 The rat pancreatic progenitor cell line B-13 is of interest for research on drug metabolism and toxicity since the cells trans-differentiate into functional hepatocyte-like cells (B-13/H) when treated with glucocorticoids.
The rat pancreatic progenitor cell line B-13 is of interest for research on drug metabolism and toxicity since the cells trans-differentiate into functional hepatocyte-like cells (B-13/H) when treated with glucocorticoids.
Abstract
The rat pancreatic progenitor cell line B-13 is of interest for research on drug metabolism and toxicity since the cells trans-differentiate into functional hepatocyte-like cells (B-13/H) when treated with glucocorticoids. In this study we investigated the trans-differentiation and liver-specific functions of B-13/H cells in a three-dimensional (3D) multi-compartment bioreactor, which has already been successfully used for primary liver cell culture. Undifferentiated B-13 cells were inoculated into the bioreactor system and exposed to dexamethasone to promote hepatic trans-differentiation (B-13/HT). In a second approach, pre-differentiated B-13 cells were cultured in bioreactors for 15 days to evaluate the maintenance of liver-typical functions (B-13/HP). During trans-differentiation of B-13 cells into hepatocyte-like cells in the 3D bioreactor system (approach B-13/HT), an increase in glucose metabolism and in liver-specific functions (urea and albumin synthesis; cytochrome P450 [CYP] enzyme activity) was observed, whereas amylase – characteristic for exocrine pancreas and undifferentiated B-13 cells – decreased over time. In bioreactors with pre-differentiated cells (approach B-13/HP), the above liver-specific functions were maintained over the whole culture period. Results were confirmed by gene expression and protein analysis showing increased expression of carbamoyl-phosphate synthase 1 (CPS-1), albumin, CYP2E1, CYP2C11 and CYP3A1 with simultaneous loss of amylase. Immunohistochemical studies showed the formation of 3D structures with expression of liver-specific markers, including albumin, cytokeratin (CK) 18, CCAAT/enhancer-binding protein beta (CEBP-β), CYP2E1 and multidrug resistance protein 2 (MRP2). In conclusion, successful culture and trans-differentiation of B-13 cells in the 3D bioreactor was demonstrated. The requirement for only one hormone and simple culture conditions to generate liver-like cells makes this cell type useful for in vitro research using 3D high-density culture systems.
Introduction
The AR42J B-13 (B-13) cell line, a subclone isolated from the rat AR42J pancreatic cell line, is related to pancreatic ductal progenitor cells1–3 and shows similarities with liver cells in their development. Upon glucocorticoid treatment, a majority (85–95%) of treated B-13 cells exhibit immunoreactivity for hepatocyte markers such as albumin, cytokeratin 8 and transferrin.2 The trans-differentiation of B-13 cells to hepatocyte-like cells (B-13/H) as a response to elevated glucocorticoids describes a pathophysiological process that can also be observed in vivo.4 Furthermore, expression of hepatic CYP enzymes and oxidative metabolism of testosterone were demonstrated in B-13/H cells.3,5,6 Since B-13 cells rapidly proliferate under standard cell culture conditions and can be trans-differentiated by glucocorticoid addition, they present a renewable, cost-effective source of functional hepatocyte-like cells and could be used in in vitro studies such as preclinical drug testing or liver disease research.
Studies on primary hepatocytes have shown an improved maintenance of cell viability and liver-specific functions like drug metabolism, disposition and toxicity when using models simulating a 3D environment, e.g. sandwich cultures (reviewed by Swift et al., 2010).7 Therefore, culture of B-13 cells in 3D culture systems might support differentiation of hepatocyte-like B-13/H cells for such studies.
In this report, a bioreactor technology for B-13 cell trans-differentiation is examined, which is based on a perfused hollow-fibre capillary network for continuous, decentralized nutrient supply and oxygenation via independent capillary systems. This approach overcomes the restrictions of static culture systems with respect to mass exchange and oxygenation.
The bioreactor system has been successfully tested in a clinical extracorporeal liver support setting with primary porcine8 or human hepatocytes.9,10 Scaled-down variants of the technology for in vitro research show stable maintenance of hepatocyte performance.11 In addition, their suitability for drug metabolism studies was demonstrated.12
In this study, the trans-differentiation of B-13 cells into hepatocyte-like cells (B-13/H) and their functional performance in the bioreactor culture system was investigated. In the first approach, progenitor B-13 cells were trans-differentiated in bioreactors by dexamethasone addition (group B-13/HT). To evaluate the maintenance of functional characteristics under 3D culture conditions, pre-differentiated B-13/H cells were also seeded and cultured in parallel bioreactors for 15 days (group B-13/HP). The efficacy of trans-differentiation and the cell performance were assessed by determination of functional parameters in the bioreactor perfusate, including synthesis of glucose, lactate, urea and albumin. Cytochrome P450 (CYP) 1A1 activity and product conjugation by Phase II enzymes were assessed by analysis of ethoxyresorufin metabolism. The reorganization grade of the cells in the bioreactor cell compartment and the distribution pattern of typical hepatocyte markers was analysed by immunofluorescence. In addition, RT-PCR analysis was performed to determine the mRNA expression of liver-typical proteins.
Results
B-13 cells are able to trans-differentiate in a 3D configuration
First, the ability of B-13 cells to differentiate into hepatocyte-like B-13/H cells in a 3D configuration was investigated. For this purpose, B-13 cells were cultured in suspension (B-13S) by seeding them into plastic dishes coated with poly(2-hydroxyethyl methacrylate) (P2HEMA), which prevents cell attachment to plastic materials13 and allows the formation of 3D aggregates by the suspended cells.
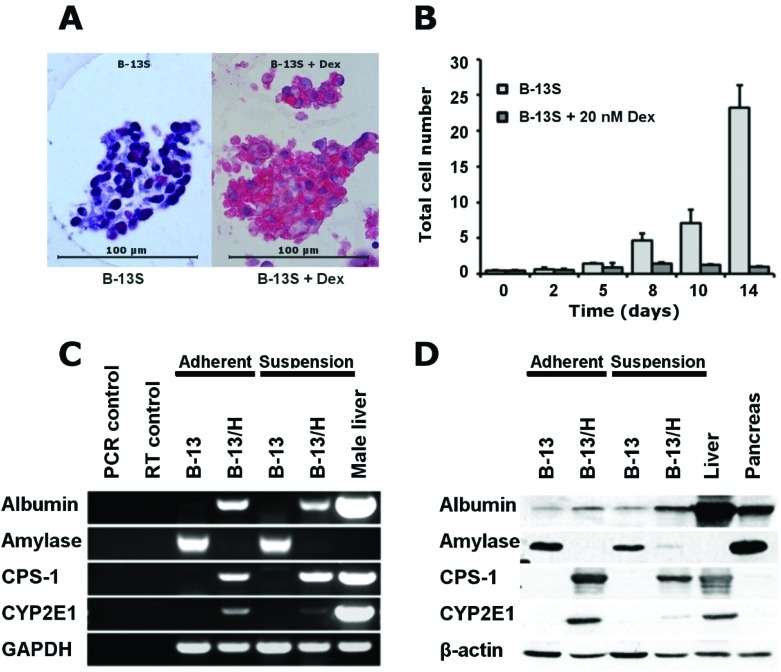
Fig. 2A demonstrates that B-13S cells formed cell aggregates (less than 100 μm in diameter), which readily excluded trypan blue and did not show any necrotic centres. Cell proliferation was suppressed upon addition of dexamethasone in B-13S cultures (Fig. 2B), however some proliferation activity was still observed (approx. 2.5 fold over 14 days). The trans-differentiation response as determined by RT-PCR and Western blotting was similar in B-13S cells after exposure to dexamethasone compared to 2D cultures of B-13 cells. Expression of amylase characteristic for pancreas exocrine tissue decreased both in mRNA and in protein expression whereas hepatocyte-specific markers (albumin, carbamoyl-phosphate synthase 1 [CPS-1] and CYP2E1) were induced (Fig. 2C and D).
Fig. 2. (A) Representative bright field microscopic picture (100-fold) of a B-13 cell aggregates cultured with or without dexamethasone obtained by suspension culture (B-13S). (B) Changes in total cell numbers in B-13S cultures treated with dexamethasone as compared with untreated control cultures. Values are shown as means ± SD. RT-PCR (C) and Western blotting (D) analysis of pancreatic or hepatic marker expression in B-13 cells maintained in adherent or suspension cultures without (B-13) or with dexamethasone (B-13/H) supplementation.
B-13 and B-13/H cells cultured in bioreactors are metabolically active and show liver-specific functions upon dexamethasone exposure
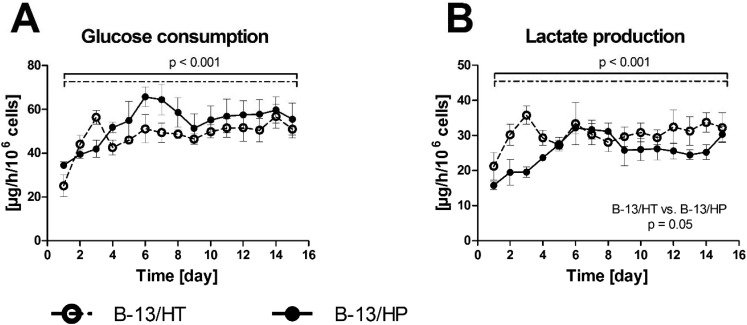
Bioreactor cultures of B-13 cells subjected to trans-differentiation (B-13/HT) showed a significant increase in glucose consumption (p = 0.029) during first three days. Those, inoculated with cells pre-differentiated prior to seeding into the bioreactor (B-13/HP) showed an increase in glucose (p = 0.005) and lactate (p = 0.009) metabolism until day 6 followed by stable rates. Both groups showed significant differences in energy metabolism considering the overall culture period (glucose consumption: p < 0.001; lactate production: p < 0.001), but no significant difference between both groups was observed at the end of culture time (glucose consumption: p = 0.609; lactate production: p = 0.691) (Fig. 3).
Fig. 3. Glucose consumption (A) and lactate production (B) in 3D multi-compartment bioreactors with B-13 cells subjected to trans-differentiation in the bioreactor (B-13/HT/dotted line) or with pre-differentiated B-13/H cells (B-13/HP/solid line) over a time period of 15 days. Values were calculated as rates per 106 cells. Comparisons concerning changes over time were performed using MANOVA (multivariate analysis of variance) with the two approaches as inter-subject variables. Two-sided p values ≤0.05 are considered significant. Significant changes over time are marked as bars enclosing respective time intervals. Graphs show means ± SEM of three independent experiments (n = 3).
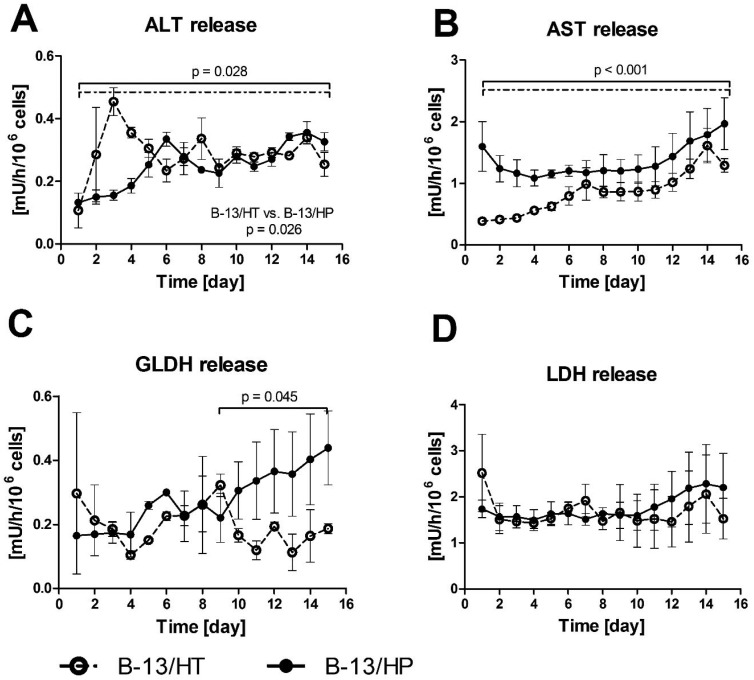
The release of intracellular enzymes (alanine transaminase [ALT], aspartate transaminase [AST], glutamate dehydrogenase [GLDH] and lactate dehydrogenase [LDH]) was analysed in bioreactor perfusates to detect potential cell damage in the cultures (Fig. 4). ALT and AST levels showed in both approaches a slight, yet significant increase during the culture period (p = 0.028 and p < 0.001), while GLDH only significantly increased in the B-13/HP group from day 9 to 15 (p = 0.045). Release rates of ALT were significant different in both groups (p = 0.026). The levels of LDH activity were constant with no significant increase (p = 0.635) over the culture period, with slightly higher enzyme release rates in B-13/HP in comparison to B-13/HT.
Fig. 4. Enzyme release of ALT (A), AST (B), GLDH (C) and LDH (D) in 3D multi-compartment bioreactors with B-13 cells subjected to trans-differentiation in the bioreactor (B-13/HT/dotted line) or with pre-differentiated B-13/H cells (B-13/HP/solid line) over time period of 15 days. Values were calculated as rates per 106 cells. Comparisons concerning changes over time were performed using MANOVA (multivariate analysis of variance) with the two approaches as inter-subject variables. Two-sided p values ≤0.05 are considered significant. Significant changes over time are marked as bars enclosing respective time intervals. Graphs show means ± SEM of three independent experiments (n = 3).
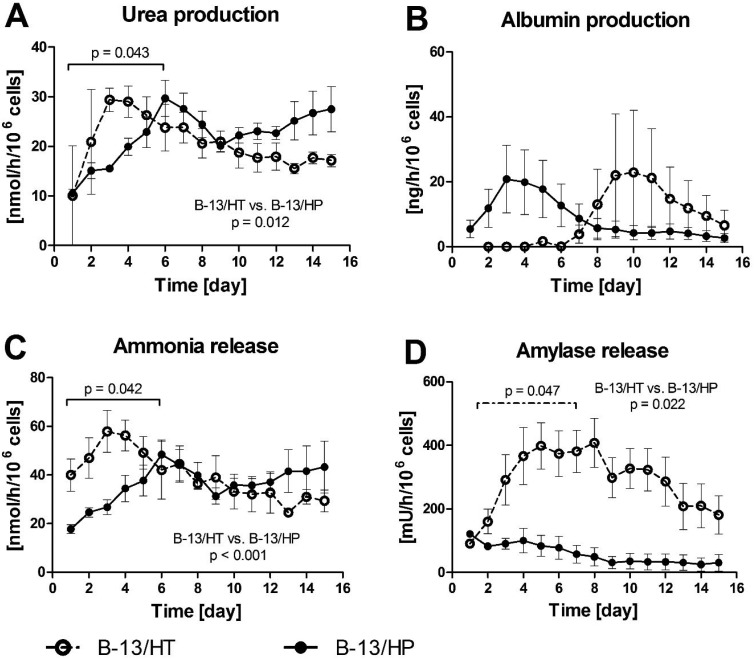
Fig. 5A and B shows the course of urea and albumin concentrations in bioreactor culture supernatants. Urea production in B-13/HT bioreactors showed an increase but not significantly until day 3 after dexamethasone induction and declined afterwards. In B-13/HP bioreactors urea production increased significantly until day 6 (p = 0.043) followed by a slight decrease to a stable level for the remaining culture time with no significant changes in production rates. In total B-13/HP showed significant higher production rates than B-13/HT (p = 0.012). Albumin synthesis showed a tendency, though not significant (p = 0.365), towards increase from day 7 in B-13/HT bioreactors with a peak on day 10, while B-13/HP showed a maximum in albumin synthesis on day 3 followed by a slight, yet not significant (p = 0.175) decline. Both groups showed different albumin production rates considering the overall culture time, although not significantly (p = 0.081).
Fig. 5. Production rates of urea (A), albumin (B) and release rates of ammonia (C) and amylase (D) in 3D multi-compartment bioreactors with B-13 cells subjected to trans-differentiation in the bioreactor (B-13/HT/dotted line) or with pre-differentiated B-13/H cells (B-13/HP/solid line) over a time period of 15 days. Values were calculated as rates per 106 cells. Comparisons concerning changes over time were performed using MANOVA (multivariate analysis of variance) with the two approaches as inter-subject variables. Two-sided p values ≤0.05 are considered significant. Significant changes over time are marked as bars enclosing respective time intervals. Graphs show means ± SEM of three independent experiments (n = 3).
Ammonia release showed a similar course as urea production in both groups, with a significant increase until day 6 in B-13/HP cultures (p = 0.042) and rather stable values thereafter with no significant changes. Similarly, ammonia levels in B-13/HT increased during the first days, although not significant (p = 0.106) and slowly declined to ammonia levels comparable to B-13/HP culture until the end of culture (Fig. 5C). B-13/HT showed significant higher ammonia release rates as B-13/HP (p < 0.001). Amylase release characteristic for pancreas exocrine cells also significantly increased over the first week of culture (p = 0.047) in the B-13/HT group and stayed stable thereafter with no significant changes. Initial values for amylase release were lower for B-13/HP compared to B-13/HT and did not significantly change over culture period (p = 0.11) (Fig. 5D). Amylase release rates were significantly different between both groups during bioreactor culture (p = 0.022).
Trans-differentiated B-13 cells show CYP1A1 activity in the bioreactor via ethoxyresorufin-O-deethylase (EROD) assay
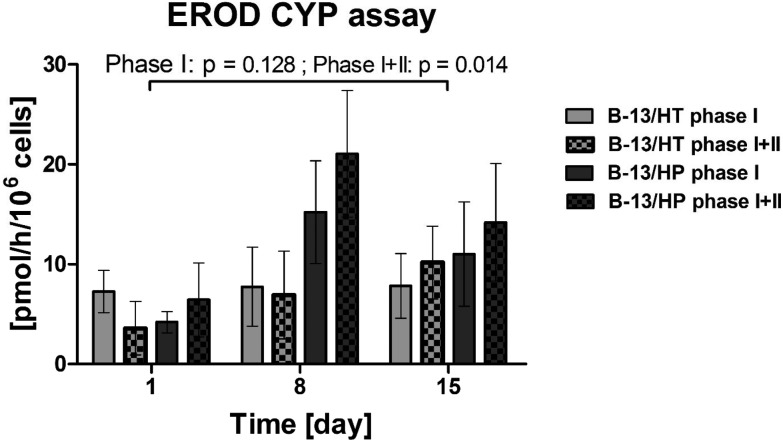
CYP1A1 dependent deethylation of ethoxyresorufin was analysed on day 1, day 8 and day 15 by detection of the fluorescent product 7-hydroxyresorufin. In both bioreactor groups (B-13/HT and B-13/HP), formation rates of non-conjugated (Phase I) and conjugated (Phase II) 7-hydroxyresorufin could be measured from day 1 to day 15 whereas Phase I + II showed significant increase (Phase I: p = 0.128; Phase I + II: p = 0.014). Values were significantly different between groups for Phase I + II (p = 0.033) (Fig. 6). B-13/HP cultures showed higher formation rates of 7-hydroxyresorufin on day 8 when compared to B-13/HT (p = 0.033), although not at a significant level for Phase I (p = 0.114), while on day 15 of culture similar formation rates were observed. Cleavage of conjugated 7-hydroxyresorufin (Phase II) resulted in the detection of higher amounts of the free product in the B-13/HP group, although not at a significant level.
Fig. 6. Biotransformation of 7-ethoxyresorufin in 3D multi-compartment bioreactors with B-13 cells subjected to trans-differentiation in the bioreactor (B-13/HT/grey) or with pre-differentiated B-13/H cells (B-13/HP/black-grey) on day 1, 8 and 15 of culture. Phase I: Formation rate of the free product, Phase I + II: Sum of formation rates of free and conjugated products. Values were calculated as rates per 106 cells. Comparisons concerning changes over time were performed using MANOVA (multivariate analysis of variance) with the two approaches as inter-subject variables. Two-sided p values ≤0.05 are considered significant. Significant changes over time are marked as bars enclosing respective time intervals. Graphs show means ± SEM of three independent experiments (n = 3).
Gene expression and protein analysis indicate an up regulation of hepatocyte-specific genes during trans-differentiation of B-13 cells
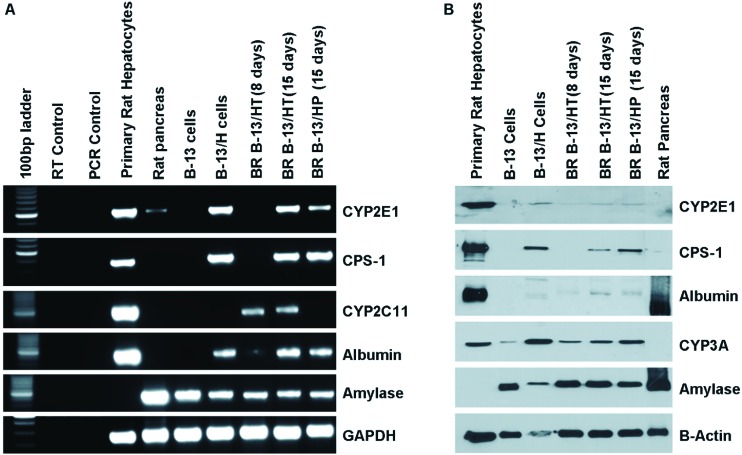
The expression of hepatic and pancreatic markers at the mRNA and protein levels was investigated in B-13/HT on day 8 and 15 and B-13/HP on day 15 in comparison with B-13 cells and B-13/H cells cultured in 2D flasks, as well as rat pancreatic tissue and primary rat hepatocytes (Fig. 7).
Fig. 7. Analysis of hepatocyte specific markers via RT-PCR and western blot in 3D multi-compartment bioreactors with B-13 cells subjected to trans-differentiation in the bioreactor (B-13/HT, day 8 and 15) or with pre-differentiated B-13/H cells (B-13/HP, day 15) compared to B-13 or B-13/H cells cultured in 2D, primary rat hepatocytes and rat pancreas tissue. (A) RT-PCR for the indicated transcripts and RT control amplification in the absence of input RNA. (B) Western blot of the indicated proteins.
The expression of genes encoding the liver-specific CPS-1 and albumin was similar in the experimental groups B-13/HT and B-13/HP as in 2D cultured B-13/H cells, but lower as compared with primary hepatocytes (Fig. 7A). The expression of CYP2E1 and also of CYP2C11 specific for male rats could be demonstrated for B-13/HT, while B-13/HP cultures showed CYP2E1 expression only, but not that of CYP2C11, similar to B-13/H cells cultured under 2D conditions. The expression of liver-specific markers increased from day 8 to day 15 in B-13/HT bioreactors. Amylase mRNA transcripts characteristic for exocrine pancreas cells were expressed in B-13/HT and B-13/HP cultures at a similar level as in B-13/H cells maintained in 2D cultures, but its expression was lower than in native rat pancreas or undifferentiated B-13 cells.
Western blot analysis (Fig. 7B) showed a weaker expression of CPS-1, albumin and CYP2E1 in B-13/HT or B-13/HP bioreactors than in primary rat hepatocytes, while CYP3A expression was comparable to that of primary cells. The expression of liver-specific markers (albumin, CPS-1, CYP2E1 and CYP3A) was similar to that of B-13/H cells generated in 2D culture. Amylase expression was comparable to B-13 and B-13/H cells, with a lower intensity than in pancreatic tissue in all approaches. In accordance with results from PCR analysis, an increase of liver-specific markers and a decline of pancreatic amylase between day 8 and day 15 of culture were observed.
Immunohistochemical staining reveals a hepatocyte-like phenotype of B-13 cells upon trans-differentiation in the 3D bioreactor
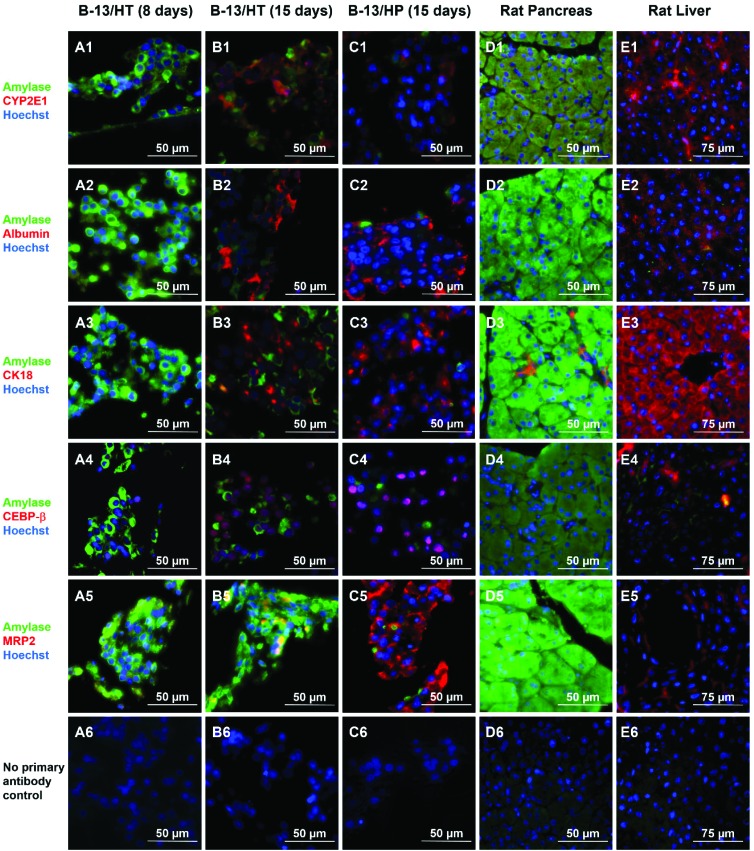
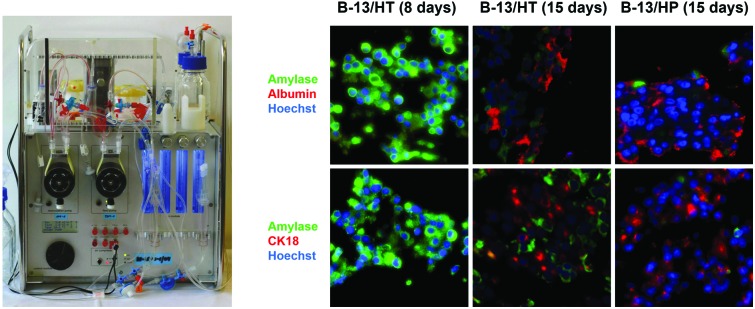
Immunofluorescence staining of liver- and pancreas-specific markers was performed in B-13/HT bioreactors on day 8 and 15 and in B-13/HP cultures on day 15 and compared to tissue of rat pancreas and liver (Fig. 8). Double-staining was performed using antibodies against pancreas-specific amylase in combination with antibodies against various liver specific marker antigens.
Fig. 8. Immunohistochemical analysis of hepatic and pancreatic markers in B-13 cells trans-differentiated in the bioreactor system (B-13/HT) over 8 (A) or 15 (B) days, B-13/H cells cultured in the bioreactor system for 15 days (B-13/HP, C), native pancreas (D), and native liver (E) in 400× magnification. Amylase positive cells are shown in green and markers characteristic for the liver are shown in red. Row (1) shows amylase and CYP2E1, row (2) amylase and albumin, row (3) amylase and CK18, row (4) amylase and CEBP-β, and row (5) amylase and MRP2 immunoreactivity. Row (6) shows negative controls (no primary antibody control). Counterstaining of nuclei was performed with Hoechst staining (blue).
Amylase was observed only in few cells in B-13/HT and B-13/HP cultures on day 15. In B-13/HT bioreactors a decrease was detected from day 8 to day 15.
The liver-specific markers albumin, CK18 and CEBP-β, a marker of early liver development, showed a similar distribution in B-13/HT (day 15) and B-13/HP bioreactors. An increase in expression of these markers between day 8 and day 15 was observed in B-13/HT, indicating progression of trans-differentiation. CYP2E1 showed a higher expression in B-13/HT cultures than in B-13/HP cultures, while positive staining for MRP2, an apical membrane transporter found in primary hepatocytes, was only observed in B-13/HP bioreactors.
Native rat and liver tissue investigated for comparison showed the typical expression of specific markers. Rat liver tissue showed albumin, CK18, CYP2E1 and MRP2 positive cells but no amylase, whereas rat pancreas tissue strongly expressed amylase, but no CYP2E1, albumin or CEBP-β. Cytokeratin 18 showed immunoreactivity in some cells of pancreatic tissue, probably associated with pancreatic duct epithelia. Negative controls (no primary antibody control) showed no unspecific staining by secondary antibodies (Fig. 8/row 6).
Discussion
The B-13 cell line is of interest because it has the potential to be readily expandable and could be used as a cost effective source of hepatocytes.1–3,5 Since the 3D multi-compartment bioreactor culture system used in this study showed successful liver cell cultivation for application in extracorporeal liver support8,9,14 or analysis of drug metabolism in small scale systems,11–12,15 we investigated the suitability of the technology to support hepatic trans-differentiation of B-13 cells.
In the first step, we investigated the influence of a 3D configuration on trans-differentiation in suspension cultures of B-13 cells (B-13S). Previous experiments with other cell types (i.e. primary human hepatocytes or human embryonic stem cells) seeded into the bioreactors had demonstrated that the cells do not only adhere to the artificial capillaries, but also to each other.11,16 Aggregation of viable cells, showing no necrotic centres and similar trans-differentiation upon dexamethasone exposure as in 2D cultures was observed in B-13S cultures, thus demonstrating the ability of B-13 cells to differentiate in a 3D aggregate configuration.
Subsequently, the suitability of the bioreactor culture system to support trans-differentiation of B-13 cells (B-13/HT) and to maintain the hepatocyte-like phenotype of pre-differentiated B-13/H cells (B-13/HP) was studied.
Monitoring of metabolic parameters in perfusates from bioreactors showed successful culture of B-13 or B-13/H cells in the system. B-13/HT and B-13/HP showed similar glucose consumption and lactate production rates with an increase in lactate production during the first days and stable metabolism afterwards. The initial lower production of lactate could be due to adaptation of the cells to the culture environment. Interestingly, glucose/lactate metabolism of B-13/HT approximated that in B-13/HP at the end of the culture period. This indicates that a comparable cell number and metabolic status was attained in both groups after 15 days supporting the estimated proliferation rate of 2.5 over the culture duration in B-13/HT used as a basis of data calculation.
The analysis of enzyme release (ALT, AST, GLDH and LDH) in bioreactor perfusates as indicators for possible cell damage,11–12,17 showed a continuous increase, although at a low level, in parallel with increasing metabolic activity, which can be interpreted as necrosis of a small proportion of cultured cells during cell growth (B-13/HT) and trans-differentiation. No drastic peaks in enzyme release, indicating harmful stress to the cells, were observed over the entire culture period.
Production of albumin, the most abundant protein produced by hepatocytes, was detected in both approaches with a maximum in the first week in the pre-differentiated cultures (B-13/HP) and in the second week in cultures trans-differentiated in the 3D bioreactor (B-13/HT), demonstrating successful induction of hepatic protein synthesis upon exposure to dexamethasone. Furthermore stable production of urea, generated by hepatocytes for nitrogen elimination via the urea cycle, was monitored over the culture period in bioreactor perfusates. The finding of lower urea concentrations in B-13/HT than in B-13/HP cultures suggests lower activity in cultures subjected to trans-differentiation within the 3D bioreactor. This is supported by the results from ammonia measurements, which show a correlation of ammonia release with urea secretion, indicating insufficient ammonia detoxification via the urea cycle. To enhance urea formation, stimulation of the urea cycle, e.g. by addition of N-carbamoyl-glutamate could be tested, which has been successfully used for improving the differentiation state of HepaRG cells.18,19
Amylase characteristic for exocrine pancreas cells is typically expressed by undifferentiated B-13 cells.1 Increasing rates of amylase as reported for B-13/H cells during trans-differentiation of B-13 cells2 could be observed in B-13/HT bioreactor cultures after addition of 10 μmol l–1 dexamethasone to the culture medium. The decrease of amylase release observed in bioreactor cultures of B-13/HP and B-13/HT in the second week of culture indicates a decrease of pancreas-specific characteristics in the cultures.
As a further functional parameter, the expression and activity of CYP isoenzymes, which are responsible for hepatic xenobiotic metabolism (Phase I reaction), was assessed. Formation of 7-hydroxyresorufin from 7-ethoxyresorufin, a known CYP1A1 substrate,20 was detected in both bioreactor groups, with B-13/HP bioreactors showing higher formation rates than B-13/HT cultures on day 8, but comparable values on day 15. Furthermore, the ongoing rise of Phase I + II metabolism between day 1 and day 15 in B-13/HT cultures indicated ongoing progression of trans-differentiation in these cultures. Thus, further prolongation of the trans-differentiation period could be useful to increase CYP activities even further.
The analysis of mRNA expression (RT-PCR) and protein expression (Western Blot) confirmed successful differentiation of B-13 cells to hepatic B-13/H cells in the bioreactor culture system. The expression of hepatocyte-specific markers previously described for B-13/H 2D cultures5,21 were maintained at both the mRNA and protein expression level. The CYP isoenzymes CYP2E1, CYP3A1, and CYP2C11, typical for rat liver22 could be detected. Furthermore, mRNA transcript levels of albumin and of CPS-1 were comparable in B-13/HT and B-13/HP to primary hepatocytes. Expression of albumin and CPS-1 proteins was also detected, though in low levels. CPS-1 is an important enzyme involved in the urea cycle and its expression in B-13/HT and B-13/HP on gene and protein levels further confirms urea production via the urea cycle. However it cannot be excluded that part of the observed urea release is produced via hydrolysis of arginine by arginase as described for human liver cell line C3A.23,24
Immunohistochemical staining showed the formation of 3D cell clusters in bioreactor cultures and confirmed the hepatic differentiation of B-13 cells in the 3D bioreactor culture system in that there was a loss in expression of amylase and an increase in hepatocyte-specific markers. CYP2E1 and albumin expression confirmed the results of gene expression and Western blot analysis. In addition, cells positive for CK18, a marker for liver epithelial cells,12 as well as expression of MRP2, a major canalicular transporter responsible for biliary elimination of drugs in hepatocytes25 indicate a liver-like phenotype of B-13/H cells in the bioreactor cultures. Further, activation of CEBP-β shows the involvement of this transcription factor in generating liver-like tissue, according to results from other studies.2
Significant improvement of hepatic differentiation in 3D as compared with 2D cultures could not be demonstrated in our study. However some liver markers, i.e. CPS-1 and albumin, while showing lower expression in suspension cultures than in 2D cultures (Fig. 2), showed at least equal expression in bioreactor cultures of pre-differentiated (B-13/HP) or trans-differentiated (B-13/HT) cells (Fig. 7). In both 2D and 3D culture systems B-13/H cells do not attain the functionality of native hepatocytes. Improvement of this cell line may need further genetic modification. For example expression of metabolically functional human CYP1A2 was achieved by introducing the corresponding gene into the cells.6
In conclusion, the studies show successful hepatic trans-differentiation of B-13 cells as well as stable maintenance of B-13/H cells over two weeks in the bioreactor. B-13 cells are cost-effective and can be generated in high amounts, which makes the cells an attractive source for larger scale in vitro studies.
Material and methods
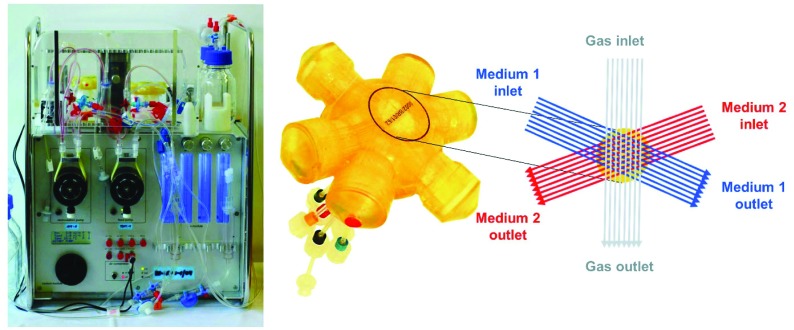
3D multi-compartment bioreactor system
The 2 ml multi-compartment bioreactor (Stem Cell Systems, Berlin, Germany, Fig. 1, right) used for 3D culture of B-13/B-13/H cells consisted of a polyurethane housing with an internal hollow space that contains a 3D network of hollow-fibre capillaries serving for counter-current medium perfusion and integral membrane oxygenation of the cells cultured in the extra-capillary space. A detailed description of the capillary structure is given elsewhere.11 The bioreactor system was operated in a perfusion device (Fig. 1, left) regulating the temperature and the medium flow rate. Gas supply (95% air, 5% CO2) and medium pH were controlled and adjusted, if required, by electronically regulated gas valves (Vögtlin Instruments, Aesch. Switzerland).
Fig. 1. Bioreactor system used for 3D high-density cultivation of B-13 cells. (A) Perfusion unit for bioreactor operation. (B) Illustration of the 2 ml bioreactor showing the capillary structure in the bioreactor with two independent bundles of hollow fibre membranes serving for counter-current medium perfusion (blue and red) and one capillary system for integral oxygenation (grey).
Preparation of B-13 cells for bioreactor culture
To investigate B-13 cells in suspension (B13S) cells were seeded into plastic dishes coated with P2HEMA as previously described.13 Cells were cultured with or without 20 nmol l–1 dexamethasone for 14 days as described elsewhere.26
B-13 cells were expanded in 2D culture as previous described26 using DMEM culture medium supplemented as shown in Table 1. B-13/H cells required for seeding in the approach B-13/HP were converted in 2D culture by adding 10 μmol l–1 dexamethasone (Sigma-Aldrich, Germany) to the culture medium five days prior inoculation. B-13 or B-13/H cells were harvested from culture plates using 0.05% trypsin/0.02% EDTA solution. Unless otherwise described reagents were obtained from Biochrom, Germany.
Table 1. Composition of B-13 culture medium.
| Final concentration |
|||
| Compound | Company | 2D culture | 3D culture |
| DMEM high glucose | Biochrom, Berlin, Germany | 88% (v/v) | 95.5% (v/v) |
| FCS | PAA, Dartmouth, MA, U.S. | 10% (v/v) | 2.5% (v/v) |
| l-Glutamine, 200 mM | Life Technologies, Carlsbad, CA, U.S. | 2 mmol l–1 | |
| Penicillin/streptomycin, 10 000 U ml–1/10 000 μg ml–1 | 100 U ml–1/100 μg ml–1 | ||
| Galactose | Sigma-Aldrich, St. Louis, MO, U.S. | 1 g l–1 | |
| Sorbitol | |||
Culture of B-13 cells or B-13/H cells in the bioreactor system
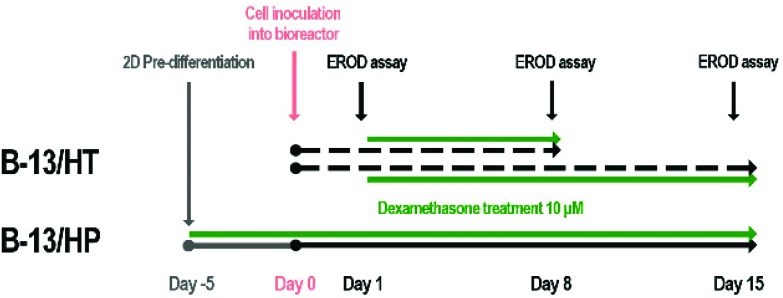
For bioreactor culture two different approaches were investigated (Fig. 9). In the first approach, 4 × 107 B-13 cells were seeded into the bioreactor and converted into B-13/H cells by treatment with 10 μmol l–1 dexamethasone from day 1 on (B-13/HT). In the second approach, 1 × 108 pre-differentiated B-13 cells (B-13/H) were seeded into the bioreactor (B-13/HP). Bioreactor cultures were perfused with DMEM culture medium as described in Table 1. In both groups dexamethasone treatment was continued over the whole culture period of 15 days. An ethoxyresorufine-O-deethylase (EROD) assay was performed on day 1, 8 and 15 in each bioreactor. Different initial cell numbers were chosen for B-13/HT and B-13/HP since previous experiments for B-13/HT (data not shown) had revealed unsuccessful trans-differentiation of B-13 cells when the bioreactor was inoculated with cells at maximum capacity.
Fig. 9. Time schedule for treatment of bioreactors cultured with B-13 (B-13/HT, dotted lines) or pre-differentiated B-13/H (B-13/HP, solid line) cells. In the approach B-13/HT, B-13 cells were seeded into the bioreactor and converted into B-13/H cells in situ over 8 or 15 days. In the approach B-13/HP, B-13 cells were converted into B-13/H cells in 2D culture prior inoculation into the bioreactor. Trans-differentiation was initiated using 10 μmol l–1 dexamethasone in both groups (green lines). Bioreactor cultures were performed over 15 days whereas on day 1, 8 and 15 an EROD CYP activity assay was performed. At the end of culture (B-13/HT on day 8 or day 15; B-13/HP on day 15) cell material was harvested and used for analysis of gene and protein expression.
Bioreactor cultures were maintained using a medium recirculation rate of 10 ml min–1, a medium feed rate of 2 ml h–1 and a gas perfusion rate of 25 ml min–1. Daily medium samples were taken from the bioreactor perfusate and analysed for ALT, AST, GLDH, LDH, amylase, urea and ammonium concentrations using an automated clinical chemistry analyser (COBAS 8000; Roche Diagnostics, Germany). Glucose and lactate concentrations and the pH of the recirculating culture medium were analysed with a blood gas analyser (ABL 700, Radiometer, Brønshøj, Denmark). Albumin concentrations were determined via an ELISA-based detection method (Bethyl Laboratories, Inc., U.S.). Values were calculated as rates per 106 cells based on initial cell numbers of pre-differentiated cells for B-13/HP and an estimated cell proliferation by the 2.5 fold over 14 days for B-13/HT. The cell proliferation rate was determined in suspension cultures (Fig. 2B).
Upon termination of the bioreactor cultures, cell material for protein and RNA analysis was withdrawn from the cell compartment on day 15. For approach B-13/HT three additional bioreactors were terminated on day 8 to characterise trans-differentiation process more in detail. For RNA analysis, cells were re-suspended in TRIZOL (Life Technologies GmbH, Germany), while cells for Western blotting were used without further processing. For immunohistochemistry, cell material was fixed in 4% formaldehyde buffer, dehydrated and embedded in paraffin.
Ethoxyresorufin-O-deethylase (EROD) assay
The CYP1A1 activity of the cultures was tested by measuring the conversion of 7-ethoxyresorufin to resorufin. 7-Ethoxyresorufin (AAT Bioquest, Inc., U.S.) was applied at a final concentration of 20 μmol l–1 to the recirculation circuit. After substrate application samples were taken from the perfusate after 0.5, 1, 1.5 and 2 hours. Samples were analysed fluorometric (extinction: 544 nm, emission: 590 nm) with an ELISA reader (FLUOstar Optima; MGB LABTECH, Germany) using authentic resorufin (Sigma-Aldrich, Germany) in a concentration range of 5–320 nmol l–1 as a standard for the free product concentration. In order to detect the absolute amount of product, including conjugated products, conjugates were cleaved by addition of β-glucuronidase/arylsulfatase (Roche Diagnostics, Germany). Therefore 150 μl of sample and 150 μl of 1 mol l–1 sodium acetate solution (pH = 5.5) were mixed with 20 μl of the enzyme solution and incubated at 37 °C over night. Resorufin formation rates were calculated from the slope of the regression line of the concentration–time curve.
Gene expression
TRIZOL (Life Technologies GmbH, Germany) was used for total RNA purification according to manufacturer's instructions and aliquots were stored at –20 °C. In addition, RNA from freshly isolated primary rat hepatocytes and pancreas (Pharmacelcus, Saarbrücken, Germany) and from B-13 and B-13/H cells cultured in 2D flasks as described before27,28 was investigated for comparison. First strand cDNA and PCR were performed using M-MLV reverse transcriptase and GoTaq polymerase respectively, following standard protocols.28 PCR reaction mixtures were incubated at 95 °C for 1 min, followed by 1 min incubation at the required annealing temperature, followed by 2 min at 73 °C for elongation for the required number of cycles (see Table 2). Amplified DNA was analysed on agarose gels containing ethidium bromide.
Table 2. DNA oligonucleotide sequences employed in RT-PCR or PCR genotyping.
| Oligo ID | 5′–3′ sequence | Annealing conditions (35 cycles) | Comments |
| RT-PCR | |||
| rmCYP2E1US | TCGACTACAATGACAAGAAGTGT | 42 °C | Will amplify a rat CYP2E (NM_031543) cDNA sequence of 525bp |
| rmCYP2EDS | CAAGATTGATGAATCTCTGGATCTC | ||
| rmhGAPDHUS | TGACATCAAGAAGGTGGTGAAG | 50 °C | Will amplify rat (NM_017008), human (NM_002046) or mouse (NM_008084) glyceraldehyde 3 phosphate dehydrogenase cDNA sequence of 243bp |
| rmhGAPDHDS2 | TCTTACTCCTTGGAGGCCATGT | ||
| rCPS1US | ATACAACGGCACGTGATGAA | 55 °C | Will amplify rat CPS (NM_017072.1) cDNA sequence of 390bp |
| rCPS1DS | GCTTAACTAGCAGGCGGATG | ||
| rmAMYLASEUS | CAAAATGGTTCTCCCAAGGA | 57 °C | Will amplify rat pancreatic amylase 2 (NM_031502.1) cDNA sequence of 224bp |
| rmAMYLASEDS | CAAAATGGTTCTCCCAAGGA | ||
| rCYP2C11 US | CTGCCATGGATCCAGTCCTAGTCC | 55 °C | Will amplify rat (NM_019184.2) cDNA sequence of 88bp |
| rCYP2C11 DS | TTCCCTCTCCCAAAGCTCTGTCTCC | ||
| rAlbumin US | CGTCAGAGGATGAAGTGCTC | 47 °C | Will amplify rat albumin (NM_134326) cDNA sequence |
| rAlbumin DS | CTTAGCAAGTCTCAGCAGCAG | Sequence of 471bp |
Western blotting
Western blotting was performed as previously described.5,26,29 In brief, protein extracts were subjected to SDS/PAGE under reducing conditions using a Bio-Rad MiniP2 electrophoresis apparatus. Protein was then transferred onto nitrocellulose and blocked overnight with 3% (w/v) dried milk/0.3% (w/v) Tween 20. Details of the primary antibodies used can be found in Table 3. After incubation with primary antibodies, blots were incubated with the appropriate horseradish-peroxidase-conjugated anti-IgG antibody. Detection was accomplished using chemiluminescence with the Fisher enhanced chemiluminescence (ECL®) kit.
Table 3. Antibodies and their concentrations employed in Western blotting and immunohistochemical staining.
| Antigen/specificity | Mwt (kDa) | Dilution | Antibodies and source |
| β-Actin | 44 | 1 : 4000 WB | Mouse monoclonal IgG (Sigma Aldrich A5441) |
| Albumin | 60 | 1 : 3000 WB | Chicken polyclonal to liver albumin (abcam ab106582) |
| 1 : 400 IHC(P) | |||
| Amylase | 57 | 1 : 3000 WB | Rabbit polyclonal to pancreatic amylase (abcam ab21156) |
| 1 : 100 IHC(P) | Mouse monoclonal (Santa Cruz sc-46657) | ||
| CEBP-β | 36 | 1 : 100 IHC(P) | Rabbit polyclonal to CEBP Beta (abcam ab53138) |
| CK18 | 48 | 1 : 100 IHC(P) | Rabbit monoclonal to Cytokeratin 18 (abcam ab133263) |
| CPS-1 | 165 | 1 : 2000 WB | Rabbit polyclonal to CPS1 – Liver Mitochondrial Marker (abcam ab3682) |
| CYP2E1 | 50 | 1 : 5000 WB | Rabbit polyclonal to CYP2E1 (abcam ab28146) |
| 1 : 100 IHC(P) | Rabbit polyclonal to CYP2E1 (abcam ab73878) | ||
| CYP3A4 | 58 | 1 : 2000 WB | Rabbit polyclonal to CYP3A4 (abcam ab155029) |
| MRP2 | 190 | 1 : 100 IHC(P) | Rabbit polyclonal to MRP2 (Sigma Aldrich M8316) |
| Anti-chicken 2° | N/A | 1 : 1000 IHC(P) | Alexa-Fluor®-594 goat anti-chicken IgG (H + L) (life technologies A-11042) |
| Anti-mouse 2° | N/A | Alexa-Fluor®-488 goat anti-mouse IgG (H + L) (life technologies A-11029) | |
| Anti-rabbit 2° | N/A | Alexa-Fluor®-594 goat anti-rabbit IgG (H + L) (life technologies A-11037) | |
| Anti-goat HRP 2° | N/A | 1 : 3000 WB | Polyclonal to goat produced in rabbit, horseradish peroxidise conjugated (Sigma Aldrich A5420) |
| Anti-rabbit HRP 2° | N/A | Polyclonal to rabbit produced in goat, horseradish peroxidise conjugated (Dako P0448) | |
| Anti-mouse HRP 2° | N/A | Polyclonal to mouse produced in goat, horseradish peroxidise conjugated (Dako P0447) |
Immunohistochemical analysis
Paraffin sections of cell material from the bioreactor, rat pancreas or rat liver tissue were deparaffinised and rehydrated. Antigen retrieval was done in citrate buffer (pH 6.0) for 15 min in a pressure cooker. Liver tissue was frozen and sections (3–4 μm in thickness) were fixed with acetone for 10 min at –20 °C. Non-specific binding sites were blocked with 2% FCS and 2.5% BSA in PBS solution for one hour at room temperature. Details of the primary and secondary antibodies used for double-staining are provided in Table 3. Counterstaining of nuclei was performed using bisBenzimide H 33342 trihydrochloride (Sigma-Aldrich, Germany). Unspecific staining was excluded by performance of negative controls omitting the primary antibody (no primary antibody control).
Statistics
Each experiment was performed three times with cells from different passages. Values are shown as means ± SEM, if not otherwise indicated. Comparisons concerning changes over time were performed using multivariate analysis of variance (MANOVA) with the two approaches as inter-subject variables. Two-sided p values ≤ 0.05 are considered significant. No Bonferroni correction was performed. SPSS 21 was used for statistical calculation.
Conclusions
The B-13 cell line in combination with the perfused 3D bioreactor system could provide a valuable tool to perform studies on drug toxicity and metabolism of the liver, enabling various modes of substance application (single or repeated injection, continuous infusion, closed circulation or open feed-mode) and regular sample-taking with the option to connect automated analytic devices. Moreover the number of animals used for research on primary cells could be reduced when using trans-differentiated B-13 cells. Recent studies show successful introduction of human genes into the B-13 cells,6 which opens the opportunity to generate cell lines with human characteristics to be used in in vitro models and in clinical liver support using the described 3D bioreactor system.
Acknowledgments
The work for this study was performed within the d-LIVER project and was funded by the European Community's FP7 Research Framework Programme under grant agreement no. 287596 (see also http://www.D-LIVER.eu/). P.M.E.P is supported by a studentship award from the National Centre for the Refinement, Reduction and Replacement of Animals in Research (NC3Rs, ; http://www.nc3rs.org.uk/) with additional support from the Alternatives Research and Development Foundation (; http://www.ardf-online.org/).
References
- Wallace K., Fairhall E. A., Charlton K. A., Wright M. C. Toxicology. 2010a;278(3):277–287. doi: 10.1016/j.tox.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Shen C. N., Slack J. M., Tosh D. Nat. Cell Biol. 2000;2(12):879–887. doi: 10.1038/35046522. [DOI] [PubMed] [Google Scholar]
- Marek C. J., Cameron G. A., Elrick L. J., Hawksworth G. M., Wright M. C. Biochem. J. 2003;370(Pt 3):763–769. doi: 10.1042/BJ20021545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace K., Flecknell P. A., Wright M. C. Am. J. Pathol. 2010c;177(3):1225–1232. doi: 10.2353/ajpath.2010.100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhall E. A., Charles M. A., Wallace K., Schwab C. J., Harrison C. J., Richter M., Hoffmann S. A., Charlton K. A., Zeilinger K., Wright M. C. Toxicol. Res. 2013;2(5):308–320. [Google Scholar]
- Probert P. M. E., Chung G. W., Cockell S. J., Agius L., Mosesso P., White S. A., Oakley F., Brown C. D. A., Wright M. C. Toxicol. Sci. 2014;137(2):350–370. doi: 10.1093/toxsci/kft258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift B., Pfeifer N. D., Brouwer K. L. R. Drug Metab. Rev. 2010;42(3):446–471. doi: 10.3109/03602530903491881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer I. M., Kardassis D., Zeillinger K., Pascher A., Grünwald A., Pless G., Irgang M., Kraemer M., Puhl G., Frank J., Müller A. R., Steinmüller T. H., Denner J., Neuhaus P., Gerlach J. C. Xenotransplantation. 2003a;10(5):460–469. doi: 10.1034/j.1399-3089.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- Sauer I. M., Zeilinger K., Pless G., Kardassis D., Theruvath T., Pascher A., Goetz M., Neuhaus P., Gerlach J. C. J. Hepatol. 2003b;39(4):649–653. doi: 10.1016/s0168-8278(03)00348-9. [DOI] [PubMed] [Google Scholar]
- Sauer I. M., Zeilinger K., Obermayer N., Pless G., Grünwald A., Pascher A., Mieder T., Roth S., Goetz M., Kardassis D., Mas A., Neuhaus P., Gerlach J. C. Int. J. Artif. Organs. 2002;25(10):1001–1005. doi: 10.1177/039139880202501015. [DOI] [PubMed] [Google Scholar]
- Zeilinger K., Schreiter T., Darnell M., Söderdahl T., Lübberstedt M., Dillner B., Knobeloch D., Nüssler A. K., Gerlach J. C., Andersson T. B. Tissue Eng., Part C. 2011;17(5):549–556. doi: 10.1089/ten.TEC.2010.0580. [DOI] [PubMed] [Google Scholar]
- Hoffmann S. A., Müller-Vieira U., Biemel K., Knobeloch D., Heydel S., Lübberstedt M., Nüssler A. K., Andersson T. B., Gerlach J. C., Zeilinger K. Biotechnol. Bioeng. 2012;109(12):3172–3181. doi: 10.1002/bit.24573. [DOI] [PubMed] [Google Scholar]
- Hodgkinson C. P., Wright M. C., Paine A. J. Mol. Pharmacol. 2000;58(5):976–981. doi: 10.1124/mol.58.5.976. [DOI] [PubMed] [Google Scholar]
- Gerlach J. C., Mutig K., Sauer I. M., Schrade P., Efimova E., Mieder T., Naumann G., Grunwald A., Pless G., Mas A., Bachmann S., Neuhaus P., Zeilinger K. Transplantation. 2003;76(5):781–786. doi: 10.1097/01.TP.0000083319.36931.32. [DOI] [PubMed] [Google Scholar]
- Lübberstedt M., Müller-vieira U., Biemel K. M., Darnell M., Hoffmann S. A., Knöspel F., Wönne E. C., Knobeloch D., Nüssler A. K., Gerlach J. C., Anderson T. B., Zeilinger K. J. Tissue Eng. Regen. Med. 2015;9(9):1017–1026. doi: 10.1002/term.1652. [DOI] [PubMed] [Google Scholar]
- Stachelscheid H., Eckert K., Jensen J., Edsbagge J., Björquist P., Rivero M., Strehl R., Jozefczuk J., Prigione A., Adjaye J., Urbaniak T., Bussmann P., Zeilinger K., Gerlach J. C. J. Tissue Eng. Regen. Med. 2013;7(9):729–741. doi: 10.1002/term.1467. [DOI] [PubMed] [Google Scholar]
- Pless G., Steffen I., Zeilinger K., Sauer I. M., Katenz E., Kehr D. C., Roth S., Mieder T., Schwartlander R., Müller C., Wegner B., Hout M. S., Gerlach J. C. Artif. Organs. 2006;30(9):686–694. doi: 10.1111/j.1525-1594.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Hoekstra R., Nibourg G. A., van der Hoeven T. V., Ackermans M. T., Hakvoort T. B., van Gulik T. M., Lamers W. H., Elferink R. P., Chamuleau R. A. Int. J. Biochem. Cell Biol. 2011;43(10):1483–1489. doi: 10.1016/j.biocel.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Ah Mew N., Payan I., Daikhin Y., Nissim I., Nissim I., Tuchman M., Yudkoff M. Mol. Genet. Metab. 2009;98(4):325–330. doi: 10.1016/j.ymgme.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith C., Scott M. P., Renwick A. B., Price R. J., Lake B. G. Xenobiotica. 2003;33(5):511–527. doi: 10.1080/0049825031000085960. [DOI] [PubMed] [Google Scholar]
- Wallace K., Marek C. J., Currie R. A., Wright M. C. J. Steroid Biochem. Mol. Biol. 2009;116(1–2):76–85. doi: 10.1016/j.jsbmb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Thangavel C., Dhir R. N., Volgin D., Shapiro B. H. Biochem. Pharmacol. 2007;74(10):1476–1484. doi: 10.1016/j.bcp.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Susando T., Lei X., Anene-Nzelu C., Zhou H., Liang L. H., Yu H. Biointerphases. 2010;5(3):FA116–FA131. doi: 10.1116/1.3521520. [DOI] [PubMed] [Google Scholar]
- Mavri-damelin D., Damelin L. H., Eaton S., Rees M., Selden C., Hodgson H. J. F. Biotechnol. Bioeng. 2008;99(3):644–651. doi: 10.1002/bit.21599. [DOI] [PubMed] [Google Scholar]
- Mottino A. D., Catania V. A. World J. Gastroenterol. 2008;14(46):7068–7074. doi: 10.3748/wjg.14.7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace K., Long Q., Fairhall E. A., Charlton K. A., Wright M. C. J. Cell Sci. 2011;124(Pt 3):405–413. doi: 10.1242/jcs.077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace K., Marek C. J., Hoppler S., Wright M. C. J. Cell Sci. 2010b;123(Pt 12):2103–2110. doi: 10.1242/jcs.070722. [DOI] [PubMed] [Google Scholar]
- Haughton E. L., Tucker S. J., Marek C. J., Durward E., Leel V., Bascal Z., Monaghan T., Koruth M., Collie-Duguid E., Mann D. A., Trim J. E., Wright M. C. Gastroenterology. 2006;131(1):194–209. doi: 10.1053/j.gastro.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Marek C. J., Tucker S. J., Konstantinou D. K., Elrick L. J., Haefner D., Sigalas C., Murray G. I., Goodwin B., Wright M. C. Biochem. J. 2005;387(Pt 3):601–608. doi: 10.1042/BJ20041598. [DOI] [PMC free article] [PubMed] [Google Scholar]