4a). Similarly, in terms of locomotion, it was noted that mir-230, mir-251, or mir-35 mutants exposed to PM2.5 had significantly less head thrash and body bend than wild-type N2, while mir-232 or mir-231 had significantly more head thrash and body bend (Fig. 4b). In contrast, nematodes with loss of function of mir-62, mir-83, or mir-234 had intestinal ROS production and locomotion approximately equal to wild-type N2 after exposure to PM2.5 (Fig. 4a and b). Under normal conditions, nematodes with a loss of function in these miRNAs displayed no significant changes in intestinal ROS production or locomotion compared to wild-type N2 (Fig. 4a and b). Therefore, the miRNAs of mir-35, mir-230, mir-231, mir-232, and mir-251 are likely involved in the control of coal combustion related PM2.5 toxicity.
Fig. 4. Confirmation of miRNAs involved in the control of coal combustion related PM2.5 toxicity. (a) Comparison of intestinal ROS production. (b) Comparison of locomotion behavior. The concentration of coal combustion related PM2.5 was 1 mg L–1. Prolonged exposure was performed from L1-larvae to young adults at 20 °C in the presence of food. Bars represent mean ± SD. **P < 0.01 vs. N2 (if not specially indicated).
SMK-1 acted as a potential target for mir-231 during regulation of coal combustion related PM2.5 toxicity
When determining the functions of specific miRNAs, the functions of the predicted target genes can provide insights.33,34 In this study, the TargetScan database was used to predict the targets of mir-231 to delineate the molecular mechanisms behind this miRNA's regulation of the response to PM2.5. Ninety-three putative targets were identified, including smk-1, which encoded an evolutionarily conserved protein ortholog of the mammalian SMEK (suppressor of MEK null) protein.35
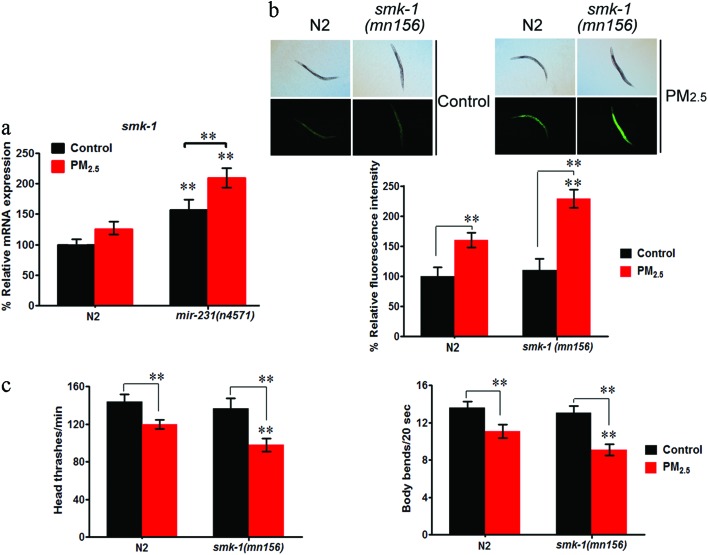
In the mir-231 mutant nematodes, it was found there was significant up-regulation of smk-1 expression, suggesting that mir-231 is a negative regulator of smk-1 (Fig. 5a). Upon exposure to PM2.5, there was an even further increase in smk-1 expression levels in mir-231 mutants compared to wild-type N2 nematodes (Fig. 5a).
Fig. 5. Effect of smk-1 mutation on coal combustion related PM2.5 toxicity in nematodes. (a) Expression pattern of smk-1 gene in mir-231 mutant nematodes. (b) Effect of smk-1 mutation on intestinal ROS production in coal combustion related PM2.5 exposed nematodes. (c) Effect of smk-1 mutation on locomotion behavior in coal combustion related PM2.5 exposed nematodes. The concentration of coal combustion related PM2.5 was 1 mg L–1. Prolonged exposure was performed from L1-larvae to young adults at 20 °C in the presence of food. Bars represent mean ± SD. **P < 0.01 vs. N2 (if not specially indicated).
When intestinal ROS production and locomotion were evaluated in the smk-1(mn156) mutant after PM2.5 exposure, it was found that this mutant had much higher levels of intestinal ROS production, as well as significantly less locomotion, compared to wild-type N2 nematodes (Fig. 5b and c). These data suggest that smk-1 mutation may induce a susceptibility of nematodes to coal combustion related PM2.5 toxicity.
Genetic interaction between mir-231 and smk-1 during the regulation of coal combustion related PM2.5 toxicity
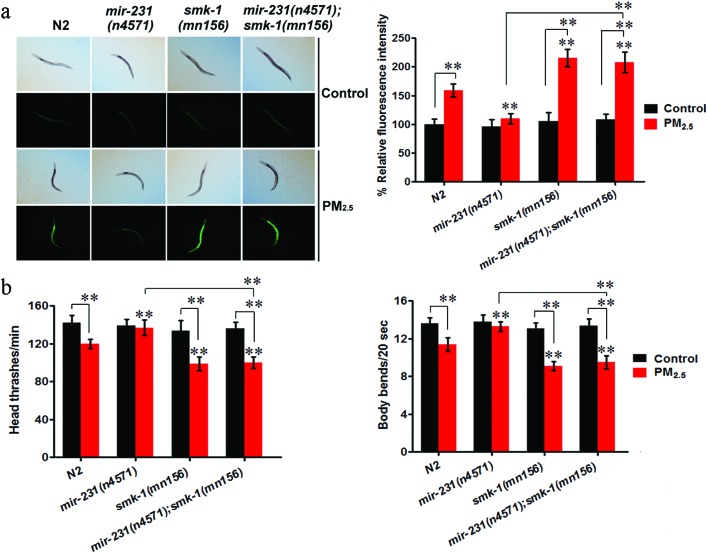
Compared to the wild-type N2 nematodes, when exposed to PM2.5, the mir-231 mutant had a significant decrease in the induction of intestinal ROS and an increase in locomotion, while the reverse phenotypes were seen in the smk-1(mn156) mutant (Fig. 6). Importantly, ROS production and locomotion in a double mutant of mir-231 (n4571);smk-1(mn156) paralleled those in the smk-1(mn156) single mutant nematodes (Fig. 6). These results suggest that the effects of a mir-231 mutation on coal combustion derived PM2.5 toxicity, in the induction of intestinal ROS production and suppression of locomotion, can be abrogated by mutating smk-1. Overall, these results suggest SMK-1 is a target of mir-231 during the regulation of coal combustion derived PM2.5 toxicity.
Fig. 6. Genetic interaction of mir-231 and smk-1 in regulating coal combustion related PM2.5 toxicity. (a) Genetic interaction of mir-231 and smk-1 in regulating coal combustion related PM2.5 toxicity in inducing intestinal ROS production. (b) Genetic interaction of mir-231 and smk-1 in regulating coal combustion related PM2.5 toxicity in decreasing locomotion behavior. The concentration of coal combustion related PM2.5 was 1 mg L–1. Prolonged exposure was performed from L1-larvae to young adults at 20 °C in the presence of food. Bars represent mean ± SD. **P < 0.01 vs. N2 (if not specially indicated).
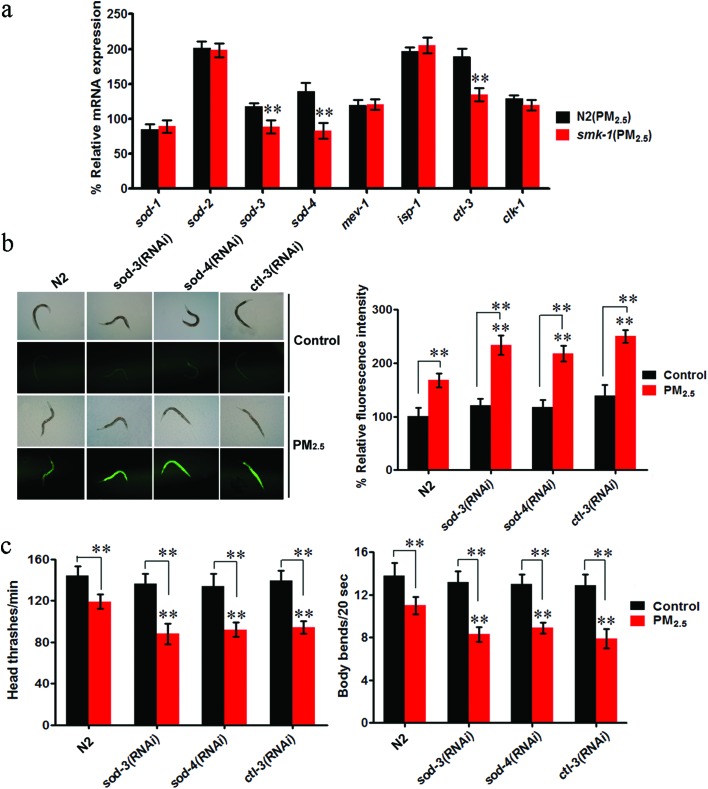
Mutation of smk-1 altered the expression of genes required for the response to oxidative stress in coal combustion related PM2.5 exposed nematodes
Due to exposure to PM2.5, the expression of some genes required for the response to oxidative stress in wild-type N2 nematodes becomes dysregulated, including sod-1, sod-2, sod-3, sod-4, ctl-3, mev-1, isp-1 and clk-1.13 Upon comparing PM2.5 exposed wild-type and smk-1 mutant nematodes, it was found that sod-3, sod-4 and ctl-3 gene expression were significantly decreased in the smk-1 mutant compared to wild-type N2 nematodes (Fig. 7a). In contrast, the expression of the sod-1, sod-2, isp-1, clk-1 and mev-1 genes was not significantly different in the smk-1 mutant compared to that in the wild-type N2 nematodes upon exposure to PM2.5 (Fig. 7a). Moreover, after PM2.5 exposure, the transcriptional expressions of sod-3, sod-4, and ctl-3 were significantly increased in mir-231 mutant nematodes, and the transcriptional expressions of sod-3, sod-4, and ctl-3 were significantly decreased in the double mutant of mir-231;smk-1 (Fig. S1†). These results imply that SOD-3, SOD-4 and CTL-3, which encode an iron/manganese superoxide dismutase, an extracellular Cu2+/Zn2+ superoxide dismutase and a catalase, respectively, may be important downstream targets of SMK-1 during PM2.5 toxicity.
Fig. 7. Oxidative stress related genes acted as downstream regulators of smk-1 in the regulation of coal combustion related PM2.5 toxicity. (a) Expression pattern of genes required for the control of oxidative stress in coal combustion related PM2.5 exposed wild-type and smk-1 mutant nematodes. (b) Effect of RNAi knockdown of sod-3, sod-4 or ctl-3 on toxicity of coal combustion related PM2.5 in inducing intestinal ROS production. (c) Effect of RNAi knockdown of sod-3, sod-4 or ctl-3 on toxicity of coal combustion related PM2.5 in decreasing locomotion behavior. The concentration of coal combustion related PM2.5 was 1 mg L–1. Prolonged exposure was performed from L1-larvae to young adults at 20 °C in the presence of food. Bars represent mean ± SD. **P < 0.01 vs. N2 (if not specially indicated).
SOD-3, SOD-4, and CTL-3 participated in the regulation of coal combustion related PM2.5 toxicity
To determine the functions of SOD-3, SOD-4, and CTL-3 during the control of PM2.5 toxicity, RNAi knockdown was performed in C. elegans for the sod-3, sod-4 and ctl-3 genes. Under normal conditions, RNAi knockdown of sod-3, sod-4, or the ctl-3 gene did not significantly alter intestinal ROS production or locomotion (Fig. 7b and c). However, after exposure to PM2.5, nematodes with RNAi knockdown of the sod-3, sod-4, or ctl-3 gene had a significantly higher induction of intestinal ROS production and a decrease in locomotion compared to the wild-type N2 (Fig. 7b and c). Therefore, SOD-3, SOD-4 and CTL-3 confer protection to nematodes against coal combustion related PM2.5 toxicity in nematodes.
Discussion
To date, the functional toxicologenomics has been widely performed to investigate the possible molecular targets for certain toxicants. However, we still know little about the molecular basis for miRNAs in the regulation of coal combustion related PM2.5 toxicity. Herein, we employed the in vivo C. elegans assay system to perform the SOLiD sequencing analysis in order to identify the possible miRNA targets for coal combustion related PM2.5 after prolonged exposure. Based on the SOLiD sequencing results, we identified 25 differentially expressed miRNAs in coal combustion related PM2.5 exposed nematodes. Some of the dysregulated miRNAs in coal combustion related PM2.5 exposed nematodes were confirmed by the qRT-PCR assay (Fig. 2c). Our results provide important in vivo miRNAs targets for coal combustion related PM2.5 in organisms.
For the identified potential miRNA targets of coal combustion related PM2.5, the gene ontology analysis suggested that they might be involved in the control of a series of important biological processes (Fig. 3a). Moreover, the KEGG analysis further demonstrated that the identified potential miRNA targets of coal combustion related PM2.5 might be associated with some important signaling pathways (Fig. 3b). These results provide important clues for further revealing the molecular mechanisms of coal combustion related PM2.5 toxicity or response of organisms to coal combustion related PM2.5.
To confirm the exact roles of candidate dysregulated miRNAs in coal combustion related PM2.5 exposed nematodes, we used the available mutants to perform the toxicity assessment using intestinal ROS production and locomotion behavior as the endpoints. After coal combustion related PM2.5 exposure, we found that 5 (mir-231, mir-232, mir-230, mir-251, and mir-35) among the examined 8 miRNA mutants had an altered toxicity in inducing intestinal ROS production and in decreasing locomotion behavior in nematodes (Fig. 4). These data confirm the possible crucial roles of mir-231, mir-232, mir-230, mir-251, and mir-35 in the regulation of coal combustion related PM2.5 toxicity. In C. elegans, mir-231 is the homologue of human miR-99 and miR-556, mir-232 is the homologue of human miR-302 and miR-519, and mir-251 is the homologue of human miR-26.36 Moreover, our results imply that the involvement of miRNAs in the regulation of PM2.5 toxicity should not be concluded only based on the expression pattern of miRNAs.
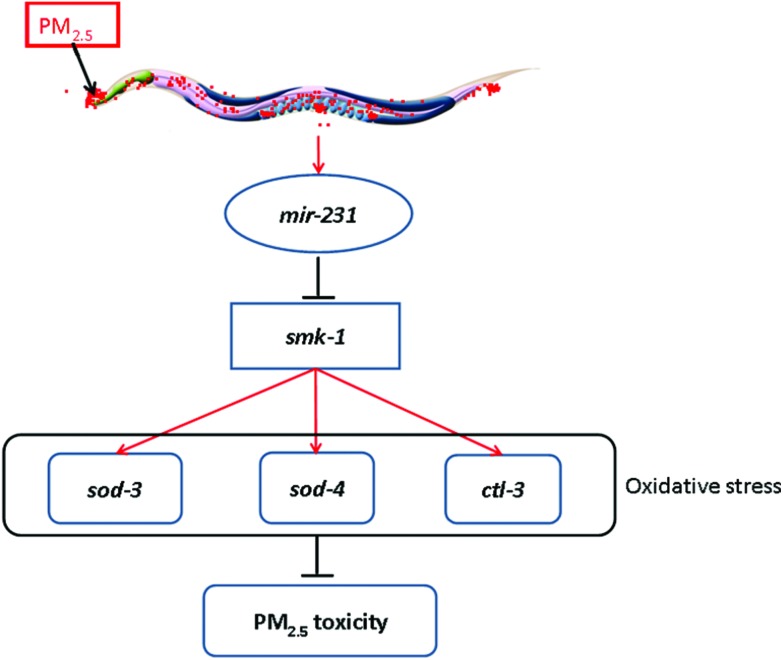
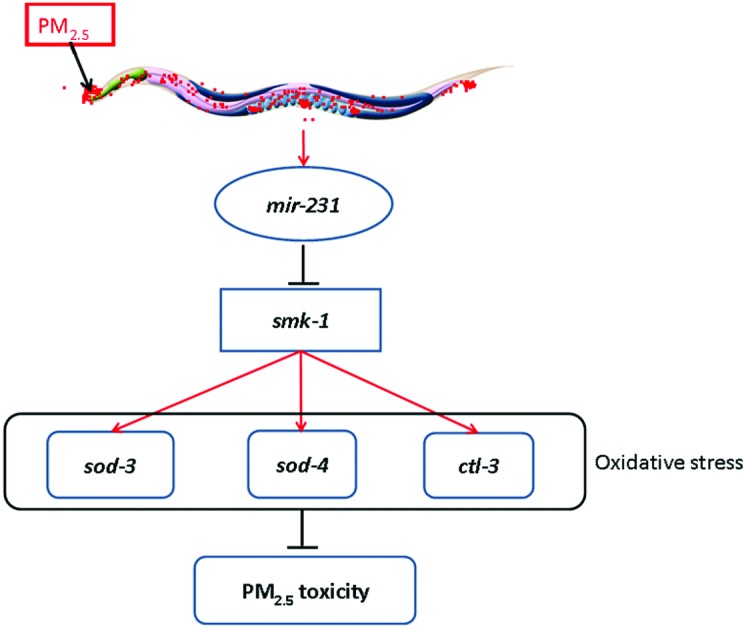
In this study, we further focused on the mir-231 to determine the underlying molecular mechanism for its involvement in the control of coal combustion related PM2.5 toxicity. At least three lines of evidence were raised to indicate the role of SMK-1 acting as the molecular target for mir-231 in the regulation of coal combustion related PM2.5 toxicity. First, after coal combustion related PM2.5 exposure, mir-231 mutation increased the expression of smk-1 (Fig. 5a). Second, the smk-1 mutant had the opposite phenotypes of intestinal ROS production and locomotion behavior to those in the mir-231 mutant after coal combustion related PM2.5 exposure (Fig. 5b and c). Third, smk-1 mutation suppressed the resistance of the mir-231 mutant to coal combustion related PM2.5 toxicity in inducing intestinal ROS production and decreasing locomotion behavior (Fig. 6). Previous study has suggested that SMK-1 activity is essential for the control of longevity and resistance to oxidative and UV-induced damage.35 Our study indicates a novel function of SMK-1 in the regulation of PM2.5 toxicity. Moreover, our data suggest that mir-231 acts as the direct upstream regulator for SMK-1 in regulating biological processes in nematodes (Fig. 8).
Fig. 8. A diagram showing the molecular mechanism for mir-231 in the regulation of coal combustion related PM2.5 toxicity.
In this study, we further identified the SOD-3, SOD-4 and CTL-3 as the downstream targets for SMK-1 in the regulation of coal combustion related PM2.5 toxicity. On one hand, after coal combustion related PM2.5 exposure, smk-1 mutation decreased the expression levels of sod-3, sod-4 and ctl-3 genes (Fig. 7a). On the other hand, RNAi knockdown of sod-3, sod-4, or ctl-3 genes induced a susceptibility to coal combustion related PM2.5 toxicity in inducing intestinal ROS production and in decreasing locomotion behavior (Fig. 7b and c). Therefore, the raised mir-231-SMK-1-SOD-3/SOD-4/CTL-3 signaling cascade may provide an important molecular basis for the role of oxidative stress in the induction of coal combustion related PM2.5 toxicity in organisms (Fig. 8). That is, after exposure, coal combustion related PM2.5 can increase the expression of mir-231. The increased mir-231 may further suppress the protection function of SOD-3, SOD-4, and CTL-4 against the activation of oxidative stress by inhibiting the SMK-1 expression.
Conclusions
In conclusion, using the SOLiD sequencing technique, we obtained the dysregulated miRNA profiling induced by coal combustion related PM2.5 in the in vivo assay system of C. elegans. Based on the determination of the expression pattern and functional analysis, we confirmed that 5 miRNAs (mir-231, mir-232, mir-230, mir-251, and mir-35) were involved in the control of coal combustion related PM2.5 toxicity. Among these 5 miRNAs, we further identified SMK-1 as the molecular target for mir-231 in the regulation of coal combustion related PM2.5 toxicity. Moreover, we raised a signaling cascade of mir-231-SMK-1-SOD-3/SOD-4/CTL-3 to explain the possible important molecular basis for the role of oxidative stress in the induction of coal combustion related PM2.5 toxicity in organisms.
Conflict of interest
None of the authors have any conflicting interests.
Supplementary Material
Acknowledgments
This study was supported by the grant from National Natural Science Foundation of China (21577016), and Fundamental Research Funds for the Central Universities in China (2242016 K41033).
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c7tx00107j
References
- Pope C. A., Burnett R. T., Thun M. J., Calle E. E., Krewski D., Ito K., Thurston G. D. J. Am. Med. Assoc. 2002;287:1132–1141. doi: 10.1001/jama.287.9.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procto S. D., Dreher K. L., Kelly S. E., Russell J. C. Toxicol. Sci. 2006;90:385–391. doi: 10.1093/toxsci/kfj100. [DOI] [PubMed] [Google Scholar]
- Lewtas J. Mutat. Res. 2007;636:95–133. doi: 10.1016/j.mrrev.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Chan J. K., Charrier J. G., Kodani S. D., Vogel C. F., Kado S. Y., Anderson D. S., Anastasio C., Van Winkle L. S. Part. Fibre Toxicol. 2013;10:34. doi: 10.1186/1743-8977-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jie Y., Houjin H., Xun M., Kebin L., Xuesong Y., Jie X. Braz. J. Med. Biol. Res. 2014;47:982–989. doi: 10.1590/1414-431X20144084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.-F., Geng C.-M., Hao W.-D., Zhao Y.-D., Li Q., Wang H.-M., Qian Y. Biomed. Environ. Sci. 2016;29:107–116. doi: 10.3967/bes2016.012. [DOI] [PubMed] [Google Scholar]
- Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoshechkin I., Sternberg P. W. Nat. Rev. Genet. 2007;8:518–532. doi: 10.1038/nrg2105. [DOI] [PubMed] [Google Scholar]
- Leung M. C., Williams P. L., Benedetto A., Au C., Helmcke K. J., Aschner M., Meyer J. N. Toxicol. Sci. 2008;106:5–28. doi: 10.1093/toxsci/kfn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.-L., Wu Q.-L., Li Y.-P., Wang D.-Y. RSC Adv. 2013;3:5741–5757. [Google Scholar]
- Wang D.-Y. Toxicol. Res. 2016;5:1003–1011. doi: 10.1039/c6tx00056h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.-L., Lin Z.-Q., Jia R.-H., Li G.-J., Xi Z.-G., Wang D.-Y. J. Hazard. Mater. 2014;274:106–114. doi: 10.1016/j.jhazmat.2014.03.064. [DOI] [PubMed] [Google Scholar]
- Sun L.-M., Lin Z.-Q., Liao K., Xi Z.-G., Wang D.-Y. Sci. Total Environ. 2015;512–513:251–260. doi: 10.1016/j.scitotenv.2015.01.058. [DOI] [PubMed] [Google Scholar]
- Yang R.-L., Zhao Y.-L., Yu X.-M., Lin Z.-Q., Xi Z.-G., Rui Q., Wang D.-Y. Toxicol. Res. 2015;4:333–343. [Google Scholar]
- Sun L.-M., Wu Q.-L., Liao K., Yu P.-H., Cui Q.-H., Rui Q., Wang D.-Y. Chemosphere. 2016;144:2392–2400. doi: 10.1016/j.chemosphere.2015.11.028. [DOI] [PubMed] [Google Scholar]
- Yang R.-L., Rui Q., Kong L., Zhang N., Li Y., Wang X.-Y., Tao J., Tian P.-Y., Ma Y., Wei J.-R., Li G.-J., Wang D.-Y. Toxicol. Res. 2016;5:1097–1105. doi: 10.1039/c6tx00022c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L. P., Lau N. C., Weinstein E. G., Abdelhakim A., Yekta S., Rhoades M. W., Burge C. B., Bartel D. P. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Zhao Y.-L., Wu Q.-L., Wang D.-Y. RSC Adv. 2015;5:92394–92405. [Google Scholar]
- Zhao Y.-L., Wu Q.-L., Wang D.-Y. Biomaterials. 2016;79:15–24. doi: 10.1016/j.biomaterials.2015.11.052. [DOI] [PubMed] [Google Scholar]
- Zhao Y.-L., Jia R.-H., Qiao Y., Wang D.-Y. Nanomedicine. 2016;12:735–744. doi: 10.1016/j.nano.2015.10.008. [DOI] [PubMed] [Google Scholar]
- Zhao Y.-L., Yang J.-N., Wang D.-Y. Sci. Rep. 2016;6:23234. doi: 10.1038/srep23234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z.-H., Li M., Liu H., Luo L.-B., Gu W.-D., Wu Q.-L., Wang D.-Y. Sci. Rep. 2016;6:32409. doi: 10.1038/srep32409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkin S., Williams P. L. Environ. Toxicol. Chem. 1995;14:2139–2147. [Google Scholar]
- Qiao Y., Zhao Y.-L., Wu Q.-L., Sun L.-M., Ruan Q.-L., Chen Y.-Y., Wang M., Duan J.-A., Wang D.-Y. PLoS One. 2014;9:e91825. doi: 10.1371/journal.pone.0091825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu C.-J., Yu X.-M., Wu Q.-L., Zhuang Z.-H., Zhang W.-M., Wang D.-Y. RSC Adv. 2015;5:8942–8951. [Google Scholar]
- Zhuang Z.-H., Zhao Y.-L., Wu Q.-L., Li M., Liu H.-C., Sun L.-M., Gao W., Wang D.-Y. PLoS One. 2014;9:e85482. doi: 10.1371/journal.pone.0085482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-P., Li Y.-X., Wu Q.-L., Ye H.-Y., Sun L.-M., Ye B.-P., Wang D.-Y. PLoS One. 2013;8:e71180. doi: 10.1371/journal.pone.0071180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.-L., Liu Q., Shakoor S., Gong J.-R., Wang D.-Y. Toxicol. Res. 2015;4:270–280. [Google Scholar]
- Liu Z.-F., Zhou X.-F., Wu Q.-L., Zhao Y.-L., Wang D.-Y. RSC Adv. 2015;5:94257–94266. [Google Scholar]
- Wu Q.-L., Zhao Y.-L., Zhao G., Wang D.-Y. Nanomedicine. 2014;10:1401–1410. doi: 10.1016/j.nano.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Miska E. A., Alvarez-Saavedra E., Abbott A. L., Lau N. C., Hellman A. B., McGonagle S. M., Bartel D. P., Ambros V. R., Horvitz H. R. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall S., Grün D., Krek A., Chen K., Wang Y.-L., Dewey C. N., Sood P., Colombo T., Bray N., Macmenamin P., Kao H.-L., Gunsalus K. C., Pachter L., Piano F., Rajewsky N. Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Bartel D. P. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S., Ma H., Burch D., Maciel G. A., Hunter T., Dillin A. Cell. 2006;124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Ibanez-Ventoso C., Vora M., Driscoll M. PLoS One. 2008;3:e2818. doi: 10.1371/journal.pone.0002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.