 As effective non-viral vectors of gene therapy, cationic lipids still have the problem of toxicity, which has become one of the main bottlenecks for their applications.
As effective non-viral vectors of gene therapy, cationic lipids still have the problem of toxicity, which has become one of the main bottlenecks for their applications.
Abstract
As effective non-viral vectors of gene therapy, cationic lipids still have the problem of toxicity, which has become one of the main bottlenecks for their applications. The toxicity of cationic lipids is strongly connected to the headgroup structures. In this article, we studied the cytotoxicity of two cationic lipids with a quaternary ammonium headgroup (CDA14) and a tri-peptide headgroup (CDO14), respectively, and with the same linker bond and hydrophobic domain. The IC50 values of CDA14 and CDO14 against NCI-H460 cells were 109.4 μg mL–1 and 340.5 μg mL–1, respectively. To determine the effects of headgroup structures of cationic lipids on cytotoxicity, apoptosis related pathways were investigated. As the lipids with a quaternary ammonium headgroup could induce more apoptotic cells than the ones with a peptide headgroup, the enzymatic activity of caspase-9 and caspase-3 increased obviously, whereas the mitochondrial membrane potential (MMP) decreased. At the same time, the reactive oxygen species (ROS) levels also increased and the cell cycle was arrested at the S phase. The results showed that the toxicity of the cationic lipid had a close relationship with its headgroup structures, and the cytotoxic mechanism was mainly via the caspase activation dependent signaling pathway and mitochondrial dysfunction. Through this study, we hope to provide the scientific basis for exploiting safer and more efficient cationic lipids for gene delivery.
Introduction
Safety is an important factor for drug and gene delivery vectors. The most extensively used delivery tools can be divided into two general categories: viral and non-viral vectors. Viral vectors can achieve high transfection efficiency in gene delivery, but the fatal defects of viruses such as their safety issues and complicated packaging process severely hinder their use in clinical trials.1,2 In contrast, cationic lipids, as a kind of non-viral vectors, are much safer than viral vectors.3 In comparison with other gene delivery materials, cationic lipids are easy to synthesize, are not as biologically hazardous as viral vectors, are readily available commercially, and can be easily adapted for specific applications.4–7 Since the first cationic lipid for gene delivery, N-[1-(2,3-dioleyloxy)propyl]-N,N,N-trimethylammonium chloride (DOTMA) was introduced by Felgner,8 a number of cationic transfection lipids with quaternary ammonium headgroups (such as DOTAP, DDAB, and CTAB) have been reported and were found to be active in a wide variety of cell types.9,10 Recently, peptide self-assembly has provided an effective approach to mimic and understand living systems due to its good biocompatibility.11 Furthermore, the rationale of these pure nanodrugs via the self-assembly approach might open an alternative avenue and give inspiration to fabricate new carrier-free nanodrugs for tumor theranostics.12,13
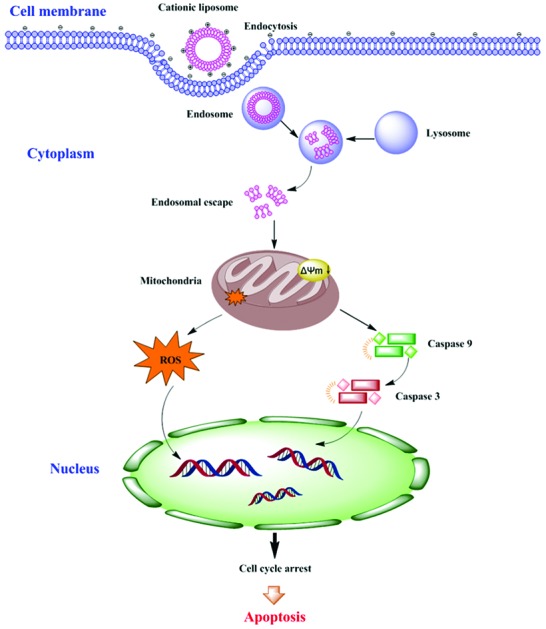
However, cationic lipids still have the problem of toxicity, as they can activate several cellular pathways like pro-apoptotic and pro-inflammatory cascades, which has become one of the main bottlenecks for their applications.14–16 Cationic lipids are positively charged amphiphiles consisting of three basic chemical functional domains: a hydrophilic headgroup, a hydrophobic domain, and a linker bond that tethers the cationic headgroup and hydrophobic tail domain.17 The cytotoxic effects are severely associated with the cationic nature of the vectors, which is mainly determined by the structure of its hydrophilic group. The hydrophilic headgroup exhibits positive charges which trigger their interaction with negatively charged DNA through electrostatic attractions, leading to the formation of complexes containing condensed DNA.18 However, the relationship between the cationic lipid headgroup structures and toxicity is rarely discussed, which greatly hinders the advancement of cationic lipids towards clinical trials. Our study showed that peptide headgroups were much superior to quaternary ammonium headgroups in terms of transfection efficiency and toxicity.19–21 Therefore, we chose quaternary ammonium and peptides as headgroups of cationic lipids for studying their cytotoxic effects (Fig. 1). Herein, the correlation of cytotoxic effects of cationic lipids with their headgroups will be elucidated based on the research on the origin of apoptosis, by analyzing caspase-3,9 enzymatic activities, reactive oxidative stress (ROS), mitochondrial membrane potential (MMP) and cell cycle arrest. This study aims at providing the scientific basis for the development of safe and efficient cationic lipids.
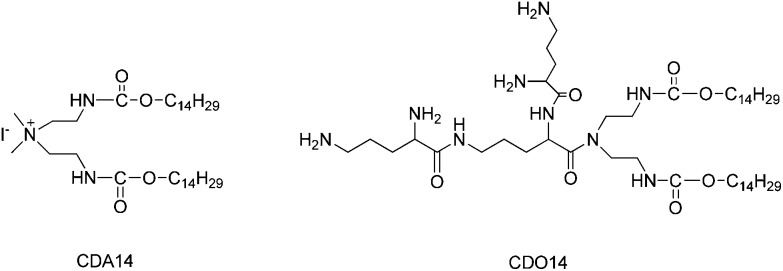
Fig. 1. The structures of CDA14 and CDO14 cationic lipids.
Materials and methods
Drugs and chemicals
CDA14 and CDO14 were synthesised by our lab. A Cell Counting Kit-8 (CCK-8), an Annexin V-FITC/PI detection kit, a caspase-3 and -9 activity assay kit, a ROS assay kit, and a cell cycle analysis kit were purchased from Beyotime Institute of Biotechnology (China). JC-1 was purchased from BD Bioscience (USA). The human non-small cell lung cancer cell line (NCI-H460 cells) was purchased from the Cell Bank of the Chinese Academy of Sciences. DMEM, RPMI-1640 medium, fetal bovine serum (FBS), and trypsin were purchased from Gibco (USA). All of the water used in this study was purified using a Milli-Q water purification system (Millipore, USA).
Liposome preparation
To prepare liposomes, 1 mg CDA14 or CDO14 was dissolved in 1 mL of chloroform in a 5 mL glass vial, respectively. The solvent was removed under a stream of nitrogen gas, followed by high vacuum desiccation. The dry lipid film was resuspended in 1 mL distilled water to give liposomes in a concentration of approximately 1 mg mL–1. The liposome solutions were subjected to several cycles of sonication in a bath sonicator and vigorous vortex mixing to form small vesicles.
Measurement of the particle size and zeta potential
For the measurement of the particle size and zeta potential, 20 μL of the liposomes were diluted in distilled water (1 mL). The particle size and zeta potential were then measured three times using a nanoparticle analyzer (HORIBA Scientific, Nanopartica, Japan).
Transmission electron microscope (TEM) observation
The morphology of the liposomes was determined using a transmission electron microscope (TEM) (H-7000; Hitachi, JEM-2100, Japan). A drop of sample solution was placed onto a 300-mesh copper grid coated with carbon. After 2 min, the grid was tapped with filter paper to remove the surface solution. After the sample was negatively stained with 2% phosphotungstic acid for 30 s, the grid was dried at room temperature and then observed by TEM.
Cell culture
NCI-H460 cells were grown in a 75 cm3 culture flask in RPMI-1640 medium supplemented with 10% FBS and antibiotics (100 U mL–1 penicillin and 100 μg mL–1 streptomycin) at 37 °C under a humidified atmosphere containing 5% CO2 in an incubator (HERAcell 150i, USA). Cells grown to confluence were subcultured every other day after being trypsinized with 0.25% trypsin and diluted to one-third in a fresh growth medium.
Cell viability assay
Cell viability was assessed with a formazan tetrazolium salt assay using a Cell Counting Kit-8 (CCK-8). NCI-H460 cells (105 cells per mL) were seeded in 96-well plates at 200 μL RPMI-1640 medium per well and were allowed to adhere for 16–24 h at 37 °C under 5% CO2 to obtain a confluence of about 80%. The cells were exposed to different concentrations of liposomes for 24 h; then the cells were incubated with 10 μL of CCK-8 reagent for 1 h at 37 °C. The optical density (OD) was determined at 450 nm using a microplate reader (Tecan, Sunrise). The cell viability was calculated as [sample/control × 100%]. All experiments were repeated six times, and the data are presented as the mean ± standard deviation (SD). The half maximal inhibitory concentration (IC50) was calculated by the Logit method.
Apoptosis analysis
NCI-H460 cells (105 cells per mL) were seeded in 6-well plates at 1 mL RPMI-1640 medium per well and were allowed to adhere for 16–24 h at 37 °C under 5% CO2 to obtain a confluence of about 80%. The medium was replaced with a fresh medium; then the cationic liposomes were added in two concentrations (15 μg mL–1 and 120 μg mL–1) for 24 h. The cells were harvested and washed twice with PBS. The cell apoptosis rate was measured using an Annexin V-FITC/PI detection kit to identify early apoptotic cells (Annexin V-FITC-positive, PI-negative) by FCM. The Cell Quest software (BD Biosciences) was used to assess the apoptosis rate. Events falling into the FITC/PI region of the lower-right quadrant were counted as apoptotic cells. The apoptosis rates of sample cells were calculated directly from the gated histograms. All experiments were repeated four times, and the data are expressed as the mean ± standard deviation.
Mitochondrial membrane potential measurements
For the assessment of mitochondrial membrane potential (MMP), NCI-H460 cells (105 cells per mL) were seeded in 6-well plates at 1 mL RPMI-1640 medium per well and allowed to adhere for 16–24 h at 37 °C under 5% CO2 to obtain a confluence of about 80%. The medium was replaced with a fresh medium; then the cationic liposomes were added in two concentrations (15 μg mL–1 and 120 μg mL–1). After 24 h, the cells were harvested and then incubated with JC-1 for 20 min at 37 °C. The excitation and emission wavelengths of fluorescence signals were 488 nm and 525 nm, respectively, and they were measured using a flow cytometer. The value of fluorescence intensity was analyzed using the Cflow Plus software (Accuri C6, FACS, BD, USA).
Caspase-3 and caspase-9 activity assay
NCI-H460 cells (105 cells per mL) were seeded in 6-well plates at 1 mL RPMI-1640 medium per well and were allowed to adhere for 16–24 h at 37 °C under 5% CO2 to obtain a confluence of about 80%. The medium was replaced with a fresh medium; then the cationic liposomes were added in two concentrations (15 μg mL–1 and 120 μg mL–1) for 24 h. The cells were collected by trypsinization, washed twice with PBS, resuspended in lysis buffer (a caspase-3 and caspase-9 activity assay kit) and incubated on ice for 15 min. The lysate was centrifuged at 4 °C and 20 000g for 15 min. The supernatant was analyzed for protein concentration by the Bradford method with BSA as a standard, and 0.1 mg of total protein was used for the caspase activity assay with Ac-DEVD-pNA (acetyl-Asp-Glu-Val-Asp p-nitroanilide) and Ac-LEHD-pNA (acetyl-Leu-Glu-His-Asp p-nitroanilide) as substrates for caspase-3 and caspase-9, respectively.22,23 The absorbance of the pNA at 405 nm was recorded using a microplate reader (Synergy H1, BioTek, USA) after 2 h.
Measurement of reactive oxygen species
The levels of intracellular mitochondrial reactive oxygen species (ROS) formation were detected with 2,7-dichlorofluorescein diacetate (DCFH-DA) according to the instructions of a ROS assay kit.24 The cells were harvested after 24 h of treatment with liposomes and then washed twice with PBS and incubated with DCFH-DA (10 mmol L–1) at 37 °C for 20 min in the dark for final analysis by flow cytometry. All measurements were performed in triplicate.
Cell cycle analysis
For the cell cycle assay, NCI-H460 cells (105 cells per mL) were seeded in 6-well plates at 1 mL RPMI-1640 medium per well and were allowed to adhere for 16–24 h at 37 °C under 5% CO2 to obtain a confluence of about 80%. The medium was replaced with a fresh medium; then cationic liposomes were added in two concentrations (15 μg mL–1 and 120 μg mL–1). After 24 h, cells were harvested and washed twice with PBS, and then fixed in 1 mL of 70% cold ethanol at 4 °C for 12 h. The fixed cells were washed with PBS and re-suspended in 1 mL Propidium Iodide (PI) staining solution (1 mg mL–1 sodium citrate, 50 μg mL–1 PI, 10 μg mL–1 RNase A, and 0.5% Triton X-100). The cells were finally incubated at 37 °C for 30 min in the dark. The distribution of cells in the cell cycle was measured by flow cytometry analysis (Accuri C6, FACS, BD, USA) with the Cflow Plus software.
Statistical analysis
Data are expressed as the mean ± standard deviation (SD). Statistical analysis was performed using a Student's t-test between two groups or single factor analysis of variance among three or more groups. Differences were judged to be significant at P < 0.05.
Results and discussion
Particle size and zeta-potential
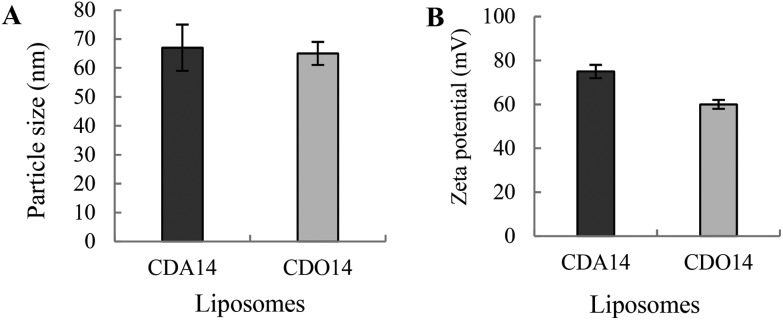
After the preparation of CDA14 and CDO14 liposomes as given in the experimental procedures, the measurement of their particle size and zeta potential was performed using dynamic light scattering (DLS) analysis. The average particle sizes of the two liposomes were around 65 nm (Fig. 2A), and the average zeta-potentials were 75 mV and 55 mV for CDA14 and CDO14 (Fig. 2B), respectively.
Fig. 2. Particle sizes and zeta potentials of the cationic liposomes. The liposomes (20 μL) were diluted in 1 mL distilled water, and their particle sizes (A) and zeta potentials (B) were measured using a nanoparticle analyzer. The PDIs of liposomes CDA14 and CDO14 were 0.203 and 0.156, respectively.
Morphological features of liposomes
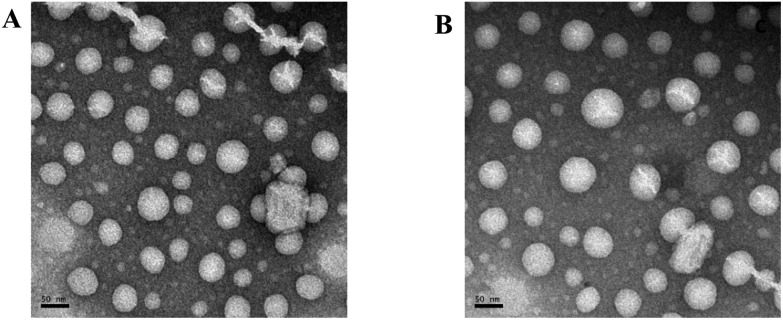
A transmission electron microscope (TEM) was used to directly visualize the size and morphology of CDA14 and CDO14 cationic liposomes. The negatively stained TEM images confirmed the formation of liposomes (Fig. 3). A smooth spherical morphology and a uniform size distribution were observed. The diameters of the CDA14 liposome (Fig. 3A) and the CDO14 liposome (Fig. 3B), ranging from 40 to 80 nm, appeared to be consistent with the results determined by using a nanoparticle analyzer.
Fig. 3. The negatively stained TEM images of the cationic liposomes. (A) The CDA14 liposome; (B) the CDO14 liposome. The scale bar is 50 nm.
Effects of cell viability and cytotoxicity
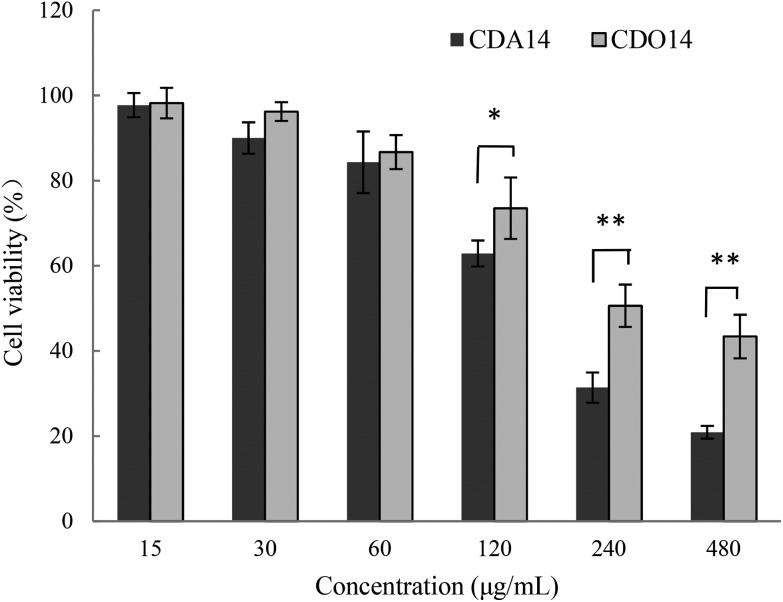
Then, we investigated the effects of CDA14 and CDO14 cationic liposomes on the cell viability of NCI-H460 cells using the CCK-8 assay with untreated cells as the control (100%). At all the concentrations measured, CDO14 showed less cytotoxicity compared with CDA14. At the concentration of 15 μg mL–1, the cell viability of CDO14 was a little higher than that of CDA14 with the cell viability rates of about 97%. With the increase of the liposome concentrations, the cell viability rates decreased. When the concentrations were more than 120 μg mL–1, the cell viability rate decreased quickly, as shown in Fig. 4, and the IC50 values of CDA14 and CDO14 were 159.4 μg mL–1 and 340.5 μg mL–1, respectively. It can be concluded that peptide lipids were much safer than quaternary ammonium lipids. Hereinafter, the concentrations of 15 μg mL–1 (transfection concentration) and 120 μg mL–1 (obvious cytotoxic concentration) were used for studying the cytotoxic mechanism.
Fig. 4. Cell viability of NCI-H460 treated with CDA14 and CDO14 liposomes. The results are expressed as the mean ± SD of triplicate experiments. *P < 0.05 and **P < 0.01, CDA14 was compared with CDO14 at the same concentrations.
Effects on the apoptosis
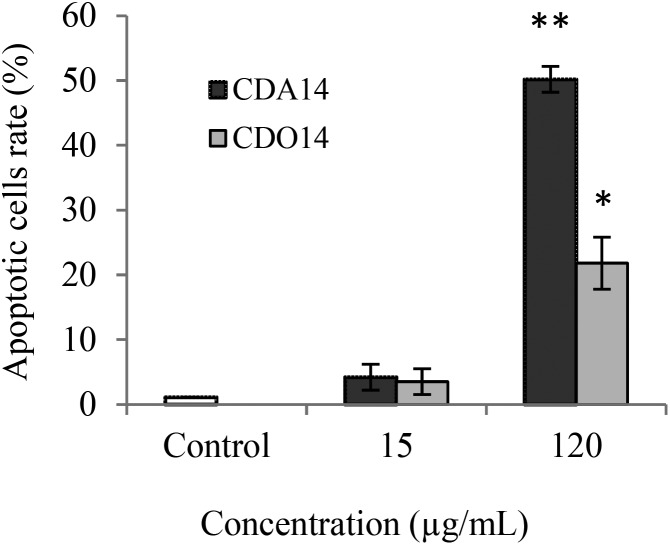
We speculated that the cytotoxicity of the liposomes may be arising from apoptosis; therefore, CDA14 and CDO14 were investigated to see if they can induce cell apoptosis. NCI-H460 cells were treated for 4 h with CDA14 and CDO14 liposomes, respectively; the apoptotic cell rate was detected using an Annexin V-FITC/PI Detection Kit by flow cytometry. The results showed that when NCI-H460 cells were treated with 15 μg mL–1 liposomes, the apoptotic cell rate increased about 5% with both CDA14 and CDO14. When NCI-H460 cells were treated with 120 μg mL–1 liposomes, the apoptotic cell rates of CDA14 and CDO14 increased about 50% and 25%, respectively, as shown in Fig. 5. Based on these results, the effects of CDA14 and CDO14 liposomes on apoptosis showed no obvious difference with 15 μg mL–1 liposome treated cells. Nevertheless, at the concentration of 120 μg mL–1, CDA14 could induce 2 times more apoptotic cells than CDO14.
Fig. 5. Effects of CDA14 and CDO14 cationic liposome induced apoptosis on NCI-H460 cells. The number of apoptotic cells was quantified by flow cytometry. Data are shown as the mean ± SD of triplicate experiments. *P < 0.05 and **P < 0.01, CDA14 and CDO14 were compared with the control.
Caspase-3, 9 enzymatic activities
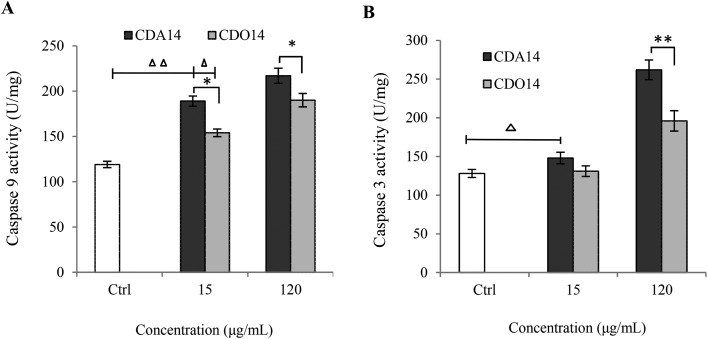
As the central regulators of apoptosis, the activities of caspase-9 and caspase-3 were measured to confirm if the apoptosis difference induced by CDA14 and CDO14 was due to the effects of headgroups.25 We examined caspase-9, 3 enzymatic activities via measuring Ac-LEHD-pNA and Ac-DEVD-pNA cleavage as substrates of NCI-H460 cell lysates collected at 24 h after treatment with CDA14 and CDO14 liposomes (15 μg mL–1 and 120 μg mL–1), respectively. As shown in Fig. 6A, the activity of caspase-9 was elevated with both CDA14 and CDO14 treatments, and CDA14 could activate this enzyme much more than CDO14. At the concentration of 15 μg mL–1, caspase-3 activity was increased with CDA14 treatment, but was not obviously changed with CDO14 treatment. At the concentration of 120 μg mL–1, caspase-3 activity was significantly increased with CDA14 treatment compared with CDO14 treatment (Fig. 6B). The results showed that CDA14-induced cell apoptosis was significantly stronger than that of CDO14, and the increase of caspase-9 and caspase-3 activity was consistent with the cell apoptosis rate in NCI-H460 cells. Because caspase-9 and caspase-3 are involved in the mitochondrial apoptotic pathway, the mechanism of CDA14 and CDO14 lipid-induced cell apoptosis may be considered to be via a caspase-dependent intrinsic mitochondrial pathway.
Fig. 6. CDA14 and CDO14 liposome induced caspase-9 and -3 activity. (A) Caspase-9 activity, (B) caspase-3 activity. Data are shown as the mean ± SD of triplicate experiments. △P < 0.05 and △△P < 0.01, CDA14 and CDO14 were compared with the control. *P < 0.05 and **P < 0.01, CDA14 was compared with CDO14 at the same concentration.
Effects on the MMP
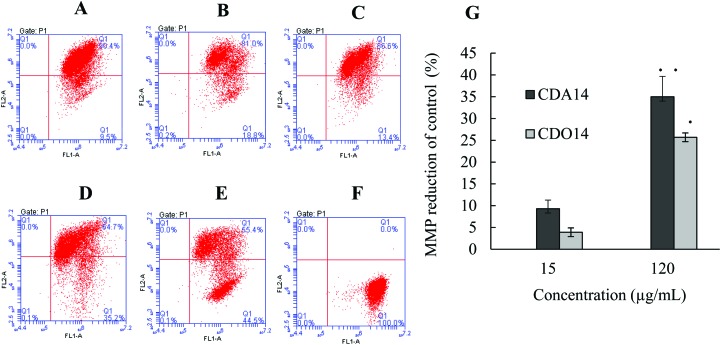
The mitochondrial membrane potential (MMP) reduction is one of the important causes leading to cell apoptosis.26 The MMP was measured using tetrachloro-tetraethyl benzimidazol carbocyanine iodide (JC-1) by flow cytometry after NCI-H460 cells were treated with CDA14 and CDO14 liposomes for 24 h, and untreated cells served as a negative control and cells treated with an apoptosis inducer as a positive control (Fig. 7A–F). The results showed that the MMP decreased about 10% and 5% at 15 μg mL–1, and decreased about 35% and 25% at 120 μg mL–1 by CDA14 and CDO14 liposome treatment compared with the control, respectively (Fig. 7G). Therefore, CDA14 and CDO14 liposomes could induce MMP reduction, causing damage to mitochondrial function. Obviously, CDA14 induced more MMP reduction than CDO14. These results were consistent with CDA14 and CDO14 induced cell apoptosis. MMP reduction is also involved in oxidative stress, and may result in reactive oxygen species (ROS) release from mitochondria.27,28
Fig. 7. Effects of the cationic liposomes on the MMP of NCI-H460 cells. (A) Negative control, (B) CDA14 liposomes at 15 μg mL–1, (C) CDO14 liposomes at 15 μg mL–1, (D) CDA14 liposomes at 120 μg mL–1, (E) CDO14 liposomes at 120 μg mL–1, (F) positive control, (G) MMP was detected based on JC-1 measurements. *P < 0.05 and **P < 0.01, CDA14 and CDO14 were compared with the control.
ROS generation in NCI-H460 cells
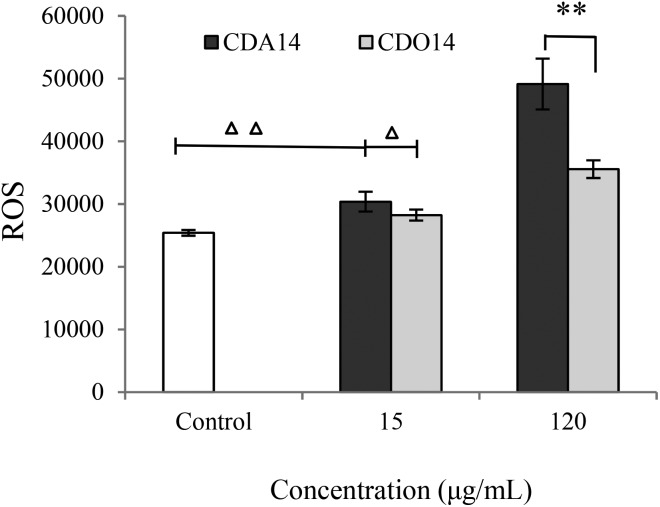
ROS is dynamically equilibrated between antioxidant defense mechanisms and cytotoxic responses leading to cell apoptosis.29–31 In particular, ROS generation could trigger a wide range of transcriptional changes as the second messengers in a signaling cascade, and it might be recovered after leading to apoptosis or cell death.32–36 In this study, the level of ROS production was detected after treatment of NCI-H460 cells with CDA14 and CDO14 liposomes for 24 h. The results showed that the intracellular ROS level was dose-dependent and increased significantly compared with the control in NCI-H460 cells. The ROS production of CDA14 was higher than that of CDO14 at the same concentrations; in particular, the ROS production of CDA14 increased remarkably at 120 μg mL–1, and was significantly higher than that of CDO14 (Fig. 8). These results showed that CDA14 and CDO14 liposomes could activate ROS, and the increase of the ROS facilitated apoptosis in NCI-H460 cells.
Fig. 8. Intracellular ROS levels by different concentrations of CDA14 and CDO14 liposomes. ROS levels were detected based on fluorescence DCF-DA measurements. Data are shown as the mean ± SD of triplicate experiments. △P < 0.05 and △△P < 0.01, CDA14 and CDO14 were compared with the control. *P < 0.05 and **P < 0.01, CDA14 were compared with CDO14 at the same concentration.
Effects on the cell cycle
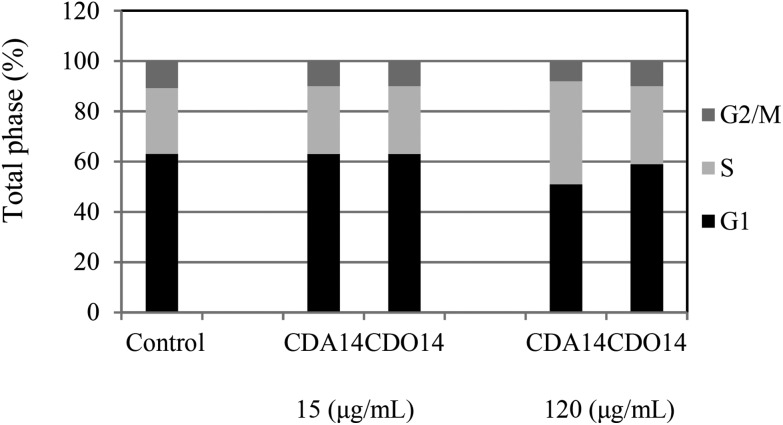
The influence of monitoring on cell cycle change is important for the development of drug delivery vectors,37–39 and a few studies have reported that cell cycle arrest may lead to the induction of apoptosis.40 Therefore, we investigated the effects of the cationic lipids on the cell cycle of NCI-H460 cells using flow cytometry after treatment with CDA14 and CDO14 for 24 h. At 15 μg mL–1 the two liposomes did not have any effects on the cell cycle phase. When the concentrations of liposomes increased to 120 μg mL–1, the sub-S population of CDA14 and CDO14 increased 1.56-fold and 1.18-fold, respectively (Fig. 9). The results showed that CDA14 and CDO14 liposomes with high concentrations could arrest more NCI-H460 cells at S phase. In contrast, at the concentration of 120 μg mL–1 CDA14 arrested about 10% more cells than CDO14. Meanwhile, G1 and G2/M phases were correspondingly shorter, leading to the inhibition of cell proliferation. The result indicated that ROS production could cause the DNA damage and the cell cycle arrest, and then induced apoptosis.
Fig. 9. Effects of CDA14 and CDO14 liposomes on the NCI-H460 cell cycle. G1 ■; S  ; G2/M
; G2/M  .
.
Conclusion
This study aims at clarifying the correlation of cytotoxic effects of cationic lipids with their headgroup structures. The results indicated that CDA14 and CDO14 showed less cytotoxicity at 15 μg mL–1 (transfection concentration); however, at higher concentrations (bigger than 120 μg mL–1), CDA14 showed obviously higher cytotoxicity than CDO14. We have proved that, compared with CDO14, CDA14 could induce more MMP reduction and ROS release, causing stronger activation of caspase enzymes and cell cycle arrest at S phase and subsequently more apoptosis of NCI-H460 cells. Our results confirmed that the cytotoxic effects of the quaternary ammonium headgroup of CDA14 are much higher compared with the tri-peptide headgroup of CDO14. This investigation suggested that the cytotoxic effects of cationic lipids are directly correlated with their headgroup structures. The peptide headgroup based cationic lipids having excellent biocompatibility showed superiority over other cationic lipids and have potential applications in improving the delivery of gene therapeutics.
Conflicts of interest
There are no conflicts of interest to declare.
Acknowledgments
The study was supported by the National Natural Science Foundation of China (21776044/21606041/21503035).
References
- Thomas C. E., Ehrhardt A., Kay M. A., Nat. Rev. Genet., 2003, 4 , 346 –358 , . [PubMed: 12728277] . [DOI] [PubMed] [Google Scholar]
- Aneed A. E., J. Controlled Release, 2004, 94 , 1 –14 , . [PubMed: 14684267] . [DOI] [PubMed] [Google Scholar]
- Sakurai F., Nishioka T., Saito H., Gene Ther., 2001, 8 , 677 –686 , . [PubMed: 11406762] . [DOI] [PubMed] [Google Scholar]
- Zhang S. B, Xu Y. M., Wang B., J. Controlled Release, 2004, 100 , 165 –180 , . [PubMed: 15544865] . [DOI] [PubMed] [Google Scholar]
- Zhi D. F., Zhang S. B., Qureshi F., Med. Chem. Lett., 2012, 22 , 3837 –3841 , . [PubMed: 22542011] . [DOI] [PubMed] [Google Scholar]
- Tokatlian T., Cam C., Segura T., Biomaterials, 2014, 35 , 825 –835 , . [PubMed: 24210142] . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foldvari M., Chen D. W., Nafissi N., J. Controlled Release, 2016, 240 , 165 –190 , . [PubMed: 26686079] . [DOI] [PubMed] [Google Scholar]
- Felgner J. H., Kumar R., Sridhar C. N., J. Biol. Chem., 1994, 269 , 2550 –2561 , . [PubMed: 8300583] . [PubMed] [Google Scholar]
- Niculescu D. D., Heyes J., Springer J. C., Curr. Med. Chem., 2003, 10 , 1233 –1261 , . [PubMed: 12678797] . [DOI] [PubMed] [Google Scholar]
- Zhou J. B., Liu J., Cheng C. J., Nat. Mater., 2012, 11 , 82 –90 , . [PubMed:22138789] . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. W., Xing R. R., Jiao T. F., ACS Appl. Mater. Interfaces, 2018, 10 , 2368 –2376 , . [PubMed: 29285927] . [DOI] [PubMed] [Google Scholar]
- Zhang R., Xing R., Jiao T., ACS Appl. Mater. Interfaces, 2016, 821 , 13262 –13269 , . [PubMed: 27176934] . [DOI] [PubMed] [Google Scholar]
- Huo S., Duan P., Jiao T., Angew. Chem., Int. Ed., 2017, 56 , 12174 –12178 , . [PubMed: 28759134] . [DOI] [PubMed] [Google Scholar]
- Bennett M. J., Aberle A. M., Balasubramaniam R. P., J. Med. Chem., 1997, 40 , 4069 –4078 , . [PubMed: 9406597] . [DOI] [PubMed] [Google Scholar]
- Lv H. T., Zhang S. B., Wan B., J. Controlled Release, 2006, 114 , 100 –109 , . [PubMed: 19699243] . [DOI] [PubMed] [Google Scholar]
- Lonez C., Vandenbranden M., Ruysschaert J. M., Adv. Drug Delivery Rev., 2012, 64 , 1749 –1758 , . [PubMed: 22634161] . [DOI] [PubMed] [Google Scholar]
- Zhi D. F., Zhang S. B., Wang B., Bioconjugate Chem., 2010, 21 , 563 –577 , . [PubMed:20121120] . [DOI] [PubMed] [Google Scholar]
- Zhi D. F., Zhang S. B., Cui S. H., Bioconjugate Chem., 2013, 24 , 487 –519 , . [PubMed:23461774] . [DOI] [PubMed] [Google Scholar]
- Zhao Y. N., Zhang S. B., Cui S. H., Expert Opin. Drug Delivery, 2012, 9 , 127 –139 , . [PubMed: 22103714] . [DOI] [PubMed] [Google Scholar]
- Zhao Y. N., Qureshi F., Zhang S. B., J. Mater. Chem. B, 2014, 2 , 2920 –2928 , . [PubMed: 25580248] . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. N., Zhang S. B., Zhang Y., J. Mater. Chem. B, 2015, 3 , 119 –126 , . [PubMed: 25580248] . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B. Z., Meng F. Y., Zhu S. L., Toxicol. Lett., 2010, 2 , 173 –178 , . [PubMed: 20079407] . [DOI] [PubMed] [Google Scholar]
- Liu X. Y., Yang Z. H., Pan X. J., Toxicol. Lett., 2010, 195 , 90 –98 , . [PubMed: 20153411] . [DOI] [PubMed] [Google Scholar]
- Xu T., Pang Q. Y., Zhou D., PLoS One, 2014, 8 , 1 –11 , . [PubMed: 25148076] . [Google Scholar]
- Brentnall M., Rodriguez-Menocal L., Guevara R. L. D., BMC Cell Biol., 2013, 14 , 32 –41 , . [PubMed: 23834359] . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W. W., Yuan L. H., Yu H. L., Life Sci., 2014, 110 , 53 –60 , . [PubMed: 25058918] . [DOI] [PubMed] [Google Scholar]
- Kongkaneramit L., Sarisuta N., Azad N., J. Pharmacol. Exp. Ther., 2008, 325 , 969 –977 , . [PubMed: 18354056] . [DOI] [PubMed] [Google Scholar]
- Gao S. Y., Gong Y. F., Sun Q. J., Molecules, 2015, 20 , 4290 –4306 , . [PubMed: 25756649] . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T., Kovochich M., Brant J., Nano Lett., 2006, 6 , 1794 –1807 , . [PubMed: 16895376] . [DOI] [PubMed] [Google Scholar]
- Kumar A., Pandey A. K., Singh S. S., Biol. Med., 2011, 51 , 1872 –1881 , . [PubMed: 21920432] . [DOI] [PubMed] [Google Scholar]
- Li J. X., Chang X. L., Chen X. X., Biotechnol. Adv., 2014, 32 , 727 –743 , . [PubMed: 24389087] . [DOI] [PubMed] [Google Scholar]
- Eom H. J., Choi J., Toxicol. Lett., 2009, 187 , 77 –83 , . [PubMed: 19429248] . [DOI] [PubMed] [Google Scholar]
- Yang X. F., He C., Li J., Toxicol. Lett., 2014, 229 , 240 –249 , . [PubMed: 24831964] . [DOI] [PubMed] [Google Scholar]
- AlGhamdi S., Leoncikas V., Plant K. E., Toxicol. Res., 2015, 46 , 1479 –1487 , . [PubMed: 26744621] . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C. S., Torres L., Miyauchi J. T., Toxicol. Res., 2016, 53 , 836 –847 , . [PubMed: 27274811] . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. M., Ma K., Jiao T. F., Sci. Rep., 2017, 7 , 42978 –42984 , . [PubMed: 5322353] . [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Huang C., Sun X., Cell. Signalling, 2012, 24 , 1923 –1930 , . [PubMed: 22735812] . [DOI] [PubMed] [Google Scholar]
- Hetheringtona A. M., Sawyez C. G., Zilberman E., Cell. Physiol. Biochem., 2016, 39 , 1648 –1662 , . [PubMed: 27626926] . [DOI] [PubMed] [Google Scholar]
- Tan H. X., Gao S.Y., Zhuang Y., Mar. Drugs, 2016, 14 , 166 –176 , . [PubMed: 27626431] . [Google Scholar]
- García V., Chica M. L., Cantarero I., Oncotarget, 2015, 7 , 4490 –4506 , . [PubMed: 26683224] . [DOI] [PMC free article] [PubMed] [Google Scholar]