Abstract
Mycobacterium africanum (Maf) causes a substantial proportion of human tuberculosis in some countries of West Africa, but little is known on this pathogen. We compared the genomes of 253 Maf clinical isolates from Ghana, including N = 175 Lineage 5 (L5) and N = 78 Lineage 6 (L6). We found that the genomic diversity of L6 was higher than in L5 despite the smaller sample size. Regulatory proteins appeared to evolve neutrally in L5 but under purifying selection in L6. Even though over 90% of the human T cell epitopes were conserved in both lineages, L6 showed a higher ratio of non-synonymous to synonymous single nucleotide variation in these epitopes overall compared to L5. Of the 10% human T cell epitopes that were variable, most carried mutations that were lineage-specific. Our findings indicate that Maf L5 and L6 differ in some of their population genomic characteristics, possibly reflecting different selection pressures linked to distinct ecological niches.
Introduction
The global phylogeography of the human-adapted Mycobacterium tuberculosis complex (MTBC) shows highest diversity in West Africa, with six out of the seven known lineages represented1,2. Two of these lineages, Lineage 5 (L5) and Lineage 6 (L6), together originally known as Mycobacterium africanum (Maf), are restricted to West Africa for unknown reasons. By contrast, MTBC lineages belonging to Mycobacterium tuberculosis sensu stricto (Mtbss), in particular Lineage 4 (L4), are more geographically widespread1. M. africanum has remained an important pathogen in West Africa since its first description in 19683, and is responsible for up to half of human tuberculosis (TB) in some regions4.
The MTBC is thought to have originally emerged in Africa and subsequently spread to other parts of the world following waves of human migrations, trade and conquests5–8. Yet the reason(s) why Maf is limited to West Africa despite, for example, centuries of the trans-Atlantic slave trade remains unknown. Some comparative studies have identified phenotypic differences between the two Maf lineages9,10, suggesting they might be fundamentally distinct and occupy different ecological niches.
Three hypotheses have been put forward to explain the restriction of Maf to West Africa. The first hypothesis proposes that Maf might have emigrated outside of Africa but was later outcompeted by Mtbss, which has been shown to be more virulent than Maf in animal models11. The second hypothesis states that the restriction of Maf to West Africa is due to its adaptation to West African human populations9,12. Finally, according to the third hypothesis, Maf might be zoonotic with an animal reservoir restricted to West Africa.
Some evidence in support of the first hypothesis is the reported association of Maf (L6) with HIV co-infection, attenuated ESAT-6 responses and delayed progression to active disease relative to Mtbss9,13–16. In addition, both Maf lineages as well as Mtbss L1, together described as “ancestral” MTBC lineages, have been shown to elicit a stronger early production of pro-inflammatory cytokines compared to the “modern” MTBC L2, L3 and L417. The delayed pro-inflammatory immune response in the “modern” MTBC lineages might allow for more rapid disease progression and transmission17. The second hypothesis is supported by the statistical association of L5 with the native West African ethnic group known as “Ewe” reported by two independent studies in Ghana9,12. The third hypothesis is mainly supported by the phylogenetic placement of Maf (L6) amidst the cluster of the animal-adapted members of the MTBC in the various phylogenies of the MTBC5,7,18.
If the first hypothesis is true, the proportion of Maf associated TB in West Africa is expected to decline over time. However, there are conflicting reports of the proportion of Maf associated TB in West Africa. Even though the report of a steady decline of Maf associated TB in some settings seems to support the first hypothesis19–21, other studies indicate that Maf remains an important cause of TB in West-Africa22–24. In Ghana for instance, a recent study showed that the proportion of TB due to Maf remained constant over the 8 year study period25. Even though, the reported statistical association of L5 with ethnicity in Ghana suggests a possible co-evolutionary scenario in favour of the second hypothesis, genetic evidence of co-evolution/co-adaptation remains to be demonstrated. In the case of the third hypothesis, the environmental or zoonotic reservoir(s) need to be identified.
In this study, we used whole genome sequencing of Ghanaian Maf clinical strains to explore population genomic differences between the two Maf lineages that might support one or more of these hypotheses.
Results
Whole genome SNP distance, average nucleotide diversity and phylogeny of Maf in Ghana
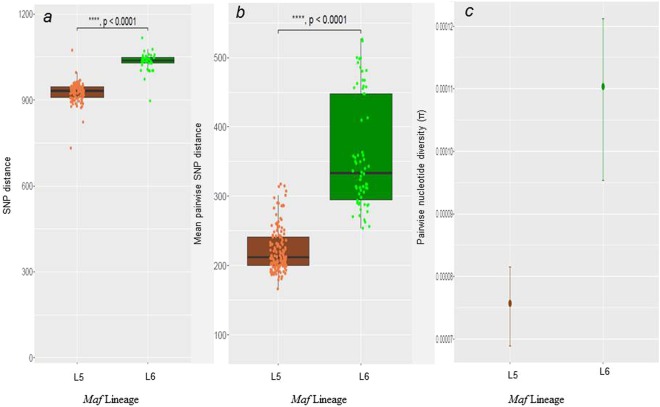
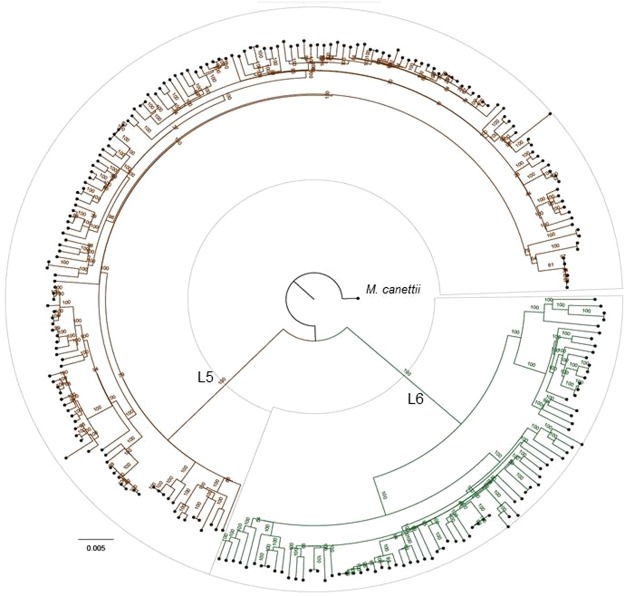
Our data set comprised Maf isolates obtained from TB patients reporting to various hospitals in Ghana. After excluding genomes that did not meet the quality criteria (Supplementary Fig. S1), 253 Maf genomes (175 L5 and 78 L6) were used for the analysis. Patients’ residential regions are provided (Supplementary Fig. S2). The upper right pie chart indicates those 97 patients (55 infected with L5 and 42 with L6) with no information on region of residence. We found the number of fixed SNPs (SNPs found in more than 95% of genomes) to be significantly higher in L6 (Median = 1,037) compared to L5 (Median = 928) (Wilcoxon rank-sum test, p < 0.0001) (Fig. 1a). Moreover, despite the larger number of L5 genomes (more than twice the number of L6 genomes) analyzed, the mean pairwise SNP distance between any two strains was significantly higher in L6 (360) compared to L5 (223) (Wilcoxon rank-sum test, p < 0.0001; Fig. 1b). Finally, the whole genome average nucleotide diversity (π) for L6 (0.000110) was significantly higher compared to L5 (0.00007) (Fig. 1c, non-overlapping 95% confidence interval (CI)). Taken together, these findings show that L6 in Ghana is significantly more genetically diverse than L5 irrespective of sample size. The whole genome-based phylogenetic tree of the Ghanaian Maf strains generated from 11,027 total polymorphic positions is shown in Fig. 2. The Maf lineages were resolved as two distinct branches of the genome-based tree with possible sub-groups (Fig. 2).
Figure 1.
Whole genome diversity of Maf Lineages (175 L5 and 78 L6 genomes). (a) Number of SNPs between Maf genomes and the hypothetical MTBC ancestor (the median fixed SNPs of L5 (934) is lower (W = 417, p-value < 2.2e-16) compared to L6 (1,039). (b) Pairwise SNPs between genomes within each lineage (the median of the pairwise SNPs is lower (W = 234, p-value < 2.2e-16) in L5 (212) compared to L6 (334). (c) Whole genome average nucleotide diversity (π) between L5 and L6 (the mean diversity of L5 (0.000076) is significantly (non-overlapping 95% confidence intervals) lower than L6 (0.000110). Error bars indicate 95% confidence intervals.
Figure 2.
Phylogeny of Ghanaian Maf strains. The maximum likelihood phylogenetic tree of 253 Ghanaian Maf isolates is based on 11,027 variable positions. The tree was rooted on M. canettii and the confidence of nodes was assessed by bootstrapping 1000 pseudo replicates. Each lineage clade is colored according to the conventional MTBC lineage color codes1.
Genetic diversity of L6 is significantly higher than L5 among T cell epitopes and genes of other functional categories
We found that the higher diversity of L6 compared to L5 was reflected across all the 8 functional categories of genes analyzed (Fig. 3). Whereas pairwise nucleotide diversity (π) for L5 was below 0.0001 across all functional categories, the estimates for L6 were all above 0.0001. The most prominent difference between L6 and L5 was within 1,226 experimentally confirmed human T cell epitopes of MTBC which we downloaded from the Immune Epitope Database (IEDB)26, for which the mean π for L5 was 0.000063 compared to the 0.000149 estimated for L6, reflecting more than a two-fold difference in diversity (non-overlapping 95% CI).
Figure 3.
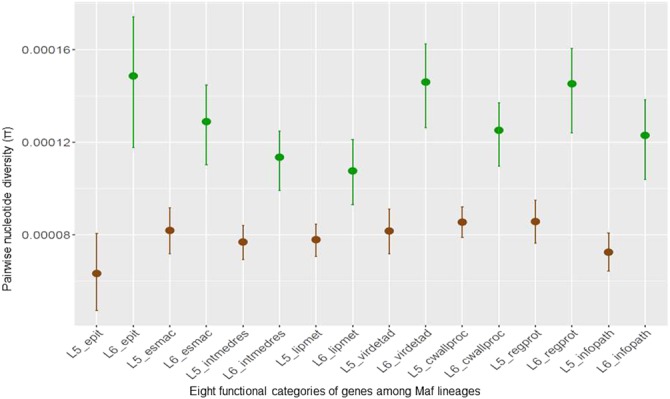
Averaged nucleotide diversity (π) of Maf within genes of eight functional categories. epit – genes encoding human T cell epitopes, esmac – genes essential for growth in macrophages, intmedres – genes involved with intermediate metabolism and respiration, lipmet – genes involved with lipid metabolism, virdetad – genes involved with virulence, detoxification and adaptation, cwallproc - genes involed with cell wall and cell processes, regprot – genes encoding regulatory proteins and infopath – genes involved with information pathways. Error bars are indications of 95% confidence intervals.
Within L5, there was no difference between the estimated π for the T cell epitopes and any of the other functionally categorized genes. However, within L6, genes encoding regulatory proteins and those involved with virulence, detoxification and adaptation were more diverse compared to those for lipid metabolism as well as intermediate metabolism and respiration (non-overlapping 95% CI). In addition, genes encoding regulatory proteins were more diverse compared to those involved with lipid metabolism (non-overlapping 95% CI).
Different selection pressures within L5 and L6 in human T cell epitopes and regulatory proteins
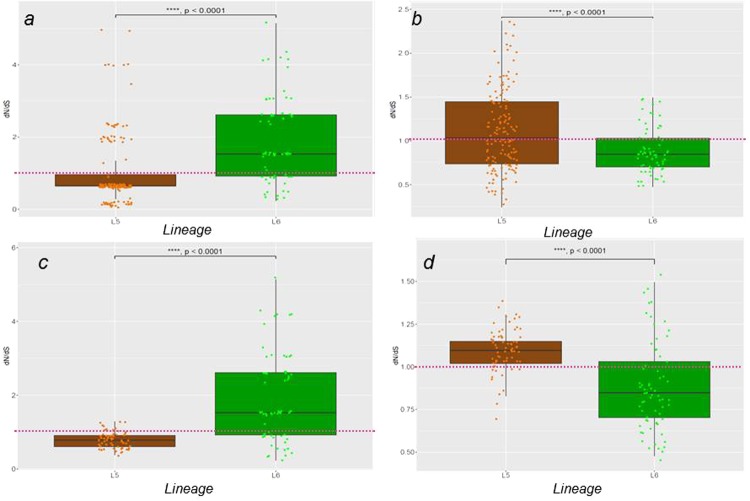
The mean pairwise dN/dS of the concatenates of T cell epitopes as well as genes of the seven other functional categories were calculated for all genomes and compared between L5 and L6. Apart from sequences encoding human T cell epitopes and regulatory proteins that had median mean pairwise dN/dS ratios greater than 1.0 in L6 and L5, respectively (Fig. 4, panel a and b), all the remaining functional categories showed a dN/dS ratio of less than 1.0 in both lineages (Supplementary Fig. S3). Human T cell epitopes of Maf L5 (median mean pairwise dN/dS = 0.64) were significantly more conserved compared to L6, which exhibited higher diversity (median mean pairwise dN/dS = 1.53) (Wilcoxon rank-sum test, W = 2265, p < 0.0001). Conversely, genes encoding regulatory proteins were more diverse among L5 genomes (with median mean pairwise dN/dS = 1.03) (Wilcoxon rank-sum test, W = 6303, p = 0.0010) compared to L6 (with median mean pairwise dN/dS = 0.85). To account for the different sample sizes; 147 L5 compared to 67 L6 genomes after excluding 43 genomes differing from others with less than 10 SNPs difference (see Methods and Supplementary Fig. S1), we repeated the analysis using mean values of 10 randomly sampled sets of L5 genomes with sample size 67 among human T cell epitopes (Fig. 4, panel c) and regulatory proteins (Fig. 4 panel d) and got similar results (Wilcoxon rank-sum test, W = 1300, p < 0.0001, W = 3466, p < 0.0001 for T cell epitopes and regulatory proteins, respectively).
Figure 4.
Pairwise dN/dS of genes encoding human T cell epitopes and regulatory proteins in L5 and L6. Estimation of pairwise dN/dS of epitopes (a) and regulatory proteins (b) using the entire 147 L5 against the 67 L6 genomes. Estimation of pairwise dN/dS of epitopes (c) and regulatory proteins (d) using the mean dN/dS values of 10 random samples (size = 67, with replacement) of L5 against the 67 L6 genomes.
Lineage-specific accumulation of mutations within human T cell epitopes
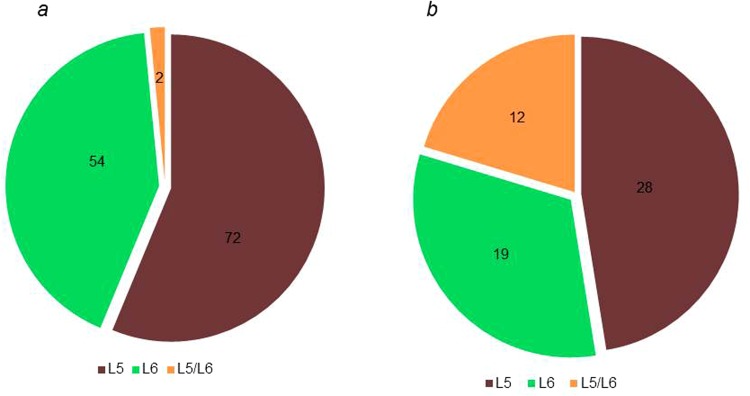
When we compared the number of epitopes with amino acid mutations between lineages, we found more epitopes mutated in L6 (N = 57) compared to L5 (N = 45), but this difference was not statistically significant (Fig. 5). In addition, we compared the number of nonsynonymous polymorphic sites between the two Maf lineages within the human T cell epitopes (Supplementary Fig S4), and found there were more frequent in L6 (N = 38) compared to L5 (N = 28) but with no statistically significant difference between L5 and L6 (X2 = 0.0055, p-value = 0.9407). We compared the identity of the mutant human T cell epitopes between the two Maf lineages (Fig. 6a) and found 72 epitopes that were uniquely mutated in L5 (among 174 genomes) compared to 54 epitopes in L6 (among 67 genomes). Only two epitopes (IEDB IDs 178644 and 178609) were mutated in both lineages. However, the mutations were at different loci with different amino acid substitutions (A183G and G278D in L5 compared to A177V and D277N in L6). In terms of T cell antigens, there were 28 uniquely mutated in L5 compared to 19 in L6 and 12 mutated in both lineages involving different epitopes within the respective antigens (Fig. 6b). The 12 T cell antigens mutated in both lineages are summarized in Table 1. All T cell epitopes and antigens with mutations among the two Maf lineages are listed in Supplementary Table S5.
Figure 5.
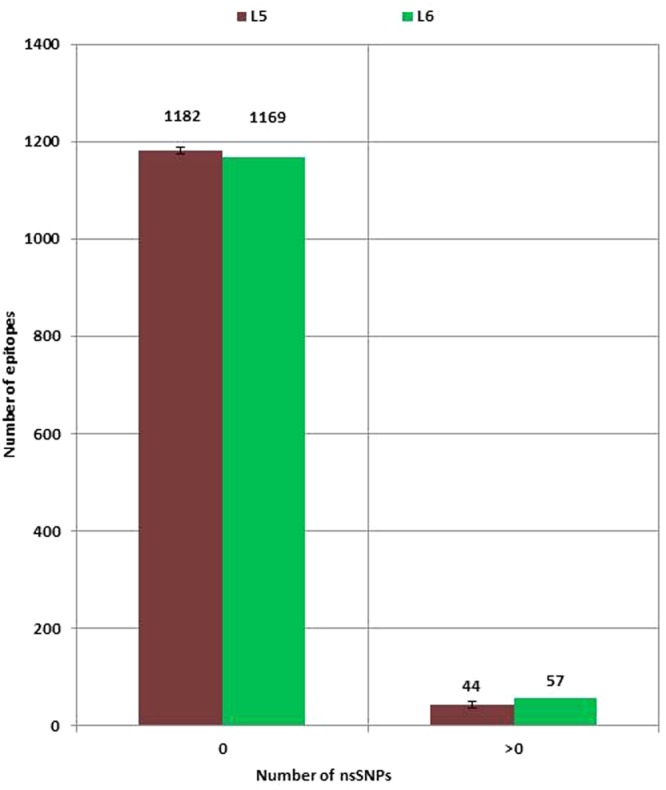
Number of human T cell epitopes with nonsynonymous SNPs (nsSNPs) stratified by Maf lineage. No significant difference (X-squared = 1.487, df = 1, p-value = 0.22) between the number of epitopes with nsSNPs among the 67 L6 genomes and L5 (mean values of 10 random samples of size = 67 with replacement).
Figure 6.
Number of human T cell epitopes (a) and human T cell antigens (b) with amino acid substitutions stratified by Maf lineage. Green represents L6-specific mutant antigens or epitopes. Brown represents L5-specific mutant antigens or epitopes. Yellow represents antigens or epitopes mutated in both L5 and L6 but at different loci with different amino acid substitutions.
Table 1.
Functions of the 12 T cell antigens mutated in both L5 and L6.
| T cell antigen | Function |
|---|---|
| Rv0288 | encodes low molecular weight antigen 7 EsxH involved with cell wall and cell processes |
| Rv0934 | encodes periplasmic phosphate-binding lipoprotein PstS1 involved with cell wall and cell processes |
| Rv2029c | encodes 6-phosphofructokinase PfkB involved with intermediate metabolism and respiration |
| Rv2627c | encoding a conserved hypothetical protein |
| Rv3003c | Encodes the large subunit of acetolactate synthase involved with valine and isoleucine biosynthesis |
| Rv3024c | encodes a probable tRNA involved with information pathways |
| Rv3763 | encodes a 19 kDa lipoprotein antigen precursor LpqH involved with cell wall and cell processes |
| Rv3804c | encodes the secreted antigen 85-a FbpA involved with lipid metabolism |
| Rv3823c | encodes conserved integral membrane transport protein MmpL8 involved with cell wall and cell processes |
| Rv3825c | encodes polyketide synthase Pks2 involved with lipid metabolism |
| Rv3879c | encodes ESX-1 secretion-associated protein EspK involved with cell wall and cell processes |
| Rv3883c | encodes membrane-anchored myosin MycP1 involved with intermediate metabolism and respiration |
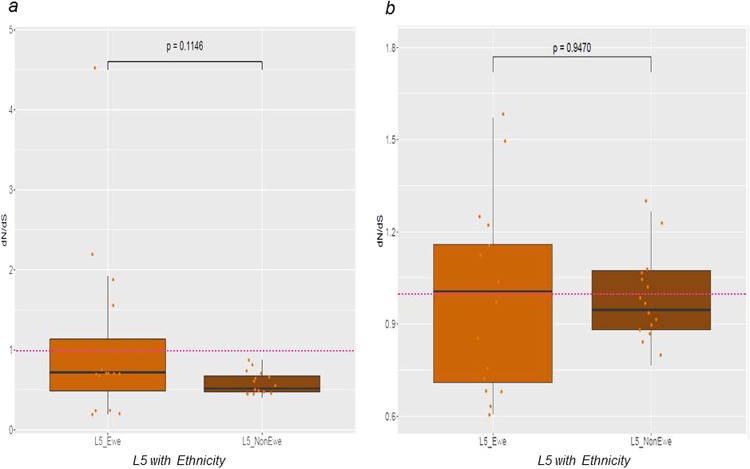
Conservation of human T cell epitopes of L5 is not affected by patient ethnicity
We previously reported an association between L5 and Ewe patient ethnicity9,12. Hence to test if conservation of T cell epitopes and/or the diversity of regulatory proteins in L5 was influenced by patient ethnicity, we estimated pairwise dN/dS for sequences encoding T cell epitopes and regulatory proteins of L5 genomes stratified by patient ethnicity (Fig. 7). The median dN/dS of T cell epitopes were all below 1.0 irrespective of patient ethnicity (Fig. 7a). However, the median dN/dS of regulatory proteins were marginally above 1.0 among L5 from Ewe TB patients and below 1.0 among L5 from non-Ewe TB patients (Fig. 7b). There was no statistically significant difference between the estimated dN/dS of either the sequences encoding T cell epitopes (Fig. 7a) or regulatory proteins (Fig. 7b) between L5 from TB patients of Ewe and non-Ewe ethnicities. In addition, there was no difference in either the number of T cell epitopes with amino acid substitutions (Supplementary Fig. S6A) or the number of non-redundant SNPs (Supplementary Fig. S6B) between L5 strains from patients of the Ewe ethnicity and those of other ethnicities.
Figure 7.
Pairwise dN/dS of sequences encoding human T cell epitopes (a) and genes encoding regulatory proteins (b) of L5 by patient ethnicity. L5 genomes from strains isolated from patients of the Ewe ethnicity (15 genomes) against, average values of 10 random samples of size 15 of L5 genomes of isolates from Non-Ewe patients.
Discussion
In this study, we analysed the largest collection of Maf genomes including both L5 and L6 reported so far. We found that (1) at the whole genome level, L6 had significantly higher pairwise nucleotide diversity, higher number of fixed SNPs as well as higher average pairwise SNPs relative to L5, (2) L6 had overall more diverse human T cell epitopes compared to L5, (3) the conservation of T cell epitopes in L5 was not influenced by patient ethnicity, and (4) genes encoding regulatory proteins of L5 had lower pairwise nucleotide diversity but a higher ratio of non-synonymous to synonymous substitution rate than L6.
Our finding that Maf L6 has a higher genetic diversity relative to L5 suggests that L6 has diversified more compared to L5 since the emergence of the two lineages5,27,28. The higher diversity of L6 could be due to either an earlier emergence, a higher mutation rate or both compared to L5. Our whole genome-based phylogenies rooted on Mycobacterium canettii showed that following the branch leading to Maf and all animal-adapted members of the MTBC defined by the characteristic deletion in RD929,30, L5 branches off earlier than L631. This designates L5 as basal to L6, hence arguing against the possibility of the higher diversity of L6 being primarily the result of earlier emergence. This notion is also supported by genomic deletion analyses showing that in addition to the deletion in RD9, L6 and all the animal-adapted members of the MTBC harbor the deletions of RD7, RD8 and RD1029. Therefore, higher intrinsic mutation rate and/or other factors are more likely to account for the higher diversity of L6 compared to L5 in Ghana.
Maf is highly restricted to West Africa, and thus could be seen as an ecological specialist compared to the other MTBC lineages. Specialists are expected to harbour less diversity across strains compared to generalists8. The observed lower genome-wide nucleotide diversity of L5 hence supports the hypothesis that L5 might be a specialist maintained in West Africa by adaptation to specific human genotypes9,12. In contrast, the higher genome-wide diversity of L6 indicates a generalist pathogen, and hence would have been expected to be globally distributed instead of displaying restriction to West Africa4,8. The observed diversity of L6 therefore may indicate a pathogen with a wider host range, supporting the hypothesis of maintenance in West Africa by possible environmental or zoonotic reservoir(s). Alternatively, it is also possible that the lower diversity of L5 observed relative to L6 could be due to a clonal expansion following a single introduction. Further studies comparing L5 and L6 from different countries in West Africa will help distinguish between these different possibilities.
Even though over 90% of T cell epitopes were conserved in both L5 and L6 (Fig. 5), which is in line with previous reports for the whole MTBC8,32,33, we found T cell epitopes in L6 overall to exhibit higher nucleotide diversity and a higher nonsynonymous to synonymous ratio compared to L5. The purifying selection of mutations within L5 is comparable to that reported for the specialist sub-lineages of L48. Interestingly, dN/dS within essential genes for survival in macrophages did not differ between L5 and L6 (Supplementary Fig. S3) supporting the notion that the genes in this category perform key functions in both L5 and L6. Since T cell responses partially drive the pathogenesis of TB34, the relative conservation of T cell epitopes in L5 indicate that it might elicit a more efficient T cell response compared to L6 in its particular host population. This therefore suggests L5 may be a more human-specific pathogen and L6, with significantly more diverse T cell epitopes, a potential opportunistic environmental or zoonotic pathogen. Even though the conserved T cell epitopes of L5 could account for geographical restriction to West Africa and the association with the Ewe ethnicity1,4,9,12, we found no difference between the diversity of L5 isolated from TB patients of Ewe and those of non-Ewe ethnic backgrounds. The limited number of L5 genomes from Ewe TB patients could possibly account for the lack of observed difference in diversity of T cell epitopes of L5 from TB patients of Ewe and non-Ewe ethnicities, and hence larger sample sizes are required to explore this further. L5 isolated from TB patients of the Ewe ethnicity were shown to be randomly distributed across the L5 clade of the Maf phylogeny instead of clustering in a particular sub-clade (Supplementary Fig. S7). This suggests that, if L5 is indeed maintained in West Africa by its co-evolution/adaptation with the Ewe ethnic group of West Africa (Cote d’Ivoire, Ghana, Nigeria, Togo and Benin)9,12, there is no specific sub-group of L5 that is responsible for this association but rather the whole of L5.
Members of the MTBC survive in the host mostly by modulation of the host immune response via the action of secretory proteins which form part of regulons controlled by specific regulatory proteins35,36. In addition, some regulatory proteins are involved in the regulation of transcription and translation of these secretory effectors as well as gene expression of other proteins involved with diverse functions36,37. Regulatory proteins in the MTBC hence play an important role in the survival and propagation of the bacteria. Therefore, our finding that regulatory proteins in L5 are under neutral selection (dN/dS = 1.03) compared to L6 in which they appear under purifying selection indicates that the mutations within regulatory proteins might be lineage-specific. This result is comparable to an earlier report comparing mutations within regulatory proteins between Mtbss and M. bovis, which found M. bovis to harbor majority of the mutations36. As mutations within some regulatory proteins have been associated with attenuated virulence38–40, our observation could account for the reported attenuated virulence of Maf relative to Mtbss14,15,17,41. This calls for further comparative studies of regulatory proteins between L5, L6 and other MTBC lineages to ascertain the effects of variation in regulatory proteins.
Our data is limited by the fact that, the number of L5 genomes was almost 3 times the number of L6 genomes; however, we used 1,000x bootstrap sampling with replacement of both L5 and L6 of equal sample size to limit any possible bias when comparing both lineages due to differences in sample size. Furthermore, a number of the L5 genomes did not have data on ethnicity and hence affected the number of L5 isolated from patients of the Ewe ethnicity for which we used average estimates of 10 random samples of L5 isolated from patients of non-Ewe origin in comparisons to account for the different sample sizes. In addition, the observed differential diversities of L5 and L6 could be due to founder effects of the lineages in Ghana which would require further studies comprising of Maf L5 and L6 from other West African countries pooled together for a sub-region-wide analysis.
In conclusion, our findings indicate that the two Maf lineages L5 and L6 are distinct in terms of population genomic diversity, and selection pressure on T cell epitopes and regulatory proteins, possibly reflecting different ecological niches. Whereas L5 may be maintained in West Africa by its co-evolution or adaptation with native West Africans, L6 may be maintained by an unknown environmental reservoir, possibly a zoonotic source. This genomic analysis of Maf from Ghana gives a glimpse of the often neglected diversity within Maf and the MTBC overall. Further studies using representative genomes of Maf from across West Africa to describe the full diversity of these members of the MTBC as well as functional assays are required to better understand the biology of Maf. Improved knowledge of Maf will have implications for our understanding of human TB and the development of better control tools.
Methods
Ethical Statement and Participant Enrollment
The study and its protocols were reviewed by the Scientific and Technical Committee (STC) and approved by the Institutional Review Board (IRB) of the Noguchi Memorial Institute for Medical Research (NMIMR), Legon-Ghana with Federal Wide Assurance number FWA00001824. All study methods were performed in accordance with the guidelines and regulations of the STC and IRB of the NMIMR.
Mycobacterium africanum Strains
Isolates used for this study were cultivated from a population based study running from July 2007 to November 2014 in Ghana9,12, West Africa, involving consecutive sputum smear positive pulmonary TB cases recruited from three geographical regions of Ghana (two from the South and one from the North) as shown in the map (Supplementary Fig. S2) constructed with the ArcGIS ArcMap tool ESRI version 10.2.2 (https://support.esri.com/en/Products/Desktop/arcgis-desktop/arcmap/10-2-2). An oral/written informed consent was sought from illiterate/literate TB patients before enrolment unto the studies. For children below 18 years, informed consent from a parent and/or legal guardian before enrolment.
Mycobacterial Sub-Culturing and Chromosomal DNA Extraction
Mycobacterium africanum strains were revived by sub-culturing on Lowenstein Jensen (LJ) slants; one supplemented with 0.4% sodium pyruvate the other with glycerol to enhance the growth of Lineage 5 and Lineage 6 strains of respectively. The cultures were incubated at 37 °C and monitored regularly until growth was observed. When confluence was achieved, five loops full of colonies were fetched into 2 mL cryo-vials containing 1 mL of sterile nuclease-free water, heat-inactivated at 98 °C for 60 minutes for DNA extraction using a hybrid DNA extraction protocol42. The isolates were confirmed MTBC by PCR amplification of IS6110, genotyped as Maf by large sequence polymorphism (LSPs) detecting region of difference (RD) 9 and 1243. Lineage identification was achieved by spoligotyping as previously described44. Strains confirmed as belonging either L5 or L6 were sequenced by the illumina platform at the Wellcome Trust Sanger Institute, United Kingdom.
DNA Sequencing, Mapping of Sequence Reads, Variance Calling and Generation of Whole Genome Fasta files
Samples were sequenced as multiplexed libraries on the Illumina HiSeq platform to produce paired end reads of 125 nt in length. Genomes provided by the Research Center Borstel was obtained by sequencing DNA libraries prepared with the Nextera XT kit and run on Illumina MiSeq (250 and 300 bp, paired end) and NextSeq (150 bp, paired end) according to the manufacturer’s instruction (Illumina, San Diego, USA). The FastQ files containing the raw paired-end reads were processed using a python pipeline developed in house as follows. The reads were first adapter- and quality- trimmed with Trimmomatic v0.3345. Reads lower than 20 bp were not kept for the downstream analysis. Overlapping paired-end reads were then merged with SeqPrep (https://github.com/jstjohn/SeqPrep). The resulting filtered reads were mapped to a hypothetical reconstructed MTBC ancestor32 with BWA v0.7.1246. Duplicated reads were marked by the MarkDuplicates module of Picard v 2.1.1 (https://github.com/broadinstitute/picard). The RealignerTargetCreator and IndelRealigner modules of GATK v.3.4.0 (https://software.broadinstitute.org/gatk/download/archive) were used to perform local realignment of reads around indels. SNPs were called with Samtools v1.2 (https://sourceforge.net/projects/samtools/files/samtools/1.2/) and VarScan v2.4.147 using the following thresholds: minimum mapping quality of 20, minimum base quality at a position of 20 and minimum read depth at a position of 7X. SNPs were considered fixed at a frequency of ≥90% and alleles were considered ancestral when the SNP frequency was ≤10%. Furthermore, SNPs were called only if the alternative basecall was supported by at least five reads and without strand bias. All variants were annotated using snpEff v4.1148, in accordance with the M. tuberculosis H37Rv reference annotation (AL123456.3). SNPs falling in regions with at least 50 bp identity to other regions in the genome were excluded from the analysis.
Generation of Variable Positions and Phylogenetic Analysis
The variable SNPs alignment was obtained by concatenating the SNP calls present in the variant calling file of each genome, using the IUPAC nucleotide ambiguity codes for heterozygous calls. A position was considered variable if at least one genome had a SNP at that position. Called deletions and positions not called according to the minimum threshold of 7 were encoded as gaps. Positions for which the proportion of gaps exceeded 50% were excluded from the alignment. Maximum likelihood phylogeny of the variable positions with 1000 bootstraps was then generated using RAxML version 8.2.349 with GTR substitution matrix and other default settings with the final tree evaluated and optimized under GAMMA with accuracy of 0.1 Log likelihood units. The best tree was then, rooted on M. canettii and annotated using figtree (http://www.webcitation.org/getfile?fileid=27177ee8dd2f34cfd254b9c5e6c6fdf4b65329f6).
Comparative genomics analysis of isolates using genes encoding proteins of 8 functional categories
Experimentally confirmed human MTBC T cell epitope (1,226 epitopes) sequences (spanning 304 antigens with some overlapping sequences) retrieved from the Immune Epitope Database (IEDB), tested in human T cell assays, with no major histocompatibility complex (MHC) restrictions and have genomic coordinates in the H37Rv reference strain8,31 were in silico extracted from the fasta whole genome files and concatenated excluding sequence redundancy using customized bash algorithms. Complementary sequences of epitopes encoded by the reversed strand were first transcribed before the concatenation to have all the sequences in the same direction. In addition, MTBC genes of other seven functional categories namely those encoding regulatory proteins (regprot; 196), genes involved with lipid metabolism (limpet; 267), genes involved with intermediate metabolism (intmedres; 917), genes involved with virulence, detoxification and adaptation (virdetad; 216), genes involved with information pathways (infopath; 234), genes involved with cell wall and cell processes (cwallproc; 768) and genes essential for growth in macrophages (esmac; 125) according to the tuberculist database50 were also retrieved and concatenated as described above excluding genes involved with drug resistance.
Estimation of Pairwise Nucleotide Diversity
Pairwise SNP distances of the whole genome excluding sites associated with drug resistance, concatenates of T cell epitopes and the genes of other seven functional categories were calculated with the dna.dist function of ape package51 of R version 3.2.352 as previously described8. Average pairwise nucleotide diversity per site (π) and confidence intervals for the π was calculated as previously described8 and plotted with ggplot2 package implemented in R. The upper and lower levels of confidence were attained by estimating the 97.5th and 2.5th quantiles of the π distribution obtained by bootstrapping (1000 replicates) as previously described8. Non-overlapping confidence intervals of π were taken as evidence of statistically significant differences53,54. Details of the algorithm for this analysis are available upon request.
Estimation of Pairwise dN/dS
The concatenates of the human T cell epitopes and the other genes of seven functional categories were also used for estimation of dN/dS ratios stratified by lineage. As a follow up, dN/dS of T cell epitopes and regulatory proteins were also estimated for 15 L5 genomes from Ewe TB patients and 77 from non-Ewe TB patients. The dN/dS estimates were calculated with all polymorphic sites within each lineage using the kaks function of the seqinr packgage55 as previously described8 and box plotted using ggplot2 package in in R version 3.2.3. Statistical difference of the estimates between the Maf lineages was accessed using the non-parametric Wilcoxon rank-sum tests with continuity correction in R version 3.4.0.
Human T cell Epitopes with Non-Synonymous SNPs and Count of Non-Redundant SNPs
Synonymous and non-synonymous mutations within the coordinates of each epitope were extracted from the variant calling file (VCF) obtained for each genome. The specific human T cell epitopes with non-synonymous SNPs were compared between the Maf lineages for lineage-specific mutated epitopes and Maf-specific mutated epitopes.
Furthermore, the number of pairwise non-redundant SNPs was estimated for the Maf lineages (67 L6 genomes and the 10 random samples of L5 of equal size as L6) as well as L5 genomes stratified by patient ethnicity (15 L5 from Ewe patients and 10 random samples of L5 from non-Ewe TB patients of size 15) using Mega656. The number of SNPs per each group was plotted and compared between the groups using the fisher’s exact test for statistical significance in R version 3.2.3.
Data availability
All the analyzed and/or generated data in this study are included in this article and its supplementary information files. Whole genome sequence reads have been submitted to the EMBL-EBI European Nucleotide Archive (ENA) Sequence Read Archive (SRA) with accession numbers provided in the supplementary document attached (Supplementary Data S8).
Electronic supplementary material
Acknowledgements
Bacterial Isolation and DNA preparations were done in the Biosafety level 3 facility at the Noguchi Memorial Institute for Medical Research, University of Ghana. Bioinformatics analyses were performed using the scientific computing core (sciCORE) at the University of Basel and the computing facility of the Wellcome Trust Sanger Institute, Genome Campus, Cambridge University. This work was supported by the Wellcome Trust Intermediate Fellowship awarded to DYM (Grant Number 097134/Z/11/Z) and by the Swiss National Science Foundation (grants 310030_166687, IZRJZ3_164171 and IZLSZ3_170834), the European Research Council (309540-EVODRTB) and SystemsX.ch.
Author Contributions
Conceived the idea: D.Y.M., S.G. Designed experiments: D.Y.M., S.G., I.D.O., M.C., S.R.H., J.P. Contributed reagents and performed experiments: I.D.O., M.C., A.A.P., L.S.B., M.C., S.O.W., A.F., C.L., G.A.A., A.I.Y., A.B., S.Y.A., P.A., C.L., D.B., S.B., F.G., P.B., T.K., S.N., M.A., S.N., C.B., B.C.d.J., J.P. and S.R.H. Analysed Data: I.D.O., M.D.C., S.R.H., L.S.B., S.G., D.Y.M. Wrote manuscript: I.D.O., M.C., S.G. and D.Y.M. All authors critically reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Isaac Darko Otchere, Mireia Coscollá, Sebastien Gagneux and Dorothy Yeboah-Manu contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29620-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sebastien Gagneux, Email: sebastien.gagneux@swisstph.ch.
Dorothy Yeboah-Manu, Email: dyboah-Manu@noguchi.ug.edu.gh.
References
- 1.Gagneux S, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 2006;103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 2007;7:328–37. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 3.Castets M, Boisvert H, Grumbach F, Brunel M, Rist N. Tuberculosis bacilli of the African type: preliminary note. Rev. Tuberc. Pneumol. (Paris). 1968;32:179–84. [PubMed] [Google Scholar]
- 4.de Jong BC, Antonio M, Gagneux S. Mycobacterium africanum-review of an important cause of human tuberculosis in WestAfrica . PLoS Negl. Trop. Dis. 2010;4:e744. doi: 10.1371/journal.pntd.0000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comas I, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 2013;45:1176–82. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comas, I. et al. Population Genomics of Mycobacterium tuberculosis in Ethiopia Contradicts the Virgin Soil Hypothesis for Human Tuberculosis in Sub-Saharan Africa. Curr. Biol. 3260–3266, 10.1016/j.cub.2015.10.061 (2015). [DOI] [PMC free article] [PubMed]
- 7.Hershberg R, et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stucki D, et al. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat. Genet. 2016;48:1535–1543. doi: 10.1038/ng.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asante-Poku A, et al. Molecular epidemiology of Mycobacterium africanum in Ghana. BMC Infect. Dis. 2016;16:385. doi: 10.1186/s12879-016-1725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeboah-manu D, et al. Genotypic diversity and drug susceptibility patterns among M. tuberculosis complex isolates from South-Western Ghana. PLoS One. 2011;6:e21906. doi: 10.1371/journal.pone.0021906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bold TD, et al. Impaired fitness of Mycobacterium africanum despite secretion of ESAT-6. J. Infect. Dis. 2012;205:984–90. doi: 10.1093/infdis/jir883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asante-Poku A, et al. Mycobacterium africanum Is Associated with Patient Ethnicity in Ghana. PLoS Negl. Trop. Dis. 2015;9:e3370. doi: 10.1371/journal.pntd.0003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Jong BC, et al. Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J. Infect. Dis. 2008;198:1037–43. doi: 10.1086/591504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Homolka S, Niemann S, Russell DG, Rohde KH. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog. 2010;6:e1000988. doi: 10.1371/journal.ppat.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tientcheu LD, et al. Differences in T-cell responses between Mycobacterium tuberculosis and Mycobacterium africanum -infected patients. Eur. J. Immunol. 2014;44:1387–1398. doi: 10.1002/eji.201343956. [DOI] [PubMed] [Google Scholar]
- 16.Jong BCD, et al. Mycobacterium africanum elicits an attenuated T cell response to early secreted antigenic target, 6 kDa, in patients with tuberculosis and their household contacts. J. Infect. Dis. 2006;193:1279–86. doi: 10.1086/502977. [DOI] [PubMed] [Google Scholar]
- 17.Portevin D, et al. Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog. 2011;7:e1001307. doi: 10.1371/journal.ppat.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niemann, S., Merker, M., Kohl, T. & Supply, P. Impact of Genetic Diversity on the Biology of Mycobacterium tuberculosis Complex Strains. Microbiol. Spectr. 4 (2016). [DOI] [PubMed]
- 19.Källenius G, et al. Evolution and clonal traits of Mycobacterium tuberculosis complex in Guinea-Bissau. J. Clin. Microbiol. 1999;37:3872–3878. doi: 10.1128/jcm.37.12.3872-3878.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niobe-Eyangoh SN, et al. Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J. Clin. Microbiol. 2003;41:2547–53. doi: 10.1128/JCM.41.6.2547-2553.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dosso M, et al. Primary resistance to antituberculosis drugs: a national survey conducted in Côte d’Ivoire in 1995–1996* for the Ivoirian Study Group on Tuberculosis Resistance ** Projet Santé Abidjan Coopération Summary. Int J Tuberc Lung Dis. 1999;3:805–809. [PubMed] [Google Scholar]
- 22.Koro FK, et al. Population dynamics of tuberculous bacilli in cameroon as assessed by spoligotyping. J. Clin. Microbiol. 2013;51:299–302. doi: 10.1128/JCM.01196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gehre F, et al. The first phylogeographic population structure and analysis of transmission dynamics of M. africanum West African 1–combining molecular data from Benin, Nigeria and Sierra Leone. PLoS One. 2013;8:e77000. doi: 10.1371/journal.pone.0077000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawson L, et al. A molecular epidemiological and genetic diversity study of tuberculosis in Ibadan, Nnewi and Abuja, Nigeria. PLoS One. 2012;7:e38409. doi: 10.1371/journal.pone.0038409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeboah-Manu D, et al. Spatio-Temporal Distribution of Mycobacterium tuberculosis Complex Strains in Ghana. PLoS One. 2016;11:e0161892. doi: 10.1371/journal.pone.0161892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vita, R. et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 43 (2015). [DOI] [PMC free article] [PubMed]
- 27.Bos KI, et al. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature. 2014;514:494–497. doi: 10.1038/nature13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kay GL, et al. Eighteenth-century genomes show that mixed infections were common at time of peak tuberculosis in Europe. Nat. Commun. 2015;6:6717. doi: 10.1038/ncomms7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brosch R, et al. A new evolutionary scenario for the Mycobacterium. Proc. Natl. Acad. Sci. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mostowy S, Cousins D, Brinkman J, Aranaz A, Behr MA. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J. Infect. Dis. 2002;186:74–80. doi: 10.1086/341068. [DOI] [PubMed] [Google Scholar]
- 31.Coscolla M, Gagneux S. Consequences of genomic diversity in mycobacterium tuberculosis. Semin. Immunol. 2014;26:431–444. doi: 10.1016/j.smim.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Comas I, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat. Genet. 2010;42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coscolla M, et al. M. tuberculosis T Cell Epitope Analysis Reveals Paucity of Antigenic Variation and Identifies Rare Variable TB Antigens Reveals Paucity of Antigenic Variation and Identifies Rare Variable TB Antigens. Cell Host Microbe. 2015;18:538–548. doi: 10.1016/j.chom.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pai M, et al. Tuberculosis. Nat. Rev. Dis. Prim. 2016;2:16076. doi: 10.1038/nrdp.2016.76. [DOI] [PubMed] [Google Scholar]
- 35.Brodin P, Rosenkrands I, Andersen P, Cole ST, Brosch R. ESAT-6 proteins: Protective antigens and virulence factors? Trends Microbiol. 2004;12:500–508. doi: 10.1016/j.tim.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 36.Bigi, M. M. et al. Polymorphisms of 20 regulatory proteins between Mycobacterium tuberculosis and Mycobacterium bovis. Microbiol. Immunol., 10.1111/1348-0421.12402 (2016). [DOI] [PubMed]
- 37.Raman, S., Hazra, R., Dascher, C. C. & Husson, R. N. Transcription regulation by the Mycobacterium tuberculosis alternative sigma factor SigD and its role in virulence. J. Bacteriol., 10.1128/JB.186.19.6605-6616.2004 (2004). [DOI] [PMC free article] [PubMed]
- 38.Gonzalo-asensio, J., Malaga, W., Pawlik, A. & Astarie-dequeker, C. Evolutionary history of tuberculosis shaped by conserved mutations in the PhoPR virulence regulator. 111 (2014). [DOI] [PMC free article] [PubMed]
- 39.Peirs P, Parmentier B, De Wit L, Content J. The Mycobacterium bovis homologous protein of the Mycobacterium tuberculosis serine/threonine protein kinase Mbk (PknD) is truncated. FEMS Microbiol. Lett. 2000;188:135–9. doi: 10.1111/j.1574-6968.2000.tb09184.x. [DOI] [PubMed] [Google Scholar]
- 40.Saïd-Salim B, Mostowy S, Kristof AS, Behr MA. Mutations in Mycobacterium tuberculosis Rv0444c, the gene encoding anti-SigK, explain high level expression of MPB70 and MPB83 in Mycobacterium bovis. Mol. Microbiol. 2006;62:1251–1263. doi: 10.1111/j.1365-2958.2006.05455.x. [DOI] [PubMed] [Google Scholar]
- 41.de Jong BC, et al. Differences between tuberculosis cases infected with Mycobacterium africanum, West African type 2, relative to Euro-American Mycobacterium tuberculosis: an update. FEMS Immunol. Med. Microbiol. 2010;58:102–5. doi: 10.1111/j.1574-695X.2009.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otchere ID, et al. Detection and characterization of drug-resistant conferring genes in Mycobacterium tuberculosis complex strains: A prospective study in two distant regions of. Tuberculosis (Edinb). 2016;99:147–154. doi: 10.1016/j.tube.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warren RM, et al. Molecular evolution of Mycobacterium tuberculosis: phylogenetic reconstruction of clonal expansion. Tuberculosis. 2001;81:291–302. doi: 10.1054/tube.2001.0300. [DOI] [PubMed] [Google Scholar]
- 44.Kamerbeek J, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 1997;35:907–14. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koboldt DC, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strainw1118; iso-2; iso-3. Fly (Austin). 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList - 10 years after. Tuberculosis (Edinb). 2011;91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 51.Popescu A-A, Huber KT, Paradis E. ape 3.0: New tools for distance-based phylogenetics and evolutionary analysis in R. Bioinformatics. 2012;28:1536–1537. doi: 10.1093/bioinformatics/bts184. [DOI] [PubMed] [Google Scholar]
- 52.R Core Team. R: A language and environment for statistical computing version 3.2.3. R Foundation for Statistical Computing, Vienna, Austria (2015). Available at: https://www.coursehero.com/file/pe12cv/Code-R-version-323-2015-12-10-Wooden-Christmas-Tree-Copyright-C-2015-The-R/. (Accessed: 26th July 2017).
- 53.Gardner MJ, Altman DG. Statistics in Medicine Confidence intervals rather than P values: estimation rather than hypothesis testing. Br. Med. J. (Clin. Res. Ed). 1986;292:746–750. doi: 10.1136/bmj.292.6522.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakagawa S, Cuthill IC. Effect size, confidence interval and statistical significance: A practical guide for biologists. Biol. Rev. 2007;82:591–605. doi: 10.1111/j.1469-185X.2007.00027.x. [DOI] [PubMed] [Google Scholar]
- 55.Charif, D. & Lobry, J. R. SeqinR 1.0-2: a contributed package to the R-project for statistical computing devoted to biological sequences retrieval and analysis. In: Bastolla U, Porto MERH, Vendruscolo M, editors. Structural approaches to sequence evolution: Molecules, networks, populations. Springer Verlag (2007).
- 56.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the analyzed and/or generated data in this study are included in this article and its supplementary information files. Whole genome sequence reads have been submitted to the EMBL-EBI European Nucleotide Archive (ENA) Sequence Read Archive (SRA) with accession numbers provided in the supplementary document attached (Supplementary Data S8).