 Sulfur mustard (SM) is a well known chemical warfare agent that poses a major threat to military personnel and also populace.
Sulfur mustard (SM) is a well known chemical warfare agent that poses a major threat to military personnel and also populace.
Abstract
Sulfur mustard (SM) is a well known chemical warfare agent that poses a major threat to military personnel and also populace. It targets multiple macromolecules, and its toxic effects are mediated by complex mechanisms. However, the sequence and manner of SM-induced cellular and molecular events underpinning the pathological processes are not fully elucidated. Effective therapeutic agents against SM poisoning are also lacking. The present study aimed to determine the dynamic cytotoxic profiles of SM in primary cultured human epidermal keratinocytes-fetal (HEK-f) and human dermal fibroblasts-adult (HDF-a) by establishing a high content analysis (HCA)-based multiparametric toxicity assay panel. SM was found to produce multiple, concentration-dependent cellular responses, including abnormal cellular morphology, cycle arrest, apoptosis, necrosis, mitochondrial membrane potential imbalance, increased membrane permeability, oxidative stress, DNA damage, and lysosome impairment. Time-course analysis indicated that the cellular and molecular responses related to the highly reactive targets of SM, such as glutathione depletion, reactive oxygen species release, DNA and lysosomal damage, and actin microfilament architecture modification, were congenerous initial events for SM injury. Moreover, this study demonstrated a novel finding that SM induced autophagy, and it was closely related to lysosome alterations in both cell types. Higher susceptibility of HEK-f cells to SM was associated with early lysosomal damage and decreased autophagy activity. Multiparametric HCA also revealed the concentration-dependent cytoprotective effect of hydroxychloroquine in HDF-a cells. The above results provided overall and objective evidence for elucidating the cytotoxic mechanism of SM, and also a good scientific base for further research on countermeasures against SM injury.
Introduction
Sulfur mustard (SM; 2,2′-bis-chloroethyl-sulfide), a powerful vesicant, has been used as a chemical warfare agent since the First World War. It still remains a serious threat to human health and public safety as it is easy to be acquired and used.1–3 Despite the great effort that has been made in the last decade to understand the toxicological mechanism of SM with some emerging technologies, such as microarray and proteomics, the mechanism for SM-induced vesication remains controversial,4–6 and effective anti-SM agents are absent.7,8
SM is a strong alkylating agent. Once exposed to the living body, it can rapidly penetrate the skin, ocular and bronchial mucous membranes to react with almost all the constituents of biological tissues and cells, causing acute and chronic lesions.9 Therefore, cells are the primary target for SM injury. DNA and membrane protein damage, oxidative stress, inflammatory reaction, cell cycle arrest, cell apoptosis, and changes in some molecules of the related pathways have been revealed as the mechanism for SM injury in different models.5,6,10,11 However, limited by the inability to measure, in a single platform, the multiple parameters that reflect a wide spectrum of function- and mechanism-related phenotypic changes, the sequence and manner of SM-induced cellular and molecular events underpinning the cyto-toxicological mechanism have not been fully elucidated.
HCA, a high-throughput screening (HTS)-based cell-imaging and multiparametric assay technology, has been developed to improve the efficiency of drug screening.12,13 HCA allows HTS and quantitative measurement of both “classical” events (such as cell and subcellular organelle size, shape, texture, cell necrosis, apoptosis, and differentiation) and molecular events (such as changes in protein level, localization, and intracellular phosphorylation) in the cellular background. The technique enables the acquisition of highly detailed and unbiased multiparametric profiles of intact cells with temporal and spatial information,14–16 which is particularly valuable for in-depth investigations of biological activities, including cytotoxicity and its associated mechanism.17–19 Nowadays, HCA-based multiparametric cytotoxic profiling has become a powerful tool in in vitro toxicity testing.19,20
The present study established a HCA-based multiparametric cytotoxicity assay panel to rapidly acquire the cellular toxic profile of SM, including information regarding the cellular morphology (nucleus, microfilament, microtubule, and whole-cell), cytotoxicity (oxidative stress, membrane permeability, mitochondrial membrane potential, and lysosomal damage), cellular death (necrosis, early apoptosis, late apoptosis, and autophagy), and cell cycle arrest (Table 1). Two dominant human skin cells, primary cultured human epidermal keratinocytes-fetal (HEK-f) and human dermal fibroblasts-adult (HDF-a), were selected because the skin is the primary target tissue of SM erosion.9–11,21 And time- and concentration-dependent cellular responses were observed to identify priming factors and to better understand the progression of SM-induced cellular toxicological pathology.
Table 1. Multi-parametric cellular phenotypic assay panel.
| Assay and parameters | Fluorescent probes |
| 1. Cellular morphology and cytoskeletal assay | |
| • Nuclear size and intensity | • Hoechst 33342 a |
| • F-actin | • Phalloidin (Alexa 488) |
| • α-Tubulin | • α-Tubulin Ab (Alexa 546) |
| • Cell morphology | • CellMask Deep Red |
| 2. Cytotoxicity assay 1 | |
| • Cell count | • Hoechst 33342 |
| • MnSOD content | • MnSODAb (Alexa 488) |
| • Mitochondrial membrane potential and mass (MMP) | • MitoTracker Red CMXRos |
| • Nuclear membrane permeability (NMP) | • TOTO-3 |
| 3. Oxidative stress assay | |
| • Cell count | • Hoechst 33342 |
| • Reduced glutathione (GSH) | • mBCI |
| • Reactive oxygen species (ROS) | • CM-H2DCFDA |
| 4. Cytotoxicity assay 2 | |
| • Cell count | • Hoechst33342 |
| • Autophagy | • LC3B Ab (Alexa 488) |
| • Lysosome membrane potential and mass | • LysoTracker Red |
| • Phosphohistone H2AX, Ser139 (DNA damage) | • pH2AXAb (Alexa 647) |
| 5. Apoptosis & necrosis assay | |
| • Cell count | • Hoechst 33342 |
| • Early apoptosis | • AnnexinV (Alexa 488) |
| • Late apoptosis | • PI |
| • Necrosis | |
| 6. Cell cycle arrest assay | |
| • Cell count | • Hoechst 33342 |
| • Cell cycle disruption | |
aThe colors of the probes represent the differential fluorescent colors of probes at the specified emission wavelength.
Materials and methods
Reagents, toxicants, and compounds
Propidiumiodide (PI) was purchased from Sigma-Aldrich (St Louis, MO, USA). Fluorescence probes (including MitoTrackerRed–CMXRos, LysoTracker Red, Alexa Fluor 488–phalloidin, Alexa Fluor 488–annexinV, TOTO-3 iodide, Cell Mask Deep Red, Hoechst 33342, CM-H2DCFDA, mBCI, and primary antibodies to LC3B, manganese superoxide dismutase [MnSOD], α-tubulin, and phosphorylated gamma-H2AX [pH2AX]) as well as all fluorescent-labeled secondary antibodies were purchased from Life Technologies (Carlsbad, CA, USA). Bovine serum albumin (BSA) was obtained from Sigma-Aldrich (St Louis, MO,USA). SM (purity ≥95%, MW: 159.08) was provided by the Institute of Chemical Defense of the Chinese People's Liberation Army (Beijing, China).
Reduced l-glutathione (GSH, purity ≥98%) was purchased from Sigma-Aldrich (St Louis, MO, USA). DSC127 (aclerastide; purity ≥99%) was synthesized by GL Biochem Ltd (Shanghai, China). Ola (olaparib), ABT-888 (veliparib), and BSI-201 (iniparib) were purchased from J&K Scientific Ltd (Beijing, China). Hydroxychloroquine (HCQ, purity ≥98%) was provided by Life Technologies (Carlsbad, CA, USA).
Cells and culture conditions
HEK-f and HDF-a cells (purchased from ScienCell, San Diego, CA, USA) were cultured and passaged according to the recommendations of the provider. Before SM treatment, HEK-f and HDF-a cells were seeded at a density of 6 × 103 cells per well in 100 μl of culture medium in 96-well assay plates (black plate, clear bottom; Costar 3603, Corning Incorporated, NY, USA) and then incubated for 18–24 h in a 5% CO2 incubator to allow cell spreading and attachment. The cultured cells were used for the SM challenge and anti-SM damage experiments.
Assay procedures
Treated manner of SM and candidate protective agents
All assays were conducted on both HEK-f and HDF-a cells. For all SM damage assays, SM was first dissolved in dimethyl sulfoxide (DMSO), and serially diluted in the cell culture medium to generate a 3× solution, which was soon added (in aliquots of 50 μl) to each well of 96-well assay plates containing pre-seeded cells. The final concentration of DMSO was strictly restricted to 0.2% across the entire assay. In the concentration-dependent experiments, three concentrations of SM (100, 300, and 450 μM) were used, and a single SM concentration of 300 μM was used in the time-course experiments. In all experiments, cell culture medium containing 0.2% DMSO was designated as the blank control, and was included in each plate to allow data normalization and plate quality control. Each test was performed in triplicate. After the addition of SM, the plates were returned to the cell culture incubator for a series of desired durations.
For anti-SM agent screening, the candidate protective agents (50 μl of 3× protective agents, including GSH, DP-7, Ola, ABT-888, BSI-201, and HCQ) were added 1 h before 50 μl of 3× SM (900 μM) was treated for 48 h. Seven different concentrations of each protective agent and three repetitions for each test were used.
Assay protocols
After SM treatments for the preset times, multiparametric assays were performed according to the assay panel shown in Table 1.This panel consisted of six assays containing almost all common cytotoxic response assays.15 Each assay was devised according to the principle of fluorescence tagging and followed the criterion of lowest interference between different fluorescence probes or markers. The detailed procedures for all six assays are described in the ESI.†
Image acquisition and analysis
The 96-well assay plates were scanned in an IN Cell Analyzer 2000 (GE HealthCare, USA) under a 20× objective lens, with a 200 W metal halide and high-pressure sodium discharge lamp, and a cooled CCD camera. For all measurements, 10 fields from each well and not less than 200 cells were observed. Excitation and emission filters were selected specially according to the specific wavelength of each fluorescent dye. DAPI filters were used for Hoechst-labeled cells; FITC filters were used for Alexa Fluor 488-labeled cells; Cy3 filters were used for cells labeled with MitoTrackerRed, LysoTracker Red, PI, or Alexa Fluor 546; and Cy5 filters were selected for cells labeled with CellMask Deep Red, TOTO-3 iodide, or Alexa Fluor 647. Exposure times that allowed the automatic optimization of fluorescent images of the control and reference wells were used.
For all assays, the acquired images were analyzed using an IN Cell Analyzer Workstation 3.5 and the Multi Target Analysis Module (GE Health Care, USA). Briefly, selecting a proper analysis mode according to each assay object, then creating an analysis protocol and optimizing segmentation parameters using control wells and various sample wells, finally, using the optimized analysis protocol to analyze the acquired images and obtain the measured values. The final measurements will be averaged over the population of cells detected in the fields of view. Specially, in assay 5, the two parameters cell divide method (Linear Discriminant 2D Filter in the Multi Target Analysis Module), which is similar to that of flow cytometry, was used to classify cells into four groups, by plotting the intensity of annexinV staining on the X axis, and the intensity of PI staining on the Y axis. The percentage of cells in four groups (survival, early apoptosis, late apoptosis, necrosis) was chosen as the output. In assay 6, a multi-parameters cell divide method (Decision Tree in the Multi Target Analysis module) was used to classify the cells into four groups (M phase, G0–G1, S, and G2 phase) according to the nuclear area, nuclear intensity, and nuclear integrated intensity.
Data processing
All results of cellular parameters were normalized and expressed as percentage changes of the control. Data from three repetitions of one test were used to generate mean, standard deviation (SD) and error (SE) values using Microsoft Excel 2010. The statistical screening parameter Z′ (Z′ factor) for each assay was calculated using the formula, Z′ = 1 – 3(σp + σn)/|(μp – μn)|, where σ and μ are the standard deviation and mean value, respectively, and ‘p’ and ‘n’ denote positive control (SM of 450 μM) and negative control (blank control solution, culture medium containing 0.2% DMSO), respectively.22 A measured difference of >3SDs was considered a significant change.23 Moreover, in the cell cycle analysis test, statistical analyses were also performed by two-tailed Welch's t-tests, one-way analysis of variance (ANOVA) followed by Tukey's multiple comparison tests, and SPSS 19.0 software was used for the statistical calculations (SPSS Inc., Chicago, IL, USA). P values <0.05 were considered statistically significant.
Results
SM is a well-known alkylating agent that rapidly reacts with numerous macromolecules in cells and produces various cytotoxic responses. In this study, a multiparametric cellular phenotypic assay panel was created to observe the concentration and time-dependent toxicological responses of SM in HEK-f and HDF-a cells. The panel consisted of 6 common combination assays involving more than 15 targets and 34 output parameters with definite biological or toxicological meaning.12,13,15,20 The Z′ values for most key assays were above 0.5 or near 0.5 (Table 2), indicating that the assays were HCS compatible and reliable in this experiment.22
Table 2. Output parameters and their implications.
| Assay targets | Parameters | Implications | Z′ factor |
| Assay 1 | |||
| Nuclear a (Nuc) | Cell count | Number of cells used in the assay | 0.66 |
| Nuc intensity | Mean nuclear intensity | 0.48 | |
| Nuc area | Area of the identified nucleus | 0.73 | |
| Nuc elongation | Ratio of the short axis to the long axis of the nucleus | — | |
| Nuc 1/(form factor) | Mean roundness index of the nucleus, perimeter 2/(4π × area) | 0.46 | |
| Nuc intensity CV | Coefficient of variation of the intensity of pixels over the population of pixels comprising the nuclear region | 0.62 | |
| F-Actin | Intensity | Average intensity of pixels for the filaments | 0.47 |
| 1/(form factor) | Mean roundness index of actins, perimeter 2/(4π × area) | — | |
| Elongation | Mean ratio of the short axis to the long axis of the filament | — | |
| Total area | Total area of filaments | 0.53 | |
| α-Tubulin | Intensity | Average intensity of pixels for tubulin | 0.46 |
| 1/(form factor) | Mean roundness index of tubulin, perimeter 2/(4π × area) | — | |
| Elongation | Mean ratio of the short axis to the long axis of tubulin | — | |
| Total area | Total area of tubulin | 0.60 | |
| Total-cell shape | Intensity × total area | Average content of tubulin in cells | — |
| 1/(form factor) | Area of cells | 0.45 | |
| Elongation | Mean intensity of pixels within the cell | — | |
| Total area | Ratio of the short axis to the long axis of the cell | 0.45 | |
| Cell 1/(form factor) | Cell roundness index, perimeter 2/(4π × area) | — | |
| Cell intensity CV | Coefficient of variation of pixel intensities over the population of pixels in the cell region | 0.56 | |
| Assay 2 | |||
| MnSOD | Intensity × total area | Average content of MnSOD in cells | 0.87 |
| MMP | Intensity × total area | Average level of MMP in cells | 0.80 |
| NMP | Intensity | Average intensity of TOTO-3 pixels within the nucleus | 0.48 |
| Assay 3 | |||
| GSH | Intensity × total area | Average content of GSH in cells | 0.46 |
| ROS | Intensity × total area | Average content of ROS in cells | 0.86 |
| Assay 4 | |||
| LC3B | Intensity × total area | Average content of LC3B in cells | 0.47 |
| Lysosome | Intensity × total area | Average number of lysosomes in cells | 0.56 |
| pH2AX | Intensity × total area | Average content of pH2AX within the nucleus | 0.65 |
| Assay 5 | |||
| mPLextro. | Intensity × total area | Average level of early apoptosis | 0.78 |
| mPLextro + Nuc(PI) | Intensity × total area | Average level of late apoptosis | 0.93 |
| Nuc(PI) | Intensity × total area | Average level of Necrosis | 0.81 |
| Assay 6 | |||
| Cell cycle | Intensity and Area | G0–G1; S; G2; M | >0.7 |
aNuclear analysis was included in each assay. mPLextro, membrane phospholipid extroversion; Nuc(PI), nuclei positively stained with propidiumiodide.
Impact of SM on cellular morphology and cytoskeleton
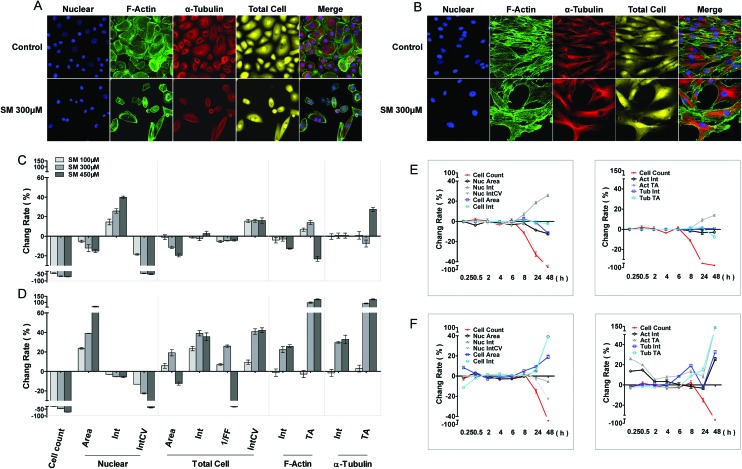
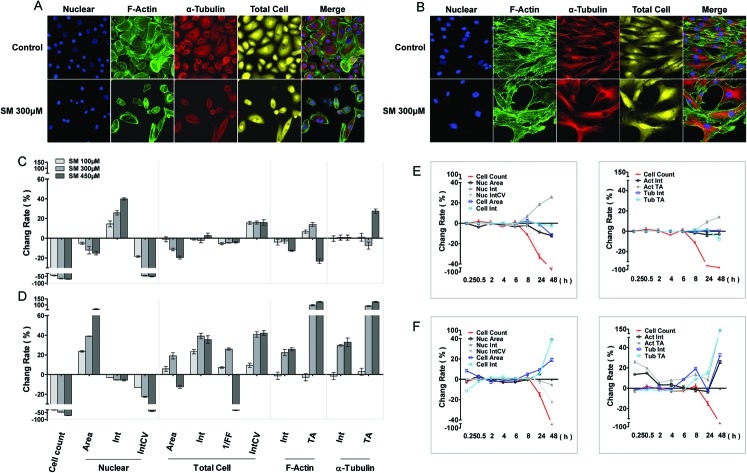
The cellular morphous texture reflects the cellular function and status, especially, under challenging conditions. After 48 h of SM treatment, the changes in cellular morphology were recorded, and the associated parameters are illustrated in Fig. 1. SM (100–450 μM) concentration dependently decreased the number of live cells and the coefficient of variation (CV) of nuclear staining intensity, and increased the CV of cellular staining intensity in both HEK-f and HDF-a cells. It also reduced the nuclear and cellular staining areas as well as the nuclear intensity in HEK-f cells (Fig. 1A and C). In contrast, it enlarged the nuclear and cellular staining areas, cellular staining intensity, and total areas of actin and α-tubulin staining in HDF-a cells (Fig. 1B and D). According to the interpretation of these output parameters described in Table 2, the above results indicated that SM induced different morphological changes in the two cell types. SM significantly reduced the nuclear and cell size, and the nucleus appeared condensed and tubulin exhibited accumulation. In HDF-a cells, SM caused nuclear and cell enlargement and cytoskeleton loosening. The time-course (0.25, 0.5, 2, 4, 6, 8, 24, and 48 h) observations (Fig. 1E and F) indicated that the earliest changes in live cell counts and nuclear morphology by SM (300 μM) exposure took place at 8 and 24 h for both HEK-f cells and HDF-a cells, respectively; then these responses worsened over time. Differently from HEK-f cells, in HDF-a cells, actin and total cell shape showed an early transient enlargement in the beginning, and an obvious area expansion took place only at 48 h. Tubulin exhibited a time-dependent increase in the area from 8 to 48 h. The aforementioned observation suggests that most of the observed cellular morphological changes were accompanied by cell death, and HEK-f cells were more sensitive to SM-induced damage.
Fig. 1. Effects of SM on the morphology of HEK-f and HDF-a cells. A and B: representative images of nuclear, actin, tubulin, and whole-cell shape in HEK-f cells and HDF-a cells, respectively; C and D: change rate (%) of morphologic parameters induced by different concentrations of SM in HEK-f cells and HDF-a cells, respectively (the treated time of SM is 48 h); E and F: change rate (%) of morphologic parameters induced by 300 μM SM for different durations of time in HEK-f cells and HDF-a cells, respectively; Elong, elongation; 1/FF, 1/(form factor); Int, staining intensity; Int CV, coefficient of variation of staining intensity; TA, total area. The nucleus was stained with Hoechst33342. Actin was stained with Alexa Fluor 488–phalloidin. Tubulin was stained with a tubulin antibody and an Alexa Fluor 546-labeled secondary antibody. Whole-cell (cytoplasm) staining was stained with CellMask Deep Red. All results of cellular parameters derived from specific cellular fluorescence images, normalized and expressed as percentage changes of the control. The data were expressed as mean ± standard error. All experiments were performed in triplicate, with no less than 200 cells per well. A measured difference of 3 SDs (standard deviation) was considered a significant change.
Mode of SM-induced cell death
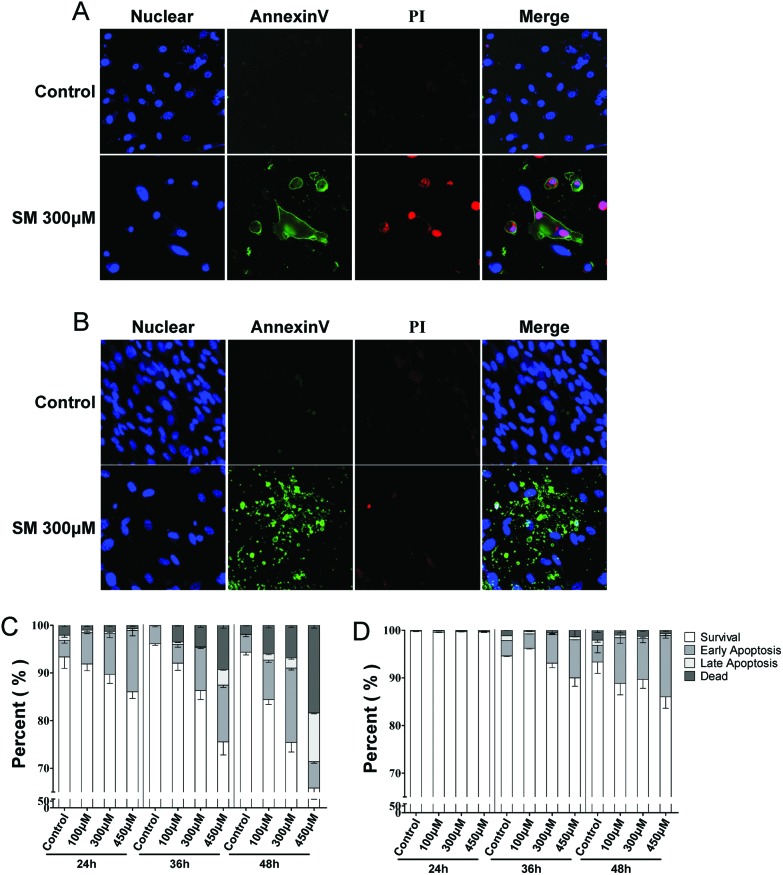
Cell death is the final event of cytotoxicity, and the mode of cell death (necrosis or apoptosis) mostly depends on the mechanism of intoxication.24 The extent of necrosis and apoptosis induced by SM (100, 300, 450 μM) was assayed at 24, 36, and 48 h in the two cell types. As shown in Fig. 2, SM induced early apoptosis in a concentration- and time-dependent manner, which was observed from 24 h onward in HEK-f cells and 48 h onward in HDF-a cells. In addition, the significantly higher rates of later apoptosis and concentration-dependent cell necrosis were observed only in HEK-f cells after 36 h of SM treatment. These results suggested that SM could induce apoptosis in both cells; however, HEK-f cells suffered a more sensitive apoptosis response. This finding was consistent with the observations mentioned earlier on the cellular and nuclear morphology.
Fig. 2. Effects of SM on mode of cell death (necrosis, apoptosis) in HEK-f and HDF-a cells. A and B: representative images of the nucleus, cell membrane eversion (annexinV), and nuclear necrosis (PI) in HEK-f cells and HDF-a cells, respectively. C and D: effects of different concentrations of SM on cell necrosis and apoptosis at different treatment times in HEK-f cells and HDF-a cells, respectively. The nucleus was stained with Hoechst 33342. Cell membrane eversion was detected using phosphatidylserine (PS)-labeled annexinV. Nuclear necrosis was stained with propidiumiodide (PI). The percentage of cells in four groups (survival, early apoptosis, late apoptosis, necrosis) was derived from the two parameters cell divide analysis method (Linear Discriminant 2D Filter in the Multi Target Analysis Module, GE Health Care, USA) according to the intensity of annexinV and PI staining on images. All the results were normalized and expressed as percentage changes of the control. The data were expressed as mean ± standard error. All experiments were performed in triplicate, with no less than 200 cells per well. A measured difference of 3 SDs (standard deviation) was considered a significant change.
Cell cycle arrest induced by SM
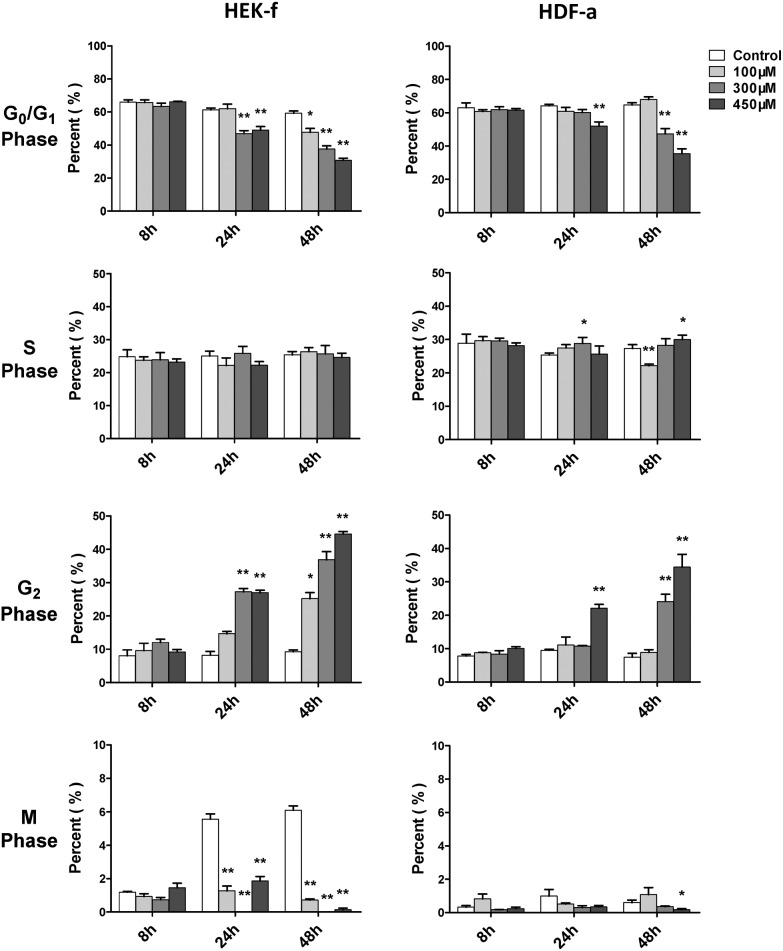
Cell cycle arrest is an endpoint event of cytotoxicity. Alkylating agents have been considered as nonspecific inhibitors of cell cycle in early studies.25 In this study, the cell cycle was examined at 8, 24, and 48 h of SM exposure, in terms of DNA content and nuclear size. The SM effect on the cell cycle could be observed only at 24 h (Fig. 3), and it became more significant at 48 h. The G2 phase arrest was the most obvious effect upon SM exposure in both cell types, meanwhile, S phase arrest was a weaker effect and only occurred in HDFa. However, more notable inhibition on M phase was noted in HEK-f cells, suggesting that the HEK-f cells seemed to be more susceptible to SM than HDF-a cells. These observations affirmed blocked normal cell growth and division by arresting the G2 and S phase.
Fig. 3. Effects of SM on the cell cycle of HEK-f cells and HDF-a cells. The percentage of cell cycle phases derived from the multiparametric cell division method (Decision Tree in the Multi Target Analysis module, GE) by classifying cells into four groups (M, G0–G1, S, and G2 phases) according to the fluorescence of nuclear area, nuclear intensity, and nuclear integrated intensity. All the results were normalized and expressed as percentage changes of the control. The data were expressed as mean ± standard error. All experiments were performed in triplicate, with no less than 200 cells per well. * and ** expressed as P < 0.5 and P < 0.01.
Changes in MnSOD, MMP, and NMP
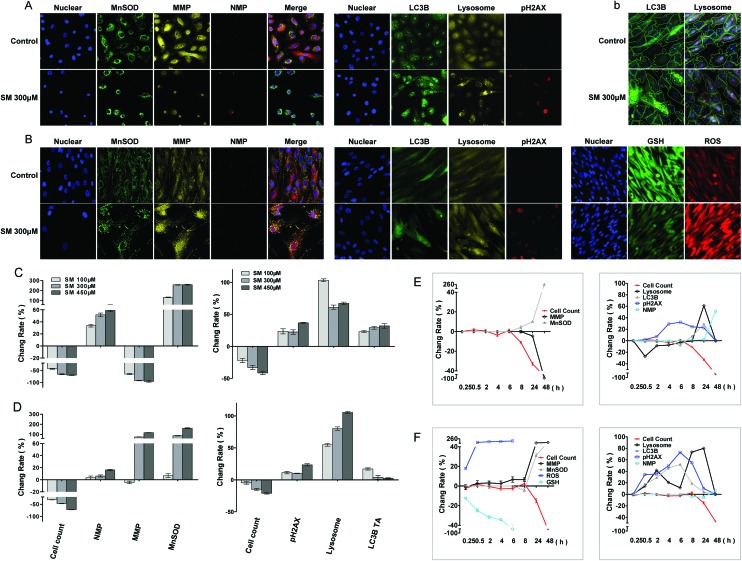
Oxidative stress, mitochondrial damage, and increased membrane permeability are common cell responses to cytotoxic agents and constitute important parameters for cytotoxicity testing. In this study, cell manganese superoxide dismutase (MnSOD) content, mitochondrial membrane potential (MMP), and nuclear membrane permeability (NMP) were measured at 8, 24, and 48 h of SM treatment. As shown in Fig. 4, SM increased the MnSOD content, MMP, and NMP in a concentration and time-dependent manner in both cells. Significant increases in MnSOD content and the change of the MMP level were observed at 24 h, and they were more obvious in the HDF-a cells than those in the HEK-f cells. These results illustrated the severe oxidative stress and mitochondrial damage caused by SM. Since the increase in MnSOD (an endogenous peroxidase for reactive oxygen species [ROS]) is an indicator of adaptive and/or protective response against oxidative stresses, these stresses must have preceded the changes in MMP and NMP in this test. Therefore, alterations in MMP and NMP were considered as the endpoint events of SM intoxication in cells.
Fig. 4. Changes in MnSOD, MMP, NMP, pH2AX, lysosomes, LC-3B, GSH, and ROS induced by SM in HEK-f cells and HDF-a cells. A and B: representative images; C and D: change rate (%) induced by different concentrations of SM treatments; E and F: change rate (%) induced by 300 μM SM for different durations of time in HEK-f cells and HDF-a cells, respectively; b: image segmentation for LC3B and lysosomes in HDF-a cells according to the Multi Target Analysis module (GE Health Care, USA). The nucleus was stained with Hoechst 33342; MnSOD was stained with a MnSOD antibody and an Alexa Fluor 546–labeled secondary antibody; MMP was stained with MitoTracker Red; NMP was stained with TOTO-3 iodide; pH2AX was stained with an anti-pH2AX primary antibody and an Alexa Fluor 647–labeled secondary antibody; lysosomes were stained with LysoTracker Red; LC-3B was stained with an anti-LC3B primary antibody and an Alexa Fluor 488–labeled secondary antibody; and GSH was stained with mBCI. ROS were indicated by CM-H2DCFDA. All the results of cellular parameters derived from specific cellular fluorescence images were normalized and expressed as percentage changes of the control. The data were expressed as mean ± standard error. All experiments were performed in triplicate, with no less than 200 cells per well. A measured difference of >3 SDs (standard deviation) was considered a significant change.
Changes in ROS and GSH in HDF-a cells
The significant increase in the cell MnSOD content upon SM exposure implied that SM could induce oxidative stress. To further probe the mechanism of oxidative stress, the intercellular levels of ROS and glutathione (GSH) were measured at 15 min, 30 min, 1 h, 2 h, 4 h, and 8 h after the SM exposure to HDF-a cells. As shown in Fig. 4, a significant decrease in the GSH level and an increase in the ROS level were detected even at 15 min. The GSH level decreased almost to 80% at 2 h, while the release of ROS reached its peak level. Thus, oxidative stress was confirmed as an initial toxic event of SM intoxication. As the SM-induced ROS release was much lower than that observed by the agents that selectively target the mitochondria in this experiment (data not shown), GSH depletion was believed to play a dominant role in SM-induced oxidative stress.
DNA damage, and alterations in lysosomes and autophagy activity
DNA is the primary target of SM. Autophagy degrades damaged organelles, cell membranes, and proteins through an autophagosomic-lysosomal pathway. The failure of autophagy is thought to be one of the main reasons for the accumulation of cell damage.26,27 Moreover, the release of lysosomes containing hydrolytic enzymes can initiate further cell damage.23 In this study, pH2AX (an indicator of DNA damage), LC3B (a marker of autophagy flux), and LysoTracker Red–stained lysosomes were assayed at 30 min, 2, 4, 6, 8 and 24 h of SM treatment. As shown in Fig. 4, with the increased SM concentration and exposure time, the aforementioned three parameters significantly changed in both cells; however, the pattern of changes differed between the two cell types. The expression of pH2AX increased in both HEK-f and HDF-a cells, peaked at 6 h, and declined from 8 h. It appeared more earlier in HDF-a cells, with the first significant change noted at 30 min, suggesting that DNA damage may occur early. The time pattern of LC3B expression changes was similar to that of pH2AX expression in HDF-a cells, while no obvious changes were observed until 8 h in HEK-f cells. The lysosome size significantly reduced at 30 min of SM exposure in HEK-f cells, then recovered to the normal level at 8 h and increased at 24 h. The late increase in lysosome size was negatively correlated with the SM concentration. In HDF-a cells, time-dependent changes were observed in the lysosome size, with the first small peak appearing at 2 h, followed by a decrease until 8 h, and then the size was increased again in a concentration dependent manner to reach the maximal size at 24 h. The above observation reflected the different features of SM-induced damage in the two cells. Specifically, DNA in HDF-a cells and lysosomes in HEK-f cells were more susceptible to SM. Changes in autophagy activity were similar to that of lysosome in both cell types.
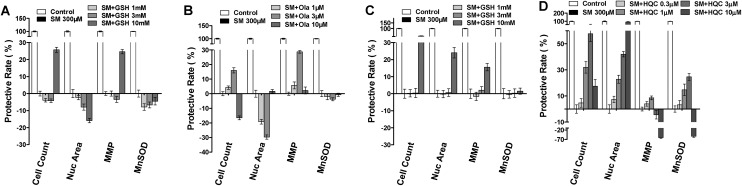
Cytoprotective agent screening
The results discussed in the preceding section revealed that SM induced concentration-dependent changes in nuclear number, MnSOD, and MMP parameters. And these parameters accurately reflected the final damage caused by SM exposure in both HEK-f and HDF-a cells. Based on the aforementioned results, the protective effects of some previously reported cytoprotective agents against SM challenge were evaluated in SM-treated HEK-f and HDF-a cells, namely, GSH and poly(ADP-ribose)polymerase (PARP) inhibitors (Ola, ABT-888, and BSI-201),28,29 DSC127 [aclerastide; active ingredient, NorLeu3-angiotensin (1–7), the only active product approved for treating diabetic ulcers],30,31 and hydroxychloroquine (HCQ, an autophagy inhibitor). All testing compounds were added to the cells prior to the SM challenge (300 μM) for 48 h. In HEK-f cells, only 10 μM GSH and 3 μM Ola demonstrated a weak protective effect (HCQ was not tested in HEK-f). The results shown in Fig. 5 indicated that the maximal recovery of cell count and MMP parameters in the GSH group was both about 25%, while that in the Ola group was about 16% and 28%, respectively. In HDF-a cells, only 10 μM GSH reversed the SM-induced decrease in cell count, nuclear area, and MMP parameters, with the maximal protective effects being about 50%, 22%, and 15%, respectively. However, GSH and Ola did not improve the MnSOD parameter. Surprisingly, HCQ reversed all the tested parameters in the testing cell in a concentration-dependent manner. But, BSI-201 and other testing compounds did not exhibit any beneficial effect in both cells, and even caused more severe cytotoxicity at higher concentrations.
Fig. 5. The cyto-protection of l-glutathione (GSH), Ola (olaparib) and hydroxychloroquine (HCQ) against SM challenge, respectively, in HEK-f cells (A and B) and HDF-a cells (C and D). Cellular phenotypic assays were carried out after SM treatment for 48 h, and the test compounds were added to the cells 1 h before the SM challenge. The nucleus was stained with Hoechst 33342, MnSOD was stained with MnSOD antibody and an Alexa Fluor 546–labeled secondary antibody, and MMP was stained with MitoTracker Red. All the results of cellular parameters derived from specific cellular fluorescence images were normalized and expressed as percentage changes of the control. The data were expressed as mean ± standard deviation (SD). All experiments were performed in triplicate, with no less than 200 cells per well. A measured difference of 3 SDs was considered a significant change.
Discussion
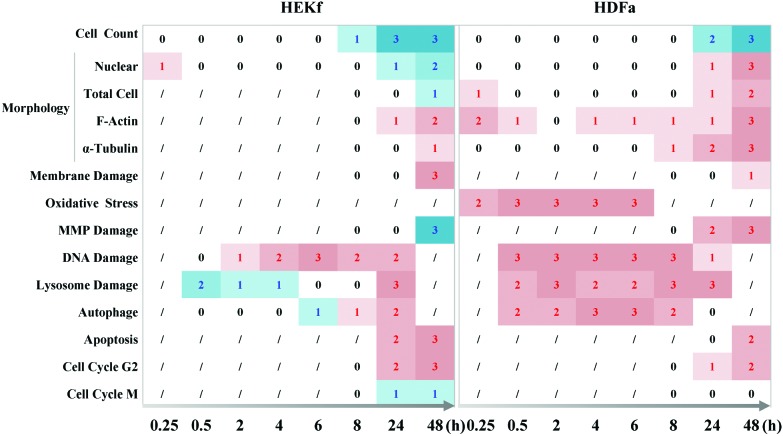
In the present work, the dynamic profile of the cytotoxicity of SM in primary cultured HEK-f and HDF-a cells was demonstrated, for the first time, using HCA-based multiparametric analysis. The main results, summarized in Fig. 6, clearly displayed the horary microscopic and molecular cellular phenotypic responses as well as the pathological processes associated with apoptosis; necrosis; mitochondria and membrane impairment; and cellular morphologic and structural injuries in the nucleus, tubulin, and whole-cell shape were the final events of SM-mediated cytotoxicity, despite the initial transient changes in actin and whole-cell shape. Meanwhile, oxidative stress, DNA damage, and lysosome challenge were early events upon SM poisoning. Moreover, cell autophagy activity was closely related to the changes observed in lysosomes. Quantitative cytotoxic profiling of SM demonstrated that HEK-f cells were more susceptible to SM damage than the HDF-a cells.
Fig. 6. Time course of cytopathological effects of SM in HEK-f and HDK-a cells. “0,1,2,3” represents ND (not detected), change, concentration-dependent change, and concentration-dependent change and maximal change were greater than 50% fold, respectively; “/” represents undetected; the red numbers represent an increased response, while the blue numbers represent a decreased response.
Quick and accurate identification of the initial molecular and cellular reaction events is the key to understanding the mechanism underlying the SM-mediated cytopathology. As a bifunctional alkylating agent, SM actively reacts with many cellular components, including lipids, proteins, and nucleic acids, especially the sulfhydryl-containing macromolecules, and N7 of guanine and the N3 of adenine in DNA/RNA, and rapidly results in thiol depletion, GSH depletion, DNA/other cellular macromolecule alkylation, and a series of related molecular and cellular fast responses.10,32–34 The present study indeed observed that α-actin microfilament (a sulfhydryl-containing macromolecule 9)-related change of cellular architecture, GSH depletion, and ROS release took place at the earliest time point of the assay, followed by the change of pH2AX expression and the lysosome. Given the fact that pH2AX (a DNA damage and repair marker) is a downstream molecule of double-stranded breaks and ataxia telangiectasia-mutated (ATM) kinase pathway,35 and together with the observation of the remarkable ROS release at the earliest time, it is reasonable to conclude that DNA damage is one of the earliest events of SM injury. The similar conclusion was also obtained with 2-chloroethyl ethyl sulfide (CEES, an analogue of SM) in HaCaT cells, which demonstrated that the phosphorylation of DNA damage sensors and checkpoint kinases, ATM at ser1981 and ataxia telangiectasia-Rad3-related (ATR) at ser428, occurred within 30 min of CEES exposure.36 In addition, some early molecular events of SM, including its analogue-mediated damage, such as p53 Ser15 phosphorylation, AKT signal pathway inhibition, nitric oxide synthase activation, and intracellular calcium level increase, have been proved to occur simultaneously with DNA repair or to be the direct results of sulfhydryl-containing macromolecule depletion (actin and Ca2+ translocase injury) or oxidative stress.37–42 These molecular changes ultimately caused cellular morphological deformities, loss of cellular integrity, cell cycle arrest, and cell apoptosis. Therefore, the molecules that are highly reactive with SM in a cell, together with their directly associated molecular pathways constitute the underlying mechanism of SM-mediated tissue injury.
In this study, we found, for the first time, that cellular autophagy is one of the earlier events induced by SM, and the change of autophagy is associated with the lysosome in two cell types. Autophagy, an evolutionarily conserved catabolic process, is a lysosome-dependent pathway for protein degradation. Among the many pathophysiological mechanisms, the lysosomes, by way of autophagy (autophagosome–lysosome pathway), degrade those damaged intracellular organelles and proteins.43,44 Generally, autophagy serves as a protective mechanism; however, persistent activation of autophagy can result in cell death. There have been ample examples of cross talk between autophagy and other modes of cell death after exposure to toxicants.26,27 In the review of toxicant-induced autophagy by Bordin,45 the authors indicated that autophagy induced by alkylating agents played a cytoprotective role in cells. For example, BO-1051, a newly synthesized nitrogen mustard analogue with high DNA affinity, induced autophagy in hepatocarcinoma. It was further demonstrated that autophagy induced by BO-1051 was a downstream target of the ATM signaling pathway.46 Taking into account the other cellular effects in the present experiment, it was deduced that SM intoxication not only leads to cell necrosis and apoptosis, but also causes cell autophagy. Since the autophagy appeared earlier than cell death and was accompanied by an increase in the lysosome size in HDF-a cells, it was believed that autophagy mainly served as a protective mechanism, at least within 8 h of SM exposure. However, the number of live cells decreased from 8 h onward in both cell types. The actual role of autophagy in the modes of cell death by SM exposure needs to be further investigated.
Dermal keratinocytes have been a focus of the in vitro studies on SM toxicology. Keratinocytes in the dermal layer of the skin are more sensitive to SM cytotoxicity because of their self-renewing nature and greater capacity for proliferation.5 This study compared the cytotoxic responses of SM between human dermal keratinocytes and fibroblasts, the two predominant cell types in the dermal layer. In vivo damage to both cell types was noted at a very early stage.5,21 In this study, despite similar SM cytotoxicity profiles, HEK-f cells exhibited more severe impairment in terms of decreased MMP, more serious NMP, stronger apoptosis activity, and earlier death. Moreover, the cells became rounder and smaller, and showed centric nuclear microtubule aggregation and nuclear shrinkage. This morphologic change may be the cause of the keratinocyte disorder, barrier damage, and enhanced skin tissue permeability observed in skin injury by SM.9 In contrast, HDF-a cells showed more earlier DNA damage, but less MMP damage, cell and nuclear swelling, and cytoskeletal loosening. More interestingly, significant lysosomal damage and associated hydrolytic enzyme release at 30 min of SM exposure were only found in HEK-f cells. While cell autophagy significantly increased from 30 min to 24 h in HDF-a cells. Since DNA damage mediated autophagy, and autophagy has a negative effect on apoptosis under most conditions, it was hypothesized that differences in cellular background regulatory machinery for autophagic lysosomes may be an important reason for the higher susceptibility of HEK-f cells to SM.
Highly efficient screening is a key step in drug discovery and development. In the present study, a four-parameter (cell count, nuclear area, MMP, and MnSOD) assay was designed based on the cytotoxic profiles of SM and technical feasibility of HCS and used to screen for the prophylactic effects of some known and/or potential protective agents against SM. GSH has been widely used in combination therapy for SM intoxication.3 PARP inhibitors are believed to be beneficial in reversing the DNA damage induced by SM.28,29 HCQ, an autophagy inhibitor, was also selected based on the finding of this study that SM induced cellular autophagy. The results showed that GSH had partial protective effects on cell count and MMP only at an adequate concentration in both cell types. Higher concentrations of GSH even caused more severe cytotoxicity (data unpublished). Surprisingly, HCQ reversed all four parameters in HDF-a cells in a concentration-dependent manner, and had a better protective effect over GSH. The PARP inhibitor showed a disappointing outcome; and GSH and Ola failed to protect against SM-induced oxidative stress. This may be due to the lower penetration ability of GSH across the cell membranes, or the limited DNA-repairing power of PARP inhibitors in the situation of acute damage by SM. In addition, the more sensitive nature of the primary cells to DNA damage may also be a cause. The screening results suggested that the GSH dose used for treating SM injury should be carefully determined. In addition to conventional strategies against SM injury, the inhibition of autophagy may also be considered as a potential prophylactic measure. The benefit of PARP inhibitors in treating chronic damage may merit further examination.
HCA-based multiparametric analysis is being increasingly used as a highly efficient approach in the cytotoxic evaluations of drugs and chemical hazards.47 By using this technology, the comprehensive and objective data were quickly acquired to describe SM cytotoxicity profiles and to evaluate protective agents in two human dermal cells in a single platform. The results accurately illustrate the sequence and manner of SM-induced cellular and molecular events, and are valuable for understanding the pathological processes of SM. However, it should be noted that the current results may not be appropriate for explaining the long-term toxic outcomes of SM in vivo due to the limitations of the two-dimensional cell culture.
Conclusions
This study is the first to report the dynamic SM cytotoxic profiling of two primary human skin cells using the HCA-based multiparametric cytotoxicity assay. The results clearly demonstrated the highly reactive target molecules of SM and their related cellular responses, such as oxidative stress, DNA and lysosomal damage, actin microfilament architecture modification. They collectively contribute to the initial events of SM cellular toxicology. In addition, this study offers a novel finding that the cellular autophagy ability is an effective element against SM intoxication. Blocking autophagy may be a direction for future research on countermeasures against SM injury.
Supplementary Material
Acknowledgments
This work was supported by the National Science and Technology Major Project of the Ministry of Science and Technology of China (grant no. 2012ZX09301-001, 2012ZX09301003, and 2015ZX09J15104) and the National Natural Science Foundation of China (grant no. 81430090).
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c5tx00305a
References
- Wattana M., Bey T. Prehosp. Disaster Med. 2009;24:19–29. doi: 10.1017/s1049023x0000649x. [DOI] [PubMed] [Google Scholar]
- C. M. Pechura and D. P. Rall, Veterans at Risk: The Health Effects of Mustard Gas and Lewisite, National Academy Press, Washington, DC, 1993. [PubMed] [Google Scholar]
- Evison D., Hinsley D., Rice P. BMJ. 2002;324:332–335. doi: 10.1136/bmj.324.7333.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerecke D. R., Chen M., Isukapalli S. S., Gordon M. K., Chang Y. C., Tong W., Androulakis I. P., Georgopoulos P. G. Toxicol. Appl. Pharmacol. 2009;234:56–165. doi: 10.1016/j.taap.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowsey P. A., Blain P. G. Toxicol. Lett. 2014;230:393–401. doi: 10.1016/j.toxlet.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Shakarjian M. P., Heck D. E., Gray J. P., Sinko P. J., Gordon M. K., Casillas R. P., Heindel N. D., Gerecke D. R., Laskin D. L., Laskin J. D. Toxicol. Sci. 2010;114:5–19. doi: 10.1093/toxsci/kfp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimfield A. A. Prog. Mol. Biol. Transl. Sci. 2012;112:209–230. doi: 10.1016/B978-0-12-415813-9.00007-6. [DOI] [PubMed] [Google Scholar]
- Thiermann H., Worek F., Kehe K. Chem. – Biol. Interact. 2013;206:435–443. doi: 10.1016/j.cbi.2013.09.015. [DOI] [PubMed] [Google Scholar]
- Sayer N. M., Whiting R., Green A. C., Anderson K., Jenner J., Lindsay C. D. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2010;878:1426–1432. doi: 10.1016/j.jchromb.2009.11.030. [DOI] [PubMed] [Google Scholar]
- Smith W. J., Dunn M. A. Arch. Dermatol. 1991;127:1207–1213. [PubMed] [Google Scholar]
- Smith K. J., Hurst C. G., Moeller R. B., Skelton H. G., Sidell F. R. J. Am. Acad. Dermatol. 1995;32:765–776. doi: 10.1016/0190-9622(95)91457-9. [DOI] [PubMed] [Google Scholar]
- Taylor D. L. Methods Mol. Biol. 2007;356:3–18. doi: 10.1385/1-59745-217-3:3. [DOI] [PubMed] [Google Scholar]
- Giuliano K. A., DeBiasio R. L., Dunlay R. T., Gough A., Volosky J. M., Zock J., Pavlakis G. N., Taylor D. L. J. Biomol. Screening. 1997;2:249–259. [Google Scholar]
- Abraham V. C., Taylor D. L., Haskins J. R. Trends Biotechnol. 2004;22:15–22. doi: 10.1016/j.tibtech.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Gasparri F., Cappella P., Galvani A. J. Biomol. Screening. 2006;11:586–598. doi: 10.1177/1087057106289406. [DOI] [PubMed] [Google Scholar]
- Li Z., Yan Y., Powers E. A., Ying X., Janjua K., Garyantes T., Baron B. J. Biomol. Screening. 2003;8:489–499. doi: 10.1177/1087057103257309. [DOI] [PubMed] [Google Scholar]
- Judson R. S., Kavlock R. J., Setzer R. W., Hubal E. A., Martin M. T., Knudsen T. B., Houck K. A., Thomas R. S., Wetmore B. A., Dix D. J. Chem. Res. Toxicol. 2011;24:451–462. doi: 10.1021/tx100428e. [DOI] [PubMed] [Google Scholar]
- Kavlock R., Chandler K., Houck K., Hunter S., Judson R., Kleinstreuer N., Knudsen T., Martin M., Padilla S., Reif D., Richard A., Rotroff D., Sipes N., Dix D. Chem. Res. Toxicol. 2012;25:1287–1302. doi: 10.1021/tx3000939. [DOI] [PubMed] [Google Scholar]
- Xu J. J., Diaz D., O'Brien P. J. Chem. – Biol. Interact. 2004;150:115–128. doi: 10.1016/j.cbi.2004.09.011. [DOI] [PubMed] [Google Scholar]
- O'Brien P. J., Irwin W., Diaz D., Howard-Cofield E., Krejsa C. M., Slaughter M. R., Gao B., Kaludercic N., Angeline A., Bernardi P., Brain P., Hougham C. Arch. Toxicol. 2006;80:580–604. doi: 10.1007/s00204-006-0091-3. [DOI] [PubMed] [Google Scholar]
- Ghanei M., Poursaleh Z., Harandi A. A., Emadi S. E., Emadi S. N. Cutan. Ocul. Toxicol. 2010;29:267–277. doi: 10.3109/15569527.2010.511367. [DOI] [PubMed] [Google Scholar]
- McGovern S. L., Caselli E., Grigorieff N., Shoichet B. K. J. Med. Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- Wolcke J., Ullmann D. Drug Discovery Today. 2001;6:637–646. doi: 10.1016/s1359-6446(01)01807-4. [DOI] [PubMed] [Google Scholar]
- Kepp O., Galluzzi L., Lipinski M., Yuan J., Kroemer G. Nat. Rev. Drug Discovery. 2011;10:221–237. doi: 10.1038/nrd3373. [DOI] [PubMed] [Google Scholar]
- Ku W. W., Bernstein I. A. Toxicol. Appl. Pharmacol. 1988;95:397–411. doi: 10.1016/0041-008x(88)90358-4. [DOI] [PubMed] [Google Scholar]
- Aki T., Funakoshi T., Unuma K., Uemura K. Toxicology. 2013;311:205–215. doi: 10.1016/j.tox.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Orrenius S., Kaminskyy V. O., Zhivotovsky B. Annu. Rev. Pharmacol. Toxicol. 2013;53:275–315. doi: 10.1146/annurev-pharmtox-011112-140210. [DOI] [PubMed] [Google Scholar]
- Balali-Mood M., Hefazi M. Fundam. Clin. Pharmacol. 2005;19:297–315. doi: 10.1111/j.1472-8206.2005.00325.x. [DOI] [PubMed] [Google Scholar]
- Smith W. J. Toxicology. 2009;263:70–73. doi: 10.1016/j.tox.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Rodgers K., Verco S., Bolton L., Dizerega G. Expert Opin. Invest. Drugs. 2011;20:1575–1581. doi: 10.1517/13543784.2011.619976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers K. E., Bolton L. L., Verco S., diZerega G. S. Adv. Wound. Care. 2015;4:339–345. doi: 10.1089/wound.2014.0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Yue L., Liu Q., Chen J., Xie J. J. Chromatog. B. 2011;879:1707–1712. doi: 10.1016/j.jchromb.2011.03.040. [DOI] [PubMed] [Google Scholar]
- Noort D., Benschop H. P., Black R. M. Toxicol. Appl. Pharmacol. 2002;184:116–126. [PubMed] [Google Scholar]
- Ghanei M., Poursaleh Z., Harandi A. A., Emadi S. E., Emadi S. N. Cutan. Ocul. Toxicol. 2010;29:269–277. doi: 10.3109/15569527.2010.511367. [DOI] [PubMed] [Google Scholar]
- Kinner A., Wu W., Staudt C., Iliakis G. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari-Singh N., Gu M., Agarwal C., White C. W., Agarwal R. Chem. Res. Toxicol. 2010;23:1034–1044. doi: 10.1021/tx100038b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbulan-Rosenthal C. M., Ray R., Benton B., Soeda E., Daher A., Anderson D., Smith W. J., Rosenthal D. S. Toxicology. 2006;227:21–35. doi: 10.1016/j.tox.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Kehe K., Raithel K., Kreppel H., Jochum M., Worek F., Thiermann H. Arch. Toxicol. 2008;82:4. doi: 10.1007/s00204-007-0265-7. [DOI] [PubMed] [Google Scholar]
- Kehe K., Balszuweit F., Steinritz D., Thiermann H. Toxicology. 2009;263:12–19. doi: 10.1016/j.tox.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Rosenthal D. S., Velena A., Chou F. P., Schlegel R., Ray R., Benton B., Anderson D., Smith W. J., Simbulan-Rosenthal C. M. J. Biol. Chem. 2003;278:8531–8540. doi: 10.1074/jbc.M209549200. [DOI] [PubMed] [Google Scholar]
- Bloch W., Elischer A., Schriek M., Bohm K., Moghbeli R., Kehe K., Szinicz L., Steinritz D. Toxicology. 2007:233. [Google Scholar]
- Inturi S., Tewari-Singh N., Agarwal C., White C. W., Agarwal R. Mutat. Res. 2014;763–764:53–63. doi: 10.1016/j.mrfmmm.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Lerner A., Bialik S., Simon H. U., Kimchi A. Cell Death Differ. 2009;16:966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- Murrow L., Debnath J. Annu. Rev. Pathol. 2013;8:107–137. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordin D. L., Lima M., Lenz G., Saffi J., Meira L. B., Mesange P., Soares D. G., Larsen A. K., Escargueil A. E., Henriques J. A. Mutat. Res. 2013;753:91–99. doi: 10.1016/j.mrrev.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Chen L. H., Loong C. C., Su T. L., Lee Y. J., Chu P. M., Tsai M. L., Tsai P. H., Tu P. H., Chi C. W., Lee H. C., Chiou S. H. Biochem. Pharmacol. 2011;81:594–605. doi: 10.1016/j.bcp.2010.12.011. [DOI] [PubMed] [Google Scholar]
- Krewski D., Acosta D., Andersen M., Anderson H., Bailar J. C., Boekelheide K., Brent R., Charnley G., Cheung V. G., Green S., Kelsey K. T., Kerkvliet N. I., Li A. A., McCray L., Meyer O., Patterson R. D., Pennie W., Scala R. A., Solomon G. M., Stephens M., Yager J., Zeise L., Comm S. J. Toxicol. Environ. Health, Part B. 2010;13:51–138. doi: 10.1080/10937404.2010.483176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.