 Recent updates on various molecular and pathophysiological aspects of the zinc transporter ZIP8 (SLC39A8).
Recent updates on various molecular and pathophysiological aspects of the zinc transporter ZIP8 (SLC39A8).
Abstract
Zinc ion (Zn2+) is essential for life; its deficiency in the human body could cause stunted growth, anemia and susceptibility to infection. The Zn transporter ZIP8 (also known as SLC39A8) is an important Zn2+ importer; aberrant Zn2+ influx mediated by ZIP8 can lead to the pathogenesis of osteoarthritis and inflammatory diseases. ZIP8 also mediates the cellular uptake of divalent metal ions including iron, manganese, and the toxic heavy metal cadmium. Individuals with SLC39A8 mutations and transgenic mouse models are starting to reveal the critical role that this gene plays in embryonic development and the metabolism of essential metal ions. Here we summarize our current understanding of ZIP8's function and regulation, at both the molecular and biological levels. We also review the association of ZIP8 with various diseases and its linkage with complex disorders like obesity, hypertension, and schizophrenia as revealed by several large genome-wide association studies.
1. Introduction
Zinc (Zn) is vital for nearly all life, it is essential for gene transcription, cell growth, differentiation and development.1 Zn is an integral component of numerous proteins, including enzymes, growth factors, cytokines, receptors, and transcription factors.2,3 The dysregulation of Zn homeostasis has been implicated in many diseases. Zn deficiency could cause stunted growth , neurodegeneration,4 immune system dysfunction, and chronic inflammation;5 while excessive Zn accumulation can be toxic to cells.1,6 In mammals, the cellular Zn level is tightly regulated by primarily two metal ion transporter families: SLC30A (ZnT) and SLC39A (ZIP).7 The ZnT family, which consists of 10 members, lowers the intracellular Zn level by shipping Zn2+ from the cytosol into the extracellular environment or intracellular organelles. In contrast, the ZIP family, which consists of 14 Zn2+ importers, transport Zn2+ into the cytoplasm from the extracellular environment or intracellular organelles, such as the endoplasmic reticulum (ER) and lysosome.8,9
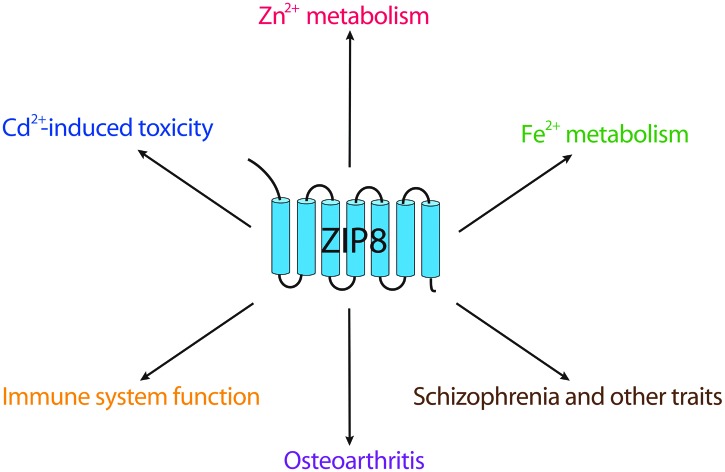
ZIP8 (also known as SLC39A8) was originally identified in monocytes by screening cDNA transcripts that were highly induced by the Mycobacterium bovis BCG cell wall, therefore it is also called as BIGM103, short for BCG-inducible gene in the monocyte clone 103.10 As a member of SLC39A transporters, ZIP8 has a high affinity for several divalent metal ions including Zn2+, iron (Fe2+), manganese (Mn2+) and the toxic heavy metal cadmium (Cd2+).10–12 ZIP14 shares a similar metal ion transport profile with that of ZIP8, however, ZIP8 is suggested to play a more important role than ZIP14 in terms of Cd2+-induced toxicity in the testis, kidney, and lung.13,14 In lung epithelia, ZIP8 is critical in Zn-mediated cytoprotection upon inflammation.15,16 In T cells, ZIP8 controls interferon (IFN)-γ expression and its immune function through the regulation of Zn release from the lysosome.8 ZIP8 also regulates innate immunity through Zn-mediated inhibition of NF-κB activity in macrophages and monocytes,17 strongly indicating that ZIP8 is a unique Zn transporter influencing immune system function during microbacterial infection and inflammation.
Recently, several renowned genome-wide association studies (GWAS) on large populations revealed the unexpected association of the genetic aberration of SLC39A8 with hypertension, obesity and schizophrenia.18–23 Close involvement of ZIP8 in osteoarthritis (OA) pathogenesis as well as direct linkage with Mn-deficiency related disorders were also reported.24,25 Although there are already several exceptional reviews covering the overall biological functions of SLC39A transporters,7,26–28 the unique roles ZIP8 play in immune system function, OA progression, and more recently Fe homeostasis haven't been well-discussed elsewhere. Here we give a review of these advances and discuss the molecular, biological, and pathological perspectives of ZIP8.
2. Structural aspects
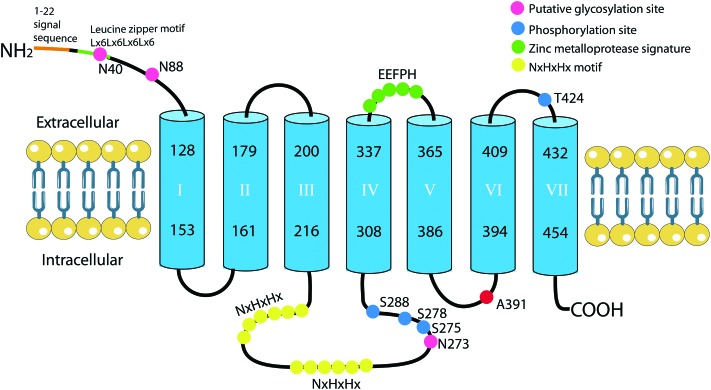
Fig. 1 illustrates the predicted transmembrane domains (TMDs) and putative functional motifs of human ZIP8.10 Most ZIPs feature three predicted TMDs at the N-terminus and four or five predicted TMDs at the C-terminus. Many ZIP members also have a long intracellular loop located between TMD III and IV, generally containing 2–5 copies of histidine-rich domains (Hx)n.26 ZIP8 is predicted to have seven TMDs with an EExxH motif localized at the extracellular loop between TMD IV and V.10 This motif fits the consensus sequence of a Zn-binding motif HExxH found in many Zn metalloproteases, except the first histidine is replaced by a glutamic acid (E). This alteration has been suggested to confer an ability to bind/transport metal ions other than Zn2+.12,29 Two copies of the NxHxHx motif exist in the long cytoplasmic loop between TMD III and IV. This histidine-rich cluster (Hx)n was proposed to interact with transition metals, especially Zn, copper and nickel, thus facilitating metal ion binding/transport,30 or participating in its ubiquitination and degradation, like in ZIP4.31
Fig. 1. Proposed structure of human Zn transporter ZIP8 (NP_001128618.1), its putative functional motifs and post-translational modifications. Four phosphorylation sites (blue dots) are verified by MS. N40, N88 and N273 (pink dots) are three putative N-glycosylation sites. The signal peptide of the first 22 amino acids is predicted using PTM code2. NxHxHx (yellow dots) indicates a putative histidine-rich region, which may play a role in metal-binding/transport, or ubiquitination and degradation. EEFPH fits the consensus sequence HEXXH of the Zn-binding motif found in metalloproteases. Non-synonymous SNP rs13107325 alters alanine to threonine at position 391 of the human ZIP8 protein (red dot), GWAS based on a large population reveals this SNP to be associated with traits related to hypertension, body mass index/obesity, and schizophrenia.
At the N-terminus, a leucine zipper motif is found on ZIP8, it consists of 28 amino acids with a periodic repetition of leucine residues at every seventh position. This motif is proposed to exist in an alpha-helical conformation, the leucine residues from two helixes could crosslink with each other, facilitating dimerization and in some cases oligomerization of proteins.32 The existence of the ZIP8 dimer was supported by two lines of evidence. First, in activated T cells, two bands of sizes of ∼75 kDa and ∼150 kDa were detected using both native and denaturing SDS gel electrophoresis; further treatment with PNGase didn't reduce the sizes of either band,8 indicating that ZIP8 is likely to exist in a dimeric form in activated T cells. Second, in HEK 293 T cells transfected with the wild type rat ZIP8, two immunoreactive bands in sizes of ∼45 kDa and ∼100 kDa were detected.12 Interestingly, when transfected with mutant rat ZIP8, in which three potential N-glycosylation sites were mutated, both bands shifted down in parallel compared to the wild type rat ZIP8, suggesting that the 100 kDa form could be the dimer of the 45 kDa protein. Although the existence of the ZIP8 protein dimer is well-appreciated, it is unclear now whether the leucine zipper motif is required for ZIP8 dimerization and the exact cellular function of the ZIP8 protein dimer.
3. Post-translational modifications
Three N-glycosylation sites have been detected in rat ZIP8 (i.e., N40, N88, and N96). Interestingly, mutation of these three sites doesn't affect its Fe or Zn transport activity, indicating that these glycans are not required for its metal ion transport.12 In agreement with rat ZIP8, human ZIP8 protein was also found to be glycosylated in tumor necrosis factor-α (TNF-α)-treated and ZIP8-transfected human BEAS-2B cells.15 The glycosylated form was exclusively detected in the membrane part, suggesting that glycosylation may play an important role for the suitable trafficking of ZIP8 to the plasma membrane or intracellular organelles. Although the existence of N-linked glycosylation on human ZIP8 is well-known, the exact glycosylation sites have not been pinpointed experimentally. Alignment between rat and human ZIP8 identifies conservation at the first two glycosylation sites i.e., N40 and N88. Another glycosylation site at N273, which is predicted with a high score using several algorithms, has been documented at the UniProtKB database (the three putative glycosylation sites on human ZIP8 protein are denoted as pink dots in Fig. 1).
So far, four mass spectrometry (MS)-verified phosphorylation sites (shown as blue dots in Fig. 1) on human ZIP8 have been documented. Prediction using NetphosK shows that S278 can be phosphorylated by casein kinase I (CKI),33 S288 by protein kinase A (PKA),34 and T424 by protein kinase C (PKC).35 Among them, PKC has been demonstrated to affect different transporters, either by directly modifying their activity or by altering their stability.36–38 In the THP-1 monocytic cell line, phorbol 12-myristate 13-acetate (PMA, an activator of PKC) treatment entirely abolished the ZIP8 protein expression.10 In line with this, PKC activation was found to decrease intracellular Cd accumulation in HEK cells,39 suggesting that PKC could be a negative modulator of ZIP8 thus protecting cells against Cd intoxication. Recently, ZIP7 was reported to be phosphorylated by casein kinase 2 (CK2) in response to the excessive extracellular Zn level and epidermal growth factor (EGF) treatment.9 ZIP7, unlike ZIP8, is primarily localized in ER. This phosphorylation could activate the transport activity of ZIP7, thereby releasing Zn2+ into the cytosol from ER, leading to subsequent activation of downstream signaling pathways that would promote cell proliferation and migration. In human proximal tubule cells, the punctate expression pattern of ZIP8 protein across the cytoplasm is consistent with its presence in ER.12,40 Besides, ZIP8 has also been observed in the mitochondria,15 endosome and lysosome.8,41 Therefore, it is tempting to speculate that the phosphorylation of ZIP8 in intracellular organelles or the plasma membrane is likely to affect its transport activity, leading to cytosolic metal ion depletion or accumulation under certain conditions.
4. Tissue distribution, subcellular localization and molecular sizes
ZIP8 expression varies markedly between different tissues and cell types. Most studies demonstrate that ZIP8 is abundantly expressed in the testis, kidney and lung while expressed at a low level in the brain and prostate.10,12,42 High expression of ZIP8 in the placenta was also noted,10,12,43 suggesting that ZIP8 could play an important role in embryo development.
Equally diverse is the subcellular localization and the sizes of ZIP8 detected in different types of cells (Table 1). The majority of studies confirmed that endogenous ZIP8 was primarily expressed at the plasma membrane,8,15,40 consistent with its proposed role as a metal ion importer. ZIP8 is localized at the apical surface of mouse renal proximal tubule cells and MDCK cells.44 However, in human proximal tubule cells, both the apical and basolateral presence were detected, reflecting the species discrepancy of ZIP8 distribution.40 Likewise, ZIP8 expression in the cytosol was also observed. In some cells, paranuclear and ER localizations were suggested,40 however, critical co-localization evidence with nuclear and ER-specific markers is required to support this statement. In addition, ZIP8 was found in the lysosome of T cells, serving as a Zn2+ intracellular store to activate its immune function upon activation.8 In transfected HEK293 T and BEAS-2B cells, endosomal and mitochondrial localizations of ZIP8 were reported,12,15 since a co-localization study using an overexpressed system can lead to artifacts, future verification of the localization of endogenous ZIP8 in these cells would be necessary.
Table 1. Summary of the molecular sizes and subcellular localizations of ZIP8.
| Protein | Localization | Molecular size | Tissue/cell line | Expression | Ref. |
| hZIP8 | Lysosome | 52 kDa | HeLa | Endogenous | 10 |
| hZIP8 | PM/cytosol | Membrane (140 kDa)/cytosol (55 kDa) | BEAS-2B/ hLECs | Endogenous | 15 |
| hZIP8-DsRed | PM/mitochondria/cytosol | PM, mitochondria (140 kDa)/cytosol (55 kDa) | BEAS-2B | Transient transfection | |
| hZIP8 | PM/lysosome | 150 kDa/75 kDa | T cells | Endogenous | 8 |
| hZIP8 | PM/cytosol/ER | 80 kDa/49 kDa | Human proximal tubule cells | Endogenous | 40 |
| hZIP8 | ND | 80 kDa/49 kDa/43 kDa | Normal proximal and distal tubule cells | Endogenous | |
| hZIP8 | PM/cytosol/ER | 80 kDa/49 kDa/43 kDa | Parental UROtsa | Endogenous | |
| hZIP8 | PM/cytosol/ER | 49 kDa | Normal urothelial cells | Endogenous | |
| hZIP8 | Membrane/cytosol | 140 kDa/55 kDa/>140 kDa (induced) | THP1/A549 /Macrophage | Endogenous | 17 |
| mZIP8-HA | PM | 86 kDa/55 kDa | MFF | Retroviral transfection | 76 |
| mZIP8-HA | Apical surface | Unknown | MDCK | Transient transfection | 44 |
| mZIP8 | PM | Unknown | Kidney | Transgenic | 104 |
| mZIP8 | PM | 120 kDa/60 kDa | RBCs | Endogenous | 41 |
| rZIP8 | PM/cytosol/endosome | 100 kDa/80 kDa/45 kDa | HEK-293T | Transient transfection | 12 |
| rZIP8 | PM/cytosol | 150 kDa/140 kDa/130 kDa | H4IIE cells | Endogenous | |
| rZIP8 | PM | 100 kDa/50 kDa | Primary hippocampal neurons | Endogenous | 78 |
| rZIP8 | Apical surface /Nucleus | ND | Hair cells in the inner ear | Endogenous | 45 |
Surprisingly, nuclear localization of ZIP8 was reported recently.45 The distribution of ZIP8 and ZIP14 is altered in rat inner ear cells during development. ZIP8 was highly expressed in the nucleus of hair cells, Hensen cells and the inner sulcus in the adult rat, while in the neonates, it was mainly localized in the cytoplasm and on the apical pole of hair cells within the organ of corti. The presence of ZIP8 in the nucleus could indicate a likely involvement in providing Zn2+ to the Zn2+-requiring transcription factor for its proper function during development. Besides, Mn2+, a substrate of both ZIP8 and ZIP14, is toxic to hair cells in the ear. Since there's no expression of the divalent metal transporter 1 (DMT1) in hair cells,46 this suggests that ZIP8 and ZIP14 may play a role in Mn2+-induced toxicity in ears.
The distribution of ZIP8 in the plasma membrane, cytosol, lysosome and other organelles strongly indicates the existence of the degradation/recycling process of ZIP8 protein. This notion was further supported by two studies. First, the expression of membrane-localized ZIP8 in polarized MDCK cells is affected by the extracellular Zn2+ level. It is induced when the Zn2+ level is low in the culturing medium, otherwise it is down-regulated when the physiological level of Zn is present,44 similar to the behavior of ZIP4.31 In ZIP4, the histidine-rich motifs between TMD III and IV are required for its ubiquitination/degradation and are highly conservative with that of ZIP8. Second, in H4IIE rat hepatoma cells, Fe loading increased the total and cell membrane level of the ZIP8 transporter.12 Similarly, ZIP14 expression on the membrane is induced by Fe overload, which results from inhibition of the endocytosis and degradation in the proteasomes.47 Intriguingly, this Fe-level-regulated degradation of ZIP14 transporter requires N-linked glycosylation, especially at the N102 site, but not the histidine-rich cluster observed in ZIP4. This N102 was not found to be conservative between human ZIP14 and ZIP8. Currently, the degradation and recycling process of ZIP8 remains unexplored, hence it would be interesting to investigate the degradation mechanism of ZIP8 and whether this process is controlled by the histidine-rich cluster and glycosylation.
Different groups have reported various molecular sizes of ZIP8 in different types of cells, we believe that ZIP8 dimerization and glycosylation could be the explanation. So far, the molecular sizes of ZIP8 discovered can be categorized into four types: heavily glycosylated ZIP8, which is generally more than 140 kDa; partially glycosylated ZIP8, which is around 100 kDa; ZIP8 dimer, which is between 100 kDa and 150 kDa; and lastly the native ZIP8, which is around 50 kDa. Usually the heavily glycosylated form is specifically localized in the membrane and exists only in exogenously transfected cells or cells induced by LPS, TNF-α or other inflammatory cytokines.12,15,17 It is also noteworthy that home-made antiserum prepared against different peptides of ZIP8 was used in most of the studies mentioned above, this may cause the disparity to some degree, and therefore independent verification of these western blot results is in great demand.
5. Regulation of ZIP8
5.1. Inflammatory cytokines
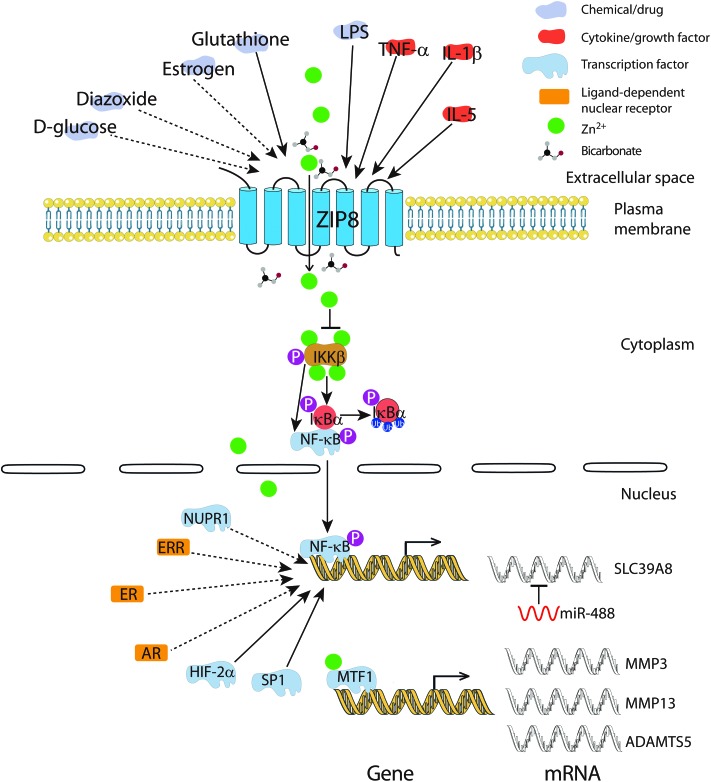
ZIP8 is highly inducible by lipopolysaccharide (LPS) and a variety of inflammatory cytokines (Fig. 2).10 LPS appears to regulate the expression of the ZIP8 transporter in a time-dependent way. In bone marrow derived macrophages, ZIP8 was induced at 4 hours of LPS treatment, however, it was suppressed at 24 hours. This suppression can be inhibited by 2-deoxyglucose, a glycolysis inhibitor.48 In lung epithelia, among the 10 ZnT and 14 ZIP transporter genes, only ZIP8 mRNA is strongly induced under TNF-α treatment, resulting in increased intracellular Zn concentration and cell survival.15 Similarly, in mouse articular chondrocytes, interleukin (IL)-1β, a proinflammatory cytokine that promotes catabolism in cartilage, markedly induced ZIP8 expression among ZnT and ZIP transporter members. In T cells, upon activation, ZIP8 is strongly upregulated, leading to Zn release from the lysosome concomitantly with increased secretion of IFN-γ and perforin (both are indicators of T cell activation).8 In mouse B cells, a comprehensive microarray analysis demonstrated that IL-5 could significantly induce the mRNA expression of ZIP8, since IL-5 is a B cell growth and differentiation factor required for its functional maturation, these results suggest that ZIP8 may contribute to B cell development through Zn2+-mediated downstream effectors.49
Fig. 2. Regulation of ZIP8. ZIP8 can be induced by LPS and various cytokines like TNF-α, IL-1β and IL-5. Glucose and estrogen can affect the mRNA expression level of ZIP8. Extracellular Zn2+ and Fe2+ concentration can regulate the expression of the plasma membrane localized ZIP8. Intracellularly, ZIP8-mediated Zn uptake can affect the activity of transcription factors directly like MTF1, or indirectly like NF-κB. Notably, NF-κB is a regulator of ZIP8, forming a negative feedback to control the intracellular Zn2+ level. Besides, the transcription factor SP1 and HIF-2α can also modulate the transcription of ZIP8. Interestingly, GSH can decrease the expression of SP1 thereby downregulating ZIP8 expression. Some evidence, though at a less sufficient level, also suggests that ligand-dependent nuclear receptors like AR, ER, and ERR control ZIP8 transcription. Solid lines represent experimentally confirmed interactions, dashed lines represent possible interactions with less-sufficient evidence.
5.2. Transcription regulators
The mechanism of ZIP8 regulation by various inflammatory cytokines was clarified recently. ZIP8 was found to be a downstream target gene of NF-κB. NF-κB controls ZIP8 expression while ZIP8 negatively regulates the NF-κB pathway and proinflammatory responses through Zn-mediated suppression of IkappaB kinase (IKK) activity in monocytes, macrophages, and lung epithelia.17 Also, the transcription factor SP1 is able to bind the promoter region of ZIP8 and is required for its expression. Elevated GSH level could reduce the expression of SP1, leading to decreased expression of ZIP8, which could attenuate Cd-induced cytotoxicity (Fig. 2).50 Microarray profiling in the MiaPaCa2 pancreas epithelial cell line also showed that ZIP8 is a nuclear protein (NUPR1)-dependent target gene.51
5.3. Chemical/drug
In mouse pancreatic β cells, ZIP8 mRNA expression was induced in response to glucose, an effect that was highly magnified by the addition of diazoxide, a pharmacologic insulin secretion inhibitor.52 As mentioned earlier, the ZIP8 transporter on the cell surface is induced by Zn depletion and Fe overload in the extracellular matrix,11,12 which can be explained by the strong competition between Zn2+ and Fe2+ for ZIP8 binding. When excessive Fe is present, the ZIP8 transporter on the membrane is highly induced due to the Zn deficiency caused by Fe competition. Although Zn depletion by treating cells with a specific Zn chelator didn't increase ZIP8 protein expression in the total lysate of H4IIE cells,12 the level of ZIP8 in the membrane remains unknown, which requires further investigation.
5.4. MicroRNA
MicroRNAs (miRNAs) have emerged as an important regulator of gene expression, estimated at regulating up to 10% of human genes.53 miRNAs are single-stranded RNA of 18–24 nucleotides,54 binding to the 7-nucleotide seed region located at 3′ UTR of the target mRNA transcripts in eukaryotes and abrogate translation by causing mRNA destabilization and degradation.55 miRNA dysregulation has been linked to many diseases, notably cancer and inflammatory cartilage disease.56–59 In chondrocytes, a study on the mechanism of OA pathogenesis discovered a novel way of ZIP8 regulation. miR-488, a single-stranded RNA of 22 nucleotides, could suppress the expression of ZIP8 and further reduce cartilage degradation through the decreased Zn content in chondrocytes.60
5.5. Hormones and hormone-related receptors
Through extensive literature mining, we noticed a significant change of ZIP8 expression can be induced by steroid hormones. Although some of the evidence is incomplete, yet, they suggest ZIP8 could be a downstream target gene of various hormones and hormone-related receptors (illustrated in Fig. 2). First, estrogen can significantly induce the expression of ZIP8 mRNA in MCF-7 cells,61 an estrogen receptor (ER) positive breast cancer cell line. Antihormones such as 4-hydroxytamoxifen and fulvestrant are routinely used for treating ER positive breast cancers.62 In an effort to examine the potential role that individual LIV-1 family members may play in the development of antihormone-resistant breast cancer, Taylor et al. discovered that the expression level of ZIP8 mRNA was highly elevated in the fulvestrant-resistant MCF-7 cell line, indicating a possible role of ZIP8 in the development of anti-hormone resistance.63 Second, using the microarray approach to screen ER and estrogen-related receptor (ERR) transcriptomes in MCF-7 cells, ER and ERR were shown to positively regulate ZIP8.64 In line with that, rosiglitazone (an activator of ERR) was also demonstrated to induce the expression of ZIP8 mRNA in the 393P-shz murine lung cancer cell line.65 Lastly, a hybridization assay using cDNA from the testis of an androgen receptor (AR)-mutated transgenic mouse showed that ZIP8 transcripts were significantly downregulated compared to the wild type,66 suggesting that AR may function as a transcriptional regulator of ZIP8 in the testis.
6. Metal ion uptake properties and its transport mechanism
ZIP8 was shown to possess high affinity for Zn2+, Fe2+, and Cd2+ followed by Mn2+ and mercury (Hg2+),12,44,67 closely matching that of ZIP14.67,68 Among these, Cd is of special interest since it is a well-known toxic and carcinogenic heavy metal. Using ZIP8 capped RNA microinjected Xenopus oocyte cultures, Nebert et al. shows that the Km value for Cd2+ is 0.48 μM, higher than that of Zn2+ and Fe2+,67 hence, it is not surprising to observe that ZIP8-mediated Cd2+ uptake demonstrated a robust inhibition by Zn2+ and Fe2+. However, Mn2+, which was a strong competitor for Cd2+ uptake in mouse fetal fibroblast cultures, didn't inhibit Cd2+ influx in the Xenopus oocyte system;11 the reason for this discrepancy remaining unclear. A further electrogenic experiment demonstrates that ZIP8 is a Zn2+/(HCO3)– symporter that moves one cation and two anions into the cells as an electroneutral complex, the optimal temperature for its metal ion transport activity is at 37 °C.12,44,67
7. Physiological function as revealed by a transgenic mouse
The generation of the SLC39A8(neo/neo) hypomorphic mouse has markedly advanced our understanding of the overall physiological function of ZIP8. This transgenic mouse line carries the SLC39A8(neo) allele where intron 3 of SLC39A8 loci was interspaced with the neo gene, thus causing significantly decreased expression of ZIP8 in the embryo, yolk sac, fetus and other tissues compared to the wild type littermate.69SLC39A8(neo/neo) homozygotes are pale, have stunted growth, and die between gestational day 18.5 and 48 hours postnatally. They exhibit hypoplasia at several organs, including the spleen, liver, lung, kidney and lower limbs, concomitant with diminished Zn2+ and Fe2+ levels. They also show symptoms of severe anemia, which results from Fe deficiency as indicated by the decreased hemoglobin level, serum Fe level, red cell number, and total Fe binding capacity. The number of erythrocytes was also strikingly affected,70 indicating that ZIP8-mediated Fe and Zn transport plays a crucial, and yet unrecognized role in multiple organ organogenesis and hematopoiesis during early embryo development.
8. The potential role of ZIP8 in metal ion metabolism
ZIP and ZnT proteins are primary regulators of cellular Zn homeostasis. Among the ZIP members, ZIP4 has the most clearly defined role; responsible for intestinal absorption of dietary Zn,71 whereas ZIP8 and ZIP14 seems to mediate Zn homeostasis in pancreatic β cells.52,72
Cd is a major component of cigarette smoke, contributing to chronic obstructive pulmonary disease and lung cancer.73–75 Because of its well-known toxicity and carcinogenicity, the mechanism of Cd2+ transport has been extensively studied. Clear evidence has identified ZIP8 as the transporter responsible for Cd2+-induced necrosis in testis, one of the most sensitive organs for Cd toxicity.76 Transgenic mice carrying 3 extra copies of the SLC39A8 gene demonstrated enhanced Cd2+-induced damage to the testis as well as the kidney, where ZIP8 is specifically localized at the apical surface of proximal tubule cells.77 In lung tissue, the research of Napolitano et al. strongly suggested ZIP8 as a key mediator of Cd2+-mediated toxicity in lung epithelia, where ZIP8 expression is highly upregulated by the inflammatory microenvironment resulting from chronic cigarette exposure.16 Additionally, their results indicate that excessive Zn can prevent Cd2+-mediated toxicity in human lung epithelial A549 cells, indisputably supporting the fact that Cd2+ enters into cells by hijacking Zn2+ transporters. The role ZIP8 plays in Zn homeostasis and Cd transport have been summarized before, for details, we refer the readers to the following reviews.26,29
As for Fe transport, given the apparent functional defects of hematopoiesis observed in SLC39A8(neo/neo) hypomorphic neonates,70 we can conclude that Fe metabolism mediated by ZIP8 plays an important role in early embryonic development. In the ZIP8 hypomorphs, the Fe level in the liver was significantly decreased probably due to the impaired Fe transfer process across the placenta. Consistent with that, in the placental cell line BeWo cells, suppression of ZIP8 expression decreased Fe uptake by 40%.12
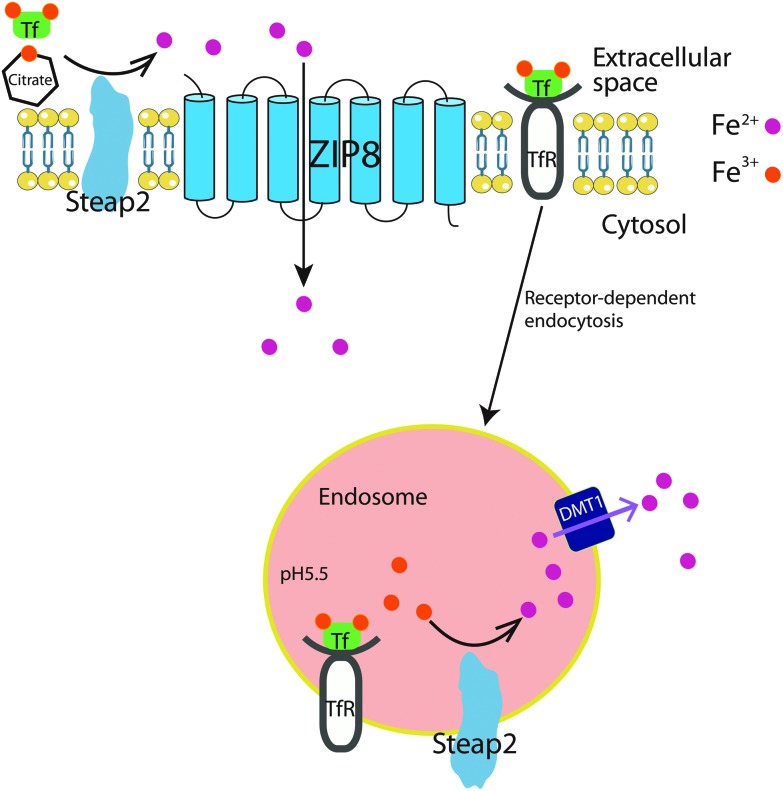
In the primary hippocampal neurons, ZIP8 was suggested to mediate both non-transferrin bound iron (NTBI) and transferrin-bound iron (TBI) uptake (shown in Fig. 3).78 First, the strong co-localization of ZIP8 and a ferrireductase Steap2 was observed on the surface of the soma and dendrites, suggesting that these two proteins may function together to support Fe3+ reduction prior to Fe2+ uptake. Secondly, ZIP8 was also shown to co-localize with the transferrin receptor/transferrin (TfR/Tf) complex on the cell membrane, suggesting that TBI uptake in neurons could also rely on a non-canonical pathway, other than the well-characterized endocytosis-dependent Fe transport. Indeed, blocking Tf internalization with an endocytosis inhibitor doesn't affect Fe uptake from TBI. Since the optimal pH of DMT1 and ZIP8 mediated Fe uptake is 5.5 and 7.5,12,79 respectively, and the pH of the brain interstitial fluid is 7.2,80 therefore, ZIP8 rather than DMT1 is a more appropriate candidate transporter for TBI uptake at the cell surface through an endocytosis-independent process. In terms of NTBI uptake in primary hippocampal neurons, whether presented as Fe2+ or Fe3+-citrate, reductase-independent Fe2+ uptake was the most efficient. On the one hand, kinetic analysis of Zn2+ inhibition of Fe2+ uptake indicated that DMT1 plays only a minor role in the uptake of NTBI. On the other hand, ZIP8 knock-down greatly impaired Fe2+ uptake, suggesting that ZIP8 plays a major role in NTBI uptake. Collectively, these observations strongly suggest that ZIP8 and Steap2 play a major role in Fe accumulation from NTBI and TBI in hippocampal neurons.
Fig. 3. Cellular mechanism of ZIP8-mediated Fe uptake in the hippocampal neuron. On one hand, Tf (transferrin)-bound Fe binds to the Tf receptor (TfR) at the cell membrane, and is transported into cells through TfR-dependent endocytosis. In the acidic environment of the endosome, Fe3+ is reduced to Fe2+ by Steap2 reductase, Fe2+ is then transported to the cytosolic plasma by DMT1, forming labile Fe pools in the cytosol for further deployment. On the other hand, Steap2 at the plasma membrane can directly reduce Fe3+, either Tf-bound or conjugated with citrate, to Fe2+, which is subsequently imported into the cytosol by ZIP8.
9. Association of ZIP8 with disease
9.1. Diseases related to infection and inflammation
As discussed previously, ZIP8 plays an important role in immune response. In accord with that, ZIP8 has been associated with diseases related to infection and inflammation. For instance, ZIP8 mRNA expression levels were found to be elevated in the monocytes from HIV-infected donors in conjunction with the increased intracellular Zn content which is linked to the resistance to apoptosis.81 Since monocytes are the targets of HIV infection and serve as a reservoir for HIV replication and assembly, this suggest that lowering the Zn content in the monocytes in HIV+ individuals could re-sensitize these cell to apoptosis and improve survival and outcome of the disease. Recently, a joint analysis of gene expression and genotypic data on malaria infected western African children identified SLC39A8 as one of the associations that are implicated in genotype-by-infection interaction.82 ZIP8, as mentioned earlier, is markedly upregulated upon primary T cell activation and affects its immune function through Zn2+ influx.8 Since supplementation of Zn has been reported to help reduce the morbidity and mortality of infectious disease,83 this interaction demonstrates a robust in vivo genotype-by-infection effect that is directly linked to the key process of ZIP8 mediated T-cell development. These observations are consistent with the results reported among different groups showing that ZIP8 was the most significantly upregulated transporter in response to cytokines, bacteria, and sepsis, suggesting its unique role in innate immunity.
9.2. Osteoarthritis
Growing evidence has shown that Zn plays a key role in the pathogenesis of OA.84,85 Zn acts as a structural component of matrix-degrading enzymes, among them, matrix metalloproteinase 3 (MMP3), MMP13 and ADAMTS5 are known to play crucial roles in OA pathogenesis.86–88 In a study to investigate the regulation of Zn2+ homeostasis during OA onset and progression, ZIP8 markedly increased, becoming the most dominantly expressed transporter among ZnT and ZIP members in chondrocytes under OA pathological conditions and in the OA cartilage.24 It is unclear why ZIP8 is markedly induced in the OA cartilage; we speculate that synovial inflammation, a common pathologic change of OA, could be one of the answers. ZIP8-mediated Zn influx increases the expression of matrix-degrading enzymes in chondrocytes. ZIP8 overexpression in mice cartilage tissue causes OA cartilage destruction whereas ZIP8 knockout inhibits OA pathogenesis. More importantly, the metal-regulatory transcription factor 1 (MTF1) was identified as a critical transcription factor in mediating Zn2+/ZIP8-induced matrix-degrading enzyme expression. What makes it more complicated is that the hypoxia-inducible factor (HIF)-2α can upregulate ZIP8, resulting in increased Zn2+ influx and MTF1 activity.89 In turn, MTF1 was revealed to be a novel transcriptional regulator of HIF-2α. In a reciprocal way, the ZIP8-Zn-MTF1 axis and HIF-2α activate each other and can synergistically promote the expression of matrix-degrading enzymes, leading to OA cartilage destruction (Fig. 2). Therefore, preventing the progression of OA, would require concomitant inhibition of the ZIP8-Zn-MTF1 catabolic cascade and HIF-2α activity in chondrocytes. The miR-488 targeting ZIP8 could also have important diagnostic and therapeutic potential, since it is significantly decreased in OA chondrocytes and could protect bone tissues from cartilage degradation by inhibiting MMP13 activity.60
9.3. Zn and Mn deficiency
During the preparation of our manuscript, Boycott et al. reported a homozygous variant, c.112G > C (p.Gly38Arg), affecting the highly conserved residue of ZIP8 protein.90 This homozygous mutation is responsible for autosomal-recessive disorders characterized by intellectual disability, developmental delay, hypotonia, strabismus, cerebellar atrophy and variable short stature. However, ZIP8 protein expression and subcellular localization have demonstrated no overt difference on lymphoblast and fibroblast cells between control and affected individuals. Whereas Zn and Mn levels in blood were variably low in all affected individuals; urine levels were high, indicating that the mutation impairs both the Zn and Mn transport of ZIP8, which can result in poor absorption of these elements through the kidney. At the same time, Park et al. identified two individuals with two different SLC39A8 variants: the first one is c.112G > C (p.Gly38Arg) and c.1019T > A (p.Ile340Asn); the second one is c.97G > A (p.Val33Met) and c.1004G > C (p.Ser335Thr) on the paternal allele and c.610G > T (p.Gly204Cys) on the maternal allele.25 These five mutations are all at highly conserved sites of ZIP8. The two affected individuals have a more severe phenotype than the case subjects described by Boycott et al. They have undetectable blood Mn levels which impairs the function of Mn-dependent enzymes, particularly β-1,4-galactosyltransferase, a Golgi enzyme essential for the glycosylation of important glycoproteins. Defective glycosylation leads to a severe disorder with a deformed skull, seizures, short limbs, profound psychomotor retardation and hearing loss.
9.4. Cancer risk and complex disorders
Recent GWAS projects have revealed surprising associations between one single nucleotide polymorphism (SNP) in SLC39A8 with schizophrenia,20,22,91 hypertension,19,23 obesity,18,21 and several metabolic traits such as body mass index and the high-density lipoprotein cholesterol (HDL-C) level (summarized in Table 2).92–94 This SNP, rs13107325, located between putative TMD V and VI, alters an alanine (major allele) to a threonine (minor allele, schizophrenia risk allele) at amino acid position 391 (denoted in Fig. 1). Alanine (A) is hydrophobic while threonine (T) is hydrophilic and polar. Ng et al. suggest that this SNP might abrogate the protein formation of ZIP8, therefore conferring an adverse effect on its metal ion transporter activity.95 Interestingly, SNP rs13107325 displays a regional distribution with high frequencies in Europe and the Middle East, followed by Central Asia and Egypt, and is totally absent in East Asia, South Asia, Southeast Asia and most regions in Africa. Based on this evolutionary analysis and other exploratory analyses, Li et al. postulate that when the ancestors of modern humans migrated out of Africa, the colder environment in Europe may have served as a selective pressure, resulting in increased SLC39A8 T allele, which enabled them to better adapt to the cold temperature and protected them from hypertension.96 Meanwhile, this T allele also increases the risk for schizophrenia, obesity and related metabolic traits (e.g., lower HDL-C and energy uptake from the nutrients). It has been suggested that ZIP8 may be involved in HDL-C metabolism through Zn-mediated inflammatory activity.29
Table 2. Summary of the association of rs13107325 (SLC39A8 T allele) with various human complex traits.
| Traits | Sample size |
Frequency | P-value | Region | Study | Ref. | |
| Initial | Replication | ||||||
| SBP, DBP, HP | 69 395 | 133 361 | 0.05 | 2.30 × 10–17 | EU | Ehret et al. (2011) | 19 |
| DBP | 87 736 | 68 368 | ND | ND | EU | Tragante et al. (2014) | 23 |
| BMI, Obesity | 123 865 | 125 931 | 0.07 | 1.50 × 10–13 | EU | Speliotes et al. (2010) | 18 |
| BMI | 168 267 | 109 703 | ND | ND | EU | Berndt et al. (2013) | 21 |
| HDL-C | 16 056 | 22 128 | 0.08 | 2.1 × 10–3 | EU | Waterworth et al. (2010) | 93 |
| HDL-C | 99 900 | — | 0.07 | <7 × 10–11 | EU | Teslovich et al. (2010) | 92 |
| HDL-C | 94 595 | 93 982 | 0.08 | 1.10 × 10–15 | EU | Willer et al. (2013) | 94 |
| Schizophrenia | 4545 cases; 15 575 controls | — | 0.08 | 2.7 × 10–6 | EU | Carrera et al. (2012) | 20 |
| Schizophrenia | 35 476 cases; 46 839 controls | — | 0.08 | 1.54 × 10–12 | EU | Ripke et al. (2014) | 22 |
| Stress | 182 | — | ND | ND | EU | Bruenig et al. (2014) | 97 |
| Gray matter volume | 1292 | — | >0.07, <0.08 | <10 × 10–4 | EU | Lajous et al. (2015) | 98 |
| Mn level in serum | 949 | — | ND | 5.1 × 10–11 | EU | Ng et al. (2015) | 95 |
It is noteworthy that rs13107325 is also associated with chronic stress,97 and gray matter volume in the subcortical region (Table 2).98 Together, these studies seem to indicate an attractive linkage between ZIP8 and cerebral functionality. Since Zn is crucial for neurotransmitter communication between neurons,99–101 we speculate that the SLC39A8 T allele might have a negative effect on neural signal transduction and contribute to psychiatric disorders through its impact on Zn homeostasis. Besides, rs13107325 associating with the Mn level in blood,95 two other SNP rs10014145 and rs233804 located at the intron of SLC39A8, have been suggested to affect the Cd concentration in human blood.102
Numerous studies have found that germline copy number variations (CNVs) can predispose individuals towards cancer occurrence. Recently, a study on the germline CNVs of renal clear cell carcinoma, the most common type of kidney cancer, revealed a significant loss of CNVs at the SLC39A8 genomic locus, occurring in 1.89% of cases (7/370) and 0.10% (3/3000) of controls.103
10. Conclusions and perspectives
We have sought here to provide detailed knowledge about the Zn transporter ZIP8, on which research has gained a lot of momentum in recent years. Although its function as a broad-spectrum divalent metal ion importer has been well-studied, it is not until recent years that we have started to appreciate its physiological and pathological implications. We believe that conditional knockout or transgenic mice of ZIP8, if possible, will continue to unravel its physiological role in different tissues, especially in the brain and liver. On the other hand, down to the molecular level, the exact role of these post-translational modifications (i.e., glycosylation and phosphorylation) sites and putative motifs mentioned in the article remains unclear; it will only be solved by site-directed mutagenesis until the crystal structure of ZIP family members is acquired. As this field moves forward, we anticipate that the insights we gained about ZIP8 will help us better understand the overall function of the ZIP protein family and improve our health.
Acknowledgments
Due to space constraints, it has been necessary to cite recent articles wherever possible; our sincere apologies to the hundreds of authors whose primary contributions are therefore not listed. This work was supported by the grants from the National Natural Science Foundation of China (no. 31170785 and 31271445), the Fund for University Talents of Guangdong Province, the Guangdong Natural Science Foundation of China (no. S2012030006289), and the Department of Education, Guangdong Government under the Top-tier University Development Scheme for Research and Control of Infectious Diseases. We would like to thank members of the Lau And Xu laboratory for critical reading of this review. The authors have declared no conflict of interest.
Biographies

Zhong-Sheng Zang
Zhong-Sheng “Eason” Zang majored in Bioengineering in 2008 and earned his Bachelor degree in 2012 at Yantai University, China. He is currently a graduate student of Profs Lau and Xu in the Department of Cell Biology and Genetics at Shantou University Medical College. His research has focused on divalent metal ion transport in human cells.

Yan-Ming Xu
Upon completion of her PhD degree (Biochemistry and Molecular Biology) in 2003 at the Fourth Military Medical University, China, Dr Xu joined the UMN in 2007 as a postdoctoral fellow in Cancer and Therapeutics Research. Since 2011, she has been working as a Principal Investigator in the Department of Cell Biology and Genetics at Shantou University Medical College. Prof. Xu's research team focuses on the action mechanism of natural compounds and drugs against cancers. Moreover, she is interested in signal transduction and regulation of protein kinases. Currently, she is an Active Member of the American Association for Cancer Research.

Andy T. Y. Lau
After obtaining his PhD degree (Molecular Biology) at the University of Hong Kong in 2005, Dr Lau joined the University of Minnesota (UMN), USA, as a postdoctoral fellow in Cancer and Epigenetics Research. Since 2011, he has been working as a Principal Investigator in the Department of Cell Biology and Genetics at Shantou University Medical College. Prof. Lau's research team focuses on the molecular basis of cancers by using “OMICS” approaches. Moreover, he is interested in post-translational modification of proteins and functions. Currently, he is an Active Member of the American Association for Cancer Research and an Academic Editor of PLoS ONE.
References
- Vallee B. L., Falchuk K. H. Physiol. Rev. 1993;73:79–118. doi: 10.1152/physrev.1993.73.1.79. [DOI] [PubMed] [Google Scholar]
- Fukada T., Yamasaki S., Nishida K., Murakami M., Hirano T. J. Biol. Inorg. Chem. 2011;16:1123–1134. doi: 10.1007/s00775-011-0797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreini C., Banci L., Bertini I., Rosato A. J. Proteome Res. 2006;5:196–201. doi: 10.1021/pr050361j. [DOI] [PubMed] [Google Scholar]
- Sensi S. L., Paoletti P., Bush A. I., Sekler I. Nat. Rev. Neurosci. 2009;10:780–791. doi: 10.1038/nrn2734. [DOI] [PubMed] [Google Scholar]
- Haase H., Rink L. Immun. Ageing. 2009;6:9. doi: 10.1186/1742-4933-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasapis C. T., Loutsidou A. C., Spiliopoulou C. A., Stefanidou M. E. Arch. Toxicol. 2012;86:521–534. doi: 10.1007/s00204-011-0775-1. [DOI] [PubMed] [Google Scholar]
- Lichten L. A., Cousins R. J. Annu. Rev. Nutr. 2009;29:153–176. doi: 10.1146/annurev-nutr-033009-083312. [DOI] [PubMed] [Google Scholar]
- Aydemir T. B., Liuzzi J. P., McClellan S., Cousins R. J. J. Leukocyte. Biol. 2009;86:337–348. doi: 10.1189/jlb.1208759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K. M., Hiscox S., Nicholson R. I., Hogstrand C., Kille P. Sci. Signaling. 2012;5:ra11. doi: 10.1126/scisignal.2002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum N. A., Kobayashi M., Moriwaki Y., Matsumoto M., Toyoshima K., Seya T. Genomics. 2002;80:630–645. doi: 10.1006/geno.2002.7000. [DOI] [PubMed] [Google Scholar]
- Liu Z., Li H., Soleimani M., Girijashanker K., Reed J. M., He L., Dalton T. P., Nebert D. W. Biochem. Biophys. Res. Commun. 2008;365:814–820. doi: 10.1016/j.bbrc.2007.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. Y., Jenkitkasemwong S., Duarte S., Sparkman B. K., Shawki A., Mackenzie B., Knutson M. D. J. Biol. Chem. 2012;287:34032–34043. doi: 10.1074/jbc.M112.367284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girijashanker K., He L., Soleimani M., Reed J. M., Li H., Liu Z., Wang B., Dalton T. P., Nebert D. W. Mol. Pharmacol. 2008;73:1413–1423. doi: 10.1124/mol.107.043588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishiro H., Ohashi T., Takuma M., Himeno S. Metallomics. 2013;5:437–444. doi: 10.1039/c3mt00003f. [DOI] [PubMed] [Google Scholar]
- Besecker B., Bao S., Bohacova B., Papp A., Sadee W., Knoell D. L. Am. J. Physiol. 2008;294:L1127–L1136. doi: 10.1152/ajplung.00057.2008. [DOI] [PubMed] [Google Scholar]
- Napolitano J. R., Liu M. J., Bao S., Crawford M., Nana-Sinkam P., Cormet-Boyaka E., Knoell D. L. Am. J. Physiol. 2012;302:L909–L918. doi: 10.1152/ajplung.00351.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. J., Bao S., Galvez-Peralta M., Pyle C. J., Rudawsky A. C., Pavlovicz R. E., Killilea D. W., Li C., Nebert D. W., Wewers M. D., Knoell D. L. Cell Rep. 2013;3:386–400. doi: 10.1016/j.celrep.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes E. K., Willer C. J., Berndt S. I., Monda K. L., Thorleifsson G., Jackson A. U., Lango Allen H., Lindgren C. M., Luan J., Magi R., Randall J. C., Vedantam S., Winkler T. W., Qi L., Workalemahu T., Heid I. M., Steinthorsdottir V., Stringham H. M., Weedon M. N., Wheeler E., Wood A. R., Ferreira T., Weyant R. J., Segre A. V., Estrada K., Liang L., Nemesh J., Park J. H., Gustafsson S., Kilpelainen T. O., Yang J., Bouatia-Naji N., Esko T., Feitosa M. F., Kutalik Z., Mangino M., Raychaudhuri S., Scherag A., Smith A. V., Welch R., Zhao J. H., Aben K. K., Absher D. M., Amin N., Dixon A. L., Fisher E., Glazer N. L., Goddard M. E., Heard-Costa N. L., Hoesel V., Hottenga J. J., Johansson A., Johnson T., Ketkar S., Lamina C., Li S., Moffatt M. F., Myers R. H., Narisu N., Perry J. R., Peters M. J., Preuss M., Ripatti S., Rivadeneira F., Sandholt C., Scott L. J., Timpson N. J., Tyrer J. P., van Wingerden S., Watanabe R. M., White C. C., Wiklund F., Barlassina C., Chasman D. I., Cooper M. N., Jansson J. O., Lawrence R. W., Pellikka N., Prokopenko I., Shi J., Thiering E., Alavere H., Alibrandi M. T., Almgren P., Arnold A. M., Aspelund T., Atwood L. D., Balkau B., Balmforth A. J., Bennett A. J., Ben-Shlomo Y., Bergman R. N., Bergmann S., Biebermann H., Blakemore A. I., Boes T., Bonnycastle L. L., Bornstein S. R., Brown M. J., Buchanan T. A., Busonero F., Campbell H., Cappuccio F. P., Cavalcanti-Proenca C., Chen Y. D., Chen C. M., Chines P. S., Clarke R., Coin L., Connell J., Day I. N., den Heijer M., Duan J., Ebrahim S., Elliott P., Elosua R., Eiriksdottir G., Erdos M. R., Eriksson J. G., Facheris M. F., Felix S. B., Fischer-Posovszky P., Folsom A. R., Friedrich N., Freimer N. B., Fu M., Gaget S., Gejman P. V., Geus E. J., Gieger C., Gjesing A. P., Goel A., Goyette P., Grallert H., Grassler J., Greenawalt D. M., Groves C. J., Gudnason V., Guiducci C., Hartikainen A. L., Hassanali N., Hall A. S., Havulinna A. S., Hayward C., Heath A. C., Hengstenberg C., Hicks A. A., Hinney A., Hofman A., Homuth G., Hui J., Igl W., Iribarren C., Isomaa B., Jacobs K. B., Jarick I., Jewell E., John U., Jorgensen T., Jousilahti P., Jula A., Kaakinen M., Kajantie E., Kaplan L. M., Kathiresan S., Kettunen J., Kinnunen L., Knowles J. W., Kolcic I., Konig I. R., Koskinen S., Kovacs P., Kuusisto J., Kraft P., Kvaloy K., Laitinen J., Lantieri O., Lanzani C., Launer L. J., Lecoeur C., Lehtimaki T., Lettre G., Liu J., Lokki M. L., Lorentzon M., Luben R. N., Ludwig B., MAGIC, Manunta P., Marek D., Marre M., Martin N. G., McArdle W. L., McCarthy A., McKnight B., Meitinger T., Melander O., Meyre D., Midthjell K., Montgomery G. W., Morken M. A., Morris A. P., Mulic R., Ngwa J. S., Nelis M., Neville M. J., Nyholt D. R., O'Donnell C. J., O'Rahilly S., Ong K. K., Oostra B., Pare G., Parker A. N., Perola M., Pichler I., Pietilainen K. H., Platou C. G., Polasek O., Pouta A., Rafelt S., Raitakari O., Rayner N. W., Ridderstrale M., Rief W., Ruokonen A., Robertson N. R., Rzehak P., Salomaa V., Sanders A. R., Sandhu M. S., Sanna S., Saramies J., Savolainen M. J., Scherag S., Schipf S., Schreiber S., Schunkert H., Silander K., Sinisalo J., Siscovick D. S., Smit J. H., Soranzo N., Sovio U., Stephens J., Surakka I., Swift A. J., Tammesoo M. L., Tardif J. C., Teder-Laving M., Teslovich T. M., Thompson J. R., Thomson B., Tonjes A., Tuomi T., van Meurs J. B., van Ommen G. J., Vatin V., Viikari J., Visvikis-Siest S., Vitart V., Vogel C. I., Voight B. F., Waite L. L., Wallaschofski H., Walters G. B., Widen E., Wiegand S., Wild S. H., Willemsen G., Witte D. R., Witteman J. C., Xu J., Zhang Q., Zgaga L., Ziegler A., Zitting P., Beilby J. P., Farooqi I. S., Hebebrand J., Huikuri H. V., James A. L., Kahonen M., Levinson D. F., Macciardi F., Nieminen M. S., Ohlsson C., Palmer L. J., Ridker P. M., Stumvoll M., Beckmann J. S., Boeing H., Boerwinkle E., Boomsma D. I., Caulfield M. J., Chanock S. J., Collins F. S., Cupples L. A., Smith G. D., Erdmann J., Froguel P., Gronberg H., Gyllensten U., Hall P., Hansen T., Harris T. B., Hattersley A. T., Hayes R. B., Heinrich J., Hu F. B., Hveem K., Illig T., Jarvelin M. R., Kaprio J., Karpe F., Khaw K. T., Kiemeney L. A., Krude H., Laakso M., Lawlor D. A., Metspalu A., Munroe P. B., Ouwehand W. H., Pedersen O., Penninx B. W., Peters A., Pramstaller P. P., Quertermous T., Reinehr T., Rissanen A., Rudan I., Samani N. J., Schwarz P. E., Shuldiner A. R., Spector T. D., Tuomilehto J., Uda M., Uitterlinden A., Valle T. T., Wabitsch M., Waeber G., Wareham N. J., Watkins H., Procardis C., Wilson J. F., Wright A. F., Zillikens M. C., Chatterjee N., McCarroll S. A., Purcell S., Schadt E. E., Visscher P. M., Assimes T. L., Borecki I. B., Deloukas P., Fox C. S., Groop L. C., Haritunians T., Hunter D. J., Kaplan R. C., Mohlke K. L., O'Connell J. R., Peltonen L., Schlessinger D., Strachan D. P., van Duijn C. M., Wichmann H. E., Frayling T. M., Thorsteinsdottir U., Abecasis G. R., Barroso I., Boehnke M., Stefansson K., North K. E., McCarthy M. I., Hirschhorn J. N., Ingelsson E., Loos R. J. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Consortium for Blood Pressure Genome-Wide Association Studies, Ehret G. B., Munroe P. B., Rice K. M., Bochud M., Johnson A. D., Chasman D. I., Smith A. V., Tobin M. D., Verwoert G. C., Hwang S. J., Pihur V., Vollenweider P., O'Reilly P. F., Amin N., Bragg-Gresham J. L., Teumer A., Glazer N. L., Launer L., Zhao J. H., Aulchenko Y., Heath S., Sober S., Parsa A., Luan J., Arora P., Dehghan A., Zhang F., Lucas G., Hicks A. A., Jackson A. U., Peden J. F., Tanaka T., Wild S. H., Rudan I., Igl W., Milaneschi Y., Parker A. N., Fava C., Chambers J. C., Fox E. R., Kumari M., Go M. J., van der Harst P., Kao W. H., Sjogren M., Vinay D. G., Alexander M., Tabara Y., Shaw-Hawkins S., Whincup P. H., Liu Y., Shi G., Kuusisto J., Tayo B., Seielstad M., Sim X., Nguyen K. D., Lehtimaki T., Matullo G., Wu Y., Gaunt T. R., Onland-Moret N. C., Cooper M. N., Platou C. G., Org E., Hardy R., Dahgam S., Palmen J., Vitart V., Braund P. S., Kuznetsova T., Uiterwaal C. S., Adeyemo A., Palmas W., Campbell H., Ludwig B., Tomaszewski M., Tzoulaki I., Palmer N. D., consortium C. A., Consortium C. K., KidneyGen C., EchoGen C., consortium C.-H., Aspelund T., Garcia M., Chang Y. P., O'Connell J. R., Steinle N. I., Grobbee D. E., Arking D. E., Kardia S. L., Morrison A. C., Hernandez D., Najjar S., McArdle W. L., Hadley D., Brown M. J., Connell J. M., Hingorani A. D., Day I. N., Lawlor D. A., Beilby J. P., Lawrence R. W., Clarke R., Hopewell J. C., Ongen H., Dreisbach A. W., Li Y., Young J. H., Bis J. C., Kahonen M., Viikari J., Adair L. S., Lee N. R., Chen M. H., Olden M., Pattaro C., Bolton J. A., Kottgen A., Bergmann S., Mooser V., Chaturvedi N., Frayling T. M., Islam M., Jafar T. H., Erdmann J., Kulkarni S. R., Bornstein S. R., Grassler J., Groop L., Voight B. F., Kettunen J., Howard P., Taylor A., Guarrera S., Ricceri F., Emilsson V., Plump A., Barroso I., Khaw K. T., Weder A. B., Hunt S. C., Sun Y. V., Bergman R. N., Collins F. S., Bonnycastle L. L., Scott L. J., Stringham H. M., Peltonen L., Perola M., Vartiainen E., Brand S. M., Staessen J. A., Wang T. J., Burton P. R., Soler Artigas M., Dong Y., Snieder H., Wang X., Zhu H., Lohman K. K., Rudock M. E., Heckbert S. R., Smith N. L., Wiggins K. L., Doumatey A., Shriner D., Veldre G., Viigimaa M., Kinra S., Prabhakaran D., Tripathy V., Langefeld C. D., Rosengren A., Thelle D. S., Corsi A. M., Singleton A., Forrester T., Hilton G., McKenzie C. A., Salako T., Iwai N., Kita Y., Ogihara T., Ohkubo T., Okamura T., Ueshima H., Umemura S., Eyheramendy S., Meitinger T., Wichmann H. E., Cho Y. S., Kim H. L., Lee J. Y., Scott J., Sehmi J. S., Zhang W., Hedblad B., Nilsson P., Smith G. D., Wong A., Narisu N., Stancakova A., Raffel L. J., Yao J., Kathiresan S., O'Donnell C. J., Schwartz S. M., Ikram M. A., Longstreth Jr. W. T., Mosley T. H., Seshadri S., Shrine N. R., Wain L. V., Morken M. A., Swift A. J., Laitinen J., Prokopenko I., Zitting P., Cooper J. A., Humphries S. E., Danesh J., Rasheed A., Goel A., Hamsten A., Watkins H., Bakker S. J., van Gilst W. H., Janipalli C. S., Mani K. R., Yajnik C. S., Hofman A., Mattace-Raso F. U., Oostra B. A., Demirkan A., Isaacs A., Rivadeneira F., Lakatta E. G., Orru M., Scuteri A., Ala-Korpela M., Kangas A. J., Lyytikainen L. P., Soininen P., Tukiainen T., Wurtz P., Ong R. T., Dorr M., Kroemer H. K., Volker U., Volzke H., Galan P., Hercberg S., Lathrop M., Zelenika D., Deloukas P., Mangino M., Spector T. D., Zhai G., Meschia J. F., Nalls M. A., Sharma P., Terzic J., Kumar M. V., Denniff M., Zukowska-Szczechowska E., Wagenknecht L. E., Fowkes F. G., Charchar F. J., Schwarz P. E., Hayward C., Guo X., Rotimi C., Bots M. L., Brand E., Samani N. J., Polasek O., Talmud P. J., Nyberg F., Kuh D., Laan M., Hveem K., Palmer L. J., van der Schouw Y. T., Casas J. P., Mohlke K. L., Vineis P., Raitakari O., Ganesh S. K., Wong T. Y., Tai E. S., Cooper R. S., Laakso M., Rao D. C., Harris T. B., Morris R. W., Dominiczak A. F., Kivimaki M., Marmot M. G., Miki T., Saleheen D., Chandak G. R., Coresh J., Navis G., Salomaa V., Han B. G., Zhu X., Kooner J. S., Melander O., Ridker P. M., Bandinelli S., Gyllensten U. B., Wright A. F., Wilson J. F., Ferrucci L., Farrall M., Tuomilehto J., Pramstaller P. P., Elosua R., Soranzo N., Sijbrands E. J., Altshuler D., Loos R. J., Shuldiner A. R., Gieger C., Meneton P., Uitterlinden A. G., Wareham N. J., Gudnason V., Rotter J. I., Rettig R., Uda M., Strachan D. P., Witteman J. C., Hartikainen A. L., Beckmann J. S., Boerwinkle E., Vasan R. S., Boehnke M., Larson M. G., Jarvelin M. R., Psaty B. M., Abecasis G. R., Chakravarti A., Elliott P., van Duijn C. M., Newton-Cheh C., Levy D., Caulfield M. J., Johnson T. Nature. 2011;478:103–109. [Google Scholar]
- Carrera N., Arrojo M., Sanjuan J., Ramos-Rios R., Paz E., Suarez-Rama J. J., Paramo M., Agra S., Brenlla J., Martinez S., Rivero O., Collier D. A., Palotie A., Cichon S., Nothen M. M., Rietschel M., Rujescu D., Stefansson H., Steinberg S., Sigurdsson E., St Clair D., Tosato S., Werge T., Stefansson K., Gonzalez J. C., Valero J., Gutierrez-Zotes A., Labad A., Martorell L., Vilella E., Carracedo A., Costas J. Biol. Psychiatry. 2012;71:169–177. doi: 10.1016/j.biopsych.2011.09.032. [DOI] [PubMed] [Google Scholar]
- Berndt S. I., Gustafsson S., Magi R., Ganna A., Wheeler E., Feitosa M. F., Justice A. E., Monda K. L., Croteau-Chonka D. C., Day F. R., Esko T., Fall T., Ferreira T., Gentilini D., Jackson A. U., Luan J., Randall J. C., Vedantam S., Willer C. J., Winkler T. W., Wood A. R., Workalemahu T., Hu Y. J., Lee S. H., Liang L., Lin D. Y., Min J. L., Neale B. M., Thorleifsson G., Yang J., Albrecht E., Amin N., Bragg-Gresham J. L., Cadby G., den Heijer M., Eklund N., Fischer K., Goel A., Hottenga J. J., Huffman J. E., Jarick I., Johansson A., Johnson T., Kanoni S., Kleber M. E., Konig I. R., Kristiansson K., Kutalik Z., Lamina C., Lecoeur C., Li G., Mangino M., McArdle W. L., Medina-Gomez C., Muller-Nurasyid M., Ngwa J. S., Nolte I. M., Paternoster L., Pechlivanis S., Perola M., Peters M. J., Preuss M., Rose L. M., Shi J., Shungin D., Smith A. V., Strawbridge R. J., Surakka I., Teumer A., Trip M. D., Tyrer J., Van Vliet-Ostaptchouk J. V., Vandenput L., Waite L. L., Zhao J. H., Absher D., Asselbergs F. W., Atalay M., Attwood A. P., Balmforth A. J., Basart H., Beilby J., Bonnycastle L. L., Brambilla P., Bruinenberg M., Campbell H., Chasman D. I., Chines P. S., Collins F. S., Connell J. M., Cookson W. O., de Faire U., de Vegt F., Dei M., Dimitriou M., Edkins S., Estrada K., Evans D. M., Farrall M., Ferrario M. M., Ferrieres J., Franke L., Frau F., Gejman P. V., Grallert H., Gronberg H., Gudnason V., Hall A. S., Hall P., Hartikainen A. L., Hayward C., Heard-Costa N. L., Heath A. C., Hebebrand J., Homuth G., Hu F. B., Hunt S. E., Hypponen E., Iribarren C., Jacobs K. B., Jansson J. O., Jula A., Kahonen M., Kathiresan S., Kee F., Khaw K. T., Kivimaki M., Koenig W., Kraja A. T., Kumari M., Kuulasmaa K., Kuusisto J., Laitinen J. H., Lakka T. A., Langenberg C., Launer L. J., Lind L., Lindstrom J., Liu J., Liuzzi A., Lokki M. L., Lorentzon M., Madden P. A., Magnusson P. K., Manunta P., Marek D., Marz W., Mateo Leach I., McKnight B., Medland S. E., Mihailov E., Milani L., Montgomery G. W., Mooser V., Muhleisen T. W., Munroe P. B., Musk A. W., Narisu N., Navis G., Nicholson G., Nohr E. A., Ong K. K., Oostra B. A., Palmer C. N., Palotie A., Peden J. F., Pedersen N., Peters A., Polasek O., Pouta A., Pramstaller P. P., Prokopenko I., Putter C., Radhakrishnan A., Raitakari O., Rendon A., Rivadeneira F., Rudan I., Saaristo T. E., Sambrook J. G., Sanders A. R., Sanna S., Saramies J., Schipf S., Schreiber S., Schunkert H., Shin S. Y., Signorini S., Sinisalo J., Skrobek B., Soranzo N., Stancakova A., Stark K., Stephens J. C., Stirrups K., Stolk R. P., Stumvoll M., Swift A. J., Theodoraki E. V., Thorand B., Tregouet D. A., Tremoli E., Van der Klauw M. M., van Meurs J. B., Vermeulen S. H., Viikari J., Virtamo J., Vitart V., Waeber G., Wang Z., Widen E., Wild S. H., Willemsen G., Winkelmann B. R., Witteman J. C., Wolffenbuttel B. H., Wong A., Wright A. F., Zillikens M. C., Amouyel P., Boehm B. O., Boerwinkle E., Boomsma D. I., Caulfield M. J., Chanock S. J., Cupples L. A., Cusi D., Dedoussis G. V., Erdmann J., Eriksson J. G., Franks P. W., Froguel P., Gieger C., Gyllensten U., Hamsten A., Harris T. B., Hengstenberg C., Hicks A. A., Hingorani A., Hinney A., Hofman A., Hovingh K. G., Hveem K., Illig T., Jarvelin M. R., Jockel K. H., Keinanen-Kiukaanniemi S. M., Kiemeney L. A., Kuh D., Laakso M., Lehtimaki T., Levinson D. F., Martin N. G., Metspalu A., Morris A. D., Nieminen M. S., Njolstad I., Ohlsson C., Oldehinkel A. J., Ouwehand W. H., Palmer L. J., Penninx B., Power C., Province M. A., Psaty B. M., Qi L., Rauramaa R., Ridker P. M., Ripatti S., Salomaa V., Samani N. J., Snieder H., Sorensen T. I., Spector T. D., Stefansson K., Tonjes A., Tuomilehto J., Uitterlinden A. G., Uusitupa M., van der Harst P., Vollenweider P., Wallaschofski H., Wareham N. J., Watkins H., Wichmann H. E., Wilson J. F., Abecasis G. R., Assimes T. L., Barroso I., Boehnke M., Borecki I. B., Deloukas P., Fox C. S., Frayling T., Groop L. C., Haritunian T., Heid I. M., Hunter D., Kaplan R. C., Karpe F., Moffatt M. F., Mohlke K. L., O'Connell J. R., Pawitan Y., Schadt E. E., Schlessinger D., Steinthorsdottir V., Strachan D. P., Thorsteinsdottir U., van Duijn C. M., Visscher P. M., Di Blasio A. M., Hirschhorn J. N., Lindgren C. M., Morris A. P., Meyre D., Scherag A., McCarthy M. I., Speliotes E. K., North K. E., Loos R. J., Ingelsson E. Nat. Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium, Ripke S., Neale B. M., Corvin A., Walters J. T., Farh K. H., Holmans P. A., Lee P., Bulik-Sullivan B., Collier D. A., Huang H., Pers T. H., Agartz I., Agerbo E., Albus M., Alexander M., Amin F., Bacanu S. A., Begemann M., Jr. Belliveau R. A., Bene J., Bergen S. E., Bevilacqua E., Bigdeli T. B., Black D. W., Bruggeman R., Buccola N. G., Buckner R. L., Byerley W., Cahn W., Cai G., Campion D., Cantor R. M., Carr V. J., Carrera N., Catts S. V., Chambert K. D., Chan R. C., Chen R. Y., Chen E. Y., Cheng W., Cheung E. F., Chong S. A., Cloninger C. R., Cohen D., Cohen N., Cormican P., Craddock N., Crowley J. J., Curtis D., Davidson M., Davis K. L., Degenhardt F., Del Favero J., Demontis D., Dikeos D., Dinan T., Djurovic S., Donohoe G., Drapeau E., Duan J., Dudbridge F., Durmishi N., Eichhammer P., Eriksson J., Escott-Price V., Essioux L., Fanous A. H., Farrell M. S., Frank J., Franke L., Freedman R., Freimer N. B., Friedl M., Friedman J. I., Fromer M., Genovese G., Georgieva L., Giegling I., Giusti-Rodríguez P., Godard S., Goldstein J. I., Golimbet V., Gopal S., Gratten J., de Haan L., Hammer C., Hamshere M. L., Hansen M., Hansen T., Haroutunian V., Hartmann A. M., Henskens F. A., Herms S., Hirschhorn J. N., Hoffmann P., Hofman A., Hollegaard M. V., Hougaard D. M., Ikeda M., Joa I., Julià A., Kahn R. S., Kalaydjieva L., Karachanak-Yankova S., Karjalainen J., Kavanagh D., Keller M. C., Kennedy J. L., Khrunin A., Kim Y., Klovins J., Knowles J. A., Konte B., Kucinskas V., Ausrele Kucinskiene Z., Kuzelova-Ptackova H., Kähler A. K., Laurent C., Keong J. L., Lee S. H., Legge S. E., Lerer B., Li M., Li T., Liang K. Y., Lieberman J., Limborska S., Loughland C. M., Lubinski J., Lönnqvist J., Jr. Macek M., Magnusson P. K., Maher B. S., Maier W., Mallet J., Marsal S., Mattheisen M., Mattingsdal M., McCarley R. W., McDonald C., McIntosh A. M., Meier S., Meijer C. J., Melegh B., Melle I., Mesholam-Gately R. I., Metspalu A., Michie P. T., Milani L., Milanova V., Mokrab Y., Morris D. W., Mors O., Murphy K. C., Murray R. M., Myin-Germeys I., Müller-Myhsok B., Nelis M., Nenadic I., Nertney D. A., Nestadt G., Nicodemus K. K., Nikitina-Zake L., Nisenbaum L., Nordin A., O'Callaghan E., O'Dushlaine C., O'Neill F. A., Oh S. Y., Olincy A., Olsen L., Van Os. J., Pantelis C., Papadimitriou G. N., Papiol S., Parkhomenko E., Pato M. T., Paunio T., Pejovic-Milovancevic M., Perkins D. O., Pietiläinen O., Pimm J., Pocklington A. J., Powell J., Price A., Pulver A. E., Purcell S. M., Quested D., Rasmussen H. B., Reichenberg A., Reimers M. A., Richards A. L., Roffman J. L., Roussos P., Ruderfer D. M., Salomaa V., Sanders A. R., Schall U., Schubert C. R., Schulze T. G., Schwab S. G., Scolnick E. M., Scott R. J., Seidman L. J., Shi J., Sigurdsson E., Silagadze T., Silverman J. M., Sim K., Slominsky P., Smoller J. W., So H. C., Spencer C. A., Stahl E. A., Stefansson H., Steinberg S., Stogmann E., Straub R. E., Strengman E., Strohmaier J., Stroup T. S., Subramaniam M., Suvisaari J., Svrakic D. M., Szatkiewicz J. P., Söderman E., Thirumalai S., Toncheva D., Tosato S., Veijola J., Waddington J., Walsh D., Wang D., Wang Q., Webb B. T., Weiser M., Wildenauer D. B., Williams N. M., Williams S., Witt S. H., Wolen A. R., Wong E. H., Wormley B. K., Xi H. S., Zai C. C., Zheng X., Zimprich F., Wray N. R., Stefansson K., Visscher P. M., Adolfsson R., Andreassen O. A., Blackwood D. H., Bramon E., Buxbaum J. D., Børglum A. D., Cichon S., Darvasi A., Domenici E., Ehrenreich H., Esko T., Gejman P. V., Gill M., Gurling H., Hultman C. M., Iwata N., Jablensky A. V., Jönsson E. G., Kendler K. S., Kirov G., Knight J., Lencz T., Levinson D. F., Li Q. S., Liu J., Malhotra A. K., McCarroll S. A., McQuillin A., Moran J. L., Mortensen P. B., Mowry B. J., Nöthen M. M., Ophoff R. A., Owen M. J., Palotie A., Pato C. N., Petryshen T. L., Posthuma D., Rietschel M., Riley B. P., Rujescu D., Sham P. C., Sklar P., St Clair D., Weinberger D. R., Wendland J. R., Werge T., Daly M. J., Sullivan P. F., O'Donovan M. C. Nature. 2014;511:421–427. [Google Scholar]
- Tragante V., Barnes M. R., Ganesh S. K., Lanktree M. B., Guo W., Franceschini N., Smith E. N., Johnson T., Holmes M. V., Padmanabhan S., Karczewski K. J., Almoguera B., Barnard J., Baumert J., Chang Y. P., Elbers C. C., Farrall M., Fischer M. E., Gaunt T. R., Gho J. M., Gieger C., Goel A., Gong Y., Isaacs A., Kleber M. E., Mateo Leach I., McDonough C. W., Meijs M. F., Melander O., Nelson C. P., Nolte I. M., Pankratz N., Price T. S., Shaffer J., Shah S., Tomaszewski M., van der Most P. J., Van Iperen E. P., Vonk J. M., Witkowska K., Wong C. O., Zhang L., Beitelshees A. L., Berenson G. S., Bhatt D. L., Brown M., Burt A., Cooper-DeHoff R. M., Connell J. M., Cruickshanks K. J., Curtis S. P., Davey-Smith G., Delles C., Gansevoort R. T., Guo X., Haiqing S., Hastie C. E., Hofker M. H., Hovingh G. K., Kim D. S., Kirkland S. A., Klein B. E., Klein R., Li Y. R., Maiwald S., Newton-Cheh C., O'Brien E. T., Onland-Moret N. C., Palmas W., Parsa A., Penninx B. W., Pettinger M., Vasan R. S., Ranchalis J. E., Rose M. R. P. L. M., Sever P., Shimbo D., Steele L., Stolk R. P., Thorand B., Trip M. D., van Duijn C. M., Verschuren W. M., Wijmenga C., Wyatt S., Young J. H., Zwinderman A. H., Bezzina C. R., Boerwinkle E., Casas J. P., Caulfield M. J., Chakravarti A., Chasman D. I., Davidson K. W., Doevendans P. A., Dominiczak A. F., FitzGerald G. A., Gums J. G., Fornage M., Hakonarson H., Halder I., Hillege H. L., Illig T., Jarvik G. P., Johnson J. A., Kastelein J. J., Koenig W., Kumari M., Marz W., Murray S. S., O'Connell J. R., Oldehinkel A. J., Pankow J. S., Rader D. J., Redline S., Reilly M. P., Schadt E. E., Kottke-Marchant K., Snieder H., Snyder M., Stanton A. V., Tobin M. D., Uitterlinden A. G., van der Harst P., van der Schouw Y. T., Samani N. J., Watkins H., Johnson A. D., Reiner A. P., Zhu X., de Bakker P. I., Levy D., Asselbergs F. W., Munroe P. B., Keating B. J. Am. J. Hum. Genet. 2014;94:349–360. doi: 10.1016/j.ajhg.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Jeon J., Shin M., Won Y., Lee M., Kwak J. S., Lee G., Rhee J., Ryu J. H., Chun C. H., Chun J. S. Cell. 2014;156:730–743. doi: 10.1016/j.cell.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Park J. H., Hogrebe M., Gruneberg M., DuChesne I., von der Heiden A. L., Reunert J., Schlingmann K. P., Boycott K. M., Beaulieu C. L., Mhanni A. A., Innes A. M., Hortnagel K., Biskup S., Gleixner E. M., Kurlemann G., Fiedler B., Omran H., Rutsch F., Wada Y., Tsiakas K., Santer R., Nebert D. W., Rust S., Marquardt T. Am. J. Hum. Genet. 2015;97:894–903. doi: 10.1016/j.ajhg.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Eide D. J. Mol. Aspects Med. 2013;34:612–619. doi: 10.1016/j.mam.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez-Peralta M., Wang Z., Bao S., Knoell D. L., Nebert D. W. Int. J. Toxicol. 2014;33:246–258. doi: 10.1177/1091581814529310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambe T. Nihon Eiseigaku Zasshi. 2013;68:92–102. doi: 10.1265/jjh.68.92. [DOI] [PubMed] [Google Scholar]
- Jenkitkasemwong S., Wang C. Y., Mackenzie B., Knutson M. D. BioMetals. 2012;25:643–655. doi: 10.1007/s10534-012-9526-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkeb L., Mukherjee I., Chatterjee I., Lahner B., Salt D. E., Connolly E. L. Plant Physiol. 2008;146:1964–1973. doi: 10.1104/pp.107.113282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Kim B. E., Wang F., Eide D. J., Petris M. J. J. Biol. Chem. 2007;282:6992–7000. doi: 10.1074/jbc.M610552200. [DOI] [PubMed] [Google Scholar]
- Alber T. Curr. Opin. Genet. Dev. 1992;2:205–210. doi: 10.1016/s0959-437x(05)80275-8. [DOI] [PubMed] [Google Scholar]
- Sharma K., D'Souza R. C., Tyanova S., Schaab C., Wisniewski J. R., Cox J., Mann M. Cell Rep. 2014;8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- Gauci S., Helbig A. O., Slijper M., Krijgsveld J., Heck A. J., Mohammed S. Anal. Chem. 2009;81:4493–4501. doi: 10.1021/ac9004309. [DOI] [PubMed] [Google Scholar]
- Kim J. E., Tannenbaum S. R., White F. M. J. Proteome Res. 2005;4:1339–1346. doi: 10.1021/pr050048h. [DOI] [PubMed] [Google Scholar]
- Krotova K. Y., Zharikov S. I., Block E. R. Am. J. Physiol. 2003;284:L1037–L1044. doi: 10.1152/ajplung.00308.2002. [DOI] [PubMed] [Google Scholar]
- Ciarimboli G., Koepsell H., Iordanova M., Gorboulev V., Durner B., Lang D., Edemir B., Schroter R., Van Le T., Schlatter E. J. Am. Soc. Nephrol. 2005;16:1562–1570. doi: 10.1681/ASN.2004040256. [DOI] [PubMed] [Google Scholar]
- Vayro S., Silverman M. Am. J. Physiol. 1999;276:C1053–C1060. doi: 10.1152/ajpcell.1999.276.5.C1053. [DOI] [PubMed] [Google Scholar]
- Martin P., Boulukos K. E., Poggi M. C., Pognonec P. FEBS J. 2009;276:1667–1679. doi: 10.1111/j.1742-4658.2009.06899.x. [DOI] [PubMed] [Google Scholar]
- Ajjimaporn A., Botsford T., Garrett S. H., Sens M. A., Zhou X. D., Dunlevy J. R., Sens D. A., Somji S. Cancer Cell Int. 2012;12:16. doi: 10.1186/1475-2867-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu M. S., Lichten L. A., Liuzzi J. P., Cousins R. J. J. Nutr. 2008;138:2076–2083. doi: 10.3945/jn.108.093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhang Y., Cui X., Yao W., Yu X., Cen P., Hodges S. E., Fisher W. E., Brunicardi F. C., Chen C., Yao Q., Li M. Curr. Mol. Med. 2013;13:401–409. [PMC free article] [PubMed] [Google Scholar]
- Wang C. Y., Jenkitkasemwong S., Sparkman B. K., Shawki A., Mackenzie B., Knutson M., FASEB J., 2012, 26 , , 641.32 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Girijashanker K., Dalton T. P., Reed J., Li H., Soleimani M., Nebert D. W. Mol. Pharmacol. 2006;70:171–180. doi: 10.1124/mol.106.024521. [DOI] [PubMed] [Google Scholar]
- Ding D., Salvi R., Roth J. A. BioMetals. 2014;27:731–744. doi: 10.1007/s10534-014-9765-0. [DOI] [PubMed] [Google Scholar]
- Ding D. L., Roth J., Salvi R. NeuroToxicology. 2011;32:233–241. doi: 10.1016/j.neuro.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N., Zhang A. S., Worthen C., Knutson M. D., Enns C. A. Proc. Natl. Acad. Sci. U. S. A. 2014;111:9175–9180. doi: 10.1073/pnas.1405355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill G. M., Curtis A. M., Adamik J., Palsson-McDermott E. M., McGettrick A. F., Goel G., Frezza C., Bernard N. J., Kelly B., Foley N. H., Zheng L., Gardet A., Tong Z., Jany S. S., Corr S. C., Haneklaus M., Caffrey B. E., Pierce K., Walmsley S., Beasley F. C., Cummins E., Nizet V., Whyte M., Taylor C. T., Lin H., Masters S. L., Gottlieb E., Kelly V. P., Clish C., Auron P. E., Xavier R. J., O'Neill L. A. J. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa K., Takatsu K. Immunology. 2006;118:497–508. doi: 10.1111/j.1365-2567.2006.02382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba I., Hossain A., Kuo M. T. Mol. Pharmacol. 2008;74:823–833. doi: 10.1124/mol.108.046862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidi T., Cano C. E., Grasso D., Garcia M. N., Sandi M. J., Calvo E. L., Dagorn J. C., Lomberk G., Urrutia R., Goruppi S., Carracedo A., Velasco G., Iovanna J. L. Clin. Cancer Res. 2012;18:5234–5246. doi: 10.1158/1078-0432.CCR-12-0026. [DOI] [PubMed] [Google Scholar]
- Bellomo E. A., Meur G., Rutter G. A. J. Biol. Chem. 2011;286:25778–25789. doi: 10.1074/jbc.M111.246082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Hannon G. J. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Kim V. N. Mol. Cells. 2005;19:1–15. [PubMed] [Google Scholar]
- Bartel D. P. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Slack F. J. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Iorio M. V., Croce C. M. EMBO Mol. Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceppi P., Peter M. E. Oncogene. 2014;33:269–278. doi: 10.1038/onc.2013.55. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D., Malizos K. N., Oikonomou P., Tsezou A. PLoS One. 2008;3:e3740. doi: 10.1371/journal.pone.0003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Kim D., Lee C. H., Lee M. S., Chun C. H., Jin E. J. J. Biomed. Sci. 2013;20:31. doi: 10.1186/1423-0127-20-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buterin T., Koch C., Naegeli H. Carcinogenesis. 2006;27:1567–1578. doi: 10.1093/carcin/bgi339. [DOI] [PubMed] [Google Scholar]
- Nicholson R. I., Johnston S. R. Breast Cancer Res. Treat. 2005;93((Suppl 1)):S3–S10. doi: 10.1007/s10549-005-9036-4. [DOI] [PubMed] [Google Scholar]
- Taylor K. M., Morgan H. E., Smart K., Zahari N. M., Pumford S., Ellis I. O., Robertson J. F., Nicholson R. I. Mol. Med. 2007;13:396–406. doi: 10.2119/2007-00040.Taylor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R. A., Chang C. Y., Kazmin D. A., Way J., Schroeder T., Wergin M., Dewhirst M. W., McDonnell D. P. Cancer Res. 2008;68:8805–8812. doi: 10.1158/0008-5472.CAN-08-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn Y. H., Yang Y. A., Gibbons D. L., Creighton C. J., Yang F., Wistuba I. I., Lin W., Thilaganathan N., Alvarez C. A., Roybal J., Goldsmith E. J., Tournier C., Kurie J. M. Mol. Cell. Biol. 2011;31:4270–4285. doi: 10.1128/MCB.05562-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhofs S., Dubois V., De Gendt K., Helsen C., Clinckemalie L., Spans L., Schuit F., Boonen S., Vanderschueren D., Saunders P. T., Verhoeven G., Claessens F. FASEB J. 2012;26:4360–4372. doi: 10.1096/fj.11-202283. [DOI] [PubMed] [Google Scholar]
- Nebert D. W., Galvez-Peralta M., Hay E. B., Li H., Johansson E., Yin C., Wang B., He L., Soleimani M. Metallomics. 2012;4:1218–1225. doi: 10.1039/c2mt20177a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinilla-Tenas J. J., Sparkman B. K., Shawki A., Illing A. C., Mitchell C. J., Zhao N. N., Liuzzi J. P., Cousins R. J., Knutson M. D., Mackenzie B. Am. J. Physiol. 2011;301:C862–C871. doi: 10.1152/ajpcell.00479.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., He L., Dong H., Dalton T. P., Nebert D. W. Biochem. Biophys. Res. Commun. 2011;410:289–294. doi: 10.1016/j.bbrc.2011.05.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez-Peralta M., He L., Jorge-Nebert L. F., Wang B., Miller M. L., Eppert B. L., Afton S., Nebert D. W. PLoS One. 2012;7:e36055. doi: 10.1371/journal.pone.0036055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews G. K. Biochem. Soc. Trans. 2008;36:1242–1246. doi: 10.1042/BST0361242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyulkhandanyan A. V., Lu H., Lee S. C., Bhattacharjee A., Wijesekara N., Fox J. E., MacDonald P. E., Chimienti F., Dai F. F., Wheeler M. B. J. Biol. Chem. 2008;283:10184–10197. doi: 10.1074/jbc.M707005200. [DOI] [PubMed] [Google Scholar]
- Wilson D. O., Weissfeld J. L., Balkan A., Schragin J. G., Fuhrman C. R., Fisher S. N., Wilson J., Leader J. K., Siegfried J. M., Shapiro S. D., Sciurba F. C. Am. J. Respir. Crit. Care Med. 2008;178:738–744. doi: 10.1164/rccm.200803-435OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado F., Bartholmai B. J., Swensen S. J., Midthun D. E., Decker P. A., Jett J. R. Chest. 2010;138:1295–1302. doi: 10.1378/chest.09-2567. [DOI] [PubMed] [Google Scholar]
- Jamal A., Homa D. M., O'Connor E., Babb S. D., Caraballo R. S., Singh T., Hu S. S., King B. A. MMWR Morb. Mortal. Wkly Rep. 2015;64:1233–1240. doi: 10.15585/mmwr.mm6444a2. [DOI] [PubMed] [Google Scholar]
- Dalton T. P., He L., Wang B., Miller M. L., Jin L., Stringer K. F., Chang X., Baxter C. S., Nebert D. W. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3401–3406. doi: 10.1073/pnas.0406085102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S. N., Liu Z., Wang B., Miller M. L., Afton S. E., Soleimani M., Nebert D. W. Internet J. Toxicol. 2014;33:14–20. doi: 10.1177/1091581813513530. [DOI] [PubMed] [Google Scholar]
- Ji C., Kosman D. J. J. Neurochem. 2015;133:668–683. doi: 10.1111/jnc.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunshin H., Mackenzie B., Berger U. V., Gunshin Y., Romero M. F., Boron W. F., Nussberger S., Gollan J. L., Hediger M. A. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- Moos T., Morgan E. H. Ann. N. Y. Acad. Sci. 2004;1012:14–26. doi: 10.1196/annals.1306.002. [DOI] [PubMed] [Google Scholar]
- Raymond A. D., Gekonge B., Giri M. S., Hancock A., Papasavvas E., Chehimi J., Kossevkov A. V., Nicols C., Yousef M., Mounzer K., Shull J., Kostman J., Showe L., Montaner L. J. J. Leukocyte. Biol. 2010;88:589–596. doi: 10.1189/jlb.0110051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idaghdour Y., Quinlan J., Goulet J. P., Berghout J., Gbeha E., Bruat V., de Malliard T., Grenier J. C., Gomez S., Gros P., Rahimy M. C., Sanni A., Awadalla P. Proc. Natl. Acad. Sci. U. S. A. 2012;109:16786–16793. doi: 10.1073/pnas.1204945109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeba A. N., Sorgho H., Rouamba N., Zongo I., Rouamba J., Guiguemde R. T., Hamer D. H., Mokhtar N., Ouedraogo J. B. Nutr. J. 2008;7:7. doi: 10.1186/1475-2891-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovesen J., Moller-Madsen B., Nielsen P. T., Christensen P. H., Simonsen O., Hoeck H. C., Laursen M. B., Thomsen J. S. J. Trace Elem. Med. Biol. 2009;23:1–8. doi: 10.1016/j.jtemb.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Roschger A., Hofstaetter J. G., Pemmer B., Zoeger N., Wobrauschek P., Falkenberg G., Simon R., Berzlanovich A., Thaler H. W., Roschger P., Klaushofer K., Streli C. Osteoarthritis Cartilage. 2013;21:1707–1715. doi: 10.1016/j.joca.2013.06.029. [DOI] [PubMed] [Google Scholar]
- Blom A. B., Lent P. L., Libregts S., Holthuysen A. E., van der Kraan P. M., van Rooijen N., van den Berg W. B. Arthritis Rheum. 2007;56:147–157. doi: 10.1002/art.22337. [DOI] [PubMed] [Google Scholar]
- Glasson S. S., Askew R., Sheppard B., Carito B., Blanchet T., Ma H. L., Flannery C. R., Peluso D., Kanki K., Yang Z., Majumdar M. K., Morris E. A. Nature. 2005;434:644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- Little C. B., Barai A., Burkhardt D., Smith S. M., Fosang A. J., Werb Z., Shah M., Thompson E. W. Arthritis Rheum. 2009;60:3723–3733. doi: 10.1002/art.25002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Won Y., Shin Y., Kim J. H., Chun J. S. Osteoarthritis Cartilage. 2016;24:134–145. doi: 10.1016/j.joca.2015.07.016. [DOI] [PubMed] [Google Scholar]
- Boycott K. M., Beaulieu C. L., Kernohan K. D., Gebril O. H., Mhanni A., Chudley A. E., Redl D., Qin W., Hampson S., Kury S., Tetreault M., Puffenberger E. G., Scott J. N., Bezieau S., Reis A., Uebe S., Schumacher J., Hegele R. A., McLeod D. R., Galvez-Peralta M., Majewski J., Ramaekers V. T., Care4Rare Canada C., Nebert D. W., Innes A. M., Parboosingh J. S., Abou Jamra R. Am. J. Hum. Genet. 2015;97:886–893. doi: 10.1016/j.ajhg.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg L., Katsel P., Roussos P., Haroutunian V., Domany E. Schizophr. Res. 2015;164:92–99. doi: 10.1016/j.schres.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., Johansen C. T., Fouchier S. W., Isaacs A., Peloso G. M., Barbalic M., Ricketts S. L., Bis J. C., Aulchenko Y. S., Thorleifsson G., Feitosa M. F., Chambers J., Orho-Melander M., Melander O., Johnson T., Li X. H., Guo X. Q., Li M. Y., Cho Y. S., Go M. J., Kim Y. J., Lee J. Y., Park T., Kim K., Sim X., Ong R. T. H., Croteau-Chonka D. C., Lange L. A., Smith J. D., Song K., Zhao J. H., Yuan X., Luan J. A., Lamina C., Ziegler A., Zhang W., Zee R. Y. L., Wright A. F., Witteman J. C. M., Wilson J. F., Willemsen G., Wichmann H. E., Whitfield J. B., Waterworth D. M., Wareham N. J., Waeber G., Vollenweider P., Voight B. F., Vitart V., Uitterlinden A. G., Uda M., Tuomilehto J., Thompson J. R., Tanaka T., Surakka I., Stringham H. M., Spector T. D., Soranzo N., Smit J. H., Sinisalo J., Silander K., Sijbrands E. J. G., Scuteri A., Scott J., Schlessinger D., Sanna S., Salomaa V., Saharinen J., Sabatti C., Ruokonen A., Rudan I., Rose L. M., Roberts R., Rieder M., Psaty B. M., Pramstaller P. P., Pichler I., Perola M., Penninx B. W. J. H., Pedersen N. L., Pattaro C., Parker A. N., Pare G., Oostra B. A., O'Donnell C. J., Nieminen M. S., Nickerson D. A., Montgomery G. W., Meitinger T., McPherson R., McCarthy M. I., McArdle W., Masson D., Martin N. G., Marroni F., Mangino M., Magnusson P. K. E., Lucas G., Luben R., Loos R. J. F., Lokki M. L., Lettre G., Langenberg C., Launer L. J., Lakatta E. G., Laaksonen R., Kyvik K. O., Kronenberg F., Konig I. R., Khaw K. T., Kaprio J., Kaplan L. M., Johansson A., Jarvelin M. R., Janssens A. C. J. W., Ingelsson E., Igi W., Hovingh G. K., Hottenga J. J., Hofman A., Hicks A. A., Hengstenberg C., Heid I. M., Hayward C., Havulinna A. S., Hastie N. D., Harris T. B., Haritunians T., Hall A. S., Gyllensten U., Guiducci C., Groop L. C., Gonzalez E., Gieger C., Freimer N. B., Ferrucci L., Erdmann J., Elliott P., Ejebe K. G., Doering A., Dominiczak A. F., Demissie S., Deloukas P., de Geus E. J. C., de Faire U., Crawford G., Collins F. S., Chen Y. D. I., Caulfield M. J., Campbell H., Burtt N. P., Bonnycastle L. L., Boomsma D. I., Boekholdt S. M., Bergman R. N., Barroso I., Bandinelli S., Ballantyne C. M., Assimes T. L., Quertermous T., Altshuler D., Seielstad M., Wong T. Y., Tai E. S., Feranil A. B., Kuzawa C. W., Adair L. S., Taylor H. A., Borecki I. B., Gabriel S. B., Wilson J. G., Holm H., Thorsteinsdottir U., Gudnason V., Krauss R. M., Mohlke K. L., Ordovas J. M., Munroe P. B., Kooner J. S., Tall A. R., Hegele R. A., Kastelein J. J. P., Schadt E. E., Rotter J. I., Boerwinkle E., Strachan D. P., Mooser V., Stefansson K., Reilly M. P., Samani N. J., Schunkert H., Cupples L. A., Sandhu M. S., Ridker P. M., Rader D. J., van Duijn C. M., Peltonen L., Abecasis G. R., Boehnke M., Kathiresan S. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth D. M., Ricketts S. L., Song K., Chen L., Zhao J. H., Ripatti S., Aulchenko Y. S., Zhang W., Yuan X., Lim N., Luan J., Ashford S., Wheeler E., Young E. H., Hadley D., Thompson J. R., Braund P. S., Johnson T., Struchalin M., Surakka I., Luben R., Khaw K. T., Rodwell S. A., Loos R. J., Boekholdt S. M., Inouye M., Deloukas P., Elliott P., Schlessinger D., Sanna S., Scuteri A., Jackson A., Mohlke K. L., Tuomilehto J., Roberts R., Stewart A., Kesaniemi Y. A., Mahley R. W., Grundy S. M., Wellcome Trust Case Control C., McArdle W., Cardon L., Waeber G., Vollenweider P., Chambers J. C., Boehnke M., Abecasis G. R., Salomaa V., Jarvelin M. R., Ruokonen A., Barroso I., Epstein S. E., Hakonarson H. H., Rader D. J., Reilly M. P., Witteman J. C., Hall A. S., Samani N. J., Strachan D. P., Barter P., van Duijn C. M., Kooner J. S., Peltonen L., Wareham N. J., McPherson R., Mooser V., Sandhu M. S. Arterioscler., Thromb., Vasc. Biol. 2010;30:2264–2276. doi: 10.1161/ATVBAHA.109.201020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Lipids Genetics Consortium, Willer C. J., Schmidt E. M., Sengupta S., Peloso G. M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M. L., Mora S., Beckmann J. S., Bragg-Gresham J. L., Chang H. Y., Demirkan A., Den Hertog H. M., Do R., Donnelly L. A., Ehret G. B., Esko T., Feitosa M. F., Ferreira T., Fischer K., Fontanillas P., Fraser R. M., Freitag D. F., Gurdasani D., Heikkila K., Hypponen E., Isaacs A., Jackson A. U., Johansson A., Johnson T., Kaakinen M., Kettunen J., Kleber M. E., Li X., Luan J., Lyytikainen L. P., Magnusson P. K., Mangino M., Mihailov E., Montasser M. E., Muller-Nurasyid M., Nolte I. M., O'Connell J. R., Palmer C. D., Perola M., Petersen A. K., Sanna S., Saxena R., Service S. K., Shah S., Shungin D., Sidore C., Song C., Strawbridge R. J., Surakka I., Tanaka T., Teslovich T. M., Thorleifsson G., Van den Herik E. G., Voight B. F., Volcik K. A., Waite L. L., Wong A., Wu Y., Zhang W., Absher D., Asiki G., Barroso I., Been L. F., Bolton J. L., Bonnycastle L. L., Brambilla P., Burnett M. S., Cesana G., Dimitriou M., Doney A. S., Doring A., Elliott P., Epstein S. E., Eyjolfsson G. I., Gigante B., Goodarzi M. O., Grallert H., Gravito M. L., Groves C. J., Hallmans G., Hartikainen A. L., Hayward C., Hernandez D., Hicks A. A., Holm H., Hung Y. J., Illig T., Jones M. R., Kaleebu P., Kastelein J. J., Khaw K. T., Kim E., Klopp N., Komulainen P., Kumari M., Langenberg C., Lehtimaki T., Lin S. Y., Lindstrom J., Loos R. J., Mach F., McArdle W. L., Meisinger C., Mitchell B. D., Muller G., Nagaraja R., Narisu N., Nieminen T. V., Nsubuga R. N., Olafsson I., Ong K. K., Palotie A., Papamarkou T., Pomilla C., Pouta A., Rader D. J., Reilly M. P., Ridker P. M., Rivadeneira F., Rudan I., Ruokonen A., Samani N., Scharnagl H., Seeley J., Silander K., Stancakova A., Stirrups K., Swift A. J., Tiret L., Uitterlinden A. G., van Pelt L. J., Vedantam S., Wainwright N., Wijmenga C., Wild S. H., Willemsen G., Wilsgaard T., Wilson J. F., Young E. H., Zhao J. H., Adair L. S., Arveiler D., Assimes T. L., Bandinelli S., Bennett F., Bochud M., Boehm B. O., Boomsma D. I., Borecki I. B., Bornstein S. R., Bovet P., Burnier M., Campbell H., Chakravarti A., Chambers J. C., Chen Y. D., Collins F. S., Cooper R. S., Danesh J., Dedoussis G., de Faire U., Feranil A. B., Ferrieres J., Ferrucci L., Freimer N. B., Gieger C., Groop L. C., Gudnason V., Gyllensten U., Hamsten A., Harris T. B., Hingorani A., Hirschhorn J. N., Hofman A., Hovingh G. K., Hsiung C. A., Humphries S. E., Hunt S. C., Hveem K., Iribarren C., Jarvelin M. R., Jula A., Kahonen M., Kaprio J., Kesaniemi A., Kivimaki M., Kooner J. S., Koudstaal P. J., Krauss R. M., Kuh D., Kuusisto J., Kyvik K. O., Laakso M., Lakka T. A., Lind L., Lindgren C. M., Martin N. G., Marz W., McCarthy M. I., McKenzie C. A., Meneton P., Metspalu A., Moilanen L., Morris A. D., Munroe P. B., Njolstad I., Pedersen N. L., Power C., Pramstaller P. P., Price J. F., Psaty B. M., Quertermous T., Rauramaa R., Saleheen D., Salomaa V., Sanghera D. K., Saramies J., Schwarz P. E., Sheu W. H., Shuldiner A. R., Siegbahn A., Spector T. D., Stefansson K., Strachan D. P., Tayo B. O., Tremoli E., Tuomilehto J., Uusitupa M., van Duijn C. M., Vollenweider P., Wallentin L., Wareham N. J., Whitfield J. B., Wolffenbuttel B. H., Ordovas J. M., Boerwinkle E., Palmer C. N., Thorsteinsdottir U., Chasman D. I., Rotter J. I., Franks P. W., Ripatti S., Cupples L. A., Sandhu M. S., Rich S. S., Boehnke M., Deloukas P., Kathiresan S., Mohlke K. L., Ingelsson E., Abecasis G. R. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng E., Lind P. M., Lindgren C., Ingelsson E., Mahajan A., Morris A., Lind L. Hum. Mol. Genet. 2015;24:4739–4745. doi: 10.1093/hmg/ddv190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Wu D. D., Yao Y. G., Huo Y. X., Liu J. W., Su B., Chasman D. I., Chu A. Y., Huang T., Qi L., Zheng Y., Group C. N. W., DietGen C., Luo X. J. Schizophr. Bull. 2016;42:178–190. doi: 10.1093/schbul/sbv070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruenig D., White M. J., Young R. M., Voisey J. Genet. Test. Mol. Biomarkers. 2014;18:683–689. doi: 10.1089/gtmb.2014.0111. [DOI] [PubMed] [Google Scholar]
- Lajous H., Da Mota B., Li J., Duchesnay E., Lemaitre H., Ducrot V., Frouin V. Act. J. RITS. 2015:32–33. [Google Scholar]
- Xie X. M., Smart T. G. Nature. 1991;349:521–524. doi: 10.1038/349521a0. [DOI] [PubMed] [Google Scholar]
- Takeda A., Minami A., Seki Y., Oku N. J. Neurosci. Res. 2004;75:225–229. doi: 10.1002/jnr.10846. [DOI] [PubMed] [Google Scholar]
- Qian J., Noebels J. L. J. Neurosci. 2006;26:6089–6095. doi: 10.1523/JNEUROSCI.0475-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentschler G., Kippler M., Axmon A., Raqib R., Skerfving S., Vahter M., Broberg K. Metallomics. 2014;6:885–891. doi: 10.1039/c3mt00365e. [DOI] [PubMed] [Google Scholar]
- Park R. W., Kim T. M., Kasif S., Park P. J. Mol. Cancer. 2015;14:25. doi: 10.1186/s12943-015-0292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Schneider S. N., Dragin N., Girijashanker K., Dalton T. P., He L., Miller M. L., Stringer K. F., Soleimani M., Richardson D. D., Nebert D. W. Am. J. Physiol. 2007;292:C1523–C1535. doi: 10.1152/ajpcell.00409.2006. [DOI] [PubMed] [Google Scholar]