 The amino polycyclic aromatic hydrocarbons (amino-PAHs) were frequently detected in PM2.5, and it was suggested that they contributed to the harmful health effects associated with PM2.5.
The amino polycyclic aromatic hydrocarbons (amino-PAHs) were frequently detected in PM2.5, and it was suggested that they contributed to the harmful health effects associated with PM2.5.
Abstract
The amino polycyclic aromatic hydrocarbons (amino-PAHs) were frequently detected in PM2.5, and it was suggested that they contributed to the harmful health effects associated with PM2.5. However, the process through which amino-PAHs induce oxidative stress responses as well as the pro-inflammatory processes along with the associated mechanisms is still not well-known. In this study, oxidative stress level, Nrf2/ARE anti-oxidative defense responses, oxidative damages and cytokine expressions were investigated in the A549 cell line after it was treated with typical airborne amino-PAHs including 1-aminopyrene (1-AP) and 3-aminofluoranthene (3-AF). The possibility of the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) pathway participating in the regulation of cytokine gene expression was also considered, and the study was conducted accordingly. The results showed that 1-AP and 3-AF in a dose-dependent manner could cause extensive damages including cell apoptosis, cell cycle arrests, and DNA damages and could up-regulate the TNF-α gene expression. In addition, the Nr2/ARE defense system was activated, as evidenced by the increased protein expression levels and nuclear translocation of Nrf2. The Nrf2/ARE binding activity was elevated and was measured with the EMSA method. Also, the protein of heme oxygenase-1 (HO-1) was up-regulated. Finally, an increase in the protein expressions of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and phosphorylation levels of Akt was observed, indicating that the PI3K/Akt pathway was activated. Furthermore, both LY294002 (an inhibitor of PI3K) and MK-2206 (an inhibitor of Akt) could significantly decrease the elevated TNF-α gene expressions, suggesting that the PI3K/Akt pathway was involved in the regulation of TNF-α expressions induced by 1-AP and 3-AF. Our results further confirmed that amino-PAHs could be a particularly important PAH derivatives contributing to the health risks caused by PM2.5.
Introduction
The fine fraction of ambient particulate matter (PM2.5, with an aerodynamic diameter of less than 2.5 μM) is becoming a serious public health problem in the world.1 The health hazards of PM2.5 at least partially result from its chemical composition.2,3 Thus, to further investigate the adverse health effects and associated biological mechanisms caused by urban PM2.5, it is necessary to identify and distinguish every toxic component in PM2.5.4 Among the chemical components of PM2.5, polycyclic aromatic hydrocarbons (PAHs) and their derivatives are first hypothesized to be the most important organic components contributing to the health risks of PM2.5.5 The derivatives of PAHs in the air usually include nitrated (nitro-PAHs), oxygenated (like quinones), hydroxylated (HO-PAHs) and amino PAHs. The sources, transformation mechanisms, and health implications of PAHs and their nitrated, hydroxylated, and oxygenated derivatives in PM2.5 have been well reviewed.6–9 In addition, there are also a few studies focusing on the adverse health outcomes generated by amino-PAHs present in PM2.5.10,11 Recently, it has been found that amino-PAHs can influence the cells in vitro through different pathways when compared with PAHs and nitro-PAHs.10–12 However, till now, amino-PAH-induced oxidative stress responses, pro-inflammatory processes, and the associated mechanisms are still not well-known.
The airborne amino-PAHs mainly originate from the direct emissions of industrial processes and the combustion process of fossil fuels and cigarettes.13 Amino-PAHs can also be produced from the reduction process of nitro-PAHs catalyzed by the catalysts applied for purifying diesel exhaust gases emitted from the vehicles.14 Additionally, amino-PAHs can be generated in vivo by the metabolism conversions of their corresponding precursors such as nitro-PAHs.11,15 In vivo, the nitro groups in nitro-PAHs can be reduced to amino groups (forming amino-PAHs) through six-electron reduction reactions. These reactions are catalyzed by cytosolic nitro-reductases and microsomal cytochrome P450s (CYPs) and/or CYP reductases in our body.16,17 The typical amino-PAHs detected in the PM2.5 samples include 1-aminopyrene (1-AP), 3-aminofluoranthene (3-AF), 3-aminobenzanthrone (3-ABA), and 9-aminoanthracene (9-AA) with the concentrations in the range of several picograms per cubic meter.13
The oxidative stress and inflammatory responses appear to be the critical biological mechanisms in the PM-induced health effects.18 Both PAHs and their derivatives have been reported to be able to cause oxidative stress and promote inflammatory responses in the cultured cells.6,19 Nuclear factor-erythroid 2-related factor 2 (Nrf2), as a redox-sensitive transcription factor, regulates the cellular antioxidant defense system. The Nrf2/antioxidant response element (ARE) pathway has been reported to be participating in regulating cellular defense mechanisms against oxidative stress caused by PM2.5.20 After bonding with ARE in the nucleus, Nrf2 can trigger the expressions of anti-oxidative genes (such as heme oxygenase-1 (HO-1)).21 In addition, the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) pathway is reported to be involved in the Nrf2/ARE-mediated defense mechanisms and HO-1 induction in response to the oxidative stress caused by PM2.5 and other environmental pollutant exposure in vitro.20
As is mentioned before, amino-PAHs can up-regulate cytokine/chemokine gene expressions in cultured cells in vitro. 1-AP, 3-AF and 3-ABA can influence the gene expressions of interleukin-6 (IL-6), Cxcl8 and Ccl5, respectively, and present pro-inflammatory potentials in the cultured cells.7,12,13 However, it is not determined whether the PI3K/Akt pathway participates in the regulation of these pro-inflammatory responses caused by amino-PAHs. The complicated compositions of urban PM2.5 create a huge challenge for the toxicological studies. To clarify the roles of amino-PAHs, in this study, 1-AP and 3-AF are used as two typical amino-PAHs that have been frequently detected in PM2.5.13 The human type II alveolar epithelial A549 cells are used as an in vitro model to explore the signaling pathway triggered by amino-PAHs. First, the oxidative stress and oxidative damage are investigated in the A549 cells after they are treated with 1-AP and 3-AF. Then, the activation of the anti-oxidative defense pathway of Nrf2/ARE and the possibility of the involvement PI3K/Akt in the regulation of cytokine gene expressions induced by amino-PAHs are examined. This information will possibly contribute to the development of targeted regulations to improve the air quality. Considering the accumulative effects of amino-PAHs in organisms, the results of the in vitro study on amino-PAHs (at several to tens of μM levels) should help in establishing regulatory control standards for these pollutants.
Materials and methods
Materials
The A549 cell line (type II pulmonary epithelial cell line) was obtained from American Tissue Type Culture Collection (ATCC, USA). 1-Aminopyrene (1-AP, 97% purity) and 3-aminofluoranthene (3-AF, 90% purity) were purchased from Sigma-Aldrich Company (USA). 2′,7′-Dichlorodihydrofluorescein diacetate (H2DCFDA), LY294002, and MK-2206 were also purchased from Sigma-Aldrich Company (USA). The cell culture medium (RPMI 1640) and fetal bovine serum (FBS) were obtained from Gibco Company (UK). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was purchased from Dojindo (Japan). All other chemicals (analytical grade purities) were obtained from Sinopharm Chemical Reagent Company (China).
Cell culture and treatment
The A549 cells were cultured in RPMI 1640 medium containing 10% FBS and 100 units per mL penicillin and streptomycin. The cells were kept in a CO2 incubator (LINDER, Germany) with a humidified atmosphere of 5% CO2 and 95% air at 37 °C. 1-AP and 3-AF were dissolved in DMSO and kept in a refrigerator at –20 °C. In each experiment, the working solutions of 1-AP and 3-AF were prepared from the storage solutions, and these solutions were freshly diluted with RPMI 1640 medium with 2% FBS. The concentrations of 1-AP and 3-NF were at 5, 10, 20, 40 and 60 μM. The cells were exposed to 1-AP and 3-AF for 24 h except for the cells to be used for ROS detection; these cells were exposed for 1 h. In all of the experiments, the concentration of DMSO was kept at 0.1% (v/v), and the cell groups treated with 0.1% (v/v) DMSO served as the control. All experiments were carried out for at least three times with three or more parallel samples.
LY294002 and MK-2206·2HCL are the inhibitors of PI3K and Akt, respectively. To determine the regulation of PI3K/Akt in the pro-inflammatory responses caused by amino-PAHs, the A549 cells were pre-treated with these two inhibitors. Briefly, in the inhibitor experiment, the A549 cells were pre-treated with LY294002 at 600 nM or MK-2206·2HCL at 15 nM for half an hour before the treatment with 1-AP and 3-AF.
MTT assay
Briefly, the cells were planted in a 96-well plate with 3000 cells per well. After adhesion, the culture medium was replaced with a fresh medium (2% FBS) containing various concentrations of 1-AP and 3-AF (0, 5, 10, 20, 40, and 60 μM). The experiment was performed at least three times with six replicates for each concentration group. After 24 h incubation, the culture medium was discarded, and freshly prepared MTT was added to each well at a final concentration of 1 mg mL–1 diluted in the culture medium and cultured for another 4 h. Then, the culture medium was replaced by 150 μL DMSO to solubilize the formazan crystals. The optical density (OD) was measured with a Multiscan Mk3 plate reader (Thermo Electron Corporation, Waltham, MA, USA) at a wavelength of 490 nm.
ROS measurement
The cells were seeded in 35 mm culture dishes with a cell density of 2–5 × 104 cells per dish. After adhesion, the cells were treated with 1-AP and 3-AF in fresh medium at different concentrations for 1 h. Then, the cells were washed with warm D-Hank's solution twice and incubated with H2DCFDA (freshly diluted in D-Hank's solution) for 30 min at 37 °C in the dark. The fluorescence intensities recorded with a fluorescent microscope (Olympus BX-51, Japan) were used to indicate the relative levels of ROS. The data were analyzed using the Image-pro plus 6 software.
Comet assay
Briefly, after the A549 cells were treated with 1-AP and 3-AF for 24 h, the cells were digested and re-suspended. The cell suspensions were mixed with low melting agarose (LMA) and pipetted on a slide coated with 1% of normal melting agarose (NMA). After this process, the third gel layer (LMA) was added to the surface to form the final “sandwich” gel. This “sandwich” gel was immersed in a cold cell lysis buffer with a pH of 10 for 1 h. This buffer solution consisted of 10 mM Tris-HCl, 2.5 M NaCl, 100 mM EDTA disodium salt, 1% Triton X-100 and 10% DMSO. Then, the slides were subjected to horizontal gel electrophoresis in a cold alkaline electrophoresis buffer (300 mM NaOH, 1 mM Na2EDTA, pH 12.5 at 4 °C) at 25 V and 300 mA for 20 min. Next, the slides were washed with the neutralization buffer (0.4 M, pH 7.5) at 4 °C for 8 min and then air-dried. The DNA was stained with propidium iodide (PI, 20 μg mL–1) and analyzed using a fluorescence microscope (Olympus BX-51, Japan). At least 300 cells were randomly captured from each sample and analyzed with the CASP software (University of Wroclaw, Poland). The DNA percentage in the tail (%Tail DNA) was selected to express the DNA damage levels.22
Flow cytometry
Flow cytometry was applied to measure the cell apoptosis ratio according to the apoptosis detection kit (BD Company, USA) with the cell staining by Annexin V-FITC and PI. After treatment with 1-AP and 3-AF for 24 h, the cells were trypsinized, washed with PBS solution twice and then re-suspended in 500 μL of a binding buffer. The cells were first incubated with Annexin-V-FITC at 37 °C for 15 min in the dark and then incubated with PI at 37 °C for 3 min in the dark. Next, the stained cells were immediately analyzed by flow cytometry. The data were analyzed with the Summit 5.2 software.
Electrophoretic mobility shift assay (EMSA)
The nuclear extracts of the A549 cells were prepared with the NE-PER Kit. Then, the DNA–protein binding reactions were conducted using the EMSA Kit (Chemiluminescent EMSA kit, Beyotime Biotechnology Company, Shanghai, China) exactly according to the manufacturer's instructions. The reaction mixtures were loaded on 4% non-denaturing polyacrylamide gels and then blotted onto nylon membranes for chemiluminescence detection.
Western blot
The Western blot method was chosen to detect the protein expression levels in the A549 cells treated with 1-AP and 3-AF for 24 h. After treatment, the cells were lysed, washed with D-Hank's balanced saline solution and collected. The total proteins and cytoplasmic and nuclear proteins were extracted using the mammalian protein extraction reagent (MPER) and cytoplasmic and nuclear protein extraction reagents (NE-PER), respectively. Then, the protein samples at an equal amount were separated in 8% or 10% sodium dodecyl sulfate-polyacrylamide gels. Subsequently, the proteins were electrophoretically transferred to nitrocellulose membranes. Then, the membranes were incubated with the primary and secondary antibodies. The blots indicating the target proteins were visualized by the chemiluminescence method. The densitometry data were evaluated based on the visible bands using the Chemi-Imager digital imaging system (Alpha Innotech, San Leandro, CA, USA).
Real time-quantitative polymerase chain reaction (RT-qPCR)
The total RNA content was isolated from the A549 cells using the TRIZOL method. The concentration of RNA was measured spectrometrically using the Gene PulserXcell Total System, and 1 μg of RNA was reverse-transcribed into cDNA with reverse transcriptase obtained from TOYOBO (Japan). The quantitative gene expression levels were measured with the Bio-Rad iQ5 Real Time PCR equipment (USA) using the THUNDERBIRD SYBR qPCR Mix (Toyobo, Japan). The il-6 (interleukin-6) and TNF-α (tumor necrosis factor-α) genes were analyzed using the following primer sequences:
GAPDH-forward primer: 5′-CCATGGAGAAGGCTGGGG-3′
GAPDH-reverse primer: 5′-CAAAGTTGTCATGGATGACC-3′
TNF-forward primer: 5′-CAGGCAGTCAGATCATCTTC-3′
TNF-reverse primer: 5′-CTTGATGGCAGAGAGGAGGT-3′
The expressions of the target gene (TG) were normalized against those of the reference gene of GAPDH. The gene expression level was expressed as fold changes and was compared with that of the control (0.1% DMSO treated cells). Also, the gene expression folds were calculated with the ΔΔCt method: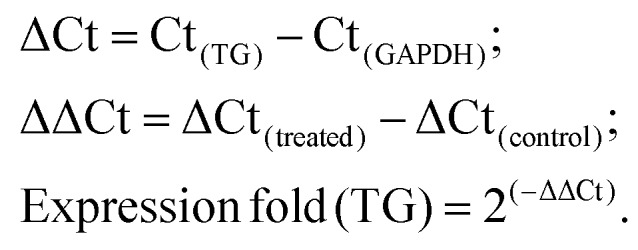
Data analysis
The data obtained are presented as mean ± standard deviation (SEM) of at least three independent experiments. A one-way ANOVA test with Bonferroni's correction was carried out to analyze the statistical difference between the groups. A p-value <0.05 was considered statistically significant.
Results
Cytotoxicity and oxidative stress responses
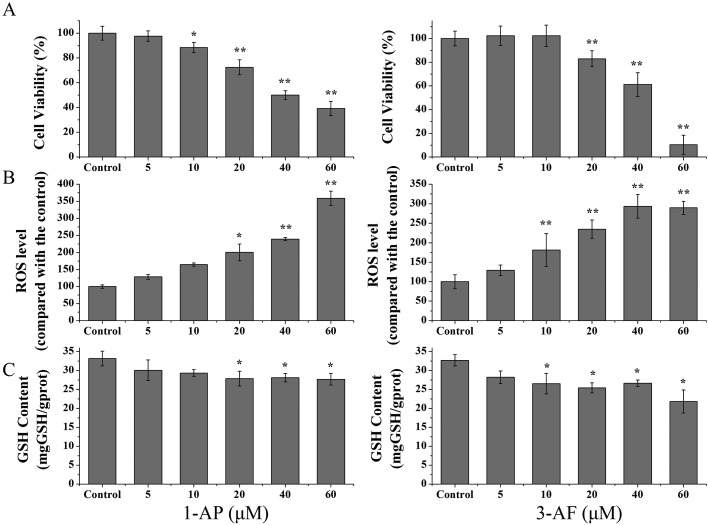
The cell viabilities of the A549 cells after they were treated with 1-AP and 3-AF were measured with the MTT method as described above. As shown in Fig. 1A, 1-AP and 3-AF caused a significant decrease in the concentration-dependent viabilities of the A549 cells (p < 0.01). The LC50 values (lethal concentration at 50%) of 1-AP and 3-AF were 44.6 and 39.0 μM for 24 h treatment, respectively. LC50 of 1-AP was a little higher than that of 3-AF but without statistical significance (p > 0.05).
Fig. 1. Cell proliferation, ROS level, and GSH content in the A549 cells after they were treated with 1-AP and 3-AF. The A549 cells were treated with different concentrations of 1-AP and 3-AF (0, 5, 10, 20, 40 and 60 μM). (A): The cell proliferation was determined by MTT assay after 24 h treatment. (B): The level of intracellular ROS was assessed by the fluorescent probe DCFH-DA after 1 h exposure. (C): The GSH content was measured according to the manufacturer's instructions after 24 h treatment. The control groups were the A549 cells treated with DMSO (0.1%, v/v). *p < 0.05 and **p < 0.01 when compared with those of the control.
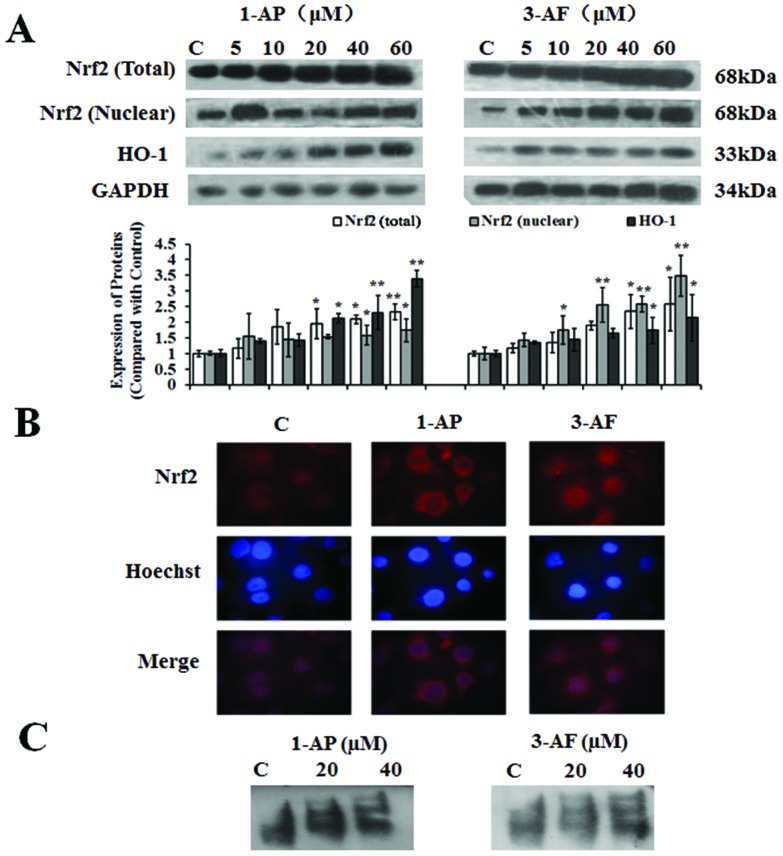
Oxidative stress in the A549 cells was evaluated by ROS generation, glutathione (GSH) consumption and HO-1 protein expression. The A549 cells were exposed to 1-AP and 3-AF for 1 h, and ROS was assessed by the DCFH-DA probe method. Fig. 1B shows that 1-AP and 3-AF could significantly increase the ROS levels in a dose-dependent manner. The ROS levels increased 3.6 ± 0.21 and 2.9 ± 0.17 times after the cells were treated with 1-AP and 3-AF, respectively, at the highest dose of 60 μM (p < 0.01). It was also observed that the cellular contents of GSH reduced significantly in the A549 cells after they were exposed to 1-AP and 3-AF for 24 h (Fig. 1C, p < 0.05). Moreover, as shown in Fig. 3A, the WB results demonstrated that both 1-AP and 3-AF could significantly increase the HO-1 protein expression when compared with the untreated cells.
Fig. 3. 1-AP and 3-AF inducing Nrf2 nuclear translocation and activation of the Nrf2/ARE pathway. The A549 cells were treated with different concentrations of 1-AP and 3-AF (0, 5, 10, 20, 40 and 60 μM) for 24 h. (A): The protein expressions of total and nuclear Nrf2 and HO-1 were measured with the Western blot assay. *p < 0.05 and **p < 0.01 when compared with those of the control. (B): Immunofluorescence staining of cells subjected to 40 μM of 1-AP and 3-AF exposure was blotted with an anti-Nrf2 antibody. Nuclear counterstaining was done with Hoechst 33342. (C): EMSA assay was performed according to the manufacturer's instructions of EMSA/Gel-Shift Kit to confirm the accumulation of Nrf2 in nuclear extracts. C: The control groups were defined as the A549 cells treated with DMSO (0.1%, v/v).
Oxidative damages
Oxidative stress and an increase in the cellular ROS levels cause oxidative damages and promote cell apoptosis. In this study, the oxidative DNA damage in the A549 cells was evaluated with the Comet assay of alkaline version after they were treated with 1-AP and 3-AF. The %Tail DNA was used as the metric for DNA migration as described in the Materials and methods section. Fig. 2A presents the %Tail DNA in the A549 cells after they were exposed to 1-AP and 3-AF for 24 h. 1-AP and 3-AF treatment caused significant DNA damage in the A549 cells in a dose-dependent manner. The %Tail DNA increased from 7.2% ± 2.7% to 45.6% ± 10.5% and 9.2% ± 3.7% to 42.3% ± 9.4% for 1-AP and 3-AF, respectively, when their concentrations were increased from 0 to 60 μM (p < 0.01).
Fig. 2. DNA damage and apoptosis of the A549 cells after they were treated with 1-AP and 3-AF. The A549 cells were treated with different concentrations of 1-AP and 3-AF (0, 5, 10, 20, 40 and 60 μM) for 24 h. (A): The DNA damage was determined by Comet assay as described in the Materials and Methods section. (B): The apoptosis was measured by flow cytometry using the Annexin V-FITC/PI probe. The control groups were the A549 cells treated with DMSO (0.1%, v/v). *p < 0.05 and **p < 0.01 when compared with those of the control.
Fig. 2B shows that 1-AP and 3-AF could significantly increase the apoptosis rate in the A549 cells as measured by the flow cytometry method. The percentages of the apoptotic cells were obviously increased in a dose-dependent manner after the 1-AP treatment (0 to 60 μM, p < 0.05). The highest apoptotic rates of 8.1% ± 1.3% were observed after the cells were treated with 1-AP at a dose of 60 μM (p < 0.01). In the 3-AF-treated A549 cells, the apoptosis rates were higher than 80% in the 20, 40 and 60 μM treated groups. After combining these results with the cell viability data, we guessed that 3-AF may probably react with the probe of Annexin V-FITC/PI and enhance the fluorescent intensity in the highest three doses, finally causing extremely high apoptosis rates.
The cell cycle distributions of the A549 cells were also measured with the flow cytometry method. The results in Table 1 presented that both 1-AP and 3-AF (40 μM, 24 h) could cause the S phase arrest, with the ratio of the A549 cells in the S phase increasing from 23.7% ± 0.8% to 29.0% ± 0.4% and 14.8% ± 3.7% to 26.4% ± 0.3%, respectively (p < 0.05).
Table 1. The distribution of cell cycle phase (%) after the 1-AP and 3-AF treatment.
| Concentration (μM) | 1-AP |
3-AF |
||||
| G0/G1 | G2/M | S | G0/G1 | G2/M | S | |
| Control | 67.8 ± 10.5 | 16.5 ± 0.5 | 23.7 ± 0.8 | 68.5 ± 0.4 | 16.4 ± 1.5 | 14.8 ± 3.7 |
| 5 | 67.0 ± 2.5 | 15.6 ± 1.5 | 25.3 ± 0.1 | 66.5 ± 3.8 | 15.0 ± 5.4 | 19.2 ± 3.9 |
| 10 | 62.9 ± 3.8 | 12.3 ± 5.0 | 27.2 ± 2.0 | 70.1 ± 5.2 | 10.0 ± 0.5 | 23.2 ± 1.6 |
| 20 | 67.4 ± 5.9 | 12.7 ± 1.2* | 27.5 ± 0.3* | 68.3 ± 4.1 | 7.5 ± 0.4* | 27.0 ± 0.1* |
| 40 | 57.4 ± 0.1 | 12.8 ± 1.0* | 29.0 ± 0.4* | 70.3 ± 4.1 | 4.4 ± 1.0* | 26.4 ± 0.3* |
| 60 | 62.5 ± 3.0 | 9.0 ± 0.7** | 28.4 ± 1.1* | 9.2 ± 6.3 | 6.2 ± 0.6* | 16.9 ± 3.4* |
The activation of Nrf2/ARE pathway
The abovementioned data summarized that 1-AP and 3-AF could induce oxidative stress and cause oxidative damages including apoptosis and DNA damage in the A549 cells. The Nrf2/ARE pathway has been shown to participate in regulating the intracellular defense mechanisms against oxidative stress induced by PM2.5. Thus, in this study, the potential roles of the Nrf2-mediated cellular defense systems against oxidative stress were further investigated. Normally, Nrf2 is presented in the form of an Nrf2-Keap 1 (Kelch-like ECH-associated protein 1) complex (without transcriptional activity) in the cytoplasm. After oxidative stress, Keap 1 is degraded and Nrf2 is translocated into the nucleus to bind with ARE. Thus, the translocation of Nrf2 from the cytoplasm into the nucleus was first measured in the A549 cells treated with 1-AP and 3-AF. As shown in Fig. 3A, both the total and nuclear Nrf2 protein expressions in the A549 cells were evaluated in a dose-dependent manner after the cells were treated with 1-AP and 3-AF, and the measurements were carried out with the Western blot method. In addition, the translocation of the Nrf2 protein from the cytoplasm to the nucleus in the A549 cells was also indicated by the pictures obtained from the immunocytochemistry method (Fig. 3B). Finally, the binding activity of the complex of Nrf2/ARE was examined by the EMSA assay as described above. As shown in Fig. 3C, 1-AP and 3-AF could induce the recruitment of Nrf2 to ARE in the A549 cells at 20- and 40 μM doses. These results together indicated that the transcriptional activity of Nrf2 was strengthened after the treatment with 1-AP and 3-AF.
Cytokine gene expressions regulated by the PI3K/Akt pathway
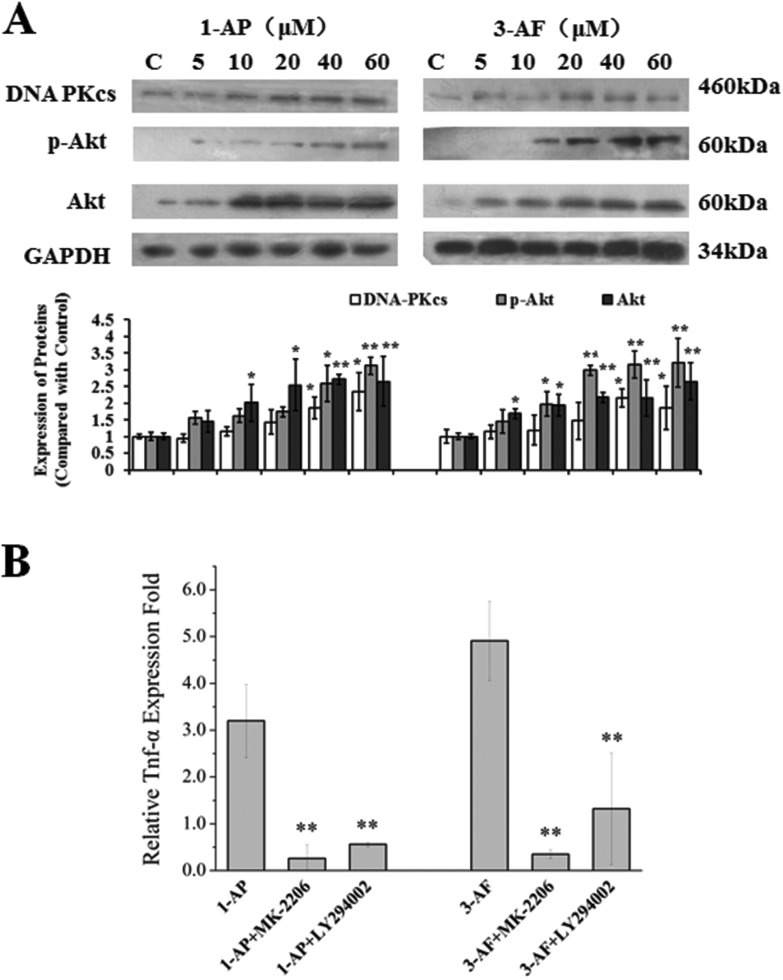
To assess whether amino-PAHs could activate the PI3K/Akt pathway in the A549 cells, we detected the expression of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs, a member of the PI3K family) and phosphorylation of Akt in the A549 cells. As shown in Fig. 4A, the DNA-PKcs protein expression and phosphorylation of Akt were markedly increased in a dose-dependent manner after 24 h exposure to 1-AP and 3-AF.
Fig. 4. The PI3K/Akt pathway-mediated cytokine expression. (A): 1-AP and 3-AF could activate the PI3K/Akt pathway. The A549 cells were treated with different concentrations of 1-AP and 3-AF for 24 h. The protein expressions of DNA-PKcs, phosph-Akt and total Akt were measured with the Western blot assay. C: The control groups were defined as the A549 cells treated with DMSO (0.1%, v/v). (B): Before the treatment with 40 μM of 1-AP and 3-AF for 6 h, the A549 cells were pre-treated with PI3K inhibitor (LY294002, 600 nM) or Akt inhibitor (MK-2206·2HCL, 15 nM) for 30 min in a serum-free medium. The expression of TNF-α was evaluated by quantitative real-time PCR. *p < 0.05 and **p < 0.01 when compared with those of the non-inhibitor groups.
The mRNA expression of TNF-α in the A549 cells was examined with the RT-qPCR method. As shown in Fig. 4B, the mRNA expression levels of TNF-α increased 3.2 ± 0.8 and 4.9 ± 0.9 folds when compared with those of the control after the cells were exposed to 1-AP and 3-AF (at 40 μM), respectively. To investigate whether the PI3K/Akt pathway participated in the regulation of TNF-α expressions induced by 1-AP and 3-AF exposure, the two inhibitors LY294002 (an inhibitor of PI3K) and MK-2206 (an inhibitor of Akt) were used. The results turned out that both LY294002 and MK-2206 could effectively suppress the gene expressions of TNF-α induced by 1-AP and 3-AF (Fig. 4B). After the treatment with MK-2206, the TNF-α levels in the A549 cells after they were exposed to 1-AP and 3-AF reduced from 3.2 ± 0.8 to 0.3 ± 0.3 folds and 4.9 ± 0.9 to 0.4 ± 0.1 folds, respectively (p < 0.01); after the treatment with LY294002, the TNF-α levels reduced from 3.2 ± 0.8 to 0.6 ± 0.1 folds and 4.9 ± 0.9 to 1.3 ± 1.2 folds, respectively (p < 0.01). It seemed that MK-2206 had higher efficiency than LY294002 on suppressing TNF-α expression in the A549 cells.
Discussions
Amino-PAHs are an important kind of PAH derivatives present in PM2.5 other than nitro-PAHs and oxygenated-PAHs. Recently, amino-PAHs have been frequently detected in urban ambient particles and are hypothesized to contribute to the adverse health effects caused by PM2.5 exposure.13,15 While several studies have focused on the cytotoxic properties and inflammatory responses of amino-PAHs, considerably less is known about the oxidative stress responses and pro-inflammatory processes caused by amino-PAHs. Ovrevik et al. have reported that amino-PAHs induce a distinctly different inflammatory response as compared with nitro-PAHs.10 Amino-PAHs can stimulate cytokine/chemokine response dominated by chemokine (C–C motif) ligand 5 (CCL5) and C–X–C motif chemokine 10 (CXCL10).11,12 In this study, we use two typical amino-PAHs present in PM2.5, 1-AP and 3-AF, to investigate their in vitro toxic effects and the associated mechanisms in the A549 cells. We have found that 1-AP and 3-AF can exert extensive cytotoxic effects and oxidative damage and can promote pro-inflammatory cytokine gene expressions in the A549 cells. These results reveal the important contributions of amino-PAHs to the health hazards of urban PM2.5. Their types and concentrations in the air might significantly influence the biological effects generated by PM2.5.
Numerous studies have reported that oxidative stress is one of the key factors correlated with the harmful effects caused by PM2.5 exposure after inhalation. Oxidative stress and the subsequent reactive oxygen species (ROS) generation can trigger redox-sensitive reactions and might finally lead to extensive harmful effects including inflammation and cell death. The Nrf2/ARE pathway is known as one of the most important anti-oxidative defense systems against oxidative stress. Commonly, the Nrf2-Keap 1 complex present in the cytoplasm has no transcriptional activity. Once translocated into the nucleus, Nrf2 can bind with ARE to protect the cells from oxidative stress-induced cellular damages. Deng et al. reported that the Nrf2/ARE/HO-1-mediated defense pathway participates in the oxidative stress induced by PM2.5 in the A549 cells.20 In this study, we have found that 1-AP and 3-AF can promote Nrf2 nuclear translocation, increase the nuclear Nrf2 protein expressions, and raise the Nrf2/ARE binding activity in the A549 cells. In addition, the HO-1 protein expression is also increased in the A549 cells after they are exposed to 1-AP and 3-AF. Analogous to amino-PAHs, other derivatives of PAHs, such as nitro-PAHs and quinones, can also increase the ROS generation and activate the Nrf2/ARE pathway.23 These results indicate that the Nrf2/ARE pathway might be a key step for protecting the pulmonary cells from oxidative damage caused by the amino-PAHs treatment. Deng et al. also reported that PI3K/Akt is involved in the signaling pathway, which leads to the PM2.5-induced Nrf2 activation and the subsequent Nrf2-mediated HO-1 transcription.20 This study is consistent with our finding that 1-AP and 3-AF can simultaneously activate the PI3K/Akt pathway.
The PI3K/Akt signaling pathway plays a role in many essential cellular physiological processes, such as survival, proliferation, migration, and differentiation, which can be activated by extracellular stimuli such as cytokines and growth factors. Intracellularly, the accumulation of oxygen species can activate the nuclear factor-κB (NF-κB) via IκB kinase (IKK) through the PI3K/Akt pathway,24 and this plays an important role in the transduction of the signals from the receptors for pro-inflammatory cytokine TNF-α. In general, the mechanisms of PAH- and their derivatives-induced DNA damage and cell death might better be investigated than their cytokine/chemokine responses. Our results in this study emphasize the notion that 1-AP and 3-AF are also potential cytokine inducers when compared with other components in PM2.5 such as bacterial lipopolysaccharide (LPS), certain metals and other organic compounds.25 In addition, the PI3K/Akt pathway is involved in regulating the TNF-α gene expressions induced by 1-AP and 3-AF exposure in the A549 cells. These findings strengthen the conclusion that amino-PAHs may be a particularly important group among the PM2.5 components.
Conclusions
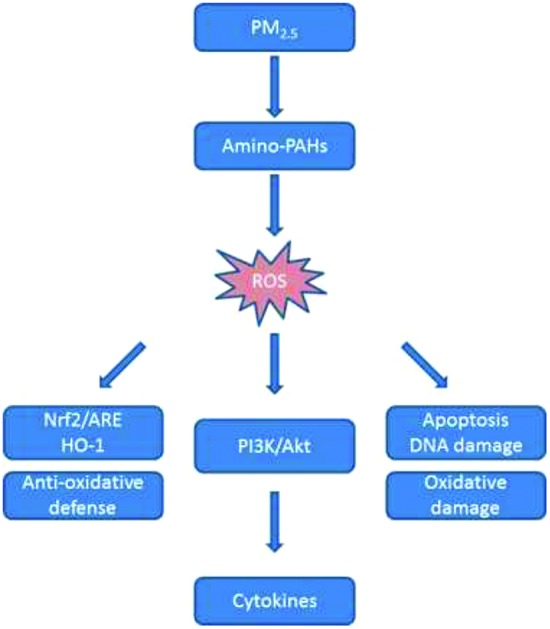
In summary, amino-PAHs (1-AP and 3-AF) can induce an increase in the ROS level, disrupt the balance between oxidation and reduction and eventually cause oxidative damages in the A549 cells. Through the PI3K/Akt pathway, ROS can activate the Nrf2/ARE antioxidant pathway to protect the cells. In addition, ROS can also regulate the intracellular inflammatory response under the mediation of the PI3K/Akt pathway. Our findings confirm and extend previous studies indicating that amino-PAHs may be a potential cytokine inducer that can promote serious oxidative damages in the A549 cells. These results emphasize that amino-PAHs can be a particularly important PAH derivative contributing to the health risks caused by ambient particles.
Conflicts of interest
There are no conflicts to declare.
Acknowledgments
The study was supported by National Natural Science Foundation of China (no. 91543123, 81771303), the Joint NSFC-ISF Research Program, jointly funded by the National Natural Science Foundation of China and the Israel Science Foundation (no. 41561144007), the Shanghai Pujiang Program (no. 17PJ1411000), and Innovative Research Team in University (no. IRT13078).
References
- Li G. X., Xue M., Zeng Q., Cai Y., Pan X. C., Meng Q. Sci. Total Environ. 2017;599:108–113. doi: 10.1016/j.scitotenv.2017.04.010. [DOI] [PubMed] [Google Scholar]
- Achilleos S., Kioumourtzoglou M. A., Wu C. D., Schwartz J. D., Koutrakis P., Papatheodorou S. I. Environ. Int. 2017;109:89–100. doi: 10.1016/j.envint.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C. R., Young L. H., Hsu H. T., Lin M. Y., Chen Y. C., Hwang B. F., Tsai P. J. Environ. Pollut. 2017;231:1085–1092. doi: 10.1016/j.envpol.2017.08.102. [DOI] [PubMed] [Google Scholar]
- Vuong N. Q., Breznan D., Goegan P., O'Brien J. S., Williams A., Karthikeyan S., Kumarathasan P., Vincent R. Part. Fibre Toxicol. 2017;14:39. doi: 10.1186/s12989-017-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besis A., Tsolakidou A., Balla D., Samara C., Voutsa D., Pantazaki A., Choli-Papadopoulou T., Lialiaris T. S. Environ. Pollut. 2017;230:758–774. doi: 10.1016/j.envpol.2017.06.096. [DOI] [PubMed] [Google Scholar]
- Kim K. H., Jahan S. A., Kabir E., Brown R. J. Environ. Int. 2013;60:71–80. doi: 10.1016/j.envint.2013.07.019. [DOI] [PubMed] [Google Scholar]
- Koike E., Yanagisawa R., Takano H. Atmos. Environ. 2014;97:529–536. [Google Scholar]
- Walgraeve C., Demeestere K., Dewulf J., Zimmermann R., Van Langenhove H. Atmos. Environ. 2010;44:1831–1846. [Google Scholar]
- Lin Y., Ma Y., Qiu X., Li R., Fang Y., Wang J., Zhu Y., Hu D. J. Geophys. Res.: Atmos. 2015;120:7219–7228. [Google Scholar]
- Ovrevik J., Arlt V. M., Oya E., Nagy E., Mollerup S., Phillips D. H., Lag M., Holme J. A. Toxicol. Appl. Pharmacol. 2010;242:270–280. doi: 10.1016/j.taap.2009.10.017. [DOI] [PubMed] [Google Scholar]
- Ovrevik J., Holme J. A., Lag M., Schwarze P. E., Refsnes M. Environ. Toxicol. Pharmacol. 2013a;35:235–239. doi: 10.1016/j.etap.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Ovrevik J., Refsnes M., Holme J. A., Schwarze P. E., Lag M. Toxicol. Lett. 2013b;219:125–132. doi: 10.1016/j.toxlet.2013.02.014. [DOI] [PubMed] [Google Scholar]
- Tang Y., Imasaka T., Yamamoto S., Imasaka T. Chemosphere. 2016;152:252–258. doi: 10.1016/j.chemosphere.2016.02.114. [DOI] [PubMed] [Google Scholar]
- Vlastimil V., Jiri B. Curr. Org. Chem. 2011;15:3059–3076. [Google Scholar]
- Totlandsdal A. I., Øvrevik J., Cochran R. E., Herseth J. I., Bølling A. K., Låg M., Schwarze P., Lilleaas E., Holme J. A., Kubátová A. J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng. 2014;49:383–396. doi: 10.1080/10934529.2014.854586. [DOI] [PubMed] [Google Scholar]
- Arlt V. M., Glatt H., Muckel E., Pabel U., Sorg B. L., Seidel A., Frank H., Schmeiser H. H., Phillips D. H. Int. J. Cancer. 2003a;105:583–592. doi: 10.1002/ijc.11143. [DOI] [PubMed] [Google Scholar]
- Arlt V. M., Stiborova M., Hewer A., Schmeiser H. H., Phillips D. H. Cancer Res. 2003b;63:2752–2761. [PubMed] [Google Scholar]
- González-Flecha B. Mol. Aspects Med. 2004;25:169–182. doi: 10.1016/j.mam.2004.02.017. [DOI] [PubMed] [Google Scholar]
- Moorthy B., Chu C., Carlin D. J. Toxicol. Sci. 2015;145:5–15. doi: 10.1093/toxsci/kfv040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Rui W., Zhang F., Ding W. Cell Biol. Toxicol. 2013;29:143–157. doi: 10.1007/s10565-013-9242-5. [DOI] [PubMed] [Google Scholar]
- Taguchi K., Motohashi H., Yamamoto M. Genes Cells. 2011;16:123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- Tice R. R., Agurell E., Anderson D., Burlinson B., Hartmann A., Kobayashi H., Miyamae Y., Rojas E., Ryu J. C., Sasaki Y. F. Environ. Mol. Mutagen. 2000;35:206–221. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Jin Y. X., Miao W. Y., Lin X. J., Pan X. H., Ye Y., Xu M. J., Fu Z. W. Environ. Toxicol. 2014;29:1399–1408. doi: 10.1002/tox.21870. [DOI] [PubMed] [Google Scholar]
- Tokuhira N., Kitagishi Y., Suzuki M., Minami A., Nakanishi A., Ono Y., Kobayashi K., Matsuda S., Ogura Y. Int. J. Mol. Med. 2015;35:10–16. doi: 10.3892/ijmm.2014.1981. [DOI] [PubMed] [Google Scholar]
- Shang Y., Zhu T., Lenz A. G., Frankenberger B., Tian F., Chen C., Stoeger T. Toxicol. In Vitro. 2013;27:2084–2093. doi: 10.1016/j.tiv.2013.08.004. [DOI] [PubMed] [Google Scholar]