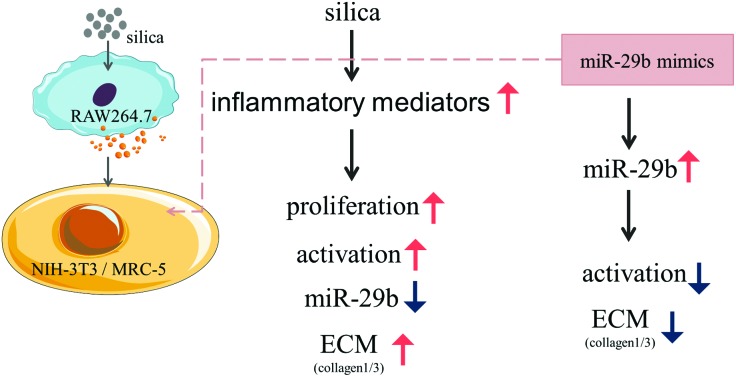
 Supernatants from silica-treated macrophages induced the lung fibroblasts proliferation, miR-29b reduced and extracellular matrix synthesis increased, which could be markedly inhibited by overexpression of miR-29b.
Supernatants from silica-treated macrophages induced the lung fibroblasts proliferation, miR-29b reduced and extracellular matrix synthesis increased, which could be markedly inhibited by overexpression of miR-29b.
Abstract
Silicosis is pathologically characterized by diffused pulmonary fibrosis and abundant deposition of extracellular matrix (ECM) components. The ECM is mainly secreted by myofibroblasts which are the activated state of fibroblasts. MicroRNA-29b (miR-29b) is one of the well-known microRNAs involved in fibrosis, but its roles in silicosis have not been specified. In this study, we hypothesized that miR-29b might play a protective role in the progression of silicosis. MTT assay, qRT-PCR, immunofluorescence and western blotting were applied. The results demonstrated that the supernatants from silica-treated macrophages not only caused the proliferation of fibroblasts (NIH-3T3 and MRC-5) but were also involved in the down-regulation of miR-29b. Meanwhile they could induce fibroblast activation, increasing the expression of ECM components such as collagen1 and collagen3, in a silica dose-dependent manner. Furthermore, overexpression of miR-29b by transfecting mimics markedly reduced the expression of ECM components and inhibited ECM synthesis. These findings indicate that miR-29b inhibits the supernatants from silica-treated macrophages from inducing extracellular matrix synthesis, thus miR-29b might have a strong anti-fibrotic capacity in silicosis and serve as a potential therapeutic agent for the treatment.
Introduction
Silicosis is a worldwide occupational lung disease caused by long-term excessive silica inhalation, which is pathologically characterized by a persistent inflammatory response and progressive pulmonary fibrosis.1,2 In this day and age, large quantities of industrial workers from all over the world have direct or indirect occupational exposure to inhalable silica particles. More than 20 000 new individuals are affected by silicosis in China every year.3 Unfortunately, there are no available effective medications to block or reverse the progression of pulmonary fibrosis induced by silica. Thus, it is significant to search for effective therapy targets in order to delay the progression of fibrosis, prolong patients’ lives and improve their quality of life.
Alveolar macrophages and pulmonary fibroblasts have been identified in a previous study as playing a central role in the progression of silicosis.4 Alveolar macrophages are the most important immune barrier against the invasion of environmental contaminants and pathogens in pulmonary innate immunity, which are key drivers of fibrogenesis. Macrophages have been found to be involved in the production of collagen by myofibroblasts.5 When silica particles are inhaled in the alveoli, they are captured and eliminated by alveolar macrophages. Once silica particles are ingested, macrophages could be activated and release some inflammatory mediators, such as reactive oxygen species, reactive nitrogen, chemokines, cytokines, and growth factors.6,7 These substances may impair pulmonary tissues, and stimulate fibroblast proliferation and differentiation into myofibroblasts. The primary pathological characteristic of silicosis is the excessive extracellular matrix (ECM) protein deposition, which is mainly secreted by myofibroblasts. Multiple genes and tissue microarrays have demonstrated that the matrix consists of the major fibrillar collagens (collagen1, collagen3 and collagen4) and fibronectin.8 Fibronectin is a macromolecular glycoprotein with high adhesion activity, which connects cells and collagens. In addition, fibronectin is one of the fibroblast chemokines that attract fibroblasts to accumulate and proliferate into the wound. Therefore, the key molecules inhibiting ECM component formation are the targets for the treatment of silicosis.
MicroRNAs (miRNAs) are endogenous small non-coding RNAs which can bind to the 3′untranslated region (3′UTR) of target mRNAs through complementary base-pairing, playing the role of post-transcriptional regulators of gene expression.9–11 MiRNAs have been involved in multiple biological processes. With the in-depth study of miRNAs, more and more evidence has indicated that deregulation of miRNAs participates in the progression of fibrosis in different tissues including the liver,12 kidney13 and myocardium.14 MicroRNA-29 (miR-29) is one of the known miRNAs involved in fibrogenesis, since many ECM related genes are the targets of miR-29, including collagen1, collagen3, collagen4, elastin and fibronectin, etc.15 In addition, miR-29b plays a protective role in angiotensin (Ang)II-mediated cardiac fibrosis by targeting the transforming growth factor (TGF)-β/Smad3 pathway.16 Furthermore, studies have shown that over-expression of miR-29b could inhibit bleomycin-induced pulmonary fibrosis in mice.17,18 However, a fundamental question as to whether or not miR-29b function is pivotal in the process of silicosis remains unexplored.
In this study, we hypothesized that miR-29b might play a protective role in the progression of silicosis. Because macrophages and fibroblasts are known as central effector cells in fibrosis, we used macrophages (Raw264.7), fibroblasts (NIH-3T3 and MRC-5) and silica to form cell models. We investigated the expression levels of ECM related genes and proteins in lung fibroblasts, which were exposed to different doses of silica-induced macrophage supernatants. Meanwhile, we used well-validated miR-29b mimics to determine whether or not miR-29b has the effect of decreasing the ECM-related mRNA and protein production.
Methods
Silica particles
The crystalline silica particles were provided by Sigma-Aldrich Co. LLC, and had a nominal size of 0.5–1.0 μm (approx. 80% between 1–5 μm). Transmission electron microscopy (TEM, JEOL, JEM-2100) and scanning electron microscopy (SEM, HITACHI, S-4800) were utilized to assess the morphology of the particles; these results have been published previously.19 The stock solution was prepared in phosphate-buffered saline (PBS), at a final silica concentration of 2 mg mL–1, by sonication. It was kept at 4 °C and used within 1 week.
Cell culture and treatment
Murine macrophage cells (Raw264.7) were purchased from Xiang Ya Central Experiment Laboratory, China, and were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Hyclone) and 1% penicillin–streptomycin solution (KeyGen Biotech, China). Mouse embryonic fibroblast cells (NIH-3T3), obtained from the China Infrastructure of Cell Line Resources, were cultured in DMEM (Hyclone) with 10% FBS (Hyclone), and 1% penicillin–streptomycin solution (KeyGen Biotech, China). Human fetal lung fibroblast cells (MRC-5), purchased from the Cell Culture Center of Peking Union Medical College China, were cultured in minimal essential medium (MEM, Hyclone) with 10% FBS (Hyclone), 1% non-essential amino acids (NEAA, Gibco) and 1% penicillin–streptomycin solution (KeyGen Biotech, China). All cells were maintained in an incubator at 37 °C with 5% CO2. All experiments were carried out using the third-passage cells.
When Raw264.7 cells had grown to 70–80% confluence, the medium was replaced with 0, 12.5, 25, 50, 100 or 200 μg mL–1 DMEM and silica solution, free of serum. The supernatants were collected and filtrated through 0.22 μm microporous membrane strainers after 24 h.
NIH-3T3 and MRC-5 cells were both grown to 70–80% confluence. They were treated with DMEM for the negative control (NC) groups. NIH-3T3 cells were also treated with the supernatants collected from Raw264.7 cells exposed to 0, 25, 50, 100 or 200 μg mL–1 silica solutions (silica 0, 25, 50, 100 and 200 μg mL–1 groups respectively), while MRC-5 cells were treated with the supernatants collected from Raw264.7 cells exposed to 0, 12.5, 25, 50 or 100 μg mL–1 silica solutions (silica 0, 12.5, 25, 50 and 100 μg mL–1 groups respectively). NIH-3T3 and MRC-5 cells were the subjects of the further analysis.
MTT assay
MTT ((4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide) assays were performed to measure the cell viability and proliferation. Labeled cells were incubated in MTT for 4 h at 37 °C, then DMSO solution was added, and the cells were shaken using a 37 °C swing bed away from light. The plates were read on a microplate reader photometer at a wavelength of 492 nm. All MTT assays were performed five times.
Immunofluorescence
NIH-3T3 and MRC-5 cells grown on chamber slides, were exposed to supernatant and (or) transfected with mimics etc. The slides were washed twice with PBS and the cells were fixed in 4% paraformaldehyde for 20 min, then 0.3% Triton X-100 for 10 min at room temperature (RT), washed again with PBS and blocked with 5% bovine serum albumin (BSA) at RT for 10 min. Then, the cells were incubated overnight at 4 °C with primary antibodies against collagen1 (1 : 500, Abcam, CA, USA) or collagen3 (1 : 100, Abcam, CA, USA). The next day the slides were washed with PBS and incubated sequentially with goat anti-rabbit antibodies (CST, 4412s) conjugated to fluorescein isothiocyanate (FITC) for 1 h at RT. 4′,6-Diamidino-2-phenylindole (DAPI) was used for nuclear staining. Immunofluorescence staining was examined by laser scanning confocal fluorescence microscopy for NIH-3T3 cells and with an Eclipse 80i microscope (Nikon, Tokyo, Japan) for MRC-5 cells.
Quantitative real-time RT-PCR
Total RNA was extracted from cells using TransZol Up (ET111, TransGen Biotech, Beijing, China), and transcribed to cDNA from mRNA using the TransScript First-Strand cDNA Synthesis SuperMix (AT301, TransGen Biotech, Beijing, China). The cDNA of miRNA was obtained using the TransScript miRNA First-Strand cDNA Synthesis SuperMix (AT351, TransGen Biotech, Beijing, China). qRT-PCR was performed with the CFX96™ real-time quantitative PCR detection system (BioRad Inc., Hercules, CA), using the SYBR Green qRT-PCR kit (AQ141, TransGen Biotech, Beijing, China). The gene expression levels were determined by normalizing to glyceraldehyde-3-phosphate dehydrogenase using the 2–ΔΔCT method. The primer sequences are listed below.
| Primers for NIH-3T3 | |
| GAPDH F | AAGAAGGTGGTGAAGCAGGC |
| GAPDH R | TCCACCACCCTGTTGCTGTA |
| Collagen1 F | GCTCCTCTTAGGGGCCACT |
| Collagen1 R | CCACGTCTCACCATTGGGG |
| Collagen3 F | CCTGGCTCAAATGGCTCAC |
| Collagen3 R | CAGGACTGCCGTTATTCCCG |
| Fibronectin F | CATTCCTGTGGGGATGGATTC |
| Fibronectin R | TACGTGCAAGCACACCGATTT |
| Collagen4 F | AGGCGAAATGGGTATGATGGG |
| Collagen4 R | CTCCCTTACCGCCCTTTTCTC |
| TIMP-1 F | GCAACTCGGACCTGGTCATAA |
| TIMP-1 R | CGGCCCGTGATGAGAAACT |
| MiR-29b F | ATCGTGGTAAACTTTAGTCACAA |
| U6 F | CTCGCTTCGGCAGCACA |
| Primers for MRC-5 | |
| GAPDH F | TCAACGACCACTTTGTCAAGCT |
| GAPDH R | CCATGAGGTCCACCACCCT |
| Collagen1 F | CCCAAGGACAAGAGGCATGT |
| Collagen1 R | CCGCCATACTCGAACTGGAA |
| Collagen3 F | TGGACAGATTCTAGTGCTGAGAAGA |
| Collagen3 R | TTGCCGTAGCTAAACTGAAAACC |
| Fibronectin F | TCTCCTGCCTGGTACAGAATATGTAGTGAG |
| Fibronectin R | GGTCGCAGCAACAACTTCCAGGT |
| Collagen4 F | CCAAGGAAGAGGTGGTGTGT |
| Collagen4 R | GTGCTTCACCAGGAGGTAGC |
| MiR-29b F | ATCGTGGTAAACTTTAGTCACAA |
| U6 F | CTCGCTTCGGCAGCACA |
MiRNA random primers used the products in the TransScript miRNA first-strand cDNA synthesis SuperMix (AT351, TransGen Biotech, Beijing, China).
Western blotting
Protein extraction was performed with Kit KGP2100 (KeyGen Biotech, China). The cells were washed three times with ice-cold PBS, and disrupted in lysis buffer. The concentration was assayed by using bicinchoninic acid (BCA, Thermo Scientific, Rockford, IL, USA). Cell extracts were separated in 15% SDS polyacrylamide gels, and transferred onto PVDF membranes (Millipore, Billerica, MA, USA), which were then blocked in 5% BSA solution and incubated overnight at 4 °C with primary antibodies against collagen1 (ab34710, Abcam; 1 : 5000), collagen3 (ab7778, Abcam; 1 : 5000), or GAPDH (#2118, CST; 1 : 1000). Anti-rabbit IgG, HRP-linked antibody (#7074, CST; 1 : 1000) was used as the secondary antibody. Proteins of interest were detected using the enhanced chemiluminescence method (ECL, Thermo). Western blots were scanned using a Tanon-5200 system (Beijing Yuan Ping Hao Biotech Co. Ltd) and the quantification was performed with Image J software, analyzing the intensity of the grey scale images.
MicroRNA transfection
NIH-3T3 and MRC-5 cells were transfected with double stranded miR-29b mimics and scramble siRNA (Sangong Biotech, Shanghai, China) using a transfection reagent and Opti-MEM Medium (Lipofectamine® RNAi MAX Reagent and Opti-MEM® Medium, Life Technologies, China) according to the manufacturer's instructions. The cells were replanted with a full-growth medium and cultured to approximately 60% confluence. Lipofectamine RNAiMAX Reagent and miRNA mimics/scramble were separately diluted in Opti-MEM Medium, after which the diluted miRNA was added to the diluted reagent (1 : 1 ratio), incubated for 5 minutes at room temperature, added to the cells (final concentration 50 nM per well) and incubated at 37 °C. The mimics were made with the chemical modification Cy3. After incubation, the cells were fixed in 4% paraformaldehyde for 20 min, and DAPI was used for nuclear staining. The transfection efficiency was detected with laser scanning confocal fluorescence microscopy for the NIH-3T3 cells. The MRC-5 cells were photographed with an Eclipse 80i microscope after incubation (Nikon, Tokyo, Japan).
Statistics
All the data were assessed by SPSS 20.0 software, using at least three independent experiments. Statistical differences between individual variables were analyzed by one-way analysis of variance (ANOVA). P < 0.05 values were considered as statistically significant for group differences. Data are presented as means ± standard deviation (means ± SD).
Results
Supernatants from silica-treated macrophages induced the proliferation of NIH-3T3 and MRC-5 cells
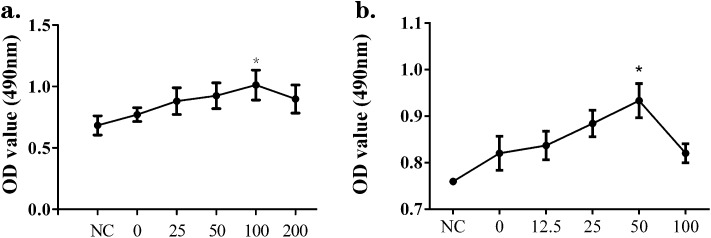
To investigate the proliferative effects of supernatants from silica-treated macrophages acting on NIH-3T3 and MRC-5 cells, MTT assays were performed. NIH-3T3 were treated with DMEM for the negative control (NC) group, or with the supernatants collected from Raw264.7 that had been exposed to 0, 25, 50, 100 or 200 μg mL–1 silica solutions (abbreviated as silica 0, 25, 50, 100 and 200 μg mL–1 groups, respectively). MRC-5 were treated with DMEM for the negative control (NC) group, or with the supernatants collected from Raw264.7 that had been exposed to 0, 12.5, 25, 50 or 100 μg mL–1 silica solutions (abbreviated as silica 0, 12.5, 25, 50 and 100 μg mL–1 groups, respectively). In the case of the NIH-3T3 cells, the peak value and most significant proliferation compared with the NC group were shown in the silica 100 μg mL–1 group, while for MRC-5, these appeared in the silica 50 μg mL–1 group (Fig. 1). Thus, the supernatants from the silica-treated macrophages had effects on both NIH-3T3 and MRC-5 cells leading to a proliferation tendency, although there was no statistical significance in many groups.
Fig. 1. Supernatants from silica-treated macrophages induced the proliferation of NIH-3T3 and MRC-5 cells. Raw264.7 cells were treated with different concentrations of silica for 24 h and the supernatants were used to culture fibroblasts. NIH-3T3 cells were cultured in DMEM (NC) or the supernatants collected from Raw264.7 cells exposed to 0, 25, 50, 100 or 200 μg mL–1 silica solution for 24 h. MRC-5 cells were cultured in DMEM (NC) or the supernatants collected from Raw264.7 cells exposed to 0, 12.5, 25, 50 or 100 μg mL–1 silica solution for 24 h. The MTT assay was performed to measure cell proliferation. In the case of NIH-3T3 (a), the peak value and significant proliferation compared with the NC group were shown in the silica 100 μg mL–1 group, while for MRC-5 (b) these appeared in the silica 50 μg mL–1 group. The results show data from five repeated tests. The data are presented as means ± SD. The significance of differences was determined by ANOVA followed by the Dunnett's test; *p < 0.05 versus the NC group.
Effects of supernatants from silica-treated macrophages on the activation of NIH-3T3 and MRC-5 cells
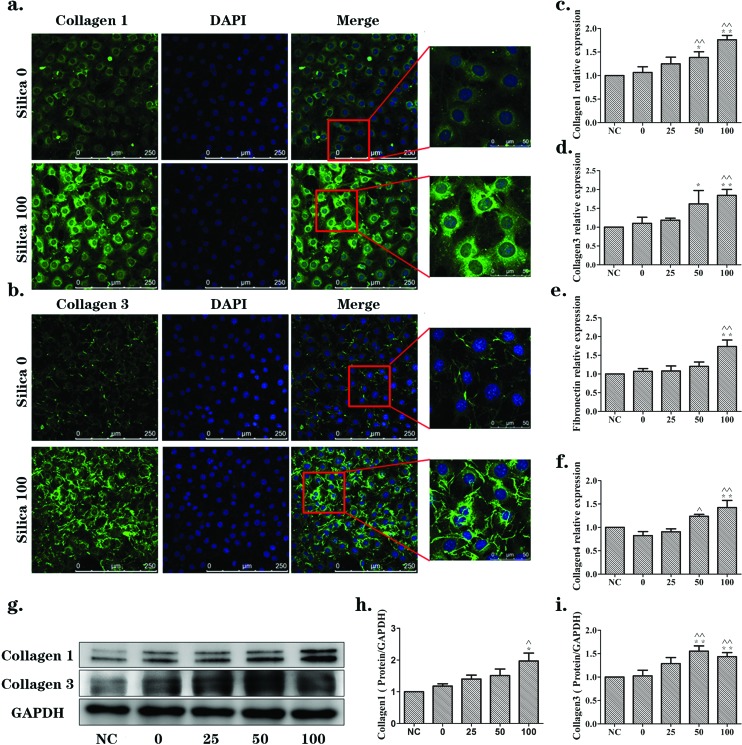
The up-regulated expression of ECM proteins is considered to be a sign of fibroblast activation. To explore whether the supernatants from silica-treated macrophages could lead to fibroblast activation, immunofluorescence, qRT-PCR and western blotting were carried out. In NIH-3T3 cells, the immunofluorescence results (Fig. 2(a) and (b)) showed that there was obviously up-regulated expression of collagen1 and collagen3 (green) in the silica 100 μg mL–1 group when compared with the silica 0 μg mL–1 group. The mRNA levels of collagen1, collagen3, fibronectin and collagen4 were evaluated by qRT-PCR assay, as shown in Fig. 2(c–f). There were increasing trends with statistically significant ascension peaks for the silica 50 and/or 100 μg mL–1 groups. Western blotting was also performed to detect the protein expression of collagen1 and collagen3. Consistent with the qRT-PCR results, collagen1 and collagen3 proteins were significantly elevated in the silica 50 and/or 100 μg mL–1 groups (Fig. 2(g–i)). The expression levels were all increased in a silica dose-dependent manner.
Fig. 2. Supernatants from silica-treated macrophages promoted changes in ECM components in a dose-dependent manner in NIH-3T3 cells. NIH-3T3 cells were cultured in DMEM (NC) or the supernatants collected from Raw264.7 cells exposed to 0, 25, 50, 100 or 200 μg mL–1 silica solution for 24 h. (a, b) Immunofluorescence staining of collagen1 and collagen3 (green), respectively, in silica 0 and silica 100 μg mL–1 groups. Collagen1 and collagen3 were mainly expressed in the cytoplasm. They were significantly increased in the silica 100 μg mL–1 group. (c, d, e, f) qRT-PCR analyses of mRNA expression levels of collagen1, collagen3, fibronectin and collagen4, respectively. mRNA levels were increased in a silica dose-dependent manner. The significant changes were observed in the silica 50 or 100 μg mL–1 groups. (g, h, i) Western blotting analyses of the protein expression levels of collagen1 and collagen3. Protein levels were increased in a silica dose-dependent manner as well. The data are presented as the means ± SD. *p < 0.05 versus the NC group, **p < 0.01 versus the NC group, ^p < 0.05 versus the silica 0 group, ^^p < 0.01 versus the silica 0 group.
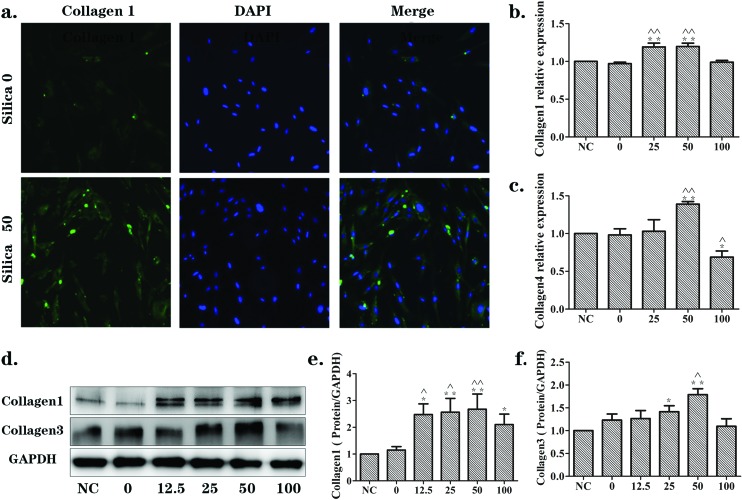
Furthermore, the results were paralleled in MRC-5 cells. The immunofluorescence results (Fig. 3(a)) showed that there was a markedly higher level of collagen1 (green) in the silica 50 μg mL–1 group when compared with the silica 0 μg mL–1 group. The mRNA levels of collagen1 and collagen4 increased in the silica 25 and/or 50 μg mL–1 groups (Fig. 3(b and c)). The protein levels of collagen1 and collagen3, discovered by western blotting, were significantly up-regulated in a silica dose-dependent manner (Fig. 3(d–f)). These results illustrate the activation effects of the supernatants from silica-treated macrophages on NIH-3T3 and MRC-5 cells.
Fig. 3. Supernatants from silica-treated macrophages promoted changes in ECM components in a dose-dependent manner in MRC-5 cells. MRC-5 cells were cultured in DMEM (NC) or the supernatants collected from Raw264.7 cells exposed to 0, 12.5, 25, 50 or 100 μg mL–1 silica solution for 24 h. (a) Immunofluorescence staining of collagen1 (green) in silica 0 and silica 50 μg mL–1 groups. Collagen1, mainly expressed in cytoplasm, was significantly increased following induction by supernatants from silica-treated macrophages. (b, c) qRT-PCR analyses of mRNA expression levels of collagen1 and collagen3, respectively. mRNA levels were up-regulated in a silica dose-dependent manner. The significant changes emerged in the silica 50 μg mL–1 group. (d, e, f) Western blotting analyses of the protein expression levels of collagen1 and collagen3. Protein levels were increased in a silica dose-dependent manner as well. The data are presented as the means ± SD. *p < 0.05 versus the NC group, **p < 0.01 versus the NC group, ^p < 0.05 versus the silica 0 group, ^^p < 0.01 versus the silica 0 group.
Effects of supernatants from silica-treated macrophages on the expression of miR-29b in NIH-3T3 and MRC-5 cells
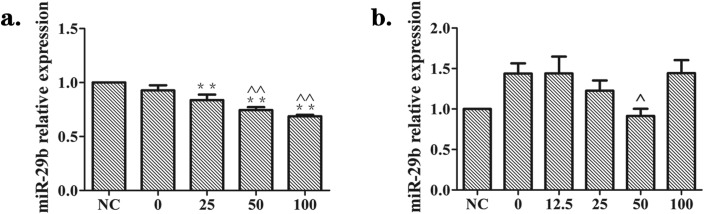
To identify whether supernatants from silica-treated macrophages regulated the level of miR-29b in NIH-3T3 and MRC-5 cells, we used miRNA qRT-PCR in repeats of the experiments described above. There was a significant and dose-dependent decrease of miR-29b in NIH-3T3 cells, and the lowest level was found in the silica 100 μg mL–1 group (Fig. 4(a)). As for MRC-5 cells, the expression of miR-29b showed a statistically significant decline in the silica 50 μg mL–1 group in comparison to the silica 0 μg mL–1 group (Fig. 4(b)). These findings showed that supernatants from silica-treated macrophages resulted in the decrease of miR-29b in NIH-3T3 and MRC-5 cells.
Fig. 4. Effects of supernatants from silica-treated macrophages on the expression of miR-29b in NIH-3T3 and MRC-5 cells. Supernatants from silica-treated macrophages suppressed the expression of miR-29b in fibroblast cells, as measured by qRT-PCR. (a) NIH-3T3 cells were cultured in DMEM (NC) or the supernatants collected from Raw264.7 cells exposed to 0, 25, 50, 100 or 200 μg mL–1 silica solution for 24 h. (b) MRC-5 cells were cultured in DMEM (NC) or the supernatants collected from Raw264.7 cells exposed to 0, 12.5, 25, 50 or 100 μg mL–1 silica solution for 24 h. There was a significant and dose-dependent decline in the level of miR-29b in NIH-3T3 (a). In MRC-5 (b), miR-29b levels showed a statistically significant decline in the silica 50 μg mL–1 group versus the silica 0 μg mL–1 group. The data are presented as the means ± SD (n = 3). The significance of differences was determined by ANOVA followed by the Dunnett's test; *p < 0.05 versus the NC group, **p < 0.01 versus the NC group, ^p < 0.05 versus the silica 0 group, ^^p < 0.01 versus the silica 0 group.
MiR-29b inhibited the activation of NIH-3T3 and MRC-5 cells induced by supernatants from silica-treated macrophages
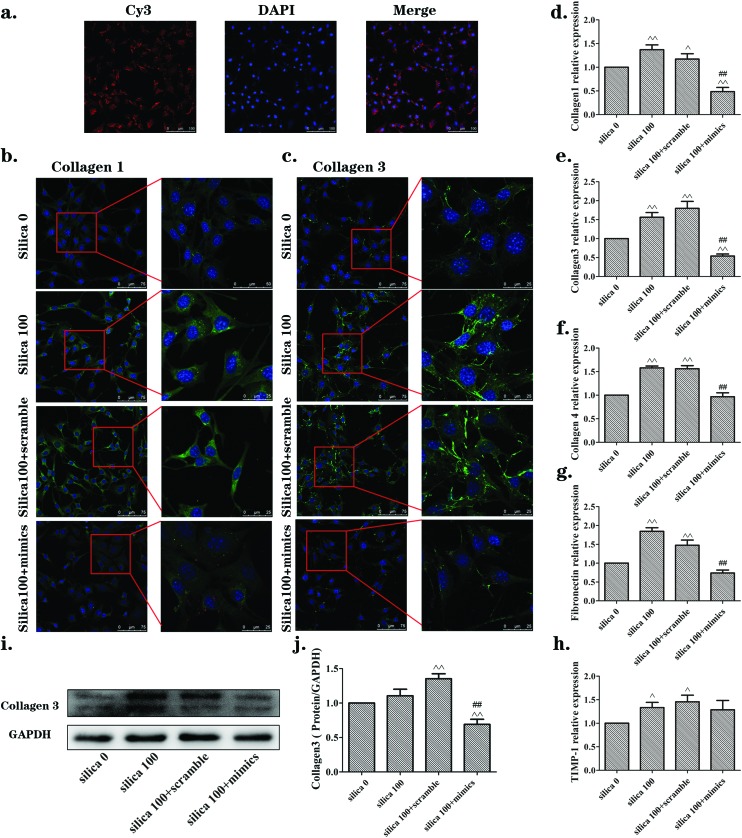
In view of the results above, the 100 μg mL–1 concentration of silica was chosen for further experiments with NIH-3T3 cells, and 50 μg mL–1 silica was chosen for MRC-5 cells. The above outcomes have demonstrated that miR-29b was reduced in NIH-3T3 and MRC-5 cells that were activated by the supernatants from silica-treated macrophages. We put forward a hypothesis that reinstating the miR-29b levels would inhibit the activation. To address this hypothesis, the cells were transfected for 24 h with either miR-29b mimics or scramble siRNA as a control, and then treated with Raw264.7 supernatants according to the protocol mentioned above. Firstly, we checked the transfection efficiency, owing to the fact that the mimics were made with the Cy3 chemical modification. NIH-3T3 cells were examined by laser scanning confocal fluorescence microscopy after 24 h incubation (Fig. 5(a)). The mimics were transfected into the NIH-3T3 cells as shown. Immunofluorescence, qRT-PCR and western blotting were carried out to examine the levels of activation of ECM components. As shown in Fig. 5(b and c), the expression of collagen1 and collagen3 were obviously up-regulated in both the silica 100 μg mL–1 and silica 100 μg/mL + scramble groups compared to those in the silica 0 μg mL–1 group, while the level of proteins in the silica 100 μg mL–1 + mimics group was markedly declined. These findings are consistent with the trends found in the qRT-PCR results for collagen1, collagen3, collagen4 and fibronectin (Fig. 5(d–g)) and in the western blotting results for collagen3 (Fig. 5(i and j)). However, the qRT-PCR result for the tissue inhibitor of metalloproteinase (TIMP)-1 was not markedly changed between silica 100 μg mL–1 group and silica 100 μg mL–1 + mimics group (Fig. 5(h)).
Fig. 5. The miR-29b inhibited the activation of NIH-3T3 cells induced by supernatants from silica-treated macrophages. (a) Transfection efficiency for NIH-3T3 cells checked by examination of Cy3 through laser scanning confocal fluorescence microscopy after 24 h incubation. (b, c) Immunofluorescence staining of collagen1 and collagen3 (green), respectively. Levels were obviously increased in silica 100 μg mL–1 and silica 100 + scramble groups, but were significantly reversed in the silica 100 + mimics group. (d, e, f, g, h) qRT-PCR results of mRNA expression levels of collagen 1, collagen3, collagen4 and fibronectin, respectively. mRNA levels were increased following induction by silica, while they were reversed in the silica 100 + mimics group. (i, j) Western blotting analyses of the protein expression levels of collagen3. The collagen3 protein levels were consistent with the qRT-PCR results. The data are presented as the means ± SD (n = 3). The significance of differences was determined by ANOVA followed by the Dunnett's test; ^p < 0.05 versus the silica 0 group, ^^p < 0.01 versus the silica 0 group, #p < 0.05 versus the silica 100 group, ##p < 0.01versus the silica 100 group.
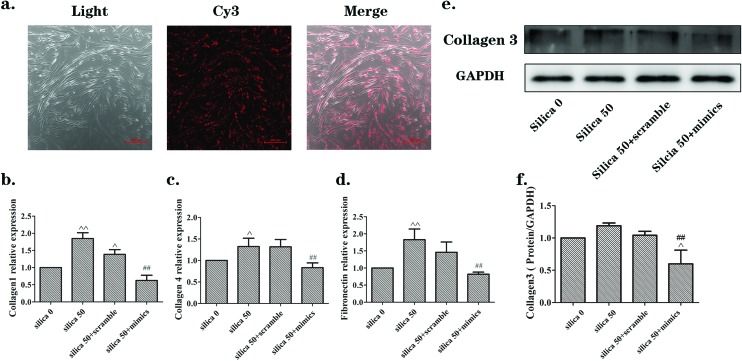
The Eclipse 80i microscope was used for MRC-5 cells (Fig. 6(a)). Cy3 indicated the mimics and was distributed in accordance with cell shape. The collagen3 protein measured by western blotting was decreased in the silica 50 μg mL–1 + mimics group, with statistical significance (Fig. 6(e and f)). The qRT-PCR results showed that the mRNA levels of collagen1, collagen4 and fibronectin increased in the silica 50 μg mL–1 group, while they decreased in the silica 50 μg/mL + mimics group (Fig. 6(b–d)). These results indicate that miR-29b inhibites the activation of NIH-3T3 and MRC-5 induced by the supernatants from silica-treated macrophages.
Fig. 6. The miR-29b inhibited the activation of MRC-5 cells induced by supernatants from silica-treated macrophages. (a) Transfection efficiency for MRC-5 cells checked by examination of Cy3 using an Eclipse 80i microscope after 24 h incubation. The Cy3 signals showed that mimics were distributed in keeping with cell shape. (b, c, d) qRT-PCR analyses of mRNA expression levels of collagen1, collagen4 and fibronectin, respectively. mRNA levels were increased in the silica 50 μg mL–1 group, while they were decreased in the silica 50 + mimics group. (e, f) Western blotting results of the protein expression levels of collagen3. The protein level was increased in the silica 50 μg mL–1 group while it was decreased in the silica 50 + mimics group. The data are presented as the means ± SD (n = 3). The significance of differences was determined by ANOVA followed by the Dunnett's test; ^p < 0.05 versus the silica 0 group, ^^p < 0.01 versus the silica 0 group, #p < 0.05 versus the silica 50 group, ##p < 0.01 versus the silica 50 group.
Discussion
Silicosis is a chronic lung disease caused by long-term silica exposure with no available effective medications. Therefore, it is critical to discover new targets against silicosis. The formation of silica-induced pulmonary fibrosis consists of a series of events: alveolar epithelial cell injury, fibroblast to myofibroblast differentiation, epithelial to mesenchymal transition, and deposition of ECM proteins in the lungs. Most of the silica particles inhaled into the pulmonary alveoli are swallowed by macrophages. These macrophages internalize the silica particles and produce pro-inflammatory cytokines, chemokines, and growth factors that are involved in the development of fibrosis, such as interleukin (IL)-1, IL-6, IL-18, tumor necrosis factor (TNF)-α, TGF-β and so on, resulting in fibroblast activation and causing recruitment of cells into the areas where silica particles are deposited.4,20,21 The macrophages and fibroblasts are the central effector cells in the progression of silicosis. Therefore, we used macrophages (Raw264.7 cells) exposed to silica solutions, and collected the supernatants of these silica-treated macrophages for culturing fibroblasts (NIH-3T3 and MRC-5 cells).
The progression of fibroblast activation and differentiation into myofibroblasts is defined by excess ECM synthesis.19 The ECM is a highly dynamic structure that is present in all organs and tissues, and interacts with cells to regulate diverse functions, including proliferation, migration and differentiation.22 Collagens are the main structural proteins of the ECM and are classified into both fibrillar (collagens1–3, 5 and 11) and non-fibrillar forms. Collagen fibrils provide tensile strength to the ECM. Fibronectin can connect cells and the ECM, but it also serves as the fibroblast chemokine, causing the cells to gather and proliferate. In our study, a cell model was successfully established in vitro. We found that NIH-3T3 and MRC-5 cells proliferated and showed increased expression of ECM component including collagen1, and collagen3, fibronectin and collagen4, in the groups which were cultured with the supernatants from silica-treated macrophages. The composition of the ECM, including collagens and fibronectin, is regulated by both transcriptional and post-transcriptional mechanisms.
In this study, we found that the level of miR-29b was reduced by exposure to the supernatants from silica-treated macrophages. MiR-29b belongs to the miR-29 family that is closely related to fibrotic diseases, and it plays an important role in the regulation of fibrosis in various organs with distinct etiologies.23–25 MiR-29b binds to target mRNA 3′UTR, and induces the suppression of target mRNA translation by post-transcriptional mechanisms. A large number of researchers have revealed that levels of miR-29b are significantly down-regulated in fibrotic disorders, while overexpression of miR-29b has an effect on restraining collagen1 and collagen3, etc.26,27 In research into hepatic fibrosis, miR-29b not only inhibited the secretion of collagens in hepatic astrocytes but was also involved in the regulation of many aspects of ECM maturation and inhibited the progression of hepatic fibrosis.27 Interestingly, in this study, we found that the miR-29b plays an inhibitory role in the progression of activation of NIH-3T3 and MRC-5 induced by supernatants of silica-treated macrophages. When we transfected cells with the miR-29b mimics, the mRNA and protein expression levels of collagens and other ECM related molecules were down-regulated. In this model, we also found that the miR-29b obstructed collagen translation by decomposing the collagen mRNA. The inhibitory effect of miR-29b on the ECM shows that miR-29b may serve as a key molecule in inhibiting the excess ECM protein formation induced by supernatants of silica-treated macrophages. Furthermore, miR-29b may play a protective role in the fibrosis progression of silicosis that will be tested and verified in a future in vivo experiment.
Extracellular matrix metalloproteinases (MMPs) play a crucial part in ECM protein degradation. Tissue inhibitors of metalloproteinase (TIMPs) are inhibitors of MMPs. ECM integrity is affected by MMPs and TIMPs that play crucial roles during matrix remodeling.28 TIMP-1, one of the four known members, can inhibit proteolytic activity of MMP.29 According to the current study, the fibrosis progression includes fibroblast generation, collagen secretion and deposition, and collagen degradation.28,30 MiR-29b-target mRNAs include collagen1, 3, 4 and so on, but not TIMP-1. We found out that the miR-29b inhibits the collagen generation caused by the supernatants from silica-treated macrophages, while the qRT-PCR result for TIMP-1 was not markedly changed (Fig. 5(h)). This might be because miR-29b does not regulate the expression of TIMP-1 in the silica-macrophage-induced activation of NIH-3T3 and MRC-5 cells. This is consistent with the fact that TIMP-1 mRNA is not a target of miR-29b according to the published papers15 and the results we got by performing Targetscan. Therefore, we have identified that the key role of miR-29b is the inhibition of the ECM synthesis rather than the promotion of collagen degradation.
Some drugs have been developed and approved for clinical use in the treatment of fibrotic disease, such as pirfenidone and nintedanib.31,32 Nevertheless, there are closely associated side effects of these drugs, such as gastrointestinal disturbances and liver function damage. In Phase III trials, pirfenidone improved the profile of disease progression and exhaustion of lung function and increased progression-free survival in patients with idiopathic pulmonary fibrosis (IPF).33 Nintedanib is a multiple tyrosine kinase inhibitor (TKI) targeting the receptor kinase(s) of vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF) and platelet-derived growth factor (PDGF).34 In a Phase III study, the small molecule led to a impairment of lung function, and inhibited physiological remodeling and the formation of blood vessels.35 The current state of affairs is that miRNA therapeutics are not yet proven.36,37 Research into the safety of miRNA intervention therapy found that delivery of miR-29 through the peripheral vasculature could increase the risk of an aneurysm.38 Therefore, in the subsequent study, we will focus on the upstream element of miR-29b to explore the upstream regulatory mechanism, with the hope of discovering a novel possibility in the treatment for silica-induced fibrosis.
In summary, we have demonstrated that miR-29b is a key molecule that can inhibit the activation of NIH-3T3 and MRC-5 cells, and the excessive ECM protein synthesis induced by supernatants from silica-treated macrophages. Furthermore, miR-29b is potentially a novel fibrosis inhibitor which could suppress the progression of silica-induced pulmonary fibrosis.
Conclusions
Our research suggests that miR-29b inhibits the supernatants from silica-treated macrophages from inducing ECM synthesis in lung fibroblasts. MiR-29b may have a strong anti-fibrotic capacity in silicosis and it may serve as a potential clinical therapeutic agent.
Conflicts of interest
There are no conflicts of interest to declare.
Acknowledgments
This work was supported by grants from the Natural Science Foundation of China (no. 81602832) and Key Projects of Science and Technology Program by Beijing Municipal Education Commission (no. KZ201610025020).
References
- Mossman B. T., Churg A. Am. J. Respir. Crit. Care Med. 1998;157:1666–1680. doi: 10.1164/ajrccm.157.5.9707141. [DOI] [PubMed] [Google Scholar]
- Yang G., Zhu Z., Wang Y., Gao A., Niu P., Tian L. Toxicol. Lett. 2013;220:103–108. doi: 10.1016/j.toxlet.2013.04.017. [DOI] [PubMed] [Google Scholar]
- Wang D., Zhang M. Chin. J. Ind. Hyg. Occup. Dis. 2012;30:801–810. [PubMed] [Google Scholar]
- Liu X., Fang S., Liu H., Wang X., Dai X., Yin Q., Yun T., Wang W., Zhang Y., Liao H., Zhang W., Yao H., Chao J. Toxicol. Appl. Pharmacol. 2015;288:152–160. doi: 10.1016/j.taap.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Wynn T., Barron L. Semin. Liver Dis. 2010;30:245–257. doi: 10.1055/s-0030-1255354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan M., Yu Z. X., Ferrans V. J., Sulciner D. J., Gutkind J. S., Irani K., Goldschmidt-Clermont P. J., Finkel T. Biochem. J. 1996;318(Pt 2):379–382. doi: 10.1042/bj3180379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura N. Curr. Opin. Pulm. Med. 2000;6:140–144. doi: 10.1097/00063198-200003000-00010. [DOI] [PubMed] [Google Scholar]
- Leppert P. C., Baginski T., Prupas C., Catherino W. H., Pletcher S., Segars J. H. Fertil. Steril. 2004;82:1182–1187. doi: 10.1016/j.fertnstert.2004.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shruti K., Shrey K., Vibha R. Biochem. Biophys. Res. Commun. 2011;407:445–449. doi: 10.1016/j.bbrc.2011.03.058. [DOI] [PubMed] [Google Scholar]
- Shukla G. C., Singh J., Barik S. Mol. Cell. Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- Chu A. S., Friedman J. R. J. Clin. Invest. 2008;118:3585–3587. doi: 10.1172/JCI36870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung A. C., Huang X. R., Meng X., Lan H. Y. J. Am. Soc. Nephrol. 2010;21:1317–1325. doi: 10.1681/ASN.2010020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum T., Gross C., Fiedler J., Fischer T., Kissler S., Bussen M., Galuppo P., Just S., Rottbauer W., Frantz S., Castoldi M., Soutschek J., Koteliansky V., Rosenwald A., Basson M. A., Licht J. D., Pena J. T., Rouhanifard S. H., Muckenthaler M. U., Tuschl T., Martin G. R., Bauersachs J., Engelhardt S. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- Cushing L., Kuang P. P., Qian J., Shao F., Wu J., Little F., Thannickal V. J., Cardoso W. V., Lü J. Am. J. Respir. Cell Mol. Biol. 2011;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Huang X.-R., Wei L.-H., Chung A. C. K., Yu C.-M., Lan H.-Y. Mol. Ther. 2014;22:974–985. doi: 10.1038/mt.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Meng X. M., Huang X. R., Chung A. C., Feng Y. L., Hui D. S., Yu C. M., Sung J. J., Lan H. Y. Mol. Ther. 2012;20:1251–1260. doi: 10.1038/mt.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery R. L., Yu G., Latimer P. A., Stack C., Robinson K., Dalby C. M., Kaminski N., van Rooij E. EMBO Mol. Med. 2014;6:1347–1356. doi: 10.15252/emmm.201303604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Wang Y., Liang D., Yang G., Chen L., Niu P., Tian L. Toxicol. Res. 2016;5:116–125. doi: 10.1039/c5tx00291e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chen J., Dong J., Liu W. Toxicol. Lett. 2005;155:353–360. doi: 10.1016/j.toxlet.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Braz N. F., Carneiro A. P., Amorim M. R., de Oliveira Ferreira F., Lacerda A. C., Silva de Miranda A., Teixeira M. M., Teixeira A. L., Mendonca V. A. J. Occup. Environ. Med. 2014;56:493–497. doi: 10.1097/JOM.0000000000000164. [DOI] [PubMed] [Google Scholar]
- Bonnans C., Chou J., Werb Z. Nat. Rev. Mol. Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Huang C., Lin X., Li J. Biochimie. 2013;95:1355–1359. doi: 10.1016/j.biochi.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Pandit K. V., Milosevic J., Kaminski N. Transl. Res. 2011;157:191–199. doi: 10.1016/j.trsl.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Patel V., Noureddine L. Curr. Opin. Nephrol. Hypertens. 2012;21:410–416. doi: 10.1097/MNH.0b013e328354e559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Iizuka M., Sekiya Y., Yoshizato K., Ikeda K., Kawada N. Biochem. Biophys. Res. Commun. 2010;391:316–321. doi: 10.1016/j.bbrc.2009.11.056. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ghazwani M., Li J., Sun M., Stolz D. B., He F., Fan J., Xie W., Li S. Biochem. Biophys. Res. Commun. 2014;446:940–944. doi: 10.1016/j.bbrc.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lin Z., Foolen J., Schoen I., Santoro A., Zenobi-Wong M., Vogel V. Matrix Biol. 2014;40:62–72. doi: 10.1016/j.matbio.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Lambert E., Dasse E., Haye B., Petitfrere E. Crit. Rev. Oncol. Hematol. 2004;49:187–198. doi: 10.1016/j.critrevonc.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Rockey D. C., Bell P. D., Hill J. A. N. Engl. J. Med. 2015;372:1138–1149. doi: 10.1056/NEJMra1300575. [DOI] [PubMed] [Google Scholar]
- Bando M. Respir. Investig. 2016;54:298–304. doi: 10.1016/j.resinv.2016.03.007. [DOI] [PubMed] [Google Scholar]
- Rogliani P., Calzetta L., Cavalli F., Matera M. G., Cazzola M. Pulm. Pharmacol. Ther. 2016;40:95–103. doi: 10.1016/j.pupt.2016.07.009. [DOI] [PubMed] [Google Scholar]
- King Jr. T. E., Bradford W. Z., Castro-Bernardini S., Fagan E. A., Glaspole I., Glassberg M. K., Gorina E., Hopkins P. M., Kardatzke D., Lancaster L., Lederer D. J., Nathan S. D., Pereira C. A., Sahn S. A., Sussman R., Swigris J. J., Noble P. W., A. S. Group N. Engl. J. Med. 2014;370:2083–2092. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- Rangarajan S., Kurundkar A., Kurundkar D., Bernard K., Sanders Y. Y., Ding Q., Antony V. B., Zhang J., Zmijewski J., Thannickal V. J. Am. J. Respir. Cell Mol. Biol. 2016;54:51–59. doi: 10.1165/rcmb.2014-0445OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richeldi L., du Bois R. M., Raghu G., Azuma A., Brown K. K., Costabel U., Cottin V., Flaherty K. R., Hansell D. M., Inoue Y., Kim D. S., Kolb M., Nicholson A. G., Noble P. W., Selman M., Taniguchi H., Brun M., Le Maulf F., Girard M., Stowasser S., Schlenker-Herceg R., Disse B., Collard H. R., I. T. Investigators N. Engl. J. Med. 2014;370:2071–2082. doi: 10.1056/NEJMoa1402584. [DOI] [PubMed] [Google Scholar]
- van Rooij E., Purcell A. L., Levin A. A. Circ. Res. 2012;110:496–507. doi: 10.1161/CIRCRESAHA.111.247916. [DOI] [PubMed] [Google Scholar]
- Michelfelder S., Trepel M. Adv. Genet. 2009;67:29–60. doi: 10.1016/S0065-2660(09)67002-4. [DOI] [PubMed] [Google Scholar]
- Boon R. A., Seeger T., Heydt S., Fischer A., Hergenreider E., Horrevoets A. J., Vinciguerra M., Rosenthal N., Sciacca S., Pilato M., van Heijningen P., Essers J., Brandes R. P., Zeiher A. M., Dimmeler S. Circ. Res. 2011;109:1115–1119. doi: 10.1161/CIRCRESAHA.111.255737. [DOI] [PubMed] [Google Scholar]